Abstract
In excitatory neurons, SCN2A (NaV1.2) and SCN8A (NaV1.6) sodium channels are enriched at the axon initial segment. NaV1.6 is implicated in several mouse models of absence epilepsy, including a missense mutation identified in a chemical mutagenesis screen (Scn8aV929F). Here, we confirmed the prior suggestion that Scn8aV929F exhibits a striking genetic background-dependent difference in phenotypic severity, observing that spike-wave discharge (SWD) incidence and severity are significantly diminished when Scn8aV929F is fully placed onto the C57BL/6J strain compared with C3H. Examination of sequence differences in NaV subunits between these two inbred strains suggested NaV1.2V752F as a potential source of this modifier effect. Recognising that the spatial co-localisation of the NaV channels at the axon initial segment (AIS) provides a plausible mechanism for functional interaction, we tested this idea by undertaking biophysical characterisation of the variant NaV channels and by computer modelling. NaV1.2V752F functional analysis revealed an overall gain-of-function and for NaV1.6V929F revealed an overall loss-of-function. A biophysically realistic computer model was used to test the idea that interaction between these variant channels at the AIS contributes to the strain background effect. Surprisingly this modelling showed that neuronal excitability is dominated by the properties of NaV1.2V752F due to “functional silencing” of NaV1.6V929F suggesting that these variants do not directly interact. Consequent genetic mapping of the major strain modifier to Chr 7, and not Chr 2 where Scn2a maps, supported this biophysical prediction. While a NaV1.6V929F loss of function clearly underlies absence seizures in this mouse model, the strain background effect is apparently not due to an otherwise tempting Scn2a variant, highlighting the value of combining physiology and genetics to inform and direct each other when interrogating genetic complex traits such as absence epilepsy.
Keywords: Absence seizures, genetic epilepsy, sodium channels, murine AE, computational analysis
Introduction
Absence epilepsy is assumed to have a complex genetic architecture, with thus far a number of rare variants revealed in a restricted number of patients. Most of these variants have been identified in the GABA receptor and calcium channels, plus a few non-ion channel related genes [1]. GABA receptors and calcium channels are both inherently important in maintaining the fine balance between excitation and inhibition in the thalamocortical circuit, necessary for proper function. Disruption to components of this circuit can result in an increased number of action potentials (APs) per cycle, which results in the 3–4Hz spike-wave discharge (SWD) on EEG, characteristic of absence seizures [1]. These brief episodes of SWDs are accompanied by a loss of consciousness.
More recently a novel mechanism of absence epilepsy was identified, with the discovery of absence seizures conferred by several mouse mutant alleles of the Scn8a gene, which encodes the voltage-gated sodium channel NaV1.6 [2]. Voltage-gated sodium channels have been heavily implicated in epilepsy, although most variants have been identified in the NaV1.1 and NaV1.2 subtypes, most associated with Dravet syndrome, Genetic Epilepsy with Febrile Seizures Plus (GEFS+) and Benign Familial Neonatal Infantile Seizures (BFNIS), reviewed in[3]. This was the first incidence of NaV1.6 being implicated in epilepsy, and the first incidence of voltage-gated sodium channels being implicated in absence epilepsy.
Voltage-gated sodium channels are integral membrane proteins essential for the initiation and propagation of APs. NaV1.6 has a relatively uniform brain distribution, with high levels of expression in hippocampus, cortex and cerebellum [4, 5]. It is localised to both excitatory and inhibitory neurons [6–11]. Hu et al (2009) [6] highlighted the important contribution of the NaV1.6 channel to the initiation of the AP in pyramidal neurons at the axon initial segment (AIS). NaV1.6 and NaV1.2 are highly concentrated in the AIS, with NaV1.6 concentrated more distally, and NaV1.2 concentrated more proximally. NaV1.6 is known to activate at a lower threshold compared with NaV1.2 [12]. The lower-threshold NaV1.6 was demonstrated to be more important for the initiation of the AP in the distal region of the AIS, and was important for forward propagation down the axon, whereas NaV1.2 was secondarily activated in the proximal region, and was more important for back-propagation to the soma and dendrites [6].
Expression of Scn8a mutation-induced absence seizures appeared to be enhanced on the C3HeB/FeJ (C3H) mouse strain background, compared with C57BL/6J (C57), the inbred mouse strain frequently used for genetic studies [2]. This preliminary observation, presumably a modifier effect due to genetic variants that differ between parent strains, was blurred by varying effects of different Scn8a mutant alleles, including Scn8a8J (NaV1.6V929F) whose biophysical properties had not been previously determined [2]. Interestingly, C3H mouse strains also harbour a private non-synonymous coding mutation in the Scn2a gene that encodes the NaV1.2 channel. Although this strain variant is one of potentially many that may modify the phenotype, it would result in spatial convergence of two altered molecules (Nav1.2, Nav1.6) in a region of the neuron critical for regulating excitability, i.e. a positive epistatic interaction between these two channel isoforms. The current study aimed to examine the functional consequences of variant NaV1.6 and NaV1.2 channels in the context of the genetic basis of the strain difference in seizure phenotype conferred by Scn8aV929F.
Materials and Methods
Animals
All animals were fed standard National Institutes of Health diet containing 6% fat and acidified water ad libitum, and maintained in a climate-controlled room, with a 12 hour light on/off cycle. All animal procedures followed Association for Assessment and Accreditation of Laboratory Animal Care guidelines and were approved by institutional Animal Care and Use Committee.
EEG
Adult mice aged between 6 and 9 weeks were anesthetised with tribromoethanol (400 mg/kg i.p.). Small burr holes were drilled (1 mm anterior to the bregma and 2 mm posterior to the bregma) on both sides of the skull 2 mm lateral to the midline. Four teflon-coated silver wires were soldered onto the pins of a microconnector (Mouser electronics, Texas). The wires were placed between the dura and the brain and an inorganic-based dental cap was then applied. The mice were given a post-operative analgesic of carprofen (5 mg/kg subcutaneous) and allowed a minimum 2 days recovery period before recordings were made. The mice were connected to the EEG Stellate Lamont Pro-36 programmable amplifier (Lamont Medical Instruments, Madison, WI) using a flexible cable with free access to all corners of the cage, including food, for 2 hr periods on 2 consecutive days (4 hr total), during the middle of the light cycle. EEG data were collected using a bipolar montage of 6 channels, representing the differential signal between each of the 4 electrodes, with the software program Stellate Harmonie (Stellate Systems, Inc., Montreal, Canada). SWD episodes were scored using the following criteria: the EEG recording showed at least 2 linked SWD discharges with amplitudes at least 2-fold higher than background and observed concurrently in the majority of the channels.
Plasmid preparation
A plasmid pcDNA3.1(+)-NaV1.2, encoding the human NaV1.2 protein, is the WT plasmid used in this study and has been previously described [13]. The variant creating the amino acid substitution in position 752 of the NaV1.2 protein was introduced into pcDNA3.1(+)-hNaV1.2 as a service by TOP Gene Technologies. Successful mutagenesis was confirmed by DNA sequencing.
A plasmid pcDNA3-NaV1.6 encoding the mouse NaV1.6 protein has been previously described [14]. The plasmid contains the Y371S substitution which converts the tetrodotoxin sensitivity of NaV1.6 from nanomolar to micromolar concentrations upon expression in mammalian cell lines [14]. This is the WT plasmid in this study. The mutation creating the amino acid substitution in position 929 of the NaV1.6 protein was introduced into pcDNA3-mNaV1.6 using the QuikChange XL II site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA, USA) and the following primers (mutated codons italicised): CCACTCCTTCCTCATCTTCTTCCGAGTGC (forward) and GCACTCGGAAGAAGATGAGGAAGGAGTGG (reverse). Successful mutagenesis was confirmed by DNA sequencing.
Cell culture and transfection
HEK293T cells (a HEK-293 cell line stably transfected with the SV40 large T antigen) were maintained in Dulbecco’s Modified Eagle’s Medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% (v/v) fetal bovine serum (Invitrogen) and 5µg/ml gentamycin for up to 30 passages. The cells were grown in T75 flasks (BD Biosciences, San Jose, CA, USA) to ~70% confluency, and then transfected with 4.5µg of the NaV1.2 WT or NaV1.2V752F plasmid using Lipofectamine-LTX™ (Invitrogen). Twenty-four hours post-transfection cells were split into T75 flasks, and at 48 hours post-transfection the cells were trypsinised and placed into suspension in extracellular solution composed of the following (mM): 140 NaCl, 4 KCl, 1 MgCl2, 2 CaCl2, 5 D-Glucose monohydrate, 10 HEPES, adjusted to pH 7.4 with NaOH. The osmolarity was adjusted to ~298mOsmol.
ND7/23 cells (Mouse neuroblastoma X Rat neuron hybrid) were maintained and passaged as above. The ND7/23 cell line produces a tetrodotoxin (TTX)-sensitive (TTX-S), but not a TTX-6 resistant (TTX-R), current [15, 16]. The cells were grown on T75 flasks (BD Biosciences) to ~70% confluency and then transfected with 19µg of the NaV1.6 WT or NaV1.6V929F plasmid using Lipofectamine-LTX™ (Invitrogen). Twenty-four hours post-transfection cells were split into T75 flasks, and at 48 hours post-transfection the cells were trypsinised and placed into suspension in extracellular solution as above.
Electrophysiological recordings
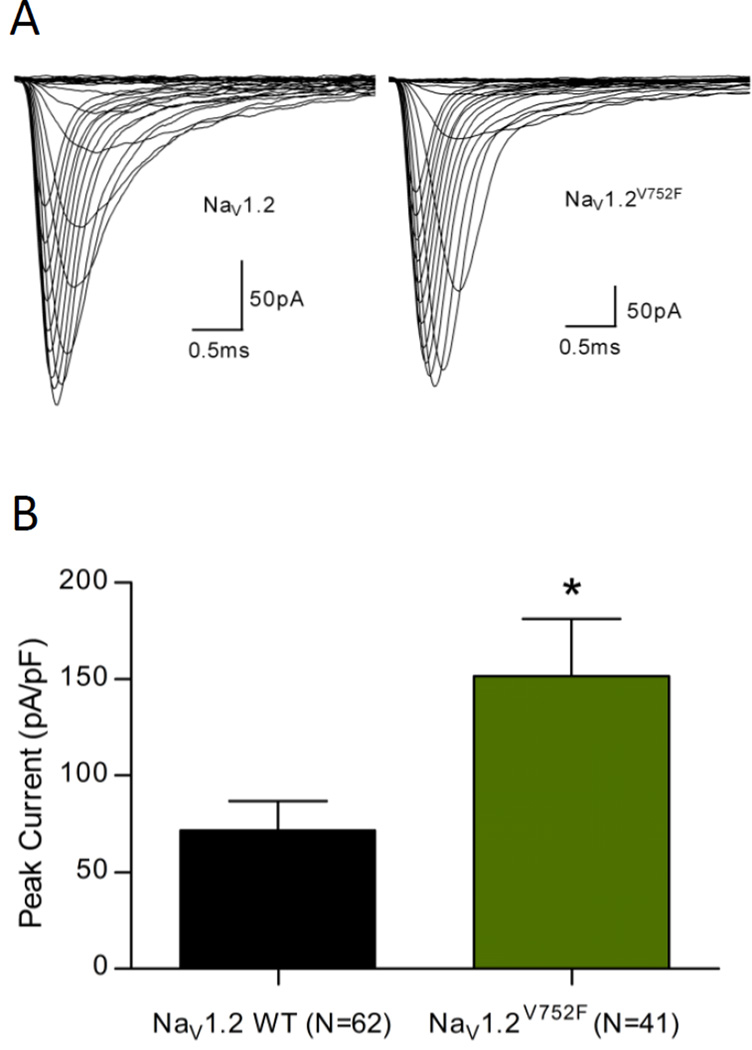
Cells suspended in extracellular solution were placed in the cell hotel of the Nanion Patchliner® (Nanion Technologies, Munich, Germany), where they were kept in suspension by gentle automatic pipetting. A medium single hole planar chip was selected for recording with an average resistance of ~2.5MΩ/hole. The intracellular solution consisted of (mM): 50 CsCl, 10 NaCl, 60 CsF, 20 EGTA, 10 HEPES, adjusted to pH 7.2 with CsOH. The osmolarity was adjusted to ~285 mOsmol. Extracellular solution was as above. Prior to recordings of the NaV1.6 channels, TTX was added to the extracellular solution at a concentration of 0.00025mM to isolate the transfected TTX-R currents. Pipette and whole cell capacitance were fully compensated and the series resistance compensation was set to 50%. Data was analysed from cells with a maximal current falling between 300pA and 5nA, so that traces were sufficiently separated from background noise, and were not subject to clamping artefacts associated with large currents. However for measures of current density all cells that produced currents were included, the results of which are shown in Fig.2B and 5B. All recordings were acquired at 50 kHz with the low pass filter set to 10–30 kHz in PATCHMASTER (HEKA Instruments Inc, NY, USA). The calculated liquid junction potential was −3.8mV and was compensated for in PATCHMASTER.
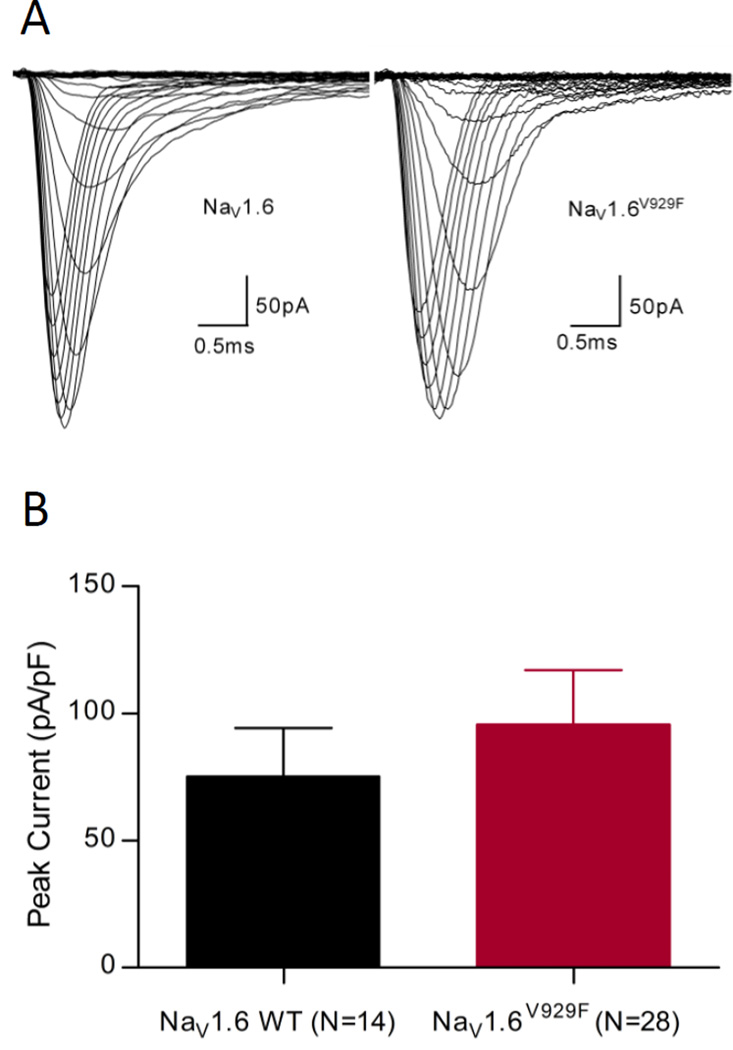
Figure 2.
NaV1.6 currents recorded in ND7/23 cells. (A) Representative traces obtained from cells expressing NaV1.6 WT and NaV1.6V929F channels using the voltage-dependence of activation protocol. (B) Comparison of average peak current densities for the two experimental groups shown with S.E.M.
Figure 5.
NaV1.2 currents recorded in HEK293T cells. (A) Representative traces obtained from cells expressing NaV1.2 WT and NaV1.2V752F channels using the voltage-dependence of activation protocol. (B) Comparison of average peak current densities for the two experimental groups shown with S.E.M. Statistical significance marked as *p<0.05.
All experiments were performed at 27°C. Data are shown as means ± S.E.M. with the number of experiments provided in the figures. Leak subtraction was performed in software before the currents were normalised. Statistical analysis was performed using Student’s t-test and differences were considered significant when p<0.05.
The voltage dependence of activation was analysed using a step protocol in which cells were depolarised to a range of potentials from −120 mV to +30mV in 5mV increments from a holding potential of −120mV. Peak currents at individual steps were measured and normalized to the maximum peak current and plotted against voltage. To calculate a reversal potential, the resulting I–V curve of each data set was individually fitted with the equation I=[1+exp(− 0.03937.z.(V−V1/2)]/.g.(V−Vr), where I is the current amplitude, z is the apparent gating charge, V is the potential of the given pulse, V1/2 is the half-maximal voltage, g is a factor related to the maximum number of open channels, and Vr is the reversal potential. Conductance was then directly calculated using the equation G=I/(V−Vr), where G is conductance. The conductance values were fit with the Boltzmann equation G=1/(1+exp[(V−V1/2)/a]), where a is the slope at half-maximum, V is the potential of the given pulse, and V1/2 is the potential for the half-maximal activation.
The voltage dependence of steady-state fast inactivation was determined using a two-step protocol composed of a conditioning pulse with depolarising potential from −120mV to +30mV in 5mV increments for 100ms from a holding potential of −120mV and a test pulse at 0mV for 20ms. The peak current amplitudes during the subsequent test pulses were normalised to the peak current amplitude during the first test pulse, plotted against the potential of the conditioning pulse, and fitted with the Boltzmann equation I=1/(1+exp[(V−V1/2)/a]), where I is equal to the test pulse current amplitude, V is the potential of the conditioning pulse, V1/2 is the voltage for the half-maximal inactivation, and a is the slope factor.
Inactivation time constants were determined using MATLAB 7.10.0 (Mathworks, MA, USA). Scripts were written that identified the peak current of each trace. A single exponential curve was used to fit each trace I=A.exp[−(t−K)/τ]+C, where I is the current, A is the relative proportion of current inactivating with the time constant τ, K is the time shift, and C is the steady-state persistent current. The time constants were plotted against voltage and the points on this graph were fit with a decaying exponential equation Y=span*exp(−K*x)+plateau, where span is the starting point of the curve, K is the decay factor and plateau is the value the curve decays to.
The recovery from inactivation protocol started with a conditioning depolarisation from a holding potential of −120mV to 0mV for 30ms to fully inactivate channels. The voltage was then returned to the holding potential of −120mV for variable intervals (every 3ms from 0 to 39ms). Finally, the voltage was stepped to 0mV for 30ms to test channel availability. The peak current amplitude during the test potentials was plotted as fractional recovery against the recovery period by normalising to the maximum current during the conditioning potentials. The recovery currents were plotted against delta time and the points on this graph were fit with a hyperbola equation Y=Imax*x/(rc+x), where Imax is the maximal current and rc is the recovery constant.
Computational model
The model was derived from that of Hu et al [17] (https://senselab.med.yale.edu/ModelDB/ accession #: 123897). It is a cortical Layer 5 pyramidal cell model designed to examine the contribution of the NaV1.2 and NaV1.6 subunits to AP initiation and backpropagation. Layer 5 cortical pyramidal cells are well characterised thalamic projection neurons [18] and, recently, also shown to receive thalamic projections [19] suggesting an intimate role in determining thalamic and cortical function and presumably participate in the generation of absence seizures. The equations for activation and inactivation gating variables for the sodium channels were identical to those used in the paper by Hu et al (2009) [6]. A voltage parameter (vshift) was used to shift the voltage dependence of activation and inactivation curves. One change that was implemented in the current study was to be able to deliver different vshift values to both activation and inactivation, so that they could be shifted in opposite directions, and by differing amounts, hence the parameter vshiin was introduced. For analysis of the variants the half maximal voltages of the NaV1.2 and NaV1.6 channels for activation and inactivation were shifted by amounts as listed in Table 7, which is based on our electrophysiological experiments (Table 1).
Table 7.
Biophysical parameters for activation and fast inactivation used for modelling
| Channel type |
Voltage-dependence of activation |
Voltage-dependence of inactivation |
Density multiplication factor (gbar) |
|---|---|---|---|
| V1/2 (mV) | V1/2 (mV) | ||
| NaV1.2 WT | −30 | −30 | 1 |
| NaV1.2V752F | −35 | −32 | 1.4 |
| NaV1.6 WT | −43 | −43 | 1 |
| NaV1.6V929F | −36 | −54 | 1 |
Table 1.
Biophysical parameters for activation and fast inactivation for NaV1.6 WT and NaV1.6V929F
| Channel type | Voltage-dependence of activation | Voltage-dependence of inactivation | ||||
|---|---|---|---|---|---|---|
| V1/2 (mV) | Slope | n | V1/2 (mV) | Slope | n | |
| NaV1.6 WT | −15.2±1.7 | 9.37±1.4 | 8 | −36.58±2.2 | −7.32±0.2 | 8 |
| NaV1.6V929F | −9.33±1.6* | 7.53±0.6 | 11 | −48.37±1.9* | −6.11±0.2* | 11 |
Values are presented as mean ± standard error.
Statistical significance is marked as *p<0.05.
Comparisons were made between the mutant channels and their WT equivalent.
Results
Profound absence seizures in C3HeB/FeJ-Scn8a8J (encoding NaV1.6V929F) congenic mutant mice
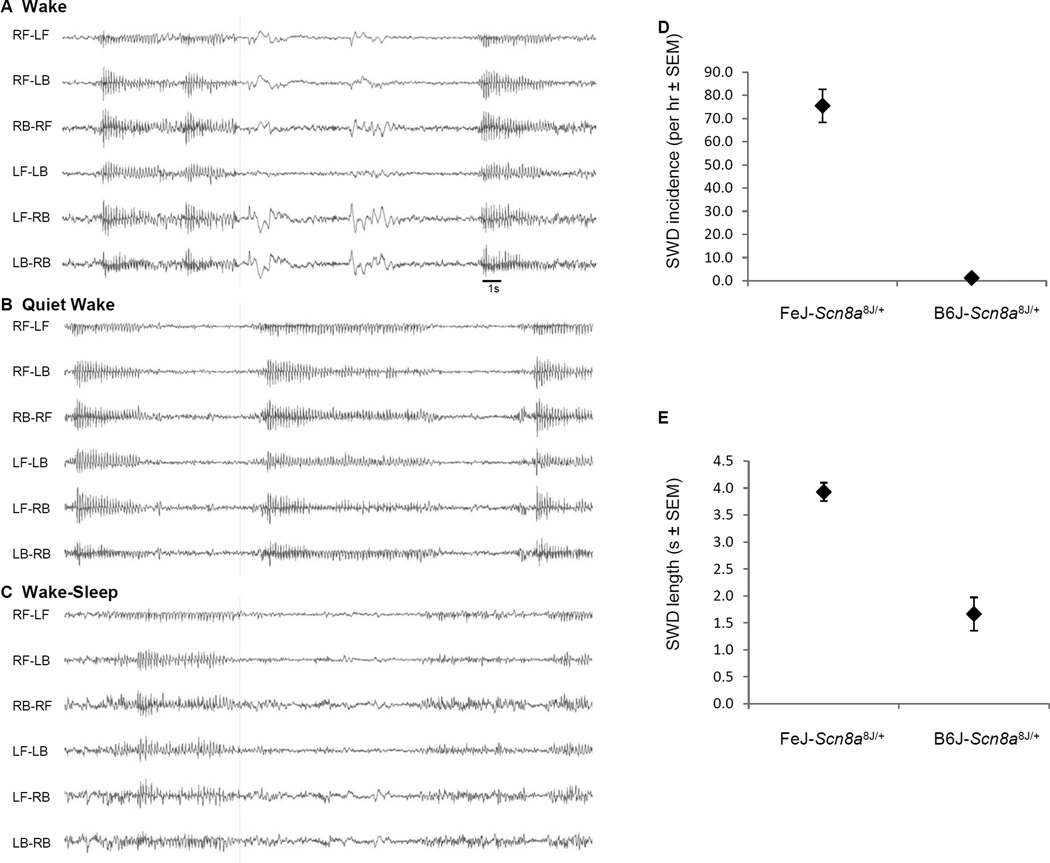
Mice heterozygous for mouse mutant alleles of Scn8a, including Scn8a8J (encoding a non-synonymous amino acid substitution in the voltage-gated sodium channel NaV1.6V929F leaving protein expression intact) were shown to exhibit modest or frequent SWDs in EEG recordings, without the severe locomotor abnormalities of Scn8a mutant homozygotes [2]. Preliminary strain background effects were also observed, as SWDs became less pronounced when NaV1.6V929F was partially backcrossed away from a mixed C3HeB/FeJ (FeJ) × C57BL/6J (B6J) and towards inbred C57BL/6J [2]. To definitively examine this effect, the mutation was backcrossed for 10 or more generations to each strain, revealing a more striking difference. In daytime EEG recordings NaV1.6V929F heterozygotes congenic on C3HeB/FeJ (N27) had an average of 76 SWDs per hour, lasting 3.9s, whereas those backcrossed to C57BL/6J (N10 or N22) had an average of 6.3 SWDs per hour, lasting 1.5s (Fig. 1). This result confirms and extends the effect, which is presumably due to one or more genetic modifier variants that differ between these parent strains.
Figure 1.
Spike-wave discharges in Scn8a8J/+ heterozygotes on B6J vs. FeJ strain backgrounds (A–C) Example of a SWD in FeJ-Scn8a8J/+ mouse in different alertness states. L- left, R-right, F-front, B-back (A) Wake – note SWD interspersed with movement artefact during interictal EEG, (B) Quiet Wake – low amplitude, low energy interictal EEG, and (C) Wake-Sleep –SWDs often occur at the end of higher energy, slower EEG bouts. (D) Histogram of SWD incidence and (E) SWD length(s). The data represent the average of six mice per genotype.
NaV1.6V929F displays a kinetic profile consistent with an overall decrease in activity of the channel
To characterise the functional consequences of the V929F mutation of the Scn8a8J allele we used a heterologous expression model and whole-cell patch clamp analysis in single cells. Expression was first attempted in HEK293T cells but expression levels were too low and inconsistent for reliable analysis as maximum current magnitude was always less than 300pA. A ND7/23 cell line based model has been developed for the study of Scn8a [14]. This model contains endogenous sodium channels that need to be blocked with TTX to enable isolation of the expressed NaV1.6 TTX-R current and typical responses are shown in Fig.2A. Simple I–V protocols were run to determine peak currents, and cell capacitances were recorded by PATCHMASTER immediately before the I–V protocol was run. There was no significant difference detected between the current density of cells expressing NaV1.6 WT or NaV1.6V929F, suggesting that NaV1.6 V929F has no impact on trafficking or expression (Fig.2B).
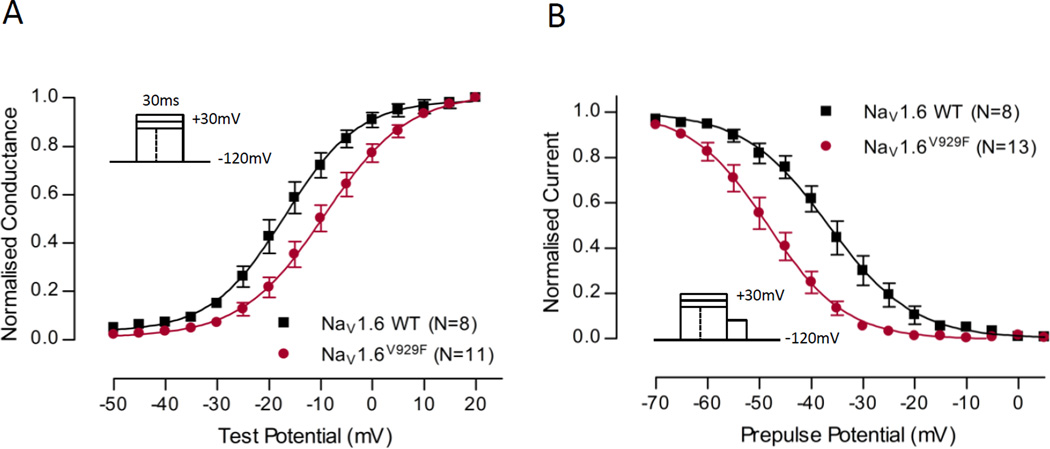
Fig.3 examines the voltage-dependence of activation and inactivation between NaV1.6 WT and NaV1.6V929F. Expression of the NaV1.6V929F mutant causes a depolarising shift in the activation curve compared with NaV1.6 WT suggesting that the mutant has a decreased number of channels open at a given voltage, consistent with a decrease in activity of the channel. The NaV1.6V929F mutant displays a large hyperpolarising shift in the fast-inactivation curve compared to NaV1.6 WT, not consistent with the depolarising shift seen in the activation curve, but consistent with a further decrease in activity of channels. The data points were fit with a Boltzmann equation and the V1/2 and slope values were obtained (Table 1). The shift in the V1/2 was significant for both activation and inactivation curves for the NaV1.6V929F mutant compared to WT, reflecting observations made by comparison of the individual data points. The slope of the inactivation curve was also significantly different compared to WT.
Figure 3.
Voltage-dependence of normalized peak conductance and steady-state inactivation. (A) Normalized conductance shown for NaV1.6 WT and NaV1.6V929F as a function of voltage. (B) Steady-state fast-inactivation shown for NaV1.6 WT and NaV1.6V929F as a function of voltage. For both graphs Boltzmann curves were fit to pooled averages and plotted (see Table 1 for V1/2 values). Protocol schemes are shown in the inset.
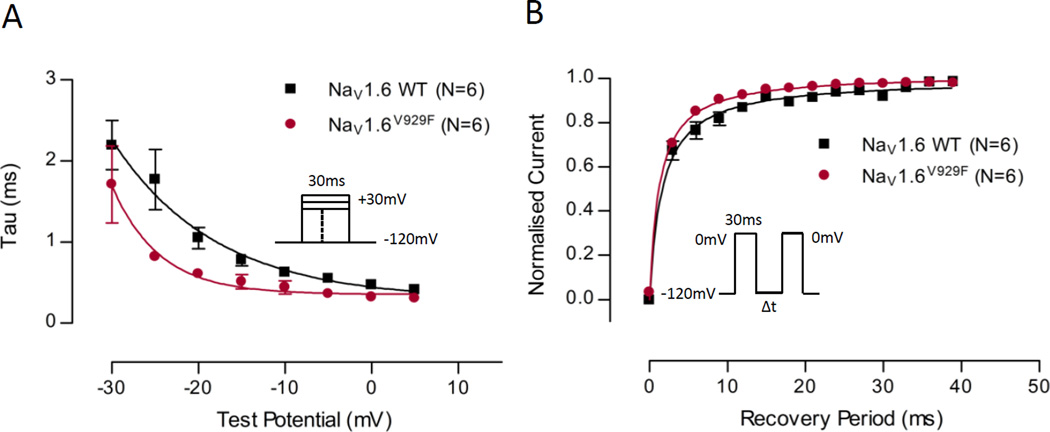
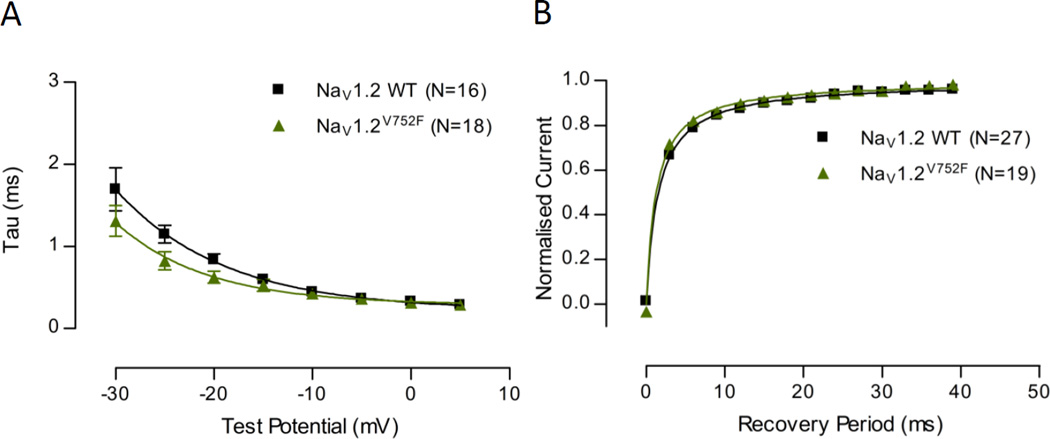
Fig.4A examines the time constant (τ) of fast inactivation at a range of voltages between NaV1.6 WT and NaV1.6V929F. The voltage range in the figure is limited to −30 to +5mV to emphasise the range in which there were differences. The time constants of the NaV1.6V929F mutant were smaller on average than the NaV1.6 WT equivalents. The data points were fit with a decaying exponential curve (Table 2), which demonstrated significant differences between the two data sets.
Figure 4.
Time constant of fast-inactivation plotted against voltage, and recovery of channel availability from fast-inactivation plotted against time. (A) Time constant of fast-inactivation shown for NaV1.6 WT and NaV1.6V929F as a function of voltage. Current for each cell was fit to a single exponential at a range of test potentials (as shown in the inset) and a time constant was determined. A one phase exponential decay was fit to pooled averages and plotted (see Table 2 for parameter values). (B) Recovery of channel availability from fast-inactivation shown for NaV1.6 WT and NaV1.6V929F as a function of time. A hyperbola was fit to pooled averages and plotted (see Table 3 for parameter values). Protocol scheme is shown in the inset.
Table 2.
Biophysical parameters for time constants of fast inactivation for NaV1.6 WT and NaV1.6V929F
| Channel type | Time constant of fast inactivation | |||
|---|---|---|---|---|
| Span | K | plateau | n | |
| NaV1.6 WT | 1.98±0.2 | 0.08±0.03 | 0.29±0.2 | 6 |
| NaV1.6V929F | 1.34±0.2* | 0.18±0.06 | 0.36±0.09 | 6 |
Values are presented as mean ± standard error.
Statistical significance is marked as *p<0.05.
Comparisons were made between the mutant channels and their WT equivalent.
Normalized current as a function of time following an inactivating voltage step is plotted in Fig.4B for NaV1.6 WT and NaV1.6V929F. NaV1.6V929F recovered more quickly on average than NaV1.6 WT. The curves were fit with a hyperbola as a means to characterise the data, for which the recovery constant was significantly quicker for the NaV1.6 V929F mutant compared to WT (Table 3), consistent with observations made by examining the individual data points.
Table 3.
Biophysical parameters for recovery of channel availability from fast inactivation for NaV1.6 WT and NaV1.6V929F
| Channel type | Recovery from inactivation | ||
|---|---|---|---|
| Imax | rc | n | |
| NaV1.6 WT | 1.00±0.01 | 1.65±0.2 | 8 |
| NaV1.6V929F | 1.02±0.01 | 1.27±0.07* | 11 |
Values are presented as mean ± standard error.
Statistical significance is marked as *p<0.05.
Comparisons were made between the mutant channels and their WT equivalent.
C3H mice harbour a NaV1.2V752F mutation
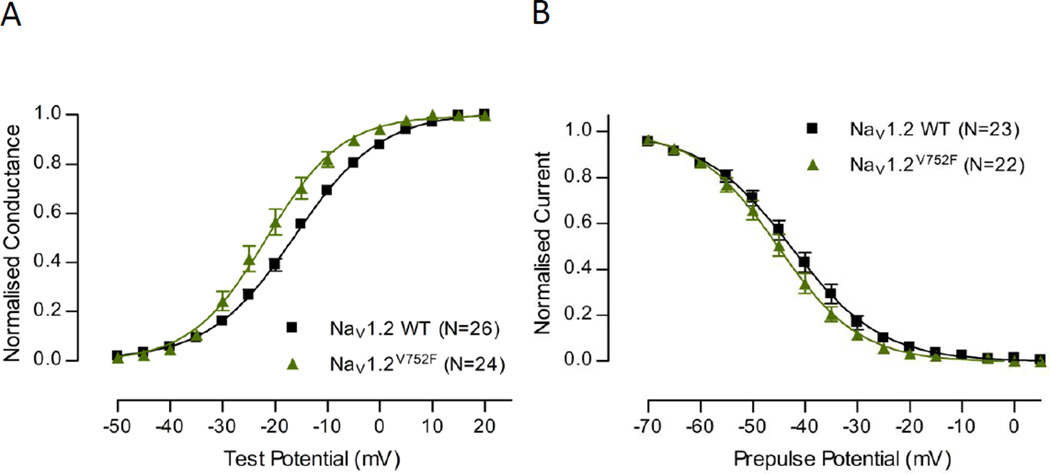
We next considered possible mechanisms by which Scn8a-associated SWDs are exacerbated on the C3H background. Scn2a (encoding NaV1.2) was identified as a potential candidate on the basis that a V752F variant had been identified in genome sequencing studies (rs27999676 in dBSNP: www.ncbi.nlm.nih.gov/SNP), in the NaV1.2 cytoplasmic linker region between domains I and II of the channel. Biophysical analysis of the NaV1.2V752F variant was undertaken and typical macroscopic currents recorded from HEK293T cells are shown in Fig.5A. The current density measures in Fig.5B show that NaV1.2V725F had a significantly increased current density compared to NaV1.2 WT. This indicates that the NaV1.2V725F variant may alter trafficking and expression of the channel. Fig.6 compares the voltage dependence of activation and inactivation in NaV1.2 WT and NaV1.2V725F. Expression of the NaV1.2V725F variant causes a hyperpolarising shift in the activation curve compared to NaV1.2 WT suggesting that the variant channel has an increased number of channels open at a given voltage, consistent with an increase in activity of the channel. The V1/2 and slope values obtained from the Boltzmann fit were both significantly different (Table 4). Expression of NaV1.2V752F displays a small hyperpolarising shift in the inactivation curve compared to NaV1.2 WT, consistent with the hyperpolarising shift seen in the activation curve, although this shift of the fitted Boltzmann curve was not significant in V1/2 change, only in slope (Table 4).
Figure 6.
Voltage-dependence of normalised peak conductance and steady-state inactivation. (A) Normalised conductance shown for NaV1.2 WT and NaV1.2V752F as a function of voltage. (B) Steady-state fast-inactivation shown for NaV1.2 WT and NaV1.2V752F as a function of voltage. For both graphs Boltzmann curves were fit to pooled averages and plotted (see Table 4 for V1/2 values).
Table 4.
Biophysical parameters for activation and fast inactivation for NaV1.2 WT and NaV1.2V752F
| Channel type | Voltage-dependence of activation | Voltage-dependence of inactivation | ||||
|---|---|---|---|---|---|---|
| V1/2 (mV) | Slope | n | V1/2 (mV) | Slope | n | |
| NaV1.2 WT | −15.65±0.8 | 8.08±0.3 | 28 | −42.84±1.4 | −7.54±0.3 | 23 |
| NaV1.2V752F | −20.82±1.4* | 6.04±0.3* | 24 | −45.22±1.4 | −6.57±0.2* | 22 |
Values are presented as mean ± standard error.
Statistical significance is marked as *p<0.05.
Comparisons were made between the variant channels and their WT equivalent.
Fig.7A examines the time constant of fast inactivation at a range of voltages between NaV1.2V752F and NaV1.2 WT. The time constants of the NaV1.2V752F were smaller on average than the NaV1.2 WT equivalents. The data points were fit with a decaying exponential curve (Table 5), which demonstrated differences between the two data sets.
Figure 7.
Time constant of fast inactivation plotted against voltage and recovery of channel availability from fast inactivation plotted against time. (A) Time constant of fast inactivation shown for NaV1.2 WT and NaV1.2V752F as a function of voltage. Current for each cell was fit to a single exponential at a range of test potentials and a time constant was determined. A one phase exponential decay was fit to pooled averages and plotted (see Table 5 for parameter values). (B) Recovery of channel availability from fast inactivation shown for NaV1.2 WT and NaV1.2V752F as a function of time. A hyperbola was fit to pooled averages and plotted (see Table 6 for parameter values).
Table 5.
Biophysical parameters for time constants of fast inactivation for NaV1.2 WT and NaV1.2V752F
| Channel type | Time constant of fast inactivation | |||
|---|---|---|---|---|
| Span | K | plateau | n | |
| NaV1.2 WT | 1.48±0.1 | 0.09±0.02 | 0.22±0.1 | 16 |
| NaV1.2V752F | 1.00±0.1* | 0.11±0.03 | 0.29±0.07 | 18 |
Values are presented as mean ± standard error.
Statistical significance is marked as *p<0.05.
Comparisons were made between the variant channels and their WT equivalent.
Normalised current as a function of time following an inactivating voltage step is plotted in Fig.7B for NaV1.2V752F and NaV1.2 WT. The curves were fit with a hyperbola as a means to characterise the data, for which the recovery constant was significantly quicker for the mutant compared to WT (Table 6), consistent with observations made by examining the individual data points.
Table 6.
Biophysical parameters for recovery of channel availability from fast inactivation for NaV1.2 WT and NaV1.2V752F
| Channel type | Recovery from inactivation | ||
|---|---|---|---|
| Imax | rc | n | |
| NaV1.2 WT | 1.00±0.01 | 1.55±0.08 | 27 |
| NaV1.2V752F | 1.00±0.01 | 1.23±0.07* | 19 |
Values are presented as mean ± standard error.
Statistical significance is marked as *p<0.05.
Comparisons were made between the variant channels and their WT equivalent.
Computational model reveals dominant effect of NaV1.2V752F variant in the pyramidal cell AIS
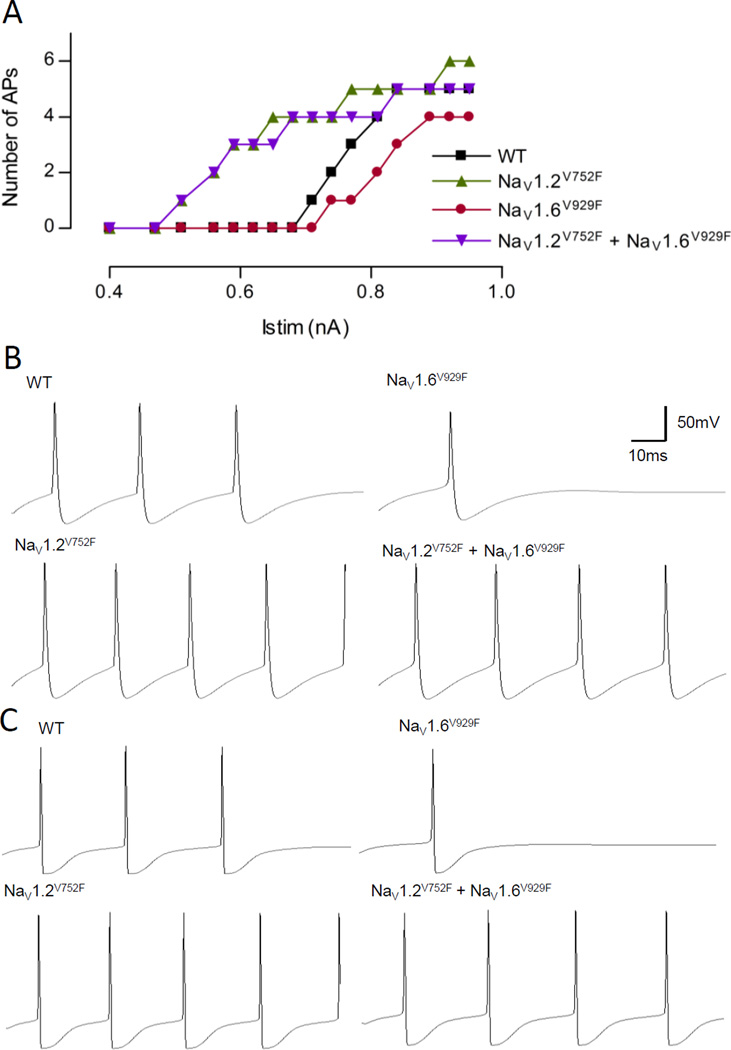
A heterozygous cell was modelled for NaV1.2V752F and NaV1.6V929F, both individually and in combination, in a model of a cortical Layer 5 pyramidal neuron, a known thalamic input and output layer. The alterations made to the steady state voltage dependence of activation and inactivation can be seen in Table 7, and the resulting input-output (i-o) curves are shown in Fig.8. This model had a proximal distribution of NaV1.2 and a distal distribution of NaV1.6 at the AIS. The sodium channel subtype modelled in the axon and nodes was NaV1.6, and the sodium channel subtype modelled in the soma, dendrites and the myelinated axon was NaV1.2. The parameters of the subunits in these regions were also altered in accordance to changes made to the subunits in the AIS. The i-o curves produced from recordings at the soma, the proximal AIS, the distal AIS, and the distal axon were identical and the curve from the soma is shown in Fig.8A. The morphology of the APs produced do differ from each compartment with example outputs shown in Fig.8B and 8C. As can be seen in Fig.8A, the model neuron harbouring the NaV1.6V929F mutation requires a larger stimulating current to initiate firing compared to WT, and fires fewer APs for any given stimulus. The NaV1.2V752F variant, however, requires a far smaller current injection to initiate firing, and fires more APs for most current injections compared to WT. When both variants are inserted into the same model, the output is dominated by the NaV1.2V752F current with the combined model largely following the i-o curve of the NaV1.2V752F variant. Contrary to our initial intuitive idea that the NaV1.6 and NaV1.2 variants could interact at the AIS or neurons where they co-localise, NaV1.6V929F channels are unable to functionally influence NaV1.2V752F containing pyramidal neurons.
Figure 8.
Results from model of a pyramidal neuron modelling the heterozygous situation for NaV1.2V752F and NaV1.6V929F, both individually and in combination. (A) The input-output curve for the WT, NaV1.2V752F, NaV1.6V929F and, NaV1.2V752F + NaV1.6V929F scenarios. (B) Example output of the four conditions at the soma for Istim = 0.77nA. (C) Example output of the four conditions in the distal axon for Istim = 0.77nA. The scale bar in B is also applicable to C.
The major modifier of Scn8a-associated SWD does not map to Scn2a
With the lack of evidence for biological interaction of NaV1.2V752F and NaV1.6V929F predicted by our pyramidal cell computational model, a more thorough search to identify the genetic basis of the strain modifier effect was undertaken. As a first step towards this, B6J-Scn8a8J heterozygotes were mated to wild-type FeJ mice and the F1 hybrid progeny mated to B6J to make the second backcross, or N2 generation, that is segregating putative modifier variants. Forty-one backcross mice heterozygous for Scn8a8J were then subjected to EEG recordings, from which SWD regions were identified and SWD incidence and duration data extracted. In the backcross, the SWD mean incidence and duration values fell between those of the parent strains, although with a wide range of non-normal distribution. These results are suggestive of genetically complex inheritance of possibly multiple trait loci contributing to the modifier effect (Fig 9A, Fig 9B – inset for SWD length and incidence, respectively).
Figure 9.
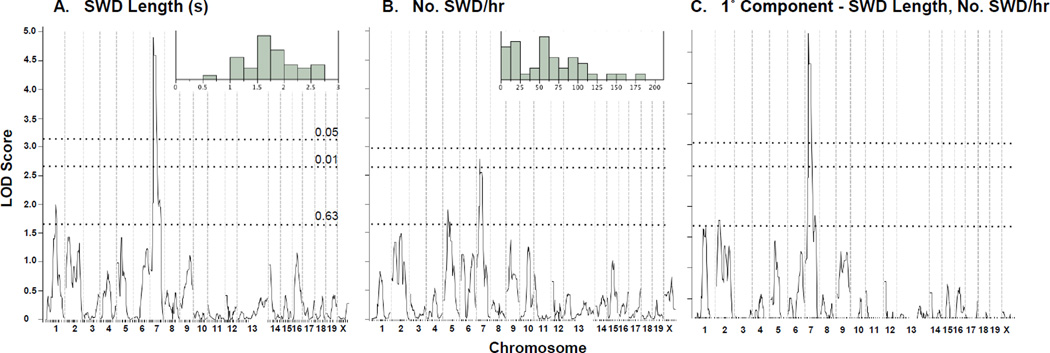
Genome scans for SWD traits in Scn8a8J/+ heterozygous (FeJ × B6J)F1 X B6J backcross mice. Genome scans for absence seizure traits SWD length (A), SWD incidence (B) and the 1° principal component between them (C). Genome-wide significance threshold p-values as determined by permutation shuffling are shown as dashed lines. The inset in A and B shows the frequency distribution of the respective raw phenotype in the backcross population.
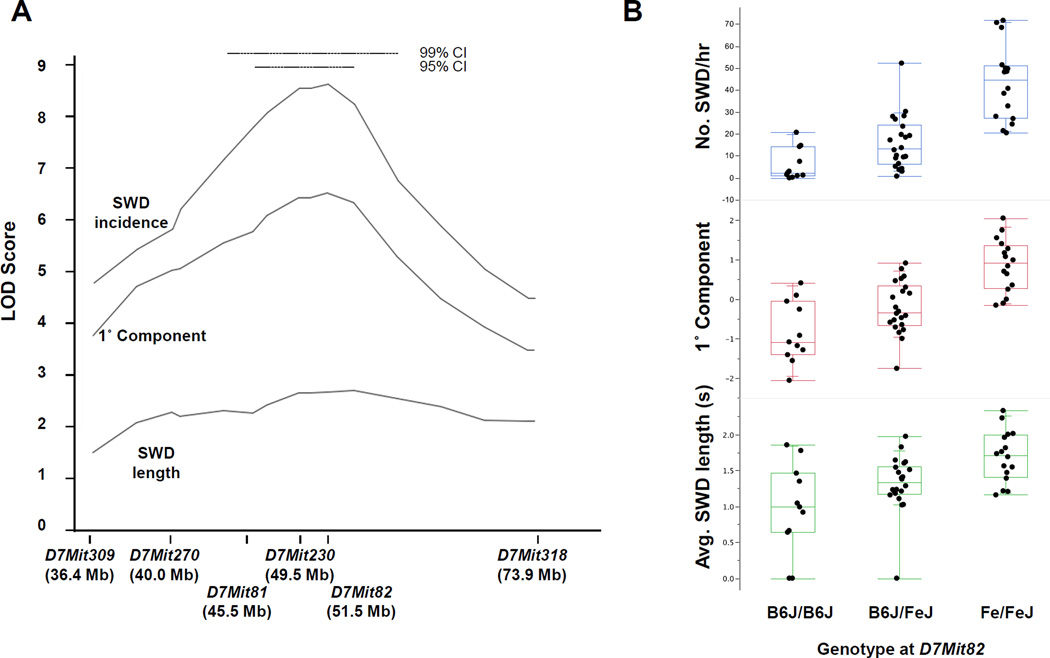
To chromosomally map modifier loci, a genome scan was done by genotyping backcross mice using 184 single nucleotide polymorphism (SNP) markers roughly evenly spaced across the autosomes and the X chromosome. By using interval mapping, we examined associations between the marker genome and SWD phenotype measures (after rank- and normal quantile-transformation). We also examined principal component phenotypes derived from the primary SWD measures to ensure fuller exploration of phenotype space and used permutation shuffling of phenotype and genotype to determine experiment-wise significance thresholds. For SWD length, a single highly significant peak was observed for Chromosome 7 (Fig 9A), with a LOD score of almost 5.0. No other significant peaks were observed, although a suggestive association was seen on Chr 1 (Fig 9A). The Chr 7 peak was not as pronounced for SWD incidence, however, although incidence did show a suggestive peak on Chr 5 (Fig 9B). Examination of the 1° principal component of SWD incidence and length, i.e. for effects that these trait measures have in common, primarily continued to support the Chr 7 association only, but this derivative trait also suggested a more minor association on Chr 2 (Fig 9C). The 2° principal component did not show any significant or suggestive associations (not shown). We also carried out pairwise marker scans to test for two-locus epistasis between segregating loci, but did not identify any significant associations – not a surprise given the small sample size of the backcross.
To confirm the effect of the Chr 7 modifier locus, provisionally named S8Jam1FeJ (Scn8a8J absence seizure modifier 1, FeJ allele), we used marker-assisted selective breeding to place the FeJ-derived 95% confidence interval onto the B6J strain background. At the ninth and tenth generation, we performed EEG on 49 Scn8a8J intercross mice (i.e. N9F1 or N10F1) segregating the S8Jam1 region of Chr 7, on a predominantly B6J genetic background – similar to the initial cross except the FeJ/FeJ homozygous genotype was represented in these test crosses. A strong association was observed between SWD traits and Chr 7 genotypes (Fig 10A), similar to that of the initial cross the effect was stronger for SWD incidence than average length on an otherwise predominantly B6J genetic background. A comparison of genotypes and SWD traits for a marker near the peak map position at 51.5 Mb showed that S8Jam1 has a clear additive effect, with FeJ/FeJ homozygous genotype having the most pronounced SWDs (Fig 10B).
Figure 10.
Location and allelic effect of Scn8a8J modifier S8Jam1 on Chr 7. (A) Chr 7 LOD score curves for SWD traits incidence, length and their 1° principal component in crosses of incipient congenic Scn8a8J/+ mice. The 95% and 99% Bayesian confidence interval (CI) for the peak location is shown at the top. (B) The allelic effect of S8Jam1 is illustrated for each trait using a marker at the peak position (D7Mit82), suggesting additivity. Each dot represents a single animal, grouped in a quartile box plot.
Even after re-examination in these more uniform congenic mice, the 95% confidence interval for S8Jam1FeJ is still large, approximately 8.8 Mb (Fig 10A), including 26 protein coding, and 43 non-coding RNA or unclassified genes (Supplemental File 1). Based on inferences from genome sequence, B6J differs from C3H/HeJ (a substrain closely-related to C3HeB/FeJ), in 13 annotated potentially functional variants (coding missense or indel mutations) spread over 9 genes, only two of which are expressed at appreciable levels in adult brain (Nav2; neuronal navigator 2, Herc2, homologous to the E6-AP domain and RCC1 domain 2; Supplemental File 1). Knockout mice for each of these have overt neurological or neurosensory phenotypes We have also performed RNAseq from thalamus (including reticular thalamus) and somatosensory cortex from FeJ and B6J parent strains. Of 30 genes in the S8Jam1FeJ 95% CI with appreciable adult brain expression, only one – the non-coding RNA gene A230056P14Rik - displayed a potentially significant expression difference (~30% decreased expression in FeJ) although its overall expression in adult brain is modest (Supplemental File 1). Three GABA receptor subunit genes Gabra5, Gabrg3 and Gabrb3 are also located in the 95% CI and are clear functional candidates for absence epilepsy, but none encode obvious deleterious variants or expression differences between C3H and B6J samples. Further work is required to examine the candidacy of Nav2, Herc2, A230056P14Rik or other candidates as the S8Jam1 modifier gene.
Discussion
Absence epilepsy has a complex genetic architecture. A number of variants have been previously identified in the GABA receptor and calcium channels associated with the disorder. In the current study we present a novel mechanism that implicates the voltage-gated sodium channel NaV1.6, as a cause of the disorder. We also investigate a potential epistatic interaction between the NaV1.6V929F mutant, and a variant in NaV1.2V752F, identified on the C3H background strain on which the seizure phenotype is strikingly enhanced.
NaV1.6V929F was identified in an ENU-induced mouse model of absence epilepsy [2]. Functional experiments in the current study suggest an overall large loss of function of the channel due to the mutation. The observation of a shift of activation in one direction and a shift of inactivation in the other is rare, suggesting a disruption of the coupling of activation and inactivation, with major loss of channel function as the net consequence [20] that contributes to the appearance of SWDs and absence.
Although an overall loss in excitation in pyramidal neurons does not seem to provide an intuitive link to an epilepsy phenotype, the functional profile of NaV1.6V929F obtained here is in keeping with previous studies looking at other NaV1.6 mutants. The classic alleles, Scn8amed and Scn8amed-jo, both associate with motor end-plate-disease and exhibit loss of function [21, 22]. Papale et al [2] described SWDs in both the Scn8amed and Scn8amed-jo mice suggesting a causative role in absence for both these alleles as well.
When the NaV1.6V929F mutation is expressed on the C3H background that carries the NaV1.2V752F allele, mice exhibit increased incidence of SWDs suggesting that interaction of NaV1.6V929F and NaV1.2V752F at the AIS of pyramidal neurons is a potential disease mechanism. Our functional experiments suggest an overall gain of function of NaV1.2V752F which does not provide an intuitive interpretation to the overall impact on excitability.
Investigating the interaction further the modelling results demonstrated a decrease in excitability of the pyramidal neuron in the presence of NaV1.6V929F, and an increase in excitability in the presence of the NaV1.2V752F, which would be expected given their individual biophysical profiles. Interestingly, when the two variants are expressed together in the model the output is dominated by the properties of NaV1.2V752F due to “functional silencing” of NaV1.6V929F. Thus, an animal that harbours NaV1.2V752F would not experience a functional change at the AIS when the disease-causing NaV1.6V929F is also present, suggesting that the AIS is not the site of biological interaction for the two genes, in direct contrast to the intuitive interpretation that two mutations in the same neuronal sub compartment are the likely site of biological epistasis. This highlights the need for caution when interpreting the results of functional studies in epilepsy using subjective analysis, especially so in the case of candidate gene approaches.
A prediction of this modelling is that there should also be no genetic interaction between NaV1.6V929F and NaV1.2V752F. The more thorough genetic analysis undertaken to map the modifiers of Scn8a-associated SWD found only a single locus on Chr 7, with a short list of candidate genes. As none of these were obviously related to Nav1.2 function or expression, genetic mapping rules out NaV1.2V752F, as a modifier of Scn8a-associated SWDs, consistent with our computational prediction.
NaV1.6 has diverse and complex roles in epilepsy. Gain of function mutations cause convulsive seizures, as shown in a patient with infantile epileptic encephalopathy and SUDEP [23] and implied by recent identification of dominant variants in patients with epileptic encephalopathy [24] and Lennox-Gastaut syndrome [25]. Such effects may well be related to function in the pyramidal neuron AIS, as NaV1.6 expression is significantly higher in epileptic mice deficient for CELF4, an RNA binding protein that binds to Scn8a mRNA [26]. Conversely, NaV1.6 loss of function mutations cause absence seizures and are protective against convulsive seizures [27]. These divergent effects between seizure types are presumably due to independent roles of Nav1.6 in separate epileptic networks. One possibility for SWD and absence seizures is NaV1.6 expression in interneurons, as shown in the spinal cord [11], Purkinje neurons [10], retinal ganglion cells [28] and the neocortex [7, 8]. A common observation is that NaV1.6 expresses in the distal AIS of inhibitory neurons, with NaV1.1 concentrated more proximally. Expression of the loss of function NaV1.6V929F mutant in interneurons would cause disinhibition, providing an intuitive explanation as to how the NaV1.6 loss of function mutations give rise to SWDs [1, 29].
If the interneuron is where NaV1.6V929F exerts its dominant effect in absence seizures, it is intriguing that pyramidal cell excitability, in the modelling results, can be pushed to a more excitable (in the presence of NaV1.6V929F and NaV1.2V752F: the C3H background), and less excitable (in the presence of NaV1.6V929F alone: the C57 background) firing pattern compared to WT with no apparent impact on the absence phenotype in vivo. One interpretation of this result is that the absence circuitry may be more sensitive to changes in interneuron properties than pyramidal cell properties, that is, there is an operational window in which we can modulate the excitability of pyramidal cells and have no affect on SWD generation. It would be interesting to look at other excitability measures and phenotypes in vivo, such as febrile seizures in the C3H animals, to try and elucidate an impact of the NaV1.2V752F variant, and the circuits that may be affected.
In conclusion, we demonstrate a loss of function mechanism in the NaV1.6 subunit that gives rise to absence seizures. Although a tempting modifier interaction with a NaV1.2 variant yielded a negative result, it gave us some clues as to how the NaV1.6V929F mutant may exert its effect. A lack of impact of NaV1.6V929F on AIS firing in pyramidal cells suggests alternative models for absence seizure generation in this model, the most attractive being an effect in inhibitory neurons leading to disinhibition. This study also reveals limitations of candidate gene predictions – even with tempting variants - without genetic mapping support, while highlighting the value of combining physiology and genetics to inform and direct each other.
Supplementary Material
Acknowledgements
We thank Verity Letts for help with EEG recordings and advice, and Joanne Smith for animal husbandry. Supported by NHMRC program grant 400121 and NMHRC fellowship 1005050 to SP. The Florey Institute of Neuroscience and Mental Health is supported by Victorian State Government infrastructure funds. WNF and scientific cores at The Jackson Laboratory was supported by grants from the NIH (R01 NS031348, P30 CA034186, respectively). SD-H and SGW are supported by grants from the Dept. of Veterans Affairs, USA.
Footnotes
Disclosure of Conflicts of Interest
None of the authors has any conflict of interest to disclose.
References
- 1.Yalcin O. Genes and molecular mechanisms involved in the epileptogenesis of idiopathic absence epilepsies. Seizure. 2012;21(2):79–86. doi: 10.1016/j.seizure.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 2.Papale LA, Beyer B, Jones JM, et al. Heterozygous mutations of the voltage-gated sodium channel SCN8A are associated with spike-wave discharges and absence epilepsy in mice. Hum. Mol. Genet. 2009;18(9):1633–1641. doi: 10.1093/hmg/ddp081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oliva M, Berkovic SF, Petrou S. Sodium channels and the neurobiology of epilepsy. Epilepsia. 2012;53(11):1849–1859. doi: 10.1111/j.1528-1167.2012.03631.x. [DOI] [PubMed] [Google Scholar]
- 4.Kress GJ, Dowling MJ, Eisenman LN, et al. Axonal sodium channel distribution shapes the depolarized action potential threshold of dentate granule neurons. Hippocampus. 2010;20(4):558–571. doi: 10.1002/hipo.20667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whitaker W, Faull R, Waldvogel H, et al. Localization of the type VI voltage-gated sodium channel protein in human CNS. Neuroreport. 1999;10(17):3703–3709. doi: 10.1097/00001756-199911260-00044. [DOI] [PubMed] [Google Scholar]
- 6.Hu W, Tian C, Li T, et al. Distinct contributions of Na(v)1.6 and Na(v)1.2 in action potential initiation and backpropagation. Nat. Neurosci. 2009;12(8):996–1002. doi: 10.1038/nn.2359. [DOI] [PubMed] [Google Scholar]
- 7.Lorincz A, Nusser Z. Cell-type-dependent molecular composition of the axon initial segment. J. Neurosci. 2008;28(53):14329–14340. doi: 10.1523/JNEUROSCI.4833-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogiwara I, Miyamoto H, Morita N, et al. Na(v)1.1 localizes to axons of parvalbuminpositive inhibitory interneurons: a circuit basis for epileptic seizures in mice carrying an Scn1a gene mutation. J. Neurosci. 2007;27(22):5903–5914. doi: 10.1523/JNEUROSCI.5270-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Wart A, Trimmer JS, Matthews G. Polarized distribution of ion channels within microdomains of the axon initial segment. J. Comp. Neurol. 2007;500(2):339–352. doi: 10.1002/cne.21173. [DOI] [PubMed] [Google Scholar]
- 10.Xiao M, Bosch MK, Nerbonne JM, et al. FGF14 localization and organization of the axon initial segment. Mol. Cell. Neurosci. 2013;56C:393–403. doi: 10.1016/j.mcn.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duflocq A, Le Bras B, Bullier E, et al. Nav1.1 is predominantly expressed in nodes of Ranvier and axon initial segments. Mol. Cell. Neurosci. 2008;39(2):180–192. doi: 10.1016/j.mcn.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 12.Rush AM, Dib-Hajj SD, Waxman SG. Electrophysiological properties of two axonal sodium channels, Nav1.2 and Nav1.6, expressed in mouse spinal sensory neurones. J Physiol. 2005;564(Pt 3):803–815. doi: 10.1113/jphysiol.2005.083089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu R, Thomas EA, Gazina EV, et al. Generalized epilepsy with febrile seizures plusassociated sodium channel beta1 subunit mutations severely reduce beta subunit-mediated modulation of sodium channel function. Neurosci. 2007;148(1):164–174. doi: 10.1016/j.neuroscience.2007.05.038. [DOI] [PubMed] [Google Scholar]
- 14.Herzog RI, Liu C, Waxman SG, et al. Calmodulin binds to the C terminus of sodium channels Nav1.4 and Nav1.6 and differentially modulates their functional properties. J. Neurosci. 2003;23(23):8261–8270. doi: 10.1523/JNEUROSCI.23-23-08261.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.John VH, Main MJ, Powell AJ, et al. Heterologous expression and functional analysis of rat Nav1.8 (SNS) voltage-gated sodium channels in the dorsal root ganglion neuroblastoma cell line ND7-23. Neuropharmacology. 2004;46(3):425–438. doi: 10.1016/j.neuropharm.2003.09.018. [DOI] [PubMed] [Google Scholar]
- 16.Zhou X, Dong XW, Crona J, et al. Vinpocetine is a potent blocker of rat NaV1.8 tetrodotoxin-resistant sodium channels. J. Pharmacol. Exp. Ther. 2003;306(2):498–504. doi: 10.1124/jpet.103.051086. [DOI] [PubMed] [Google Scholar]
- 17.Wimmer VC, Reid CA, Mitchell S, et al. Axon initial segment dysfunction in a mouse model of genetic epilepsy with febrile seizures plus. J Clin Investig. 2010;120(8):2661–2671. doi: 10.1172/JCI42219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paz JT, Bryant AS, Peng K, et al. A new mode of corticothalamic transmission revealed in the Gria4(−/−) model of absence epilepsy. Nat. Neurosci. 2011;14(9):1167–1173. doi: 10.1038/nn.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Constantinople CM, Bruno RM. Deep cortical layers are activated directly by thalamus. Science. 2013;340(6140):1591–1594. doi: 10.1126/science.1236425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rhodes TH, Vanoye CG, Ohmori I, et al. Sodium channel dysfunction in intractable childhood epilepsy with generalized tonic-clonic seizures. J Physiol. 2005;569(Pt 2):433–445. doi: 10.1113/jphysiol.2005.094326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kohrman DC, Harris JB, Meisler MH. Mutation detection in the med and medJ alleles of the sodium channel Scn8a. Unusual splicing due to a minor class AT-AC intron. J. Biol. Chem. 1996;271(29):17576–17581. doi: 10.1074/jbc.271.29.17576. [DOI] [PubMed] [Google Scholar]
- 22.Kohrman DC, Smith MR, Goldin AL, et al. A missense mutation in the sodium channel Scn8a is responsible for cerebellar ataxia in the mouse mutant jolting. J. Neurosci. 1996;16(19):5993–5999. doi: 10.1523/JNEUROSCI.16-19-05993.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Veeramah KR, O'Brien JE, Meisler MH, et al. De Novo Pathogenic SCN8A Mutation Identified by Whole-Genome Sequencing of a Family Quartet Affected by Infantile Epileptic Encephalopathy and SUDEP. Am. J. Hum. Genet. 2012;90(3):502–510. doi: 10.1016/j.ajhg.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carvill GL, Heavin SB, Yendle SC, et al. Targeted resequencing in epileptic encephalopathies identifies de novo mutations in CHD2 and SYNGAP1. Nat. Genet. 2013;45(7):825–830. doi: 10.1038/ng.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allen AS, Berkovic SF, Cossette P, et al. De novo mutations in epileptic encephalopathies. Nat. 2013;501(7466):217–221. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wagnon JL, Briese M, Sun W, et al. CELF4 regulates translation and local abundance of a vast set of mRNAs, including genes associated with regulation of synaptic function. PLoS Genet. 2012;8(11):e1003067. doi: 10.1371/journal.pgen.1003067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin MS, Tang B, Papale LA, et al. The voltage-gated sodium channel Scn8a is a genetic modifier of severe myoclonic epilepsy of infancy. Hum. Mol. Genet. 2007;16(23):2892–2899. doi: 10.1093/hmg/ddm248. [DOI] [PubMed] [Google Scholar]
- 28.Van Wart A, Trimmer JS, Matthews G. Polarized distribution of ion channels within microdomains of the axon initial segment. J. Comp. Neurol. 2007;500(2):339–352. doi: 10.1002/cne.21173. [DOI] [PubMed] [Google Scholar]
- 29.Beyer B, Deleuze C, Letts VA, et al. Absence seizures in C3H/HeJ and knockout mice caused by mutation of the AMPA receptor subunit Gria4. Hum. Mol. Genet. 2008;17(12):1738–1749. doi: 10.1093/hmg/ddn064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.