Abstract
Pseudallescheria boydii KNU13-2 was isolated from crop field soil and identified by analysis of internal transcribed spacer regions of rDNA and morphological characteristics. In the literature, P. boydii has been mentioned as a human pathogen. This is the first record of P. boydii isolated from crop field soil in Korea.
Keywords: Fungi, Molecular identification, Morphology, Pathogenic fungi, Pseudallescheria
The fungal pathogen Pseudallescheria boydii (Shear) McGinnis et al., 1982 [anamorph, Scedosporium apiospermum (Sacc.) Sacc.] has special public health importance because it causes localized as well as disseminated infections in both immunocompromised and immunocompetent hosts [1, 2, 3]. The increasing number of P. boydii infections could be associated with the increasing occurrence of this fungus in the environment. It has been shown that P. boydii is much more abundant in urban and industrial areas and in agricultural soils than in habitats with low human activity [4]. A few studies have shown that different strains of P. boydii can degrade aromatic hydrocarbons [5, 6] and produce stable fungistatic substances [7].
During an investigation of the fungal community in crop field soil, P. boydii was encountered and identified by its morphological and molecular characteristics. This finding is of particular interest because this species is a potential human and animal pathogen and has not previously been reported in Korea. In this report, we describe the isolation and identification of P. boydii from soil, discuss the significance of this discovery, and provide an explanation for the occurrence of this fungus in soil.
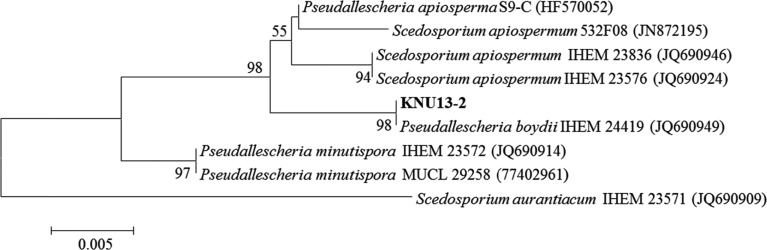
Fungi were isolated from crop field soil collected at a depth of 0~15 cm in Jeongseon province, South Korea. The fungi were isolated by conventional dilution and grown for 7 days at 25℃ on potato dextrose agar (PDA; Difco Laboratories, Detroit, MI, USA) supplemented with chloramphenicol (100 µg/mL). Pure cultures were maintained in sterile distilled water at 4℃. Genomic DNA of strain KNU13-2 was extracted using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) following the manufacturer's instructions. The internal transcribed spacer (ITS) regions, including the 5.8S rRNA gene, were amplified with the primers ITS1 and ITS4 [8]. The amplified PCR product was purified using a QIAquick PCR purification Kit (Qiagen, Valencia, CA, USA) following the manufacturer's instructions. The PCR product was sequenced with an ABI Prism 3730 DNA analyzer (Applied Biosystems, Foster City, CA, USA). The sequence was compared with reference ITS1~ITS4 rDNA sequences in GenBank using BLAST analysis (http://www.ncbi.nlm.nih.gov/Blast). The nucleotide sequence reported here has been deposited in GenBank (accession No. KJ921604). The sequences of closely related strains were aligned using the MultAlin program. The phylogenetic analysis was carried out by the neighbor-joining method using MEGA software [9] with the Kimura 2-parameter model. The robustness of the tree shown in Fig. 1 was evaluated using 1,000 bootstrap replications. Scedosporium aurantiacum IHEM 23571 was used as the outgroup.
Fig. 1.
Neighbor-joining phylogenetic analysis of the partial 18S-ITS1-5.8S-ITS2-28S rDNA sequence of Pseudallescheria boydii KNU13-2 obtained from crop field soil in Korea. The sequence obtained in the study is shown in boldface. Numerical values (> 50) on branches are the percentage of 1,000 bootstrap replicates that support the branch. Scedosporium aurantiacum IHEM 23571 was used as the outgroup. The scale represents the number of substitutions per site.
The ITS sequence of KNU13-2 matched that of P. boydii IHEM 23827 (GenBank accession No. JQ690949) with 100% similarity. The distribution of P. boydii IHEM 23827 in French patients with cystic fibrosis has been described [10]. P. boydii encountered in the context of cystic fibrosis is capable of chronically colonizing the respiratory tract of patients [10]. Based on similarity analysis, the fungus KNU13-2 was identified as P. boydii. Phylogenetic analysis revealed that the isolate was grouped with reference isolates of P. boydii with 98% bootstrap support (Fig. 1). These results indicate that isolate KNU13-2 is P. boydii.
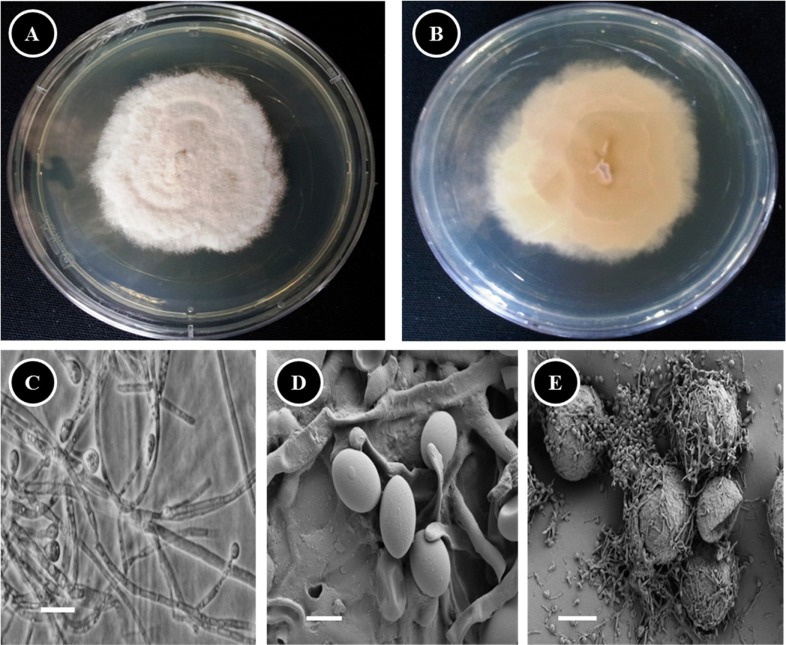
One of the most typical features of P. boydii is its ability to develop sexual structures on routine culture media. The presence of spherical ascomata (cleistothecia) and fusiform or ellipsoidal ascospores allows easy differentiation of this species from other species of Scedosporium, including Scedosporium prolificans [11]. To confirm the molecular result, the morphology of isolate KNU13-2 was compared with previous descriptions of P. boydii [12, 13]. Photomicrographs were taken with a Kodak 14n digital camera (Tokyo, Japan) attached to a compound microscope or a scanning electron microscope. A dense conidial suspension of isolate KNU13-2 was inoculated on PDA in 9-cm plastic petri dishes and grown for 2 wk at 25℃. The fungus produced two synanamorphs, P. boydii and S. apiospermum, which fit the description of P. boydii and S. apiospermum on PDA. The morphological characteristics of the identified species are summarized in Table 1. Colonies on PDA attained diameters of 13~16 (~18) mm after 7 days growth at 25℃. Colonies were wooly to cottony, with moderate sporulation, white to greyish or dark brown, reverse pale with brownish zones (Fig. 2A and 2B). Hyphae were hyaline and septate. No exudates were observed on the upper or lower surface. Conidiophores were simple, bearing annellides of varying lengths, and exhibited little differentiation from the vegetative hyphae (Fig. 2C). Conidia were ovoid with truncate bases, born singly or in small clusters at the ends of the conidiophores or from short annellidic necks (Fig. 2C and 2D). Large (50 to 230 µm) multicellular sexual fruiting bodies (cleistothecia) were observed after 2 wk of incubation (Fig. 2E). The morphological characteristics of the isolate agreed with the description of P. boydii. In conclusion, based on the phylogenetic analysis and the morphological characteristics, strain KNU13-2 was identified as P. boydii.
Table 1.
Comparison of morphological characteristics of the study isolate with respect to reported Pseudallescheria boydii characteristics
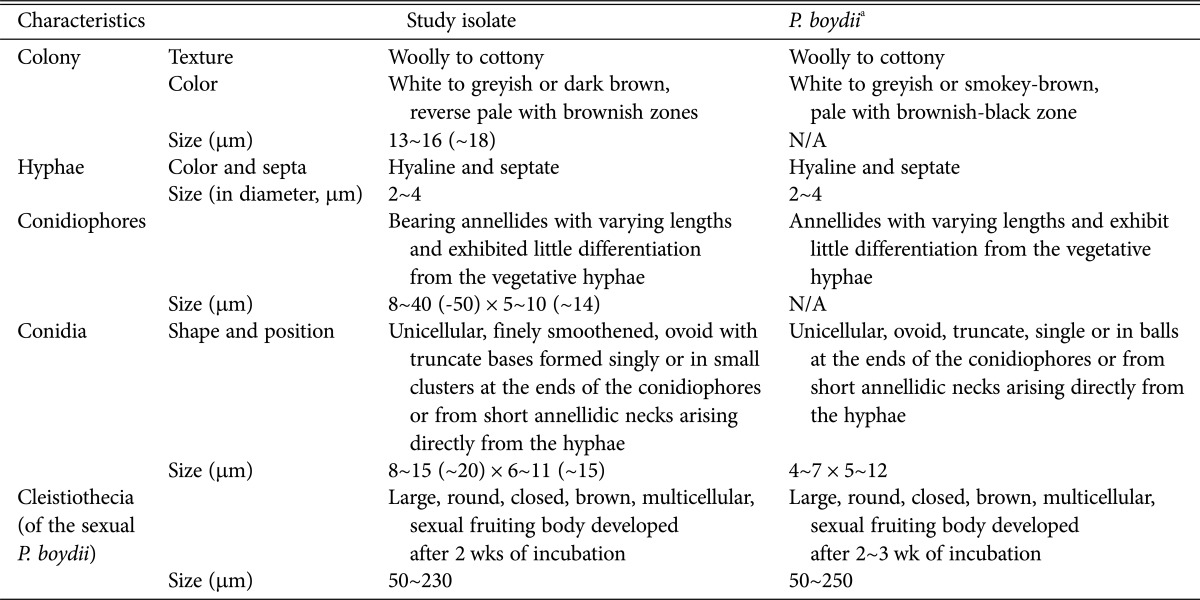
Fig. 2.
Morphology of Pseudallescheria boydii KNU13-2 observed using a compound microscope and scanning electron microscope (SEM). A, Front side of colony; B, Reverse side of colony; C, Apical part of hyphae of the Scedosporium apiospermum anamorph producing conidia (compound microscope images); D, Conidia (SEM image); E, Ascoma (cleistothecium) (SEM image) (scale bars: C = 30 µm, D = 3 µm, E = 40 µm).
Ko et al. [14] reported that P. boydii possesses a strong competitive saprophytic ability and is widespread in soil. Thus, the occurrence of P. boydii in crop field soils shows the importance of monitoring fungi in these soils to evaluate the hygienic quality of organic amendments and to establish soil management recommendations for farmers. Though it is known that P. boydii has the ability to degrade aromatic hydrocarbons [5, 6] and produce stable fungistatic substances [7], more studies on the pathogenicity of this fungus are needed.
ACKNOWLEDGEMENTS
This work was supported by the National Institute of Biological Resources under the Ministry of Environment, Republic of Korea, as part of a project to survey and excavate Korean indigenous fungal species.
References
- 1.McGinnis MR, Padhye AA, Ajello L. Pseudallescheria boydii (Shear) McGinnis, A.A. Padhye & Ajello, 1982. Mycotaxon. 1982;14:97. [Google Scholar]
- 2.Rainer J, de Hoog GS, Wedde M, Gräser Y, Gilges S. Molecular variability of Pseudallescheria boydii, a neurotropic opportunist. J Clin Microbiol. 2000;38:3267–3273. doi: 10.1128/jcm.38.9.3267-3273.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janda-Ulfig K, Ulfig K, Cano J, Guarro J. A study of the growth of Pseudallescheria boydii isolated from sewage sludge and clinical sources on tributyrin, rapeseed oil, biodiesel oil and diesel oil. Ann Agric Environ Med. 2008;15:45–49. [PubMed] [Google Scholar]
- 4.Rainer J, de Hoog GS. Molecular taxonomy and ecology of Pseudallercheria, Petriella and Scedosporium prolificans (Microascaceae) containing opportunistic agents on humans. Mycol Res. 2006;110(Pt 2):151–160. doi: 10.1016/j.mycres.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 5.April TM, Abbott SP, Foght JM, Currah RS. Degradation of hydrocarbons in crude oil by the ascomycete Pseudallescheria boydii (Microascaceae) Can J Microbiol. 1998;44:270–278. doi: 10.1139/w97-152. [DOI] [PubMed] [Google Scholar]
- 6.Prenafeta-Boldú FX, Summerbell R, Sybren de Hoog G. Fungi growing on aromatic hydrocarbons: biotechnology's unexpected encounter with biohazard? FEMS Microbiol Rev. 2006;30:109–130. doi: 10.1111/j.1574-6976.2005.00007.x. [DOI] [PubMed] [Google Scholar]
- 7.Nirma C, Eparvier V, Stien D. Antifungal agents from Pseudallescheria boydii SNB-CN73 isolated from a Nasutitermes sp. termite. J Nat Prod. 2013;76:988–991. doi: 10.1021/np4001703. [DOI] [PubMed] [Google Scholar]
- 8.Govinda Rajulu MB, Thirunavukkarasu N, Babu AG, Aggarwal A, Suryanarayanan TS, Reddy MS. Endophytic Xylariaceae from the forests of Western Ghats, southern India: distribution and biological activities. Mycology. 2013;4:29–37. [Google Scholar]
- 9.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 10.Zouhair R, Rougeron A, Razafimandimby B, Kobi A, Bouchara JP, Giraud S. Distribution of the different species of the Pseudallescheria boydii/Scedosporium apiospermum complex in French patients with cystic fibrosis. Med Mycol. 2013;51:603–613. doi: 10.3109/13693786.2013.770606. [DOI] [PubMed] [Google Scholar]
- 11.Gilgado F, Cano J, Gené J, Guarro J. Molecular phylogeny of the Pseudallescheria boydii species complex: proposal of two new species. J Clin Microbiol. 2005;43:4930–4942. doi: 10.1128/JCM.43.10.4930-4942.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fun with microbiology (What's buggin' you?) Short descriptions and photographs of some photogenic microorganisms overview [Internet] Yuri; [cited 2011 Nov 17]. Available from: http://thunderhouse4-yuri.blogspot.ca/2011/01/pseudallescheria-boydiiscedospori-um.html. [Google Scholar]
- 13.Doctor Fungus. Fungus overview [Internet] Doctor Fungus; [cited 2001 Jan 3]. Available from: http://www.doctorfungus.org/Thefungi/pseudallescheria.php. [Google Scholar]
- 14.Ko WH, Tsou YJ, Ju YM, Hsieh HM, Ann PJ. Production of a fungistatic substance by Pseudallescheria boydii isolated from soil amended with vegetable tissues and its significance. Mycopathologia. 2010;169:125–131. doi: 10.1007/s11046-009-9237-1. [DOI] [PubMed] [Google Scholar]