Abstract
The synthesis and evaluation of two cathepsin S-specific probes is described. For long-term retention of the probe at the target site and a high signal-to-noise ratio, we introduced a lipidation approach via the simple attachment of palmitoic acid to the reporter. After cathepsin S-specific cleavage in cultured cells and in a grafted tumor mouse model, fluorescence increased owing to dequenching and we observed an intracellular accumulation of the fluorescence in the target tissue. The lipidated probe provided a prolonged and strongly fluorescent signal in tumors when compared to the very similar non-lipidated probe, demonstrating that non-invasive tumor identification is feasable. The homing principle by probe lipidation might also work for selective administration of cytotoxic compounds to specifically reduce tumor mass.
Keywords: fluorescence probes, FRET, homing, lipidation, tumor diagnosis
It is the ultimate goal in cancer diagnostics to locate tumors in humans early and with minimal invasion.[1] In the past decade, optical molecular imaging technologies have become a powerful addition to both tumor detection and therapeutic treatment evaluation owing to its high selectivity, good spatial resolution, and non-invasive mode of action.[2] However, the limitation of this method over techniques such as MRI and CT is that fluorescent signals are attenuated by body mass, skin, and hair, mostly through light absorption and scattering. Additionally, high autofluorescence in animal tissue reduces the signal-to-noise ratio.[3] One way of mitigating these problems is the use of tumor-specific enzyme substrates in combination with dyes emitting in the near infrared (NIR). Proteases are typical enzymes secreted into the tumor microenvironment by both tumor and tumor-associated cells. They are required for promoting tumor growth, tissue invasion, and the altered metabolism of tumor cells.[4] A number of probes targeting proteases have been developed in the past and several are on the way to the clinic, among them several substrates.[5] A common problem with these probes is a lack of local accumulation of the signal generated, often referred to as “homing”. Therefore, we hypothesized that probes with specific cancerous tissue accumulation would enhance the signal-to-noise ratio after being cleaved by the enzyme.
Among the proteases that have a major role in cancer are cysteine cathepsins, in particular cathepsins B, L, and S, as judged on the basis of pharmacological and genetic approaches.[6] In cancer, the primary source of these cathepsins are tumor and tumor-associated cells, and in particular macrophages.[4, 7] A number of groups have studied the mechanistic role of cathepsin S in cancer using in vitro and in vivo models.[7b] Genetic ablation of cathepsin S in the pancreatic islet model revealed that the enzyme plays a role in tumor invasion, resistance to apoptosis, and also in tumor angiogenesis.[7b] Moreover, Kwok et al. have shown that cathepsin S is expressed on the surface of carcinoma cells and stays associated with the cell membrane.[8]
Based on the reverse design principle, we equipped a highly specific non-peptidic substrate for cathepsin S with an NIR fluorophore and a suitable quencher and used a lipidation approach to target the fluorescent product of the enzymatic reaction to the region of enzymatic activity, that is, the tumor (Scheme 1). Reverse design for protease substrates is based on the idea that a non-peptidic high affinity (or covalently binding) inhibitor provides optimized interaction with the protease of interest and is turned into a high turnover substrate by placing a peptide bond close to the catalytic center of the protease.[9] We attached a long wavelength fluorophore (Cy5.5 for in vivo near-infrared imaging) and a suitable quencher (BHQ-3) by reactive amino groups at the molecule termini. Another orthogonally protected amino group was used to attach palmitic acid as the lipid moiety (Supporting Information, Scheme S1). Such lipidations were previously used to locate peptidic FRET reporters to the outer surface of inflammatory macrophages or neutrophils that secrete elastases enabling the hydrolytic enzyme activity to reside exclusively on the cell surface.[10] In some cases, a local memory effect is enforced by internalization of the fluorescent fragment suggesting the possibility of successful homing.[10a] As cathepsin S is secreted and relocalized to the cell surface during tumor development,[7b] we expected the signal to be confined to the tumor and its microenvironment thereby generating a strong localized signal. Provided that the reverse design approach prevents unspecific cleavage, all other areas in the specimen should show very low fluorescence. This should result in largely increased signal-to-noise ratios and improved tumor detection compared to non-lipidated fluorescent substrates.[11]
Scheme 1.

Highly specific cathepsin S inhibitor 1 was turned into a non-lipidated (2) and a lipidated substrate (3) for cathepsin S (reverse design) and equipped with a long wavelength fluorophore and a suitable quencher. The cleavage site of the probe is marked with a red asterisk.
Cathepsin S-sensitive activity-based probes have been designed as suicide substrates that label the enzyme covalently and inactivate it upon binding.[12] While such probes are a powerful way to localize cathepsins, turnover-based probes are particularly useful for the detection of activities of low-abundance proteases.[9b] Compound 1 has a high affinity for cathepsin S while exhibiting excellent selectivity against the related cathepsins, which provided an attractive scaffold for the reverse design of a near-infrared (NIR) quenched fluorescent probe for cathepsin S.[9b] For extension beyond the reporter unit itself, the P1 moiety was equipped with a branched linker for attaching palmitoic acid as the lipid moiety (Scheme 1). The synthesis started from the non-natural amino acid Fmoc-DAB(Boc)-OH using standard solid-phase peptide chemistry, followed by further derivatization of orthogonally protected amino groups in solution phase (Supporting Information, Scheme S1).
The resulting lipidated probe 3 was synthesized with the fluorophore Cy5.5 and the quencher BHQ-3 (see the Supporting Information for details). For comparison, the non-lipidated probe 2 was synthesized by using the same linker with maleimide as a chemical tag, which may potentially be used to facilitate the conjugation with macromolecular delivery systems in the future. A sample of probe 2 was reacted with cysteine to quench the Michael acceptor (probe 2 b). Cleavage of the probes was tested using recombinant enzymes. Despite the use of bulky fluorophores, 3 and 2 were good substrates for cathepsin S, with turnover rate constants (kcat/Km) values of 2700 and 41 700 L mol−1 s−1, respectively. The probes showed a pronounced selectivity for cathepsin S over the related cysteine cathepsins B, K, L, and V, all of which share very high sequence homology. No turnover was detected for cathepsins B, K, L, and V except for probe 2, that was cleaved by cathepsin V with kcat/Km value of about 500 L mol−1 s−1 with> 80-fold selectivity for cathepsin S (Figure 1). The kinetic profiling further showed that lipidation did not have a significant effect on the Km value (Km≈2 μm for both probes), but decreased the turnover efficiency by a factor of about 15, which is most likely due to reduced water solubility and/or aggregation effects induced by the fatty acid moiety. Moreover, both compounds 2 and 3 were cleaved by cathepsin S at exactly the same site as determined by mass spectrometry (Supporting Information, Figures S1 and S2), indicating that the specificity was conserved. As lipidation of the probes was shown previously to confine them to the membrane of inflammatory cells,[10a] we next tested 3 in the presence of liposomes as model membrane vesicles for the effect on turnover kinetics. A slight but rapid self-dequenching of 3 was observed, indicative of insertion of the probe into the membrane, whereas no changes were seen for 2 (Supporting Information, Figure S3). Moreover, in the presence of liposomes, the Kcat/Km value for hydrolysis of 3 increased by a factor of about 7 to a value of 19 500 L mol−1 S−1, whereas no significant effect was seen for 2, suggesting that the insertion of the lipid moiety into the liposome membrane facilitated the hydrolysis of 3 by cathepsin S. Very similar results were obtained also in the presence of BSA, suggesting that the probe may bind to serum albumines.
Figure 1.
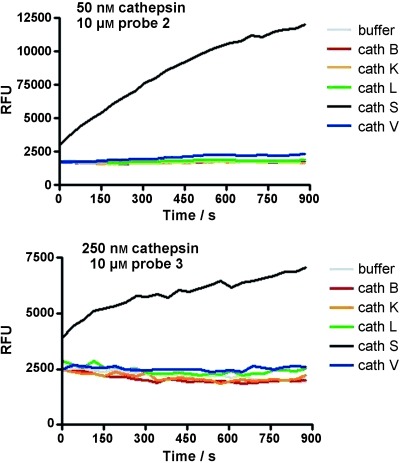
Cleavage of cathepsin S reporters in vitro. Change in emission intensity at λem=695 nm (λex=670 nm) of 2 (a) and lipidated probe 3 (b) after addition of various cathepsins. RFU=relative fluorescence units.
As macrophages are known as a major source of cathepsin S in the tumor microenvironment,[7a] we next tested enzyme activity in THP-1 cells differentiated into macrophages as these cells express both intracellular and extracellular cathepsin S. Cells were incubated with probes 2 or 3, respectively, and reporter cleavage was analyzed by live cell microscopy. Both probes were readily cleaved and taken up by the cells, although longer incubation time was needed for efficient cleavage of 3 (Figure 2 a). Pre-treatment of cells with the broad-spectrum membrane-impermeant cathepsin inhibitor E-64 had very little effect on the hydrolysis of 2, suggesting that the probe was largely cleaved intracellularly after internalization. In contrast, E-64 significantly protected the lipidated probe 3 against hydrolysis, suggesting that the lipidated probe was cleaved on the surface of the cells by extracellular cathepsin S prior to internalization of the lipidated fragment carrying the fluorophore. The cell-permeant form of the inhibitor, E-64d, essentially abolished processing of both probes, suggesting that cathepsins and not other proteases are responsible for probe hydrolysis (Figure 2 b). Furthermore, blocking endocytosis by cooling the cells to 4 °C for 30 min abolished hydrolysis of 2, whereas a signal for 3 was observed only on the cell membrane, confirming the above conclusions (Supporting Information, Figure S4).
Figure 2.

Bright-field (left) and NIR fluorescence (right, (λex=635–675 nm, λem=696–736 nm) images of PMA-differentiated THP-1 cells incubated with probe 2 (t=1 h) or lipidated probe 3 (t=4 h). Longer incubation time for probe 3 was required to overcome the effect of self-dequenching. Cells were incubated with the cell-impermeant cathepsin inhibitor E-64 or its cell-permeant form E-64d for 1 h prior to addition of the probe. Scale bar=200 μm. PMA=phorbol ester.
We then investigated if the homing of the lipidated probe 3 was sufficient to allow accumulation in an animal tumor model and if the difference in the probe distribution would affect the duration of the signal. We therefore applied probes 2 and 3 to visualize cathepsin S activity in vivo in a grafted tumor mouse model (4T1 cells, probe hydrolysis in single cells shown in the Supporting Information, Figure S5 a,b). Whole body fluorescence images were obtained with a fluorescence imager (Cri Maestro 500, Wolburn, MA, USA) after intravenous injection of probe 3 or 2, respectively. A substantial fluorescence signal was detected in the tumor in vivo as early as 30 min after injection for both probes. The Cy5.5 signal in tumors significantly increased in the next 6 h indicating probe cleavage. All animals showed steady and pronounced fluorescence signals at 24 h postsystemic injection (Figure 3 a) and a decrease thereafter (Supporting Information, Figures S6–S8). The cysteine-quenched probe 2 b behaved identical to probe 2 (Supporting Information, Figure S6).
Figure 3.
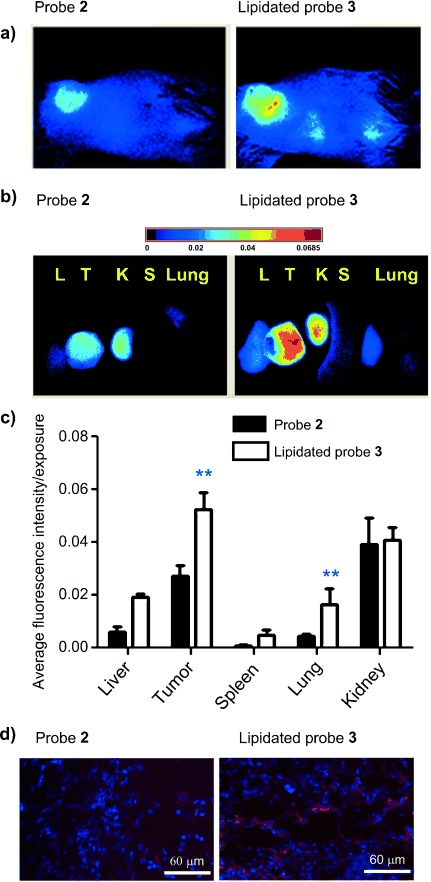
a) Whole-mouse optical images of 4T1 tumor-bearing mice at 24 h post i.v. injection of 2 (left) and 3 (right), respectively (n=5). b), c) Distribution of 2 and 3 in key organs at 5 days post injection. d) Cy5.5 fluorescence images of deep-frozen tumor sections of mice injected with 2 (left) and 3 (right), respectively. DAPI (4′,6′-diamidino-2-phenylindole; blue) was applied by the mounting media. (λex=405/680 nm; λem=425/700 nm, scale bar=60 μm). ** p<0.003.
Over time, the lipidated probe 3 showed increased tumor-specific fluorescence, resulting in a substantially better contrast at all-time points compared to the non-lipidated probe 2. The tumor-to-healthy tissue signal peaked after 5 days post-injection of probe 3 with Cy5.5 intensities in tumors about 6 times higher than that of surrounding skin in vivo and up to 18 times higher than in muscle ex vivo (Supporting Information, Table S2). Moreover, the lipidated probe 3 signal was twice as high after 24 h (Figure 3 a) and allowed effective tumor imaging after 5 and 8 days, when the signal from probe 2 was reduced close to the limit of quantification (Supporting Information, Figures S7 and S8). These significant differences demonstrate the advantage of the lipidated probe 3 for the detection of tumor tissue in vivo. Because the grafted tumor is located close to the skin, accumulation of the fluorescence in key organs cannot be easily detected at comparable levels owing to the poor light penetration in deeper tissue. Therefore, ex vivo imaging of excised tumors and other organs was performed after 5 days to investigate the probe fate in tumor versus organs. As shown in Figure 3 b, the biodistribution study indicated that the non-lipidated probe 2 was cleaved predominantly in the tumor, to a lesser extend found in the kidney and hardly at all in other organs, while the lipidated probe 3 was activated predominantly in the tumor and to some extent in the kidney (Figure 3 a–c) with almost no other organs being affected. To confirm that the cleaved probes entered the cell and accumulated intracellularly, tumors were removed from the mouse, fixed, frozen, and sectioned. Slices were evaluated by fluorescence microscopy. NIR excitation showed strong intracellular fluorescence signals in the tumor sections. The signal from mice treated with the lipidated probe 3 were much stronger than those from the non-lipidated probe 2, consistent with the images obtained by in vivo imaging (Figure 3 d).
In summary, the synthesis and evaluation of two cathepsin S-specific probes is described based on the concept of reverse design. To achieve long-term retention of the probe at the target site and a high signal-to-noise ratio, we introduced a lipidation approach by simply attaching palmitoic acid to the reporter. Lipidation was expected to locate the probe predominantly on the cell surface or bound to serum albumins. The successful application to mice by tail vein injection was a surprise as we expected predominantly staining of the endothelium close to the injection site. We speculate that albumins in the blood stream are able to prevent instantaneous targeting to cells and keep the lipidated probe in circulation for extended periods of time. This might also be the reason that the successful homing of the fluorescent probe fragment lasted days rather than hours. After cathepsin S-specific cleavage, fluorescence increased due to dequenching and we observed an accumulation of the fluorescence intracellularly in the target tissue. It appears that only a fraction of the fluorescent probe is leaving the tumor mass and partially stains liver (probe 3) or kidneys (probe 2). The lipidated probe 3 clearly retained a prolonged and strongly fluorescent signal in tumors when compared to the very similar non-lipidated probe 2 (Supporting Information, Figure S7), demonstrating that non-invasive tumor identification with 3 might be possible in the future. We suggest that the homing principle via probe lipidation might also work for selective administration of cytotoxic compounds to specifically reduce tumor mass.
Supporting Information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
miscellaneous_information
References
- 1.O’Farrell AC, Shnyder SD, Marston G, Coletta PL, Gill JH. Br. J. Pharmacol. 2013;169:719–735. doi: 10.1111/bph.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drummen G. Molecules. 2012;17:14067–14090. doi: 10.3390/molecules171214067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chudakov DM, Lukyanov S, Lukyanov KA. Trends Biotechnol. 2005;23:605–613. doi: 10.1016/j.tibtech.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 4.Mohamed MM, Sloane BF. Nat. Rev. Cancer. 2006;6:764–775. doi: 10.1038/nrc1949. [DOI] [PubMed] [Google Scholar]
- 5a.Nguyen QT, Olson ES, Aguilera TA, Jiang T, Scadeng M, Ellies LG, Tsien RY. Proc. Natl. Acad. Sci. USA. 2010;107:4317–4322. doi: 10.1073/pnas.0910261107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5b.Edgington LE, Verdoes M, Bogyo M. Curr. Opin. Chem. Biol. 2011;15:798–805. doi: 10.1016/j.cbpa.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5c.Verdoes M, Bender KO, Segal E, van der Linden WA, Syed S, Withana NP, Sanman LE, Bogyo M. J. Am. Chem. Soc. 2013;135:14726–14730. doi: 10.1021/ja4056068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Lecaille F, Kaleta J, Brömme D. Chem. Rev. 2002;102:4459–4488. doi: 10.1021/cr0101656. [DOI] [PubMed] [Google Scholar]
- 6b.Reiser J, Adair B, Reinheckel T. J. Clin. Invest. 2010;120:3421–3431. doi: 10.1172/JCI42918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6c.Vasiljeva O, Reinheckel T, Peters C, Turk D, Turk V, Turk B. Curr. Pharm. Des. 2007;13:387–403. doi: 10.2174/138161207780162962. [DOI] [PubMed] [Google Scholar]
- 6d.Gocheva V, Joyce JA. Cell Cycle. 2007;6:60–64. doi: 10.4161/cc.6.1.3669. [DOI] [PubMed] [Google Scholar]
- 6e.Mikhaylov G, Mikac U, Magaeva AA, Itin VI, Naiden EP, Psakhye I, Babes L, Reinheckel T, Peters C, Zeiser R, Bogyo M, Turk V, Psakhye SG, Turk B, Vasiljeva O. Nat. Nanotechnol. 2011;6:594–602. doi: 10.1038/nnano.2011.112. [DOI] [PubMed] [Google Scholar]
- 7a.Gocheva V, Wang HW, Gadea BB, Shree T, Hunter KE, Garfall AL, Berman T, Joyce JA. Genes Dev. 2010;24:241–255. doi: 10.1101/gad.1874010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7b.Gocheva V, Zeng W, Ke D, Klimstra D, Reinheckel T, Peters C, Hanahan D, Joyce J. Genes Dev. 2006;20:543–556. doi: 10.1101/gad.1407406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwok HF, Buick R, Kuehn D, Gormley J, Doherty D, Jaquin T, McClurg A, Ward C, Byrne T, Jaworski J, Leung KL, Snoddy P, McAnally C, Burden R, Gray B, Lowry J, Sermadiras I, Gruszka N, Courtenay-Luck N, Kissenpfennig A, Scott C, Johnston J, Olwill S. Mol. Cancer. 2011;10:147. doi: 10.1186/1476-4598-10-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Watzke A, Kosec G, Kindermann M, Jeske V, Nestler H-P, Turk V, Turk B, Wendt KU. Angew. Chem. 2008;120:412–415. doi: 10.1002/anie.200702811. [DOI] [PubMed] [Google Scholar]
- Angew. Chem. Int. Ed. 2008;47:406–409. doi: 10.1002/anie.200702811. [DOI] [PubMed] [Google Scholar]
- 9b.Caglič D, Globisch A, Kindermann M, Lim N-H, Jeske V, Juretschke H-P, Bartnik E, Weithmann KU, Nagase H, Turk B, Wendt KU. Bioorg. Med. Chem. 2011;19:1055–1061. doi: 10.1016/j.bmc.2010.10.028. [DOI] [PubMed] [Google Scholar]
- 10a.Cobos-Correa A, Trojanek JB, Diemer S, Mall MA, Schultz C. Nat. Chem. Biol. 2009;5:628–630. doi: 10.1038/nchembio.196. [DOI] [PubMed] [Google Scholar]
- 10b.Gehrig S, Mall MA, Schultz C. Angew. Chem. 2012;124:6363–6366. doi: 10.1002/anie.201109226. [DOI] [PubMed] [Google Scholar]
- Angew. Chem. Int. Ed. 2012;51:6258–6261. doi: 10.1002/anie.201109226. [DOI] [PubMed] [Google Scholar]
- 11.Jiang T, Olson ES, Nguyen QT, Roy M, Jennings PA, Tsien RY. Proc. Natl. Acad. Sci. USA. 2004;101:17867–17872. doi: 10.1073/pnas.0408191101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verdoes M, Edgington LE, Scheeren FA, Leyva M, Blum G, Weiskopf K, Bachmann MH, Ellman JA, Bogyo M. Chem. Biol. 2012;19:619–628. doi: 10.1016/j.chembiol.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
miscellaneous_information


