Abstract
In the past two decades, European ash trees (Fraxinus spp.) have been severely damaged due to ash dieback disease, which is caused by the fungal species Hymenoscyphus fraxineus (Chalara fraxinea in the anamorphic stage). Recent molecular phylogenetic and population genetic studies have suggested that this fungus has been introduced from Asia to Europe. During a fungal survey in Korea, H. fraxineus-like apothecia were collected from fallen leaves, rachises, and petioles of Korean ash and Manchurian ash trees. The morphological and ecological traits of these materials are described with the internal transcribed spacer rDNA sequence comparison of H. fraxineus strains collected from Korea, China and Japan.
Keywords: Ash dieback, Chalara fraxinea, Hymenoscyphus fraxineus, Korean ash, Manchurian ash
Ash dieback is a highly infectious fungal disease that is threatening various ash trees (Fraxinus spp.) in Europe. Since ash dieback was first observed in North-East Poland in the early 1990s, it has throughout the continent [1]. Chalara fraxinea T. Kowalski was described as the causal agent of ash dieback [2]. Hymenoscyphus albidus (Gillet) W. Phillips, a common helotialean species in Europe, was suggested to be a teleomorph of C. fraxinea based on their morphological similarity [3]. However, a recent molecular study [4] showed that H. albidus is a species complex that can be subdivided into pathogenic and non-pathogenic groups. In the pathogenic group, a new species, H. pseudoalbidus Queloz, Grünig, Berndt, T. Kowalski, T. N. Sieber & Holdenr, has been designated and suggested as a teleomorph of C. fraxinea [4]. Further cultural and population genetic studies have demonstrated the distinctions between H. albidus and H. pseudoalbidus [5, 6, 7, 8]. Zhao et al. [9] described that the presence or absence of croziers at the base of asci can be a taxonomic key to distinguish these two species. It was recently hypothesized that the causal agent of the sudden outbreak of European ash dieback may have been introduced from East Asia [4, 9]. Zhao et al. [9] found that Japanese materials of Lambertella albida are morphologically and genetically similar to H. pseudoalbidus. Zheng and Zhuang [10] also reported the occurrence of H. pseudoalbidus in China. According to the recent amendment of fungal nomenclature, the correct name for H. pseudoalbidus is H. fraxineus (T. Kowalski) Baral, Queloz & Hosoya [11]. During a fungal biodiversity survey in Korea, Hymenoscyphus species on fallen leaves, rachises and petioles of Korean ash (Fraxinus rhynchophylla Hance, also known as F. chinensis subsp. rhynchophylla (Hance) Hemsl.) and Manchurian ash (F. mandshurica Rupr.) were collected. We performed a detailed morphological examination and sequence comparison with other Hymenoscyphus spp. including the close relatives H. albidus and H. fraxineus (= H. pseudoalbidus).
Apothecia collected on fallen leaves, rachises, and petioles were pressed with clean papers and dried on the laboratory bench. All collected materials have been deposited at the Korea University Herbarium (KUS). Fresh materials were primarily mounted with distilled water for the confirmation of the natural colors of their microstructures. Dried materials were rehydrated in 3~10% aqueous KOH. Amyloid reactions were conducted with Melzer's reagent or Lugol's solution (IKI) without KOH pretreatment. Line drawings were made with the aid of an Olympus BX50 microscope equipped with an Olympus U-DA drawing tube. Measurements were made in distilled water, Congo red, lactophenol-cotton blue, IKI, or Melzer's reagent and are reported as follows: minimum-maximum (length) ×minimum-maximum (width) [mean length ± standard deviation × mean width ± standard deviation, Q (length/width ratio) = average ± standard deviation].
Genomic DNA was extracted from dried apothecia using the methodology described by Lee and Taylor [12]. The concentration and purity of extracted nucleic acids were assessed using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA). PCR was performed in 50 µL reactions that consisted of 39 µL of sterile water, each primer at 0.4 µM, 1 unit of ExTaq DNA Polymerase (TaKaRa, Tokyo, Japan), a dNTP mixture consisting of all four nucleotides, each at a concentration of 2.5 mM, 10× ExTaq buffer containing 20 mM Mg2+, and approximately 100 ng of template DNA. ITS1 and ITS4 primers [13] were used to amplify the internal transcribed spacer (ITS) region of rDNA, including the 5.8S gene. The cycling parameters for ITS amplification included an initial denaturation at 95℃ for 5 min, followed by 35 cycles of denaturation at 95℃ for 1 min, annealing at 58℃ for 1min, and extension at 72℃ for 2min, with a final extension at 72℃ for 10 min. After confirming the PCR products on an agarose/TBE electrophoretic gel (Certified Molecular Biology Agarose; Bio-Rad Laboratories, Madrid, Spain), they were purified using a LaboPass PCR Kit (COSMO Genetech, Seoul, Korea) following the manufacturer's protocols. Sequencing was performed on an automatic sequencer (ABI Prism 377 DNA Sequencer) using the BigDye Cycle Sequencing Kit (version 3.1; Applied Biosystems, Foster City, CA, USA) with the same primers used for the PCR amplifications. The obtained ITS sequences were deposited in GenBank with the accession Nos. KF830851, KF830852, KF830850, and KF830853.
In addition to the newly generated sequence data, 37 fungal ITS sequences were retrieved from GenBank to determine the phylogenetic placement of Korean Hymenoscyphus specimens (Table 1). The obtained sequences were aligned using ClustalW [14] and manually edited using BioEdit ver. 7.0.5.2 [15] when necessary. Ambiguously aligned positions were excluded from the subsequent analysis. Phylogenetic analyses using neighbor-joining (NJ), maximum likelihood (ML), and maximum parsimony (MP) were performed with MEGA ver. 6.06 [16]. In the NJ analysis, the distances between ITS sequences were calculated using the Kimura 2-parameter model. In the ML analysis, the general time-reversible model with a gamma-distributed substitution rate and proportion of invariant sites (GTR + I + G) was separately applied to each gene for the nucleotide substitution model. An MP heuristic search was performed with 5,000 replications, each with 500 random sequence additions, and branch swapping by tree bisection-reconnection. All nucleotide substitutions were equally weighted and unordered with gaps treated as missing data. The nodal support for the individual branches was estimated by bootstrapping (BS) using 1,000 replicates [17].
Table 1.
Sequences used in this study
Previously, Hymenoscyphus albidus and H. fraxineus were regarded as morphologically indistinguishable until H. O. Baral (personal communication) and Zhao et al. [9] demonstrated differences in their ascal bases. In Korea, the apothecial discomycetes collected on the fallen leaves, rachises, and petioles of Fraxinus spp. were confirmed as H. albidus based on earlier descriptions [18, 19]. However, during the re-examination of Korean materials, we observed that all samples possessed distinct croziers at the ascal bases (Fig. 1). We report H. fraxineus for the first time in Korea and describe its morphological and ecological traits.
Fig. 1.
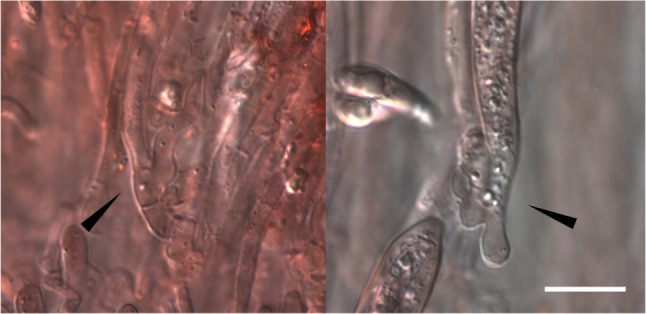
Distinct croziers observed at the ascal bases of Hymenoscyphus fraxineus material (KUS-F52255). Mounted in Congo red solution (scale bar = 10 µm).
Hymenoscyphus fraxineus (T. Kowalski) Baral, Queloz & Hosoya (Figs. 1, 2, 3)
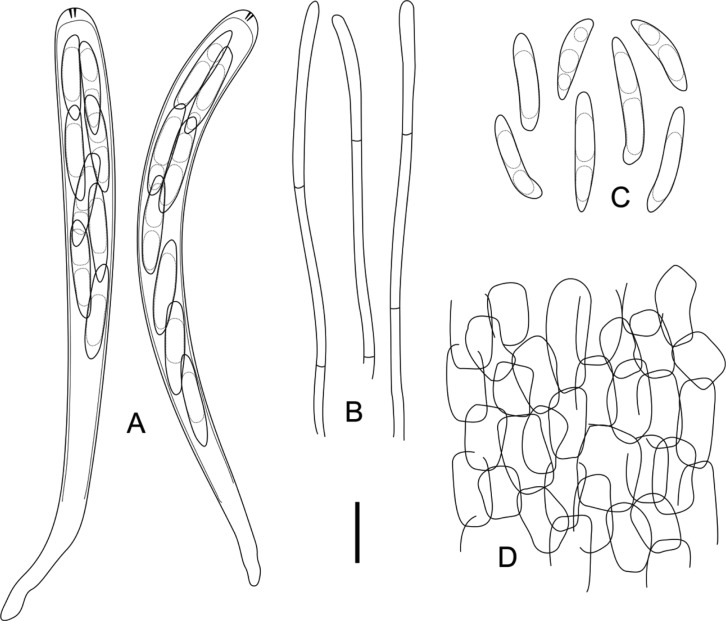
Fig. 2.

Hymenoscyphus fraxineus. A, Fruiting bodies on fallen rachis of F. rhynchophylla; B, C, Upper and side views of cupulate apothecia (scale bars: A = 10 mm, B, C = 1 mm).
Fig. 3.
Hymenoscyphus fraxineus. A, 8-spored asci with MLZ+ apical pores; B, Cylindric paraphyses; C, Ellipticfusoid ascospores containing several oil drops; D, Ectal excipulum composed of prismatic to angular cells (scale bar: A~D = 10 µm).
≡ Chalara fraxinea T. Kowalski, For. Pathol. 36: 264 (2006). = Hymenoscyphus pseudoalbidus Queloz, Grünig, Berndt, T. Kowalski, T. N. Sieber & Holdenr., For. Pathol. 41: 140 (2011). Apothecia gregarious, superficial, seated on a distinct stipe. Receptacle shallow cupulate to discoid, ivory to white or cream when fresh, turning light brown when dry. Disc up to 2 mm in diameter, plano-concave, concolorous with the receptacle. Stipe cylindric, slightly tapering towards the base, concolorous with the receptacle, blackened near the base, up to 1 mm long. Ectal excipulum composed of prismatic to angular cells, individual cells hyaline to light yellowish, thin-walled, 8~18 × 6~10 µm, decreasing in size near the apothecial margin. Asci arising from distinct croziers, cylindric, hyaline, 8-spored, apex conical, apical pore blued in IKI without KOH pretreatment, 85~105 × 7~9 µm (95.6 ± 3.21 × 8.5 ± 0.33 µm, n = 35). Ascospores elliptic-fusoid to clavate, scutuloid, hyaline, biseriate, occupying the upper 1/2~3/4 of the entire asci length, 15~21 × 3~ 4 µm (19.3 ± 0.75 × 3.6 ± 0.13 µm, Q = 5.03 ± 0.13, n = 80). Paraphyses cylindric, hyaline, septate, unbranched, 2~2.5 µm wide, tips slightly swollen up to 3 µm, slightly exceeding the asci.
Specimens examined
On leaves, rachises, and petioles of Korean ash (Fraxinus rhynchophylla)-Korea: Danyang, Darian Valley in Mt. Sobaek National Park, 36°57'46'' N, 128°25'24'' E, alt. 400 m, 14 Jul 2007, KUS-F51679 and KUS-F51684; Pyeongchang, Mt. Taegi, 37°35'25'' N, 128°16'47'' E, alt. 1050 m, 14 Aug 2008, KUS-F52255; Yangpyeong, Seolmaejae Recreation Forest, 37°33'14'' N, 127°30'12'' E, alt. 300 m, 15 Jul 2009, KUS-F52542; Yangpyeong, Saneum Recreation Forest, 37°36'9'' N, 127°35'54'' E, alt. 200m, 15 Jul 2009, KUS-F52552; Hoengseong, Cheongtaesan Recreation Forest, 37°31'32'' N, 128°17'34'' E, alt. 850 m, 19 Aug 2009, KUS-F52610; Hoengseong, Supchaewon, 37°23'25'' N, 128°17'34'' E, alt. 1,050 m, 20 Aug 2009, KUS-F52613; on damp rotting leaves of Manchurian ash (Fraxinus mandshurica)-Korea: Hoengseong, Supchaewon, 37°23'24'' N, 128°17'39'' E, alt. 1,100 m, 23 Sep 2008, KUS-F52355.
Morphological characteristics of the Korean materials were in agreement with the previous descriptions of H. albidus [18, 19]. Based on the croziers at the ascal bases that were first noticed by Zhao et al. [9], this fungus was identified as H. fraxineus. Korf [20] and Hosoya et al. [19] regarded it as a member of Lambertella (Rutstroemiaceae, Helotiales) based on the existence of stromatized patches on the substrates. In a recent molecular study, however, the genus Lambertella was determined to be a polyphyletic group, and Hymenoscyphus was suggested to be the most appropriate genus for this taxon [9]. Hymenoscyphus caudatus (P. Karst.) Dennis is also similar to H. fraxineus due to its white stipitate apothecia and scutuloid ascospores as well as its foliicolous habit, but differs in the small size of its apothecia (up to 1.5 mm diameter) [18, 21]. Hymenoscyphus ginkgonis J. G. Han & H. D. Shin is another similar species possessing ascospores of a similar shape and size, but is distinguished by its substrate-specificity on ginkgo (Ginkgo biloba L.) leaves and the presence of deep violet pigments in its paraphyses [22]. Hymenoscyphus albidoides H. D. Zheng & W. Y. Zhuang recently described from China is distinct from this fungus owing to an outer covering composed of 1~3 parallel hyphal layers, cuboid crystals in the basal tissue of the stipe, and its occurrence on the veins of rotten leaves of Picrasma quassioides (D. Don) Benn. [10].
Korean materials of H. fraxineus were usually isolated from Korean ash (F. rhynchophylla), which is a new host for this fungus. However, ash dieback symptoms were not detected when we collected these materials, indicating the possibility that Korean ash has an inherited resistance to this pathogen. Korean ash is phylogenetically closely related to manna ash (F. ornus L.) [23], which is known to be less susceptible to ash dieback than other European ash species [24]. In China and Japan, H. fraxineus has been reported on Manchurian ash (F. mandshurica), which is widely distributed in Eastern Asia [9, 10].
The analyzed dataset included 41 Hymenoscyphus spp. and two outgroup taxa, Cudoniella clavus (Helotiaceae) and Sclerotinia sclerotiorum (Sclerotiniaceae) (Table 1). The final alignment contained 477 nucleotide positions, of which 333 were invariant and 94 were parsimony-informative characters. The ML analysis yielded an optimal tree with a best log-likelihood of -1,795.14. Ten equally most-parsimonious trees of 251 steps were obtained by the MP analysis, with a consistency index of 0.6972 and an retention index of 0.8403. Since the inferred NJ, ML, and MP trees were topologically congruent, the NJ tree is represented with the bootstrap values of NJ, ML, and MP analyses above the corresponding branches (Fig. 4). In the inferred trees, the Hymenoscyphus spp. formed a single group with moderate support values (NJ BS = 77, ML BS = 78, and MP BS = 72), reflecting the broad circumscription of the genus as well as the possibility of the heterogeneous assemblage of diverse fungal species. As shown in previous studies, H. albidus and H. fraxineus were morphologically similar but clearly separated in the phylogeny [4, 9, 10]. Korean materials (KUS-F52255, KUS-F52355, KUS-F52610, and KUS-F52613) were grouped with other H. fraxineus (= Chalara fraxinea) strains with strong nodal supports (NJ BS = 99, ML BS = 99, and MP BS = 95). Hymenoscyphus albidoides formed a sister-group with H. albidus and H. fraxineus.
Fig. 4.
Neighbor-joining (NJ) tree of Hymenoscyphus spp. based on the internal transcribed spacer rDNA sequences. The bootstrap values greater than 60% are indicated above the corresponding branches (NJ BP/ML BP/MP BP). The newly generated sequences in this study are shown in bold. The scale bar indicates the proportion of sites changing along each branch. BP, bootstrap proportion; ML, maximum likelihood; MP, maximum parsimony.
Zhao et al. [9] investigated the intraspecific variation in H. fraxineus (= H. pseudoalbidus), and found that the ITS sequence of Asian strains concurred with that of European strains except for two nucleotide positions, 83 (G instead of a gap) and 124 (C instead of T) (Table 2), reflecting their close relation. Among the Asian strains, positions 155, 379, and 443 showed some variations (Table 2). The obtained ITS sequences of Korean isolates were nearly identical, irrespective of the host. KUS-F52255 showed a degenerate base R (indicating A or G bases) instead of G at the nucleotide position 258 (Table 2), but it is likely to be an incorrect base call since all known ITS sequences of H. fraxineus, H. albidus, and even the close relative H. albidoides show the same nucleotide G at this position.
Table 2.
Comparison of ITS sequences from Hymenoscyphus albidus and H. fraxineus strains used in this study
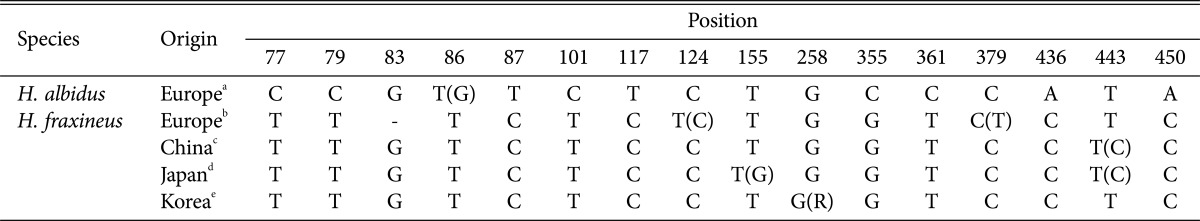
Numbering from H. albidus 90803.16 (GU586879). The second dominant base is indicated by parentheses in case there is a variation. '-' indicates gap.
aGU586877, GU586879, GU586880, GU586882, GU586884, GU586887, GU586893, GU586895, and GU586899.
bFJ429386, FJ597975, GU586901, GU586904, GU586905, and GU586917.
cKF188725, KF188726, KF188727.
Based on the field observations and artificial inoculation studies showing Asian ash species tend to be immune to ash dieback disease [24, 25], Asia is considered a possible origin of the ash dieback pathogen [4, 9]. This hypothesis was verified by subsequent population genetic analyses using polymorphic microsatellite markers showing that European H. fraxineus has limited genetic diversity among and within populations [5, 6]. In contrast, Zhao et al. [9] found that Japanese H. fraxineus possess more genetic variation compared to European populations. The ITS sequences of Japanese H. fraxineus is most variable among the Asian materials, while those of Chinese and Korean materials are relatively conserved (Table 2) although more population sampling from various sites is necessary.
ACKNOWLEDGEMENTS
This work was supported by a research grant for a research program for agricultural science and technology of the National Institute of Horticultural and Herbal Science (PJ008418 and PJ010279), Rural Development Administration, Republic of Korea.
References
- 1.Timmermann V, Børja I, Hietala AM, Kirisits T, Solheim H. Ash dieback: pathogen spread and diurnal patterns of ascospore dispersal, with special emphasis on Norway. EPPO Bull. 2011;41:14–20. [Google Scholar]
- 2.Kowalski T. Chalara fraxinea sp. nov. associated with dieback of ash (Fraxinus excelsior) in Poland. For Pathol. 2006;36:264–270. [Google Scholar]
- 3.Kowalski T, Holdenrieder O. The teleomorph of Chalara fraxinea, the causal agent of ash dieback. For Pathol. 2009;39:304–308. [Google Scholar]
- 4.Queloz V, Grünig CR, Berndt R, Kowalski T, Sieber TN, Holdenrieder O. Cryptic speciation in Hymenoscyphus albidus. For Pathol. 2011;41:133–142. [Google Scholar]
- 5.Bengtsson SB, Vasaitis R, Kirisits T, Solheim H, Stenlid J. Population structure of Hymenoscyphus pseudoalbidus and its genetic relationship to Hymenoscyphus albidus. Fungal Ecol. 2012;5:147–153. [Google Scholar]
- 6.Gross A, Grünig CR, Queloz V, Holdenrieder O. A molecular toolkit for population genetics investigations of the ash dieback pathogen Hymenoscyphus pseudoalbidus. For Pathol. 2012;42:252–264. [Google Scholar]
- 7.Kirisits T, Dämpfle L, Kräutler K. Hymenoscyphus albidus is not associated with an anamorphic stage and displays slower growth than Hymenoscyphus pseudoalbidus on agar media. For Pathol. 2013;43:386–389. [Google Scholar]
- 8.Husson C, Scala B, Caël O, Frey P, Feau N, Ioos R, Marçais B. Chalara fraxinea is an invasive pathogen in France. Eur J Plant Pathol. 2011;130:311–324. [Google Scholar]
- 9.Zhao YJ, Hosoya T, Baral HO, Hosaka K, Kakishima M. Hymenoscyphus pseudoalbidus, the correct name for Lambertella albida reported from Japan. Mycotaxon. 2012;122:25–41. [Google Scholar]
- 10.Zheng HD, Zhuang WY. Hymenoscyphus albidoides sp. nov. and H. pseudoalbidus from China. Mycol Prog. 2014;13:625–638. [Google Scholar]
- 11.Baral HO, Queloz V, Hosoya T. Hymenoscyphus fraxineus, the correct scientific name for the fungus causing ash dieback in Europe. IMA Fungus. 2014;5:79–80. doi: 10.5598/imafungus.2014.05.01.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee S, Taylor J. Isolation of DNA from fungal mycelia and single spores. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego: Academic Press; 1990. pp. 282–287. [Google Scholar]
- 13.White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. New York: Academic Press; 1990. pp. 315–322. [Google Scholar]
- 14.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 16.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 18.Dennis RW. A revision of the British Helotiaceae in the herbarium of the Royal Botanic Gardens, Kew, with notes on related European species. Mycol Pap. 1956;62:1–216. [Google Scholar]
- 19.Hosoya T, Otani Y, Furuya K. Materials for the fungus flora of Japan (46) Trans Mycol Soc Jpn. 1993;34:429–432. [Google Scholar]
- 20.Korf RP. New combination and a new name for discomycetes illustrated by Boudier in the Icones Mycologicae. Mycotaxon. 1982;14:1–2. [Google Scholar]
- 21.Lizoň P. The genus Hymenoscyphus (Helotiales) in Slovakia, Czechoslovakia. Mycotaxon. 1992;45:1–59. [Google Scholar]
- 22.Han JG, Shin HD. Hymenoscyphus ginkgonis sp. nov. growing on leaves of Ginkgo biloba. Mycotaxon. 2008;103:189–195. [Google Scholar]
- 23.Wallander E. Systematics of Fraxinus (Oleaceae) and evolution of dioecy. Plant Syst Evol. 2008;273:25–49. [Google Scholar]
- 24.Kirisits T, Matlakova M, Mottinger-Kroupa S, Cech TL, Halmschlager E. The current situation of ash dieback caused by Chalara fraxinea in Austria. In: Doğmuş-Lehtijarvi T, editor. Proceedings of the Conference of IUFRO Working Party 7.02.02; 2009 May 11-16; Eğirdir, Turkey. SDU Faculty of Forestry Journal, Serial A, Special Issue; 2009. pp. 97–119. [Google Scholar]
- 25.Drenkhan R, Hanso M. New host species for Chalara fraxinea. New Dis Rep. 2010;22:16. [Google Scholar]