Abstract
The phytohormone abscisic acid (ABA) regulates plant growth, development, and abiotic stress responses. ABA signaling is mediated by a group of receptors known as the PYR1/PYL/RCAR family, which includes the pyrabactin resistance 1–like protein PYL8. Under stress conditions, ABA signaling activates SnRK2 protein kinases to inhibit lateral root growth after emergence from the primary root. However, even in the case of persistent stress, lateral root growth eventually recovers from inhibition. We showed that PYL8 is required for the recovery of lateral root growth, following inhibition by ABA. PYL8 directly interacted with the transcription factors MYB77, MYB44, and MYB73. The interaction of PYL8 and MYB77 increased the binding of MYB77 to its target MBSI motif in the promoters of multiple auxin-responsive genes. Compared to wild-type seedlings, the lateral root growth of pyl8 mutant seedlings and myb77 mutant seedlings was more sensitive to inhibition by ABA. The recovery of lateral root growth was delayed in pyl8 mutant seedlings in the presence of ABA, and the defect was rescued by exposing pyl8 mutant seedlings to the auxin IAA (3-indoleacetic acid). Thus, PYL8 promotes lateral root growth independently of the core ABA-SnRK2 signaling pathway by enhancing the activities of MYB77 and its paralogs, MYB44 and MYB73, to augment auxin signaling.
INTRODUCTION
Abscisic acid (ABA) is an important phytohormone that regulates many growth and developmental processes, including seed longevity, dormancy and germination, vegetative growth, root architecture, and abiotic stress responses (1–4). ABA, together with auxin and other hormones, regulates root growth and architecture to acclimate to the variable growth environment, such as drought and salt stress, nutrient availability, gravity, and light (5–12). The root system of higher plants consists of primary, lateral, and adventitious roots. High concentrations of ABA inhibit both primary and lateral root growth (5, 9, 13, 14). Osmotic stress and salt stress inhibit primary and lateral root growth in a partially ABA-dependent manner (1, 6, 9, 15). During salt stress, both primary roots and lateral roots that have emerged from the primary root undergo a quiescent phase followed by a recovery phase (9, 15). ABA signaling functions in growth suppression during the quiescent phase of primary roots and lateral roots that have emerged from the primary root (post-emergence), although the quiescent phases for primary and lateral roots last for different periods and show different sensitivities to salt (9, 15). A low concentration of ABA is required for primary root elongation in response to water deficit and salt stress (15, 16). ABA is required for the growth recovery of primary root from inhibition and for long-term growth of primary roots during salt stress (15). In addition, the phytohormone gibberellic acid promotes growth recovery of both primary and lateral roots (9, 15).
In contrast to the inhibitory role of ABA on lateral root initiation and growth, auxin stimulates lateral root initiation and promotes lateral root growth (8, 12). The repression of lateral root formation under osmotic stress can be overcome by exogenous application of the synthetic auxin 1-naphthaleneacetic acid (6), implying that the balance between the repression by ABA and promotion by auxin may determine the fate of the lateral root primordium. As reported previously, ABA antagonizes auxin by promoting production of reactive oxygen species (ROS), leading to the inhibition of primary root growth (17–20). ABA also reduces auxin transport in roots by suppressing the expression of PIN-FORMED 1 through ABA-insensitive (ABI) 4, resulting in suppression of lateral root formation and elongation (21). Furthermore, ABA greatly reduces the auxin reporter ProDR5:GUS expression in roots, indicating a reduction in auxin concentration or response (20, 22). In contrast, ABA can enhance auxin signaling without increasing the amount of auxin by activating auxin-responsive promoters to repress embryonic axis elongation (23). Similarly, auxin has both antagonistic and synergistic effects on responses to ABA. Auxin response factor (ARF) 2 directly suppresses the expression of HOMEOBOX PROTEIN 33, which is a positive regulator in ABA repression of primary root growth (22). However, auxin enhances ABA signaling through ARF10-and ARF16-mediated activation of ABI3 expression in the regulation of lateral root formation and seed dormancy (24, 25). Plants with mutations in the auxin resistant (AXR) genes, axr2-1 and axr3-1, and auxin transport mutants aux1 and pin2 are insensitive to the effects of ABA and auxin on embryonic axis elongation and root growth (23, 25, 26).
The core ABA signaling pathways consist of ABA-specific receptors and their downstream protein phosphatases and kinases. The pyrabactin resistance 1 (PYR1) and PYR1-like proteins (PYLs), also known as the regulatory component of ABA receptor (RCAR) family proteins, are ABA receptors (27, 28). PYR1/PYL/RCARs interact with and inhibit clade-A protein phosphatase type 2Cs (PP2Cs) in the presence of ABA and release the SnRK2 protein kinases from inhibition by the PP2Cs (27, 28). Activated SnRK2s phosphorylate transcription factors such as ABA-responsive element binding factors (29), ABI5 (30), enhanced late embryogenesis abundant level (30), and flowering basic helix-loop-helix–type transcription factor 3 (30) to alter gene expression. Activated SnRK2s also phosphorylate ion channels such as slow anion channel 1 (31–36), K+ channel in Arabidopsis 1 (37), and mechanosensitive channel of small conductance– like 9 (38) to stimulate stomatal closure. SnRK2s also phosphorylate the nicotinamide adenine dinucleotide phosphate oxidase RBOHF, leading to the production of ROS (17–19). In addition, several other substrates of SnRK2s include proteins related to RNA binding, microRNA and epi-genetic regulation, chloroplast function, and many other processes based on phosphoproteomics analyses (38, 39). Thus, PYR1/PYL/RCARs are thought to primarily regulate cellular functions using the PP2C-SnRK2 pathway.
Here, we found a previously uncharacterized function of PYL8 (also known as RCAR3) in the regulation of lateral root growth of Arabidopsis thaliana seedlings exposed to ABA treatment. PYL8 has a high affinity for ABA (40) and was recently shown to mediate the ability of ABA to inhibit primary root growth through the PP2C-SnRK2 pathway (7). We found that PYL8 mediates a synergistic action of ABA and auxin in promoting the growth recovery of post-emergence lateral roots. In addition, we found evidence for the function of the core ABA signaling components, PYR1/PYL/RCARs, PP2Cs, and SnRK2s, in the inhibition of lateral root growth. Our results suggest a signaling pathway in which the ABA receptor PYL8 directly interacts with a group MYB transcription factors to mediate crosstalk between ABA and auxin signaling to promote the growth recovery of lateral root from inhibition.
RESULTS
PYL8 promotes recovery from lateral root growth inhibition by ABA
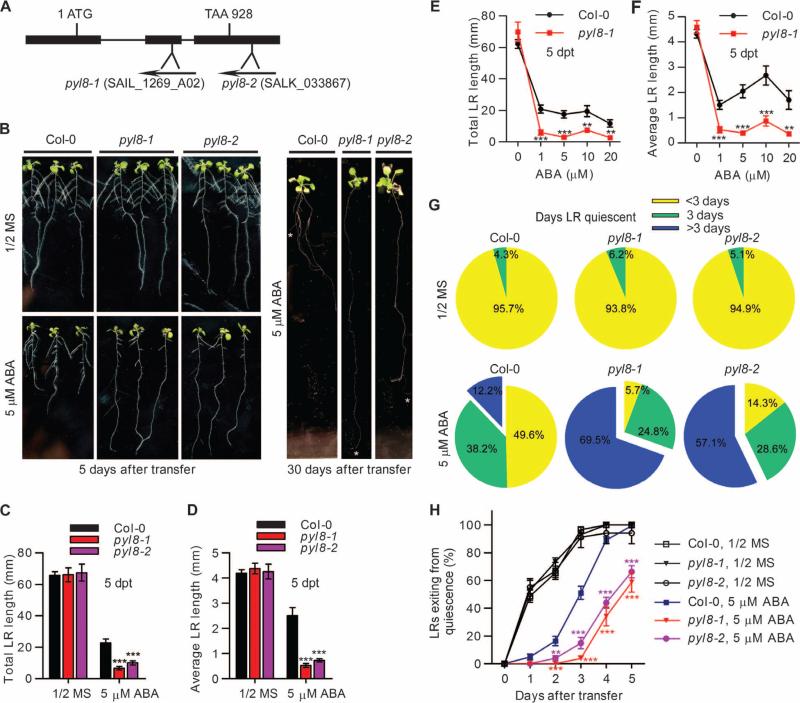
ABA regulates the architecture of plant root systems during abiotic stress. To investigate how ABA controls root architecture, we analyzed the lateral root growth of A. thaliana plants with mutations in components of the ABA signaling pathway. To assess the role of PYL8 in ABA signaling, we analyzed two plants with transferred DNA (T-DNA) insertion mutations in PYL8 (pyl8-1 and pyl8-2) (Fig. 1A) that result in loss of function of PYL8 (7). We found that pyl8 mutant seedlings showed more suppression of lateral root growth than did wild-type seedlings when exposed to ABA for 5 days (Fig. 1B). Seedlings of pyl8 mutant seedlings grown on ABA-containing media for 30 days exhibited altered root architecture compared to wild-type seedlings (Fig. 1B, right panel). Lateral root growth was severely suppressed, whereas primary root growth was less inhibited, in pyl8 mutant seedlings exposed to ABA (Fig. 1B, right panel). Both the total and average lateral root lengths of pyl8 mutant seedlings were reduced compared to the wild-type seedlings when grown on ABA-containing media for 5 days (Fig. 1, C and D), suggesting that the lateral root growth of pyl8 mutant seedlings is more sensitive to ABA. To confirm this observation, we examined lateral root growth with different concentrations of ABA. The lateral root growth of pyl8 mutant seedlings was more sensitive to ABA inhibition at all concentrations tested (Fig. 1, E and F). Thus, PYL8 promotes lateral root growth in response to ABA.
Fig. 1. pyl8 mutant seedlings have enhanced sensitivity to ABA-induced inhibition of lateral root growth.
(A) Diagram of T-DNA insertions in the PYL8 gene. The T-DNA insertions in the two SALK lines are inserted in exons, which are presented as closed boxes. (B) The root architecture of wild-type (Col-0) or pyl8 mutant seedlings was documented at 5 dpt (days post-transfer) (left panel) or 30 dpt (right panel) to media (1/2 MS, 1% sucrose) with or without ABA. The primary root tips are marked by white asterisks. (C and D) Lateral root (LR) length of Col-0 and pyl8 mutant seedlings at 5 dpt to media with or without ABA. n ≥ 12 seedlings. Data are means ± SEM. ***P < 0.001, Student's t test. (E and F) Lateral root length of Col-0 and pyl8-1 mutant seedlings at 5 dpt to media with varying concentrations of ABA. n = 9 seedlings. Data are means ± SEM. **P < 0.01, ***P < 0.001, Student's t test. (G) Pie charts showing the percentage of quiescent lateral roots for the indicated number of days for seedlings grown in media with or without ABA media. n = 10 to 24 seedlings per condition. (H) Percentage of quiescent lateral roots (<0.5 mm at 0 dpt) that exited quiescence within 5 dpt to the media with or without ABA. Percentage was calculated as total lateral roots exiting quiescence (>0.5 mm) at the indicated days divided by total number of quiescent lateral roots at 0 dpt. n = 10 to 24 seedlings per condition. Data are means ± SEM. **P < 0.01, ***P < 0.001, analysis of variance (ANOVA).
A previous study suggests that after being shifted to salt stress, post-emergence lateral roots undergo a quiescent phase followed by a recovery phase, and their growth is spatially and temporally regulated (9). To test whether the retarded lateral root growth of pyl8 mutants exposed to ABA resulted from an extended quiescent phase, we monitored lateral roots for 5 days after emergence. Lateral roots that were less than 0.5 mm in length were considered quiescent, which is defined as stage C in the development of lateral root (5, 9). We found that the quiescent phase was extended in pyl8 mutant compared to wild-type seedlings when grown on ABA-containing media, whereas the quiescent phase of pyl8 mutant seedlings was comparable to that of wild-type seedlings when grown on control media (Fig. 1G). As a result, growth recovery was delayed in pyl8 mutant compared to wild-type seedlings when exposed to ABA (Fig. 1H). Thus, inhibition of lateral root growth by ABA in pyl8 mutant seedlings may be caused by prolonged quiescence, implying that PYL8 may function in promoting lateral root growth recovery in the presence of ABA.
Because salt or osmotic stress is known to induce ABA accumulation (41, 42), we analyzed the lateral and primary root growth of pyl8 seedlings in media supplemented with various concentrations of NaCl or mannitol. However, we did not observe significant differences in lateral root growth between pyl8 mutant and wild-type seedlings (fig. S1). Plant responses to these stressors are more complicated than responses to ABA because NaCl causes both osmotic stress and ion toxicity (43) and mannitol causes osmotic stress and other effects due to mannitol entry into cells (44). These differences may explain the differences in the response of lateral root growth in pyl8 mutant plants to ABA and these other stressors.
PYLs, PP2Cs, and SnRK2s are required for lateral root growth suppression by ABA
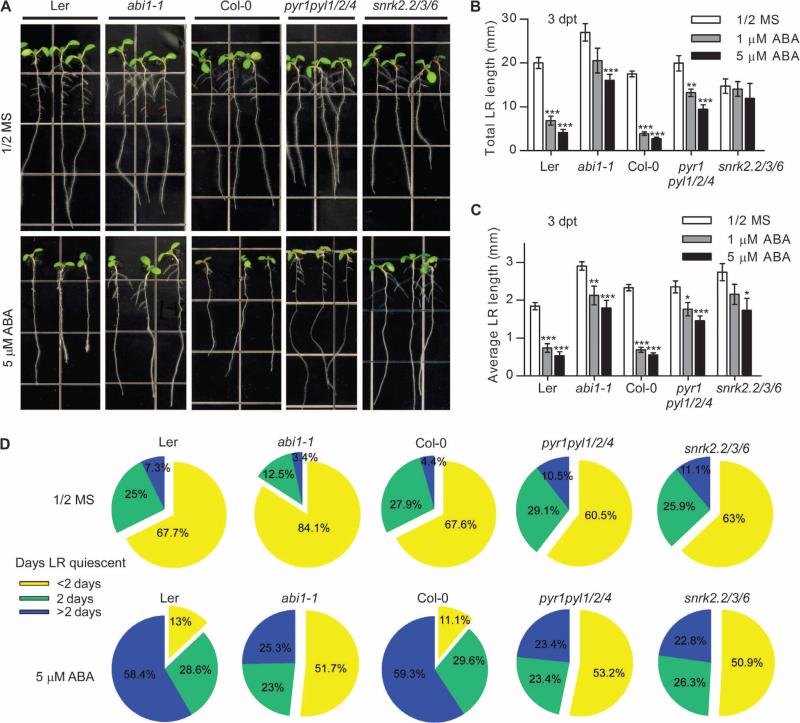
To test whether the lateral root phenotype of pyl8 mutant seedlings is due to reduced response to ABA, we analyzed lateral root growth of seedlings with mutations in the core ABA signaling pathway, including pyr1, pyl1, pyl2, and pyl4 (pyr1pyl1/2/4); snrk2.2, snrk3, and snrk6 (snrk2.2/3/6); or abi1-1, each of which is resistant to ABA-induced inhibition of seed germination and seedling growth (13, 27, 45). Unlike pyl8 mutant seedlings, the lateral root growth of abi1-1, pyr1pyl1/2/4, or snrk2.2/3/6 mutant seedlings was less sensitive than that of wild-type seedlings to inhibition by ABA (Fig. 2, A to C). The lateral roots of wild-type and abi1-1, pyr1pyl1/2/4, or snrk2.2/3/6 mutant seedlings had similar length of quiescent phases when grown in control media. However, the quiescent phase was shorter in abi1-1, pyr1pyl1/2/4, or snrk2.2/3/6 mutant compared to wild-type seedlings when grown on ABA-containing media (Fig. 2D). Thus, the core ABA signaling pathway promotes lateral root quiescence in plants exposed to ABA.
Fig. 2. Lateral root growth is not suppressed by exposure to ABA in ABA-resistant mutant seedlings.
(A) The root architecture of wild-type (Col-0 or Ler) or abi1-1, pyr1pyl1/2/4, or snrk2.2/3/6 mutant seedlings was documented at 3 dpt to media (1/2 MS, 1% sucrose) with or without ABA. (B and C) Lateral root (LR) length of seedlings at 3 dpt to media with varying concentrations of ABA. n ≥ 11 seedlings. Data are means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, Student's t test. (D) Pie charts for the percentage of quiescent lateral roots for the indicated number of days on seedlings grown in media with or without ABA. n = 10 to 12 seedlings per condition.
Suppression of primary root growth and lateral root formation by ABA is reduced in pyl8 and ABA-resistant mutants
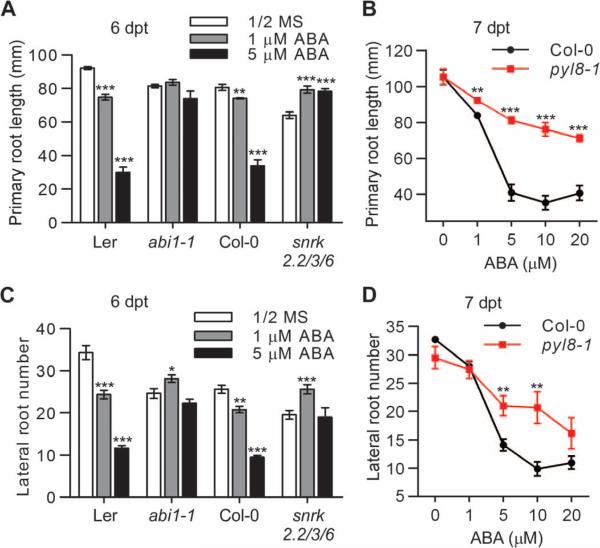
Primary and lateral roots use common and distinct molecular mechanisms to initiate and grow (5, 6, 9, 46, 47). We compared primary root elongation and the number of lateral roots in pyl8 or ABA-resistant mutant seedlings. As reported previously (7, 13, 14, 48), the primary root growth of pyl8-1, abi1-1, or snrk2.2/3/6 mutant seedlings was less sensitive to ABA exposure compared to that of wild-type seedlings (Fig. 3, A and B). In wild-type seedlings, exposure to ABA reduced the number of lateral roots. In contrast, in pyl8-1, abi1-1, or snrk2.2/3/6 mutant seedlings, the number of lateral roots was less affected by exposure to ABA (Fig. 3, C and D). We found that the expression of ABA-responsive genes was similar in the roots of pyl8 mutant and wild-type plants exposed to ABA (fig. S2), suggesting that PYL8 has functional redundancy with other PYLs in mediating ABA-responsive gene expression. Thus, PYL8 likely plays a similar role to other PYLs in primary root growth and lateral root initiation.
Fig. 3. Suppression of primary root growth and lateral root formation by exposure to ABA is reduced in pyl8 and ABA-resistant mutant seedlings.
(A and C) The primary root length and lateral root number of wild-type (Col-0 or Ler) or abi1-1 or snrk2.2/3/6 mutant seedlings were documented at 6 dpt to media (1/2 MS, 1% sucrose) with varying concentrations of ABA. n ≥ 10 seedlings. Data are means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, Student's t test. (B and D) The primary root length and lateral root number of Col-0 and pyl8-1 mutant were documented at 7 dpt to media with varying concentrations of ABA. n = 9 seedlings. Data are means ± SEM. **P < 0.01, ***P < 0.001, Student's t test.
We also assessed whether exposing wild-type seedlings to ABA could affect the expression of PYL8. We found that growing seedlings for 12 hours on ABA-containing media reduced the abundance of PYL8 transcripts to about 33% of that in seedlings grown on control media (fig. S3). However, because ABA stabilizes PYL8 protein by reducing ubiquitination (49), the abundance of PYL8 protein may not substantially decrease in response to ABA.
Application of exogenous auxin promotes lateral root growth in pyl8 mutant seedlings exposed to ABA
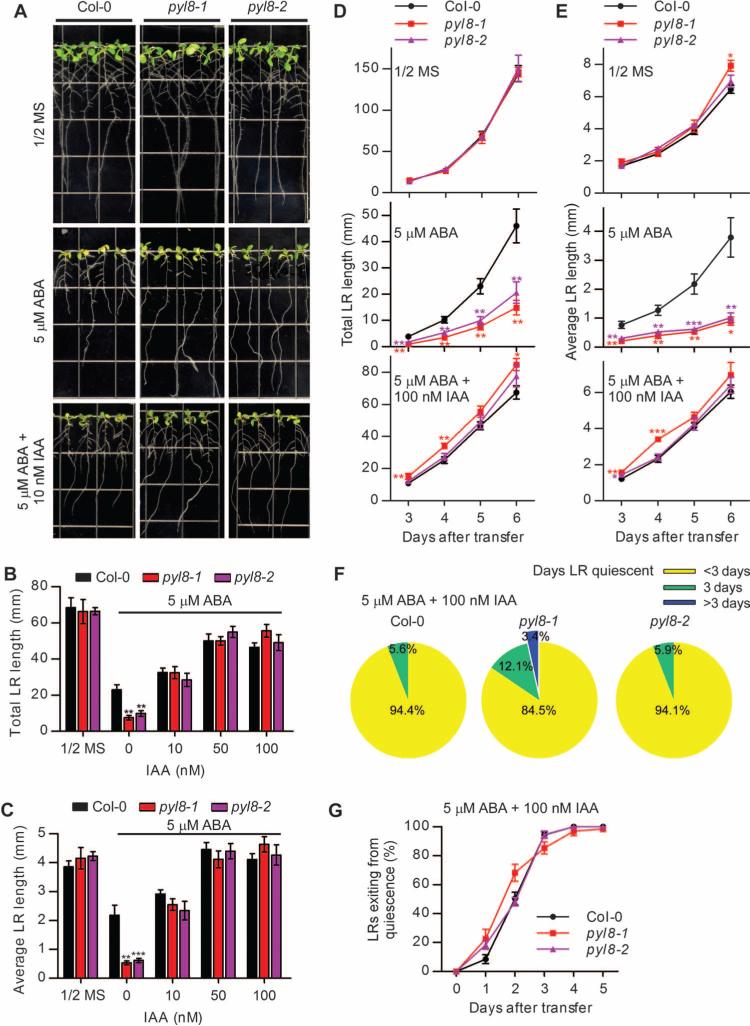
Suppression of lateral root development by osmotic stress can be overcome by exogenous application of auxin (6). To test whether the inhibition of lateral root growth recovery in pyl8 mutants could be rescued by auxin, we analyzed the lateral root growth of pyl8 seedlings grown on ABA-containing media supplemented with different concentrations of the auxin IAA (3-indoleacetic acid). We found that 10 nM IAA was sufficient to rescue the inhibition of lateral root growth in pyl8 mutant seedlings exposed to ABA for 5 days (Fig. 4, A to C). We also analyzed lateral root growth over time and found that wild-type and pyl8 mutant seedlings showed similar lateral root growth on control media, and that lateral root elongation of pyl8 mutant seedlings was suppressed by exposure to ABA (Fig. 4, D and E). However, in pyl8 mutant seedlings exposed to both IAA and ABA, lateral root growth was enhanced compared to that of wild-type seedlings (Fig. 4, D and E).
Fig. 4. Exogenous IAA rescues the lateral root growth defects of pyl8 mutant seedlings.
(A) The root architecture of wild-type (Col-0) or pyl8 mutant seedlings was documented at 5 dpt to media (1/2 MS, 1% sucrose) with or without ABA or ABA plus IAA. (B and C) Lateral root (LR) length of seedlings at 5 dpt to media with or without ABA or ABA plus IAA. n ≥ 11 seedlings for Col-0 and pyl8-2, and n = 6 seedlings for pyl8-1. Data are means ± SEM. **P < 0.01, ***P < 0.001, Student's t test. (D and E) The lateral root length of seedlings was measured at the indicated days after transfer to media with or without ABA or ABA plus IAA. n ≥ 11 seedlings for Col-0 and pyl8-2, and n = 6 seedlings for pyl8-1. Data are means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, Student's t test. (F) Pie charts for the percentage of quiescent lateral roots for the indicated number of days for seedling grown in media with or without ABA or ABA plus IAA. n = 6 to 18 seedlings per treatment. (G) Percentage of quiescent lateral roots (<0.5 mm at 0 dpt) that exited within 5 dpt to the media with ABA plus IAA. n = 6 to 18 seedlings per treatment. Data are means ± SEM. *P < 0.05, ANOVA.
To test whether IAA treatment alters the quiescent or recovery phases of lateral roots, we monitored the lateral roots of wild-type and pyl8 mutant seedlings for 5 days. The pyl8 mutant seedlings had a similar length of quiescent phase as wild-type seedlings in the presence of both IAA and ABA (Fig. 4F). The recovery of post-emergence lateral root growth in pyl8 mutant seedlings exposed to ABA treatment was accelerated by exposure to IAA compared to exposure to ABA alone, resulting in comparable lengths of lateral roots to wild-type seedlings (Figs. 1H and 4G). Thus, the prolonged quiescent phase in lateral roots of pyl8 mutant seedlings exposed to ABA was shortened by exposure to IAA, suggesting that the delayed recovery of lateral root growth may be due to auxin deficiency or reduced response to auxin.
PYL8 enhances MYB77 activity to promote lateral root growth
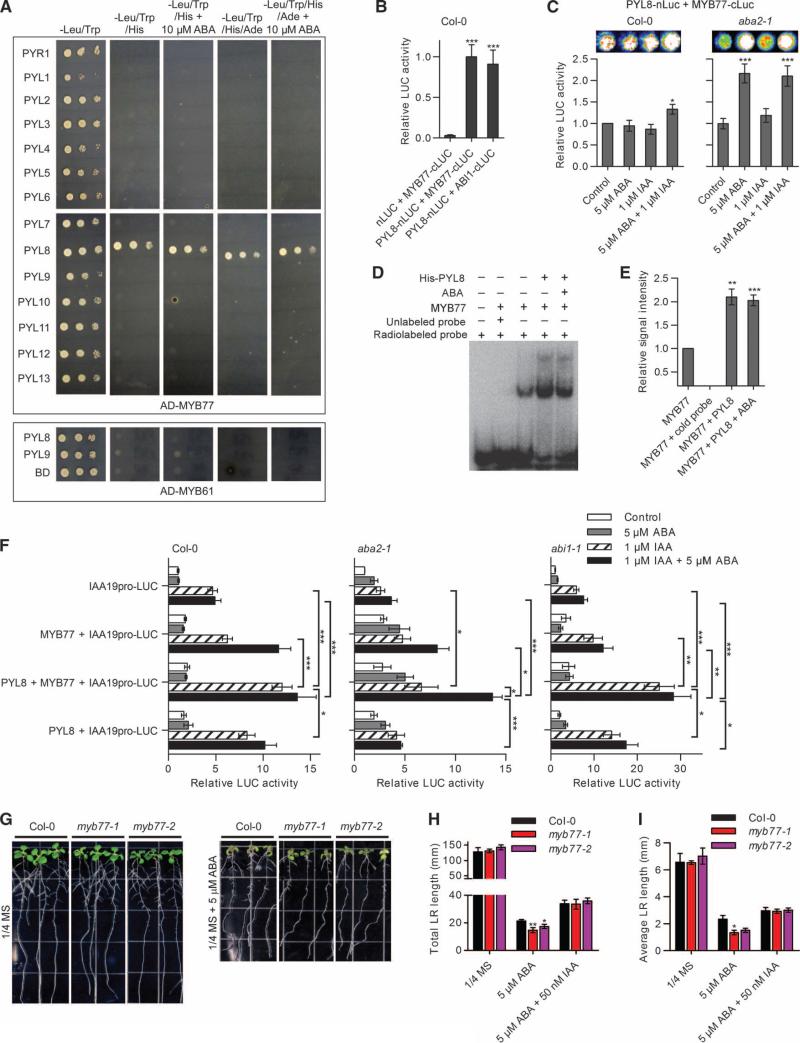
Auxin signaling can activate the transcription factor MYB77 to promote lateral root growth (50). Recently, systematic yeast two-hybrid screens found MYB77 as an interacting protein of PYL8 (51). To test whether the interaction between PYL8 and MYB77 was specific, we fused several PYLs to the GAL4 DNA binding domain (BD) and MYB77 or MYB61 to the GAL4-activating domain (AD) and performed yeast two-hybrid assays. We found that PYL8, but not other PYLs, interacted with MYB77, but not MYB61, in an ABA-independent manner (Fig. 5A). To further characterize the interaction between PYL8 and MYB77 in plant cells, we fused PYL8 to the N-terminal domain of firefly luciferase (LUC) (PYL8-nLUC) and MYB77 to the C-terminal domain of LUC (MYB77-cLUC) and co-expressed the two constructs in A. thaliana protoplasts. Coexpression of PYL8-nLUC, but not nLUC, with MYB77-cLUC produced measurable luciferase activity, comparable to coexpression of MYB77-cLUC with the positive control ABI1-nLUC (7) (Fig. 5B). To test whether the interaction between PYL8 and MYB77 was altered in the presence of ABA and auxin in vivo, we treated wild-type protoplasts coexpressing PYL8-nLUC and MYB77-cLUC with different combinations of ABA and IAA.
Fig. 5. PYL8 enhances auxin signaling through MYB77.
(A) PYL-MYB77 interactions in the yeast two-hybrid assay. Interaction was determined by yeast growth in media lacking His or His and Ade in the presence and absence of ABA. Dilutions (10−1, 10−2, and 10−3) of saturated cultures were spotted onto the plates, which were photographed after 5 days. (B) PYL8-MYB77 interaction in LUC complementation as-says in wild-type Col-0 protoplasts. n = 4 experiments. Data are means ± SEM. ***P < 0.001, Student's t test. (C) PYL8-MYB77 interaction in Col-0 wild-type (left panel) and aba2-1 mutant protoplasts (right panel) with or without ABA in LUC complementation assays. n ≥ 7 experiments. Data are means ± SEM. *P < 0.05, ***P < 0.001, Student's t test. (D and E) Binding between MYB77 and the MBSI motif–containing DNA oligonucleotides with or without PYL8 in EMSA assay. n = 4 experiments. Data are means ± SEM. **P < 0.01, ***P < 0.001, Student's t test. (F) IAA19pro-LUC expression in Col-0 wild-type (left panel), aba2-1 mutant (middle panel), and abi1-1 mutant (right panel) protoplasts cotransformed with the indicated combinations of PYL8, MYB77, and IAA19pro-LUC with varying concentrations of ABA and IAA. n ≥ 3 experiments. Data are means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, Student's t test. (G) The root architecture of Col-0 or myb77 mutant seedlings was documented at 5 dpt to media (1/4 MS, 1% sucrose) with or without ABA. (H and I) Lateral root (LR) length of Col-0 and myb77 mutant seedlings at 5 dpt to media with or without ABA. n ≥ 11 seedlings. Data are means ± SEM. *P < 0.05, **P < 0.01, Student's t test.
We found that LUC activity was enhanced in cells exposed to both ABA and IAA, but not in cells exposed to either ABA or IAA alone (Fig. 5C, left panel). To investigate the effect of endogenous ABA, we performed LUC complementation assays in protoplasts derived from the ABA-deficient mutant aba2-1 (52) and found that ABA exposure promoted the interaction between PYL8 and MYB77 in the absence or presence of IAA (Fig. 5C, right panel). The interaction between PYL8 and MYB77 was not reduced by overexpression of ABI1 or ABI2 in aba2-1 mutant protoplasts (fig. S4), suggesting that MYB77 and the PP2Cs may interact with different regions of PYL8. Thus, PYL8 specifically interacts with MYB77, and this interaction may be enhanced by ABA and IAA.
We asked whether the interaction between PYL8 and MYB77 affected the ability of MYB77 to bind to DNA and activate transcription. A previous study found that MYB77ΔC1 (amino acids 1 to 200) recognizes several DNA motifs, including MBSI (CNGTTR) and MBSII (GTTAGTTA), and preferentially binds to MBSI motif–containing promoters (53). Using an electrophoretic mobility shift assay (EMSA), we found that full-length MYB77 bound to MBSI motif–containing DNA oligonucleotides (CRGTTA), but not to oligonucleotides containing M1 (CTGTTC), M2 (CAGTGA), or M3 (CCATTA) motifs (fig. S5, A and B). To test whether PYL8 affects the ability of MYB77 to bind to MBSI motifs, we preincubated recombinant MYB77 and PYL8 with or without ABA before performing the EMSA and found that PYL8 enhanced the binding of MYB77 to MBSI motifs in an ABA-independent manner (Fig. 5, D and E). PYL8 alone did not bind to MBSI motifs (fig. S5C). We identified putative MBSI and MBSII motifs in the 500–base pair (bp) promoter regions of multiple auxin-responsive genes, including IAA1, IAA7, IAA17, IAA19, GH3.2, PIN1, and HAT2 (fig. S5, D and E). To test whether PYL8 can enhance MYB77 transcriptional activity, we fused a 439-bp fragment of IAA19 promoter to the gene encoding LUC (IAA19pro-LUC, fig. S5D) and used it as an auxin-responsive reporter. We expressed PYL8, MYB77, IAA19pro-LUC, or combinations thereof in wild-type protoplasts. As expected, IAA, but not ABA, enhanced LUC activity in control protoplasts expressing only the reporter (Fig. 5F, left panel). However, co-overexpression of PYL8 and MYB77 increased LUC activity in the presence of IAA (Fig. 5F, left panel). To investigate the effects of endogenous ABA, we repeated the experiments in protoplasts from aba2-1 mutant plants and found that ABA increased the ability of PYL8 and MYB77 overexpression to enhance LUC activity in the presence of IAA (Fig. 5F, middle panel). Because ABA can inhibit auxin-responsive gene expression (20, 22), we performed reporter assays in protoplasts of abi1-1 mutant plants (54), which show inhibition of the core ABA signaling pathway (45), and found that cooverexpression of PYL8 and MYB77 increased LUC activity with or without ABA in the presence of IAA in these cells (Fig. 5F, right panel). Thus, PYL8 enhances the binding of MYB77 to its target motif MBSI, leading to increased response to auxin in an ABA-enhanced manner.
To further understand whether MYB77 can activate auxin responses to control lateral root growth in response to ABA, we analyzed the lateral root growth of two plants with T-DNA insertion mutations in MYB77 (50). We found that lateral root growth in myb77 mutant seedlings was more sensitive to inhibition by ABA than that in wild-type seedlings and that exposure to IAA could reverse ABA-induced inhibition of lateral root growth in myb77 mutant seedlings (Fig. 5, H and I). Thus, MYB77, like PYL8, promotes lateral root growth in seedlings exposed to ABA.
PYL8 interacts with paralogs of MYB77 and enhances their transcriptional activity
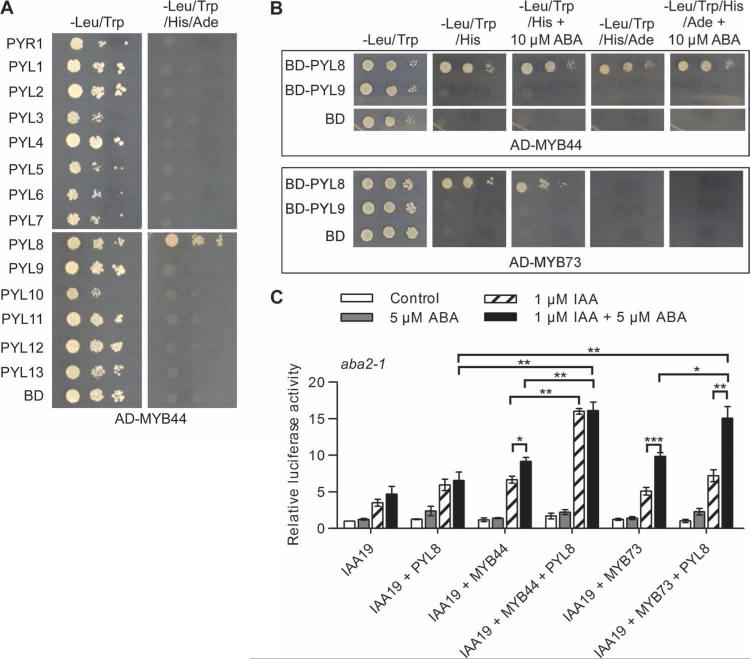
We asked whether PYL8 could bind paralogs of MYB77 and modify their transcriptional activity. MYB77, MYB73, MYB70, and MYB44 belong to the R2R3 MYB family subgroup 22, which is defined by two conserved motifs, TGLYMSPxSP and GxFMxVVQEMIxxEVRSYM (fig. S6) (55). PYL8 is a putative interacting protein with MYB44 (56). To test whether MYB73 and MYB44 could specifically bind PYL8, we performed yeast two-hybrid assays and found that PYL8, but not other PYLs, bound to both MYB44 and MYB73 in an ABA-independent manner (Fig. 6, A and B). PYL8 showed a much stronger interaction with MYB77 and MYB44 than with MYB73 (Figs. 5A and 6B). To test whether MYB44 and MYB73 could enhance the transcriptional response to auxin, we performed LUC assays in IAA19pro-LUC–expressing protoplasts from aba2-1 and abi1-1 mutant plants. We found that co-overexpression with PYL8 enhanced the ability of MYB44 or MYB73 to increase LUC activity in the presence of IAA (Fig. 6C and fig. S7) and that exposure to ABA enhanced the ability of PYL8 to activate MYB73 (Fig. 6C). Thus, PYL8 likely binds to and enhances the activities of paralogs of MYB77, which may function redundantly with MYB77 to enhance auxin signaling.
Fig. 6. PYL8 interacts with and activates MYB44 and MYB73.
(A and B) PYL-MYB44 and PYL-MYB73 interactions in the yeast two-hybrid assay. Interaction was determined by yeast growth in media lacking His or His and Ade in the presence and absence of ABA. Dilutions (10−1, 10−2, and 10−3) of saturated cultures were spotted onto the plates, which were photographed after 5 days. (C) IAA19pro-LUC expression in aba2-1 mutant protoplasts cotransformed with the indicated combinations of PYL8, MYB44, MYB73, and IAA19pro-LUC with varying concentrations of ABA and IAA. n = 3 experiments. Data are means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, Student's t test.
DISCUSSION
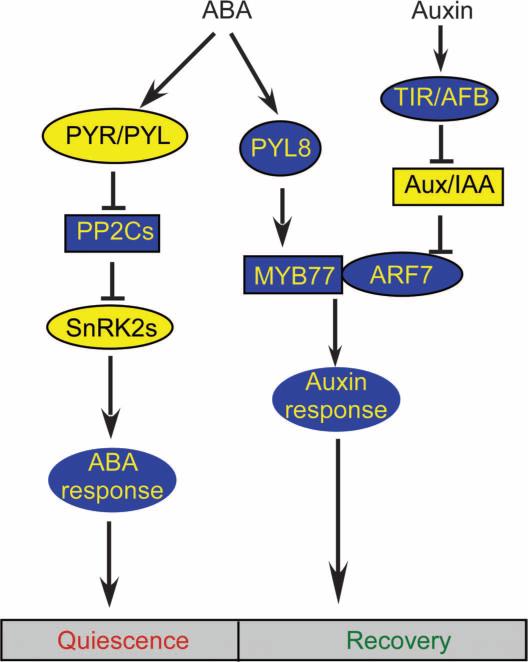
The root system of plants is one of the most sensitive organs in sensing the availability of water and nutrients or other adverse soil conditions. As a critical part of plant responses to environmental cues, phytohormone signaling finely controls the growth and development of roots for acclimating to environmental changes. Here, we found a synergistic action of ABA and auxin in controlling the growth of plant lateral roots. Our study reveals that ABA promotes lateral root growth recovery through a pathway mediated by a previously uncharacterized action of ABA receptor PYL8. PYL8 directly interacts with a group of transcription factors, MYB77, MYB44, and MYB73, leading to the enhancement of auxin-dependent transcription. Together with evidence from previous studies (5, 6, 9), our results suggest that ABA has dual roles in controlling lateral root growth via two different pathways (Fig. 7). In this model, ABA signaling inhibits lateral root growth during the quiescent phase and promotes lateral root growth during the recovery phase of post-emergence lateral root growth during abiotic stress.
Fig. 7. Proposed model of ABA response pathways in regulating lateral root growth.
ABA has dual roles in controlling lateral root growth in Arabidopsis. ABA exposure promotes periods of quiescence and growth recovery in post-emergence lateral roots. The inhibitory effect of ABA on lateral root growth is mediated by the core ABA signaling pathway through the PYL receptors, PP2C co-receptors, and SnRK2 protein kinases. PYL8 has a previously undescribed function in accelerating lateral root growth during the recovery phase through activating MYB77. PYL8 directly interacts with MYB77 and enhances the activity of MYB77 to up-regulate the expression of multiple auxin-responsive genes at the time when lateral roots enter the recovery phase. MYB77 forms heterodimers with ARF7 and promotes the transcription of ARF7 target genes. At high concentrations of auxin, Aux/IAA is degraded by transport inhibitor response 1 (TIR1)– and auxin signaling F-box (AFB)–mediated ubiquitination to release the inhibition on the ARFs and induce auxin-responsive gene expression, bypassing PYL8.
The growth of post-emergence lateral roots under salt stress undergoes a quiescent phase followed by a recovery phase. Endogenous ABA stimulated by salt stress promotes lateral root quiescence, and application of exogenous ABA treatment mimics this effect (9). Therefore, together with our results, it is likely that lateral root growth undergoes quiescent and recovery phases when exposed to exogenous ABA. We found that lateral root growth was less sensitive to ABA in ABA-resistant mutant seedlings including pyr1pyl1/2/4, abi1-1, and snrk2.2/3/6 (Fig. 2, A to C). Consistent with this result, lateral root growth is less sensitive to salt stress in transgenic seedlings expressing ABI1-1 in the root endodermis (9). The ABI4 gene encodes an ABA-regulated AP2 domain transcription factor, and lateral root growth of abi4 mutant seedlings is insensitive to ABA (21). Similarly, the double mutant seedlings abi4-1 and fus3-3 are less sensitive to salt stress (9, 21), suggesting that ABI4 may be the downstream regulator in controlling lateral root quiescence during exposure to ABA or salt stress. Thus, lateral root quiescence is likely controlled by core ABA signaling through the PYL receptors, PP2C co-receptors, SnRK2 protein kinases, ABI4 transcription factor, and downstream ABA-responsive genes, and our results suggest that PYL8-mediated lateral root growth recovery is independent of the ABA core signaling pathway. Instead, PYL8 interacts directly with the MYB77 paralogs to enhance auxin-dependent gene expression.
In support of the proposed model, our data showed that the recovery of post-emergence lateral root growth was delayed in pyl8 mutant seedlings exposed to ABA and that the application of exogenous IAA accelerated the lateral root growth of pyl8 mutant seedlings exposed to ABA (Figs. 1 and 4). The direct interaction of PYL8 with the MYB transcription factors bridges ABA and auxin signaling in controlling lateral root growth. We showed that ABA enhances the effects of PYL8 on the activities of MYB73 and MYB77 in the presence of IAA (Figs. 5F and 6C), suggesting a synergistic action of ABA and IAA on MYB44, MYB73, and MYB77 activities through their interactions with PYL8. It is known that MYB77 positively regulates auxin signaling to promote lateral root formation (50). MYB77 forms a heterodimer with ARF7 and increases the transcription of ARF7 target genes (50), as indicated in our model (Fig. 7). In addition, both ARF7 and ARF19 increase the expression of early auxin response genes such as LBD16 (also known as ASL18) and LBD29 (also known as ASL16) in lateral roots, promoting lateral root formation and elongation (57–59).
ABA signaling can enhance auxin-dependent responses by activating an auxin-responsive promoter (23). Truncated MYB77 and MYB44 proteins bind to the motifs of MBSI and MBSII (53, 60) that were identified in the 500-bp fragments of the promoters of multiple auxin-responsive genes (fig. S5, D and E). Auxin-responsive genes were highly induced by salt stress in plants overexpressing MYB44 (61), suggesting that salt stress or ABA can enhance auxin signaling when MYB44 abundance is high. MYB73 and MYB77 are expressed in lateral roots (50, 62), whereas PYL8 is expressed extensively in roots (7). The spatial expression patterns of MYBs and PYL8 indicate that the interaction of PYL8 and MYBs may occur specifically in roots. In addition, we observed that the inhibition of lateral root growth in myb77 mutant seedlings was not as severe as that in pyl8 mutant seedlings when exposed to ABA, suggesting that MYB44 and MYB73 may be functionally redundant with MYB77. In protoplasts, ABA enhanced the ability of IAA and PYL8 overexpression to increase the ability of MYB73 and MYB77 to activate an auxin reporter gene (Figs. 5F and 6C). This result implies that the synergistic action of ABA and IAA can be mediated by PYL8-MYB73 and PYL8-MYB77. Furthermore, the expression of MYB44, MYB73, and MYB77 is strongly induced by dehydration, salt, cold, ABA, or auxin (50, 61, 63).
PYL8 has been shown to mediate ABA inhibition of primary root growth (7). This role is likely mediated through the PP2C-SnRK2 pathway because primary root elongation in abi1-1 mutant seedlings and snrk2.2/3/6 mutant seedlings was not sensitive to ABA exposure (Fig. 3, A and B). Our results show that PYL8 also has a role in promoting lateral root elongation exposed to ABA, and suggest that this function is mediated through the previously uncharacterized PYL8-MYB77 pathway. Therefore, PYL8 has different roles mediated through distinct pathways in the ABA-dependent regulation of different aspects of root growth and development.
MATERIALS AND METHODS
Plant materials
The pyl8-1 (SAIL_1269_A02) (7, 14), pyl8-2 (SALK_033867) (7), pyr1pyl1/2/4 quadruple mutant (27), snrk2.2/3/6 triple mutant (13), myb77-1 (50), and myb77-2 (50) were in Col-0 background, except pyl2 (27) and abi1-1 (45), which were in the Landsberg erecta (Ler) background. The pyl8 and myb77 mutants were obtained from the Arabidopsis Biological Resource Center (ABRC).
Plant growth conditions
Seeds were surface-sterilized for 10 min in 20% bleach and then rinsed in sterile deionized water four times. Sterilized seeds were grown on 0.6% Phytagel (Sigma) media containing 1/2 MS nutrients or 1/4 MS nutrients as indicated (catalog no. M524, PhytoTechnology Laboratories), 1% sucrose adjusted to pH 5.7 (control media), and kept at 4° to 8°C for 3 days. Seedlings were grown vertically for 3 days before transfer to control media or media with the indicated concentrations of ABA (Sigma, A1049) or IAA (Sigma, I2886). Seedlings were grown in a Percival CU36L5 incubator at 23°C under a 16-hour light, 8-hour dark photoperiod. Plates were sealed with micropore tape (3M).
For protoplast analysis, seedlings were grown on Jiffy 7 peat soil (42mm Pellets) in a Percival chamber with a relatively short photoperiod (12 hours of light at 23°C, 12 hours of dark at 20°C) under low light (about 100 μE m−2 s−1) and 50 to 70% relative humidity under well-watered conditions (no drought and no flooding). Protoplasts were isolated as described (64, 65).
Phenotype analysis
The length of all lateral roots was quantified using a Leica EZ4HD stereo microscope. The total and average lateral root lengths of individual plants were quantified by summing or averaging the lengths of all the lateral roots on each plant. Lateral roots shorter than 0.5 mm were categorized as quiescent (9).
Plasmid constructs
All primer sequences are shown in table S1. To generate IAA19pro-LUC construct, the 439-bp IAA19 promoter fragment amplified from Col-0 genomic DNA with primers IAA19proF and IAA19proR was cloned into the Bam HI and Nco I of the RD29B-LUC vector (65). To generate pET28a-PYL8 construct, the coding sequence of PYL8 was amplified from His-PYL8 (65) using PYL8 primers and cloned into pET28a vectors. To generate pENTR-MYB77 construct, MYB77 CDS was amplified using pENTR-MYB77 primers and cloned into pENTR vector using the pENTR/D-TOPO Cloning Kit (Invitrogen). MYB77 was recombined into the pEarley-nLUC and pEarley-cLUC binary vectors using an LR reaction (Invitrogen). MYB44, MYB73, and MYB77 were cloned into pHBT95 using transfer polymerase chain reaction with pHBT-MYBs primers, cloned into pGADT7 with AD-MYBs primers, and cloned into pGEX-6P1 with 6P1-MYB77 primers. ABI1, ABI2, BD-PYLs, and PYL8-nLUC were the same as reported (64). ABI1 was cloned into pHBT95-cLUC using cLUC-ABI1 primers. His-PYL8 was the same as reported (65). ZmUBQ::GUS was provided by J. Sheen.
Yeast two-hybrid assay
Yeast two-hybrid assays were performed as described (66). pGBKT7-PYLs were the same as reported (64). PYLs fused to the GAL4 DNA binding domain were used as baits. MYB44, MYB73, MYB77, and MYB61 fused to the GAL4-activating domain were used as preys. Interaction was determined by growth assay on media lacking His or His and Ade in the presence and absence of 10 μM ABA. Dilutions (10−1, 10−2, and 10−3) of saturated cultures were spotted onto the plates and photographed after 5 days.
Transient expression assay in Arabidopsis
Assays for transient expression in protoplasts were performed as described (64). All steps were kept at room temperature. IAA19pro-LUC was used as the auxin-responsive reporter. ZmUBQ-GUS was used as the internal control. After transfection, protoplasts were incubated in washing and incubation solution (0.5 M mannitol, 20 mM KCl, 4 mM MES, pH 5.7) with or without ABA and IAA at the indicated concentrations with 12 hours of light. LUC complementation assays were performed as described (64).
Electrophoretic mobility shift assay
GST-MYB77 and His-PYL8 were purified as described (67). EMSA assay was performed as described (64, 68). The radiolabeled DNA probes were labeled with [γ-32P]adenosine 5′-triphosphate using T4 polynucleotide kinase and purified using Micro Bio-Spin 6 columns (Bio-Rad, 732-6221). EMSAs were performed with radiolabeled oligonucleotides (0.6 nM) and recombinant GST-MYB77 protein (17 nM) with or without the indicated unlabeled oligonucleotides (40 nM). Recombinant His-PYL8 protein (17 nM) was preincubated with GST-MYB77 (17 nM) for 10 min with or without ABA (0.2 mM) on ice before the radiolabeled oligo-nucleotide was added. The DNA binding reactions were performed in binding buffer containing 10 mM Hepes-KOH (pH 7.5), 0.1 mM EDTA, 75 mM KCl, 1.25 mM MgCl2, 0.2 mM dithiothreitol, bovine serum albumin (2 μg/ml), and 5% glycerol. The reaction was incubated with the radio-labeled oligonucleotide for 30 min on ice and then loaded onto 5% polyacrylamide native gels, which had been pre-run at 180 V in 0.5× tris-borate EDTA buffer for 30 min at 4°C, and run at 180 V for 35 min at 4°C. After electrophoresis, the gel was dried under vacuum at 80°C for 15 min on a filter paper and exposed to a phosphorimager for 2 to 10 hours.
Supplementary Material
Acknowledgments
We thank J. Sheen for the ZmUBQ::GUS construct and S. R. Cutler for the pyr1pyl1/2/4 quadruple mutant. Funding: This work was supported by NIH grant R01GM059138 and by the Chinese Academy of Sciences.
Footnotes
Author contributions: Y.Z., P.W., and J.-K.Z. designed the research; Y.Z., L.X., X.W., Y.-J.H., J.G., and C.-G.D. performed the experiments and analyzed the data; and Y.Z., X.Z., and J.-K.Z. wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
Citation: Y. Zhao, L. Xing, X. Wang, Y.-J. Hou, J. Gao, P. Wang, C.-G. Duan, X. Zhu, J.-K. Zhu, The ABA receptor PYL8 promotes lateral root growth by enhancing MYB77-dependent transcription of auxin-responsive genes. Sci. Signal. 7, ra53 (2014).
REFERENCES AND NOTES
- 1.Zhu JK. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 3.Raghavendra AS, Gonugunta VK, Christmann A, Grill E. ABA perception and signalling. Trends Plant Sci. 2010;15:395–401. doi: 10.1016/j.tplants.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 4.Ooms J, Leon-Kloosterziel KM, Bartels D, Koornneef M, Karssen CM. Acquisition of desiccation tolerance and longevity in seeds of Arabidopsis thaliana (a comparative study using abscisic acid-insensitive abi3 mutants). Plant Physiol. 1993;102:1185–1191. doi: 10.1104/pp.102.4.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Smet I, Signora L, Beeckman T, Inzé D, Foyer CH, Zhang H. An abscisic acid-sensitive checkpoint in lateral root development of Arabidopsis. Plant J. 2003;33:543–555. doi: 10.1046/j.1365-313x.2003.01652.x. [DOI] [PubMed] [Google Scholar]
- 6.Deak KI, Malamy J. Osmotic regulation of root system architecture. Plant J. 2005;43:17–28. doi: 10.1111/j.1365-313X.2005.02425.x. [DOI] [PubMed] [Google Scholar]
- 7.Antoni R, Gonzalez-Guzman M, Rodriguez L, Peirats-Llobet M, Pizzio GA, Fernandez MA, De Winne N, De Jaeger G, Dietrich D, Bennett MJ, Rodriguez PL. PYRABACTIN RESISTANCE1-LIKE8 plays an important role for the regulation of abscisic acid signaling in root. Plant Physiol. 2013;161:931–941. doi: 10.1104/pp.112.208678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lavenus J, Goh T, Roberts I, Guyomarc'h S, Lucas M, De Smet I, Fukaki H, Beeckman T, Bennett M, Laplaze L. Lateral root development in Arabidopsis: Fifty shades of auxin. Trends Plant Sci. 2013;18:450–458. doi: 10.1016/j.tplants.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 9.Duan L, Dietrich D, Ng CH, Chan PM, Bhalerao R, Bennett MJ, Dinneny JR. Endodermal ABA signaling promotes lateral root quiescence during salt stress in Arabidopsis seedlings. Plant Cell. 2013;25:324–341. doi: 10.1105/tpc.112.107227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Overvoorde P, Fukaki H, Beeckman T. Auxin control of root development. Cold Spring Harb. Perspect. Biol. 2010;2:a001537. doi: 10.1101/cshperspect.a001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uga Y, Sugimoto K, Ogawa S, Rane J, Ishitani M, Hara N, Kitomi Y, Inukai Y, Ono K, Kanno N, Inoue H, Takehisa H, Motoyama R, Nagamura Y, Wu J, Matsumoto T, Takai T, Okuno K, Yano M. Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 2013;45:1097–1102. doi: 10.1038/ng.2725. [DOI] [PubMed] [Google Scholar]
- 12.Smith S, De Smet I. Root system architecture: Insights from Arabidopsis and cereal crops. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012;367:1441–1452. doi: 10.1098/rstb.2011.0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujii H, Zhu JK. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. U.S.A. 2009;106:8380–8385. doi: 10.1073/pnas.0903144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzalez-Guzman M, Pizzio GA, Antoni R, Vera-Sirera F, Merilo E, Bassel GW, Fernández MA, Holdsworth MJ, Perez-Amador MA, Kollist H, Rodriguez PL. Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell. 2012;24:2483–2496. doi: 10.1105/tpc.112.098574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geng Y, Wu R, Wee CW, Xie F, Wei X, Chan PM, Tham C, Duan L, Dinneny JR. A spatio-temporal understanding of growth regulation during the salt stress response in Arabidopsis. Plant Cell. 2013;25:2132–2154. doi: 10.1105/tpc.113.112896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharp RE, Poroyko V, Hejlek LG, Spollen WG, Springer GK, Bohnert HJ, Nguyen HT. Root growth maintenance during water deficits: Physiology to functional genomics. J. Exp. Bot. 2004;55:2343–2351. doi: 10.1093/jxb/erh276. [DOI] [PubMed] [Google Scholar]
- 17.Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell. 2002;14:3089–3099. doi: 10.1105/tpc.007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwak JM, Mori IC, Pei ZM, Leonhardt N, Torres MA, Dangl JL, Bloom RE, Bodde S, Jones JD, Schroeder JI. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 2003;22:2623–2633. doi: 10.1093/emboj/cdg277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sirichandra C, Gu D, Hu HC, Davanture M, Lee S, Djaoui M, Valot B, Zivy M, Leung J, Merlot S, Kwak JM. Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Lett. 2009;583:2982–2986. doi: 10.1016/j.febslet.2009.08.033. [DOI] [PubMed] [Google Scholar]
- 20.He J, Duan Y, Hua D, Fan G, Wang L, Liu Y, Chen Z, Han L, Qu LJ, Gong Z. DEXH box RNA helicase-mediated mitochondrial reactive oxygen species production in Arabidopsis mediates crosstalk between abscisic acid and auxin signaling. Plant Cell. 2012;24:1815–1833. doi: 10.1105/tpc.112.098707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shkolnik-Inbar D, Bar-Zvi D. ABI4 mediates abscisic acid and cytokinin inhibition of lateral root formation by reducing polar auxin transport in Arabidopsis. Plant Cell. 2010;22:3560–3573. doi: 10.1105/tpc.110.074641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L, Hua D, He J, Duan Y, Chen Z, Hong X, Gong Z. Auxin Response Factor2 (ARF2) and its regulated homeodomain gene HB33 mediate abscisic acid response in Arabidopsis. PLOS Genet. 2011;7:e1002172. doi: 10.1371/journal.pgen.1002172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Belin C, Megies C, Hauserová E, Lopez-Molina L. Abscisic acid represses growth of the Arabidopsis embryonic axis after germination by enhancing auxin signaling. Plant Cell. 2009;21:2253–2268. doi: 10.1105/tpc.109.067702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brady SM, Sarkar SF, Bonetta D, McCourt P. The ABSCISIC ACID INSENSITIVE 3 (ABI3) gene is modulated by farnesylation and is involved in auxin signaling and lateral root development in Arabidopsis. Plant J. 2003;34:67–75. doi: 10.1046/j.1365-313x.2003.01707.x. [DOI] [PubMed] [Google Scholar]
- 25.Liu X, Zhang H, Zhao Y, Feng Z, Li Q, Yang HQ, Luan S, Li J, He ZH. Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2013;110:15485–15490. doi: 10.1073/pnas.1304651110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson AK, Pickett FB, Turner JC, Estelle M. A dominant mutation in Arabidopsis confers resistance to auxin, ethylene and abscisic acid. Mol. Gen. Genet. 1990;222:377–383. doi: 10.1007/BF00633843. [DOI] [PubMed] [Google Scholar]
- 27.Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, Alfred SE, Bonetta D, Finkelstein R, Provart NJ, Desveaux D, Rodriguez PL, McCourt P, Zhu JK, Schroeder JI, Volkman BF, Cutler SR. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 29.Fujii H, Verslues PE, Zhu JK. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell. 2007;19:485–494. doi: 10.1105/tpc.106.048538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakashima K, Fujita Y, Kanamori N, Katagiri T, Umezawa T, Kidokoro S, Maruyama K, Yoshida T, Ishiyama K, Kobayashi M, Shinozaki K, Yamaguchi-Shinozaki K. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 2009;50:1345–1363. doi: 10.1093/pcp/pcp083. [DOI] [PubMed] [Google Scholar]
- 31.Brandt B, Brodsky DE, Xue S, Negi J, Iba K, Kangasjärvi J, Ghassemian M, Stephan AB, Hu H, Schroeder JI. Reconstitution of abscisic acid activation of SLAC1 anion channel by CPK6 and OST1 kinases and branched ABI1 PP2C phosphatase action. Proc. Natl. Acad. Sci. U.S.A. 2012;109:10593–10598. doi: 10.1073/pnas.1116590109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geiger D, Maierhofer T, Al-Rasheid KA, Scherzer S, Mumm P, Liese A, Ache P, Wellmann C, Marten I, Grill E, Romeis T, Hedrich R. Stomatal closure by fast abscisic acid signaling is mediated by the guard cell anion channel SLAH3 and the receptor RCAR1. Sci. Signal. 2011;4:ra32. doi: 10.1126/scisignal.2001346. [DOI] [PubMed] [Google Scholar]
- 33.Vahisalu T, Puzõrjova I, Brosché M, Valk E, Lepiku M, Moldau H, Pechter P, Wang YS, Lindgren O, Salojärvi J, Loog M, Kangasjärvi J, Kollist H. Ozone-triggered rapid stomatal response involves the production of reactive oxygen species, and is controlled by SLAC1 and OST1. Plant J. 2010;62:442–453. doi: 10.1111/j.1365-313X.2010.04159.x. [DOI] [PubMed] [Google Scholar]
- 34.Geiger D, Scherzer S, Mumm P, Marten I, Ache P, Matschi S, Liese A, Wellmann C, Al-Rasheid KA, Grill E, Romeis T, Hedrich R. Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc. Natl. Acad. Sci. U.S.A. 2010;107:8023–8028. doi: 10.1073/pnas.0912030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee SC, Lan W, Buchanan BB, Luan S. A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc. Natl. Acad. Sci. U.S.A. 2009;106:21419–21424. doi: 10.1073/pnas.0910601106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geiger D, Scherzer S, Mumm P, Stange A, Marten I, Bauer H, Ache P, Matschi S, Liese A, Al-Rasheid KA, Romeis T, Hedrich R. Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc. Natl. Acad. Sci. U.S.A. 2009;106:21425–21430. doi: 10.1073/pnas.0912021106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sato A, Sato Y, Fukao Y, Fujiwara M, Umezawa T, Shinozaki K, Hibi T, Taniguchi M, Miyake H, Goto DB, Uozumi N. Threonine at position 306 of the KAT1 potassium channel is essential for channel activity and is a target site for ABA-activated SnRK2/OST1/SnRK2.6 protein kinase. Biochem. J. 2009;424:439–448. doi: 10.1042/BJ20091221. [DOI] [PubMed] [Google Scholar]
- 38.Wang P, Xue L, Batelli G, Lee S, Hou YJ, Van Oosten MJ, Zhang H, Tao WA, Zhu JK. Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc. Natl. Acad. Sci. U.S.A. 2013;110:11205–11210. doi: 10.1073/pnas.1308974110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Umezawa T, Sugiyama N, Takahashi F, Anderson JC, Ishihama Y, Peck SC, Shinozaki K. Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci. Signal. 2013;6:rs8. doi: 10.1126/scisignal.2003509. [DOI] [PubMed] [Google Scholar]
- 40.Szostkiewicz I, Richter K, Kepka M, Demmel S, Ma Y, Korte A, Assaad FF, Christmann A, Grill E. Closely related receptor complexes differ in their ABA selectivity and sensitivity. Plant J. 2010;61:25–35. doi: 10.1111/j.1365-313X.2009.04025.x. [DOI] [PubMed] [Google Scholar]
- 41.Pierce M, Raschke K. Correlation between loss of turgor and accumulation of abscisic acid in detached leaves. Planta. 1980;148:174–182. doi: 10.1007/BF00386419. [DOI] [PubMed] [Google Scholar]
- 42.Jia W, Wang Y, Zhang S, Zhang J. Salt-stress-induced ABA accumulation is more sensitively triggered in roots than in shoots. J. Exp. Bot. 2002;53:2201–2206. doi: 10.1093/jxb/erf079. [DOI] [PubMed] [Google Scholar]
- 43.Niu X, Bressan RA, Hasegawa PM, Pardo JM. Ion homeostasis in NaCl stress environments. Plant Physiol. 1995;109:735–742. doi: 10.1104/pp.109.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tholakalabavi A, Zwiazek J, Thorpe T. Effect of mannitol and glucose-induced osmotic stress on growth, water relations, and solute composition of cell suspension cultures of poplar (Populus deltoides var.Occidentalis) in relation to anthocyanin accumulation. In Vitro Cell Dev. Biol. 1994;30:164–170. [Google Scholar]
- 45.Gosti F, Beaudoin N, Serizet C, Webb AA, Vartanian N, Giraudat J. ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell. 1999;11:1897–1910. doi: 10.1105/tpc.11.10.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roychoudhry S, Del Bianco M, Kieffer M, Kepinski S. Auxin controls gravitropic setpoint angle in higher plant lateral branches. Curr. Biol. 2013;23:1497–1504. doi: 10.1016/j.cub.2013.06.034. [DOI] [PubMed] [Google Scholar]
- 47.Rosquete MR, von Wangenheim D, Marhavý P, Barbez E, Stelzer EH, Benková E, Maizel A, Kleine-Vehn J. An auxin transport mechanism restricts positive orthogravitropism in lateral roots. Curr. Biol. 2013;23:817–822. doi: 10.1016/j.cub.2013.03.064. [DOI] [PubMed] [Google Scholar]
- 48.Leung J, Merlot S, Giraudat J. The Arabidopsis ABSCISIC ACID–INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell. 1997;9:759–771. doi: 10.1105/tpc.9.5.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Irigoyen ML, Iniesto E, Rodriguez L, Puga MI, Yanagawa Y, Pick E, Strickland E, Paz-Ares J, Wei N, De Jaeger G, Rodriguez PL, Deng XW, Rubio V. Targeted degradation of abscisic acid receptors is mediated by the ubiquitin ligase substrate adaptor DDA1 in Arabidopsis. Plant Cell. 2014;26:712–728. doi: 10.1105/tpc.113.122234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shin R, Burch AY, Huppert KA, Tiwari SB, Murphy AS, Guilfoyle TJ, Schachtman DP. The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell. 2007;19:2440–2453. doi: 10.1105/tpc.107.050963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arabidopsis Interactome Mapping Consortium Evidence for network evolution in an Arabidopsis interactome map. Science. 2011;333:601–607. doi: 10.1126/science.1203877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.González-Guzmán M, Apostolova N, Bellés JM, Barrero JM, Piqueras P, Ponce MR, Micol JL, Serrano R, Rodríguez PL. The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell. 2002;14:1833–1846. doi: 10.1105/tpc.002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Romero I, Fuertes A, Benito MJ, Malpica JM, Leyva A, Paz-Ares J. More than 80 R2R3-MYB regulatory genes in the genome of Arabidopsis thaliana. Plant J. 1998;14:273–284. doi: 10.1046/j.1365-313x.1998.00113.x. [DOI] [PubMed] [Google Scholar]
- 54.Koornneef M, Reuling G, Karssen CM. The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol. Plant. 1984;61:377–383. [Google Scholar]
- 55.Stracke R, Werber M, Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001;4:447–456. doi: 10.1016/s1369-5266(00)00199-0. [DOI] [PubMed] [Google Scholar]
- 56.Jaradat MR, Feurtado JA, Huang D, Lu Y, Cutler AJ. Multiple roles of the transcription factor AtMYBR1/AtMYB44 in ABA signaling, stress responses, and leaf senescence. BMC Plant Biol. 2013;13:192. doi: 10.1186/1471-2229-13-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilmoth JC, Wang S, Tiwari SB, Joshi AD, Hagen G, Guilfoyle TJ, Alonso JM, Ecker JR, Reed JW. NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J. 2005;43:118–130. doi: 10.1111/j.1365-313X.2005.02432.x. [DOI] [PubMed] [Google Scholar]
- 58.Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D, Onodera C, Quach H, Smith A, Yu G, Theologis A. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell. 2005;17:444–463. doi: 10.1105/tpc.104.028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell. 2007;19:118–130. doi: 10.1105/tpc.106.047761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jung C, Kim YK, Oh NI, Shim JS, Seo JS, Choi YD, Nahm BH, Cheong JJ. Quadruple 9-mer-based protein binding microarray analysis confirms AACnG as the consensus nucleotide sequence sufficient for the specific binding of AtMYB44. Mol. Cells. 2012;34:531–537. doi: 10.1007/s10059-012-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jung C, Seo JS, Han SW, Koo YJ, Kim CH, Song SI, Nahm BH, Choi YD, Cheong JJ. Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 2008;146:623–635. doi: 10.1104/pp.107.110981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim JH, Nguyen NH, Jeong CY, Nguyen NT, Hong SW, Lee H. Loss of the R2R3 MYB, AtMyb73, causes hyper-induction of the SOS1 and SOS3 genes in response to high salinity in Arabidopsis. J. Plant Physiol. 2013;170:1461–1465. doi: 10.1016/j.jplph.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 63.Fowler S, Thomashow MF. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell. 2002;14:1675–1690. doi: 10.1105/tpc.003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao Y, Chan Z, Xing L, Liu X, Hou YJ, Chinnusamy V, Wang P, Duan C, Zhu JK. The unique mode of action of a divergent member of the ABA-receptor protein family in ABA and stress signaling. Cell Res. 2013;23:1380–1395. doi: 10.1038/cr.2013.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK. In vitro reconstitution of an abscisic acid signalling pathway. Nature. 2009;462:660–664. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bai G, Yang DH, Zhao Y, Ha S, Yang F, Ma J, Gao XS, Wang ZM, Zhu JK. Interactions between soybean ABA receptors and type 2C protein phosphatases. Plant Mol. Biol. 2013;83:651–664. doi: 10.1007/s11103-013-0114-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao Y, Zhao S, Mao T, Qu X, Cao W, Zhang L, Zhang W, He L, Li S, Ren S, Zhao J, Zhu G, Huang S, Ye K, Yuan M, Guo Y. The plant-specific actin binding protein SCAB1 stabilizes actin filaments and regulates stomatal movement in Arabidopsis. Plant Cell. 2011;23:2314–2330. doi: 10.1105/tpc.111.086546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao J, Zhang W, Zhao Y, Gong X, Guo L, Zhu G, Wang X, Gong Z, Schumaker KS, Guo Y. SAD2, an importin β-like protein, is required for UV-B response in Arabidopsis by mediating MYB4 nuclear trafficking. Plant Cell. 2007;19:3805–3818. doi: 10.1105/tpc.106.048900. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.