Abstract
Since their initial discovery in the early 1990s, microRNAs have now become the focus of a multitude of lines of investigation ranging from basic biology to translational applications in the clinic. Previously believed to be of no biological relevance, microRNAs regulate processes fundamental to human health and disease. In diseases of the lung, microRNAs have been implicated in developmental programming, as drivers of disease, potential therapeutic targets, and clinical biomarkers; however, several obstacles must be overcome for us to fully realize their potential therapeutic use. Here, we provide for the clinician an overview of microRNA biology in selected diseases of the lung with a focus on their potential clinical application.
Keywords: microRNA, epigenetics, gene, biomarkers
In the last several years, noncoding RNAs (ncRNAs) have emerged as potential disease-modifying molecules in lung disease. Discovered more than 20 years ago, this family of RNAs, previously considered to be nonfunctional members of the human genome, have proven to be important to fundamental biological processes, including growth, differentiation, programmed cell death, and inflammation (1, 2). Among the large family of ncRNAs, microRNAs (miRs) represent the best-studied member. Often localized to fragile chromosomal regions, these small, 18- to 25-nucleotide (nt), molecules can either induce mRNA degradation or inhibit translation (3). In fact, it is estimated that up to 60% of the human genome may be under the regulation of microRNAs (4). Until recently, the majority of microRNA-based studies had been conducted in cancers; however, we are now observing a rapidly growing body of investigation focused on nonmalignant diseases of many organs, including the lung. The parallels between microRNA deregulation in lung disease and their deregulation during development of the lung are becoming apparent. Several microRNAs critical to lung embryogenesis appear to resurface in expression later in life, thus suggesting their biological role in the development of diseases of the lung. The molecular triggers for this observed deregulation remain a mystery. Increased investigation of fundamental microRNA biology during lung development may serve to inform our understanding of lung disease. We have now seen several microRNA-based studies in lung cancer, pulmonary fibrosis, chronic obstructive pulmonary disease (COPD), asthma, and pulmonary hypertension (PH) all implicating microRNAs in the molecular pathogenesis of these diseases.
Although our knowledge of microRNAs in lung disease is increasing, we are met with several questions regarding both their biological complexity and potential as both biomarkers and targeted therapies. We are still challenged to identify their eventual targeted therapeutic purpose. We are also challenged with determining how these small molecules integrate with components of the human genome and proteome to promote or prevent disease. Last, long noncoding RNAs (lncRNAs) are another form of ncRNA whose regulatory role is only beginning to be realized. Among the lncRNAs, both long intervening noncoding RNAs (lincRNAs) and ultraconserved regions appear to have regulatory properties that may also be of biological relevance to disease (5). Given the rapidly advancing field of ncRNA investigation in lung disease, herein we provide for the clinician an overview of the current state of primarily microRNA application across lung disease with a focus on their role in several diseases of the lung, novel applications for detection, and the challenges to translating microRNA to the clinic.
MicroRNA Biogenesis and Regulation
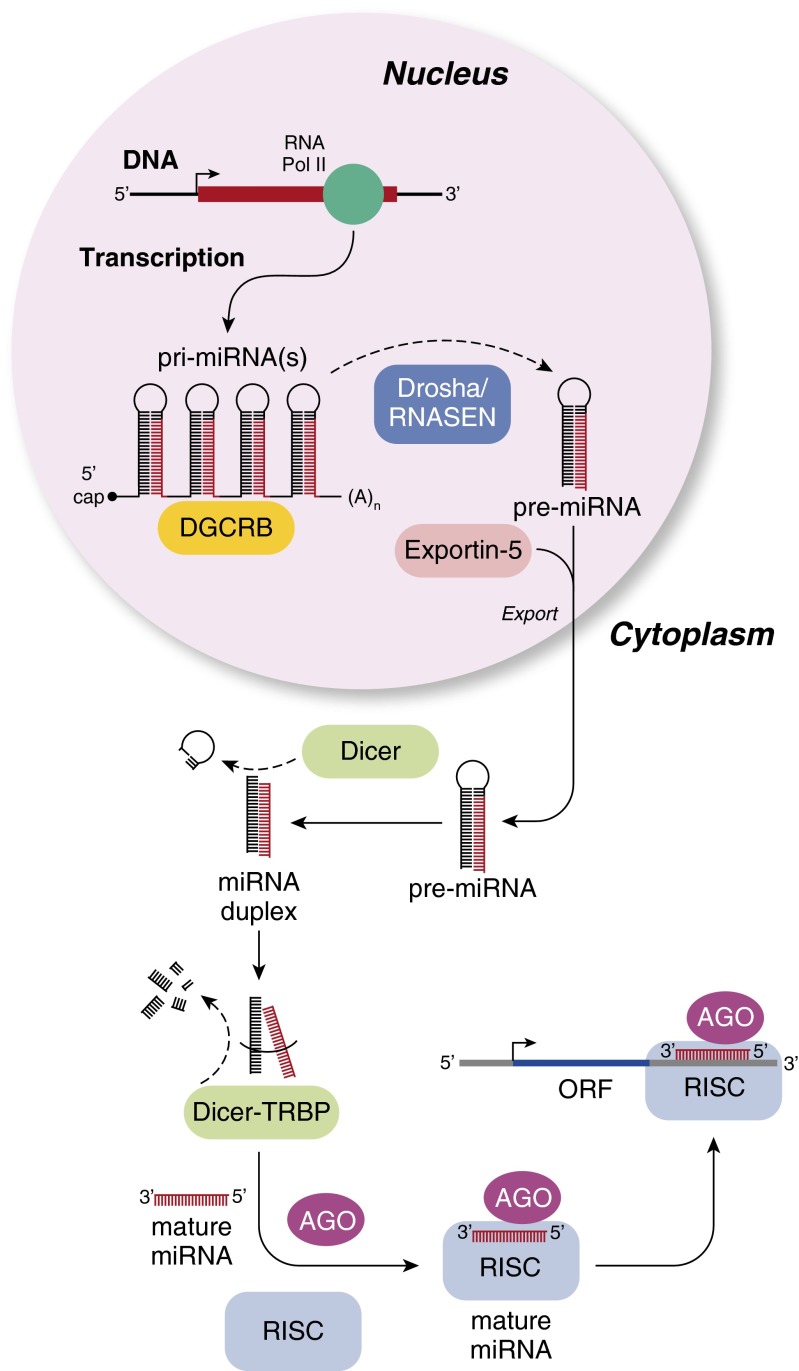
The process of microRNA biogenesis involves a series of well-orchestrated steps, each of which is complex (Figure 1). A long primary transcript (pri)-microRNA undergoes transcription by RNA polymerase II leading to the generation of what are termed pri-microRNAs. These pri-microRNAs are then bound to the double-stranded RNA-binding domain (dsRBD) protein known as DiGeorge syndrome critical region gene 8 (DGCR8) for vertebrates (6). The pri-microRNA is then enzymatically converted by RNASEN/Drosha to a smaller stem-loop ∼70-nt precursor microRNA (pre-microRNA) (7). Exportin, which is a double-stranded binding protein, transports the pre-microRNA from the nucleus to the cytoplasm, after which Dicer generates a mature microRNA by cleavage. It is at this point that the mature microRNA may form complementarity to either the 3′ or 5′ untranslated region (UTR) of a target gene to induce either RNA degradation or inhibition of translation. Several studies have demonstrated that miRNAs tend to be globally deregulated in many diseases, including cancers. Although the mechanisms for this deregulation have yet to be fully elucidated, we now know that microRNAs are susceptible to several mechanisms for deregulation, including epigenetic silencing, chromosomal amplification, chromosomal deletion, and environmental stressors (3).
Figure 1.
MicroRNA processing. AGO = Argonaute proteins; DGCR8 = DiGeorge syndrome critical region gene 8; ORF = open reading frame; RISC = RNA-induced silencing complex; TRBP = transactivating response RNA-binding protein.
MicroRNAs and Lung Development
Lung organogenesis is an inherently complex course, regulated by a multitude of genes and signaling molecules. Evidence continues to support the importance that microRNAs play in lung development. MicroRNAs have long been implicated in the organogenesis innate to lung development. Lung development in mammalian species has been divided into a series of six stages: embryonic, glandular, canalicular, saccular, alveolar, and vascular maturation. An evaluation for microRNA expression patterns during the various stages of lung development identified 21 microRNAs that were differentially expressed (8). Williams and colleagues compared microRNA expression patterns between fetal and adult human lungs, using real-time polymerase chain reaction, and concluded that 13 microRNAs were up-regulated in human fetal lungs and 8 were up-regulated in human adult lungs (9). The significance of these changes remains poorly understood.
The importance of microRNA function to lung development and disease is further underscored by investigations focused on the miR-17-92 cluster This cluster is a polycistronic gene localized to chromosome 13 and includes six microRNA genes (miR-17, -18a, -19a, -20a, 19b-1, and 92-1) (10). This particular cluster has been implicated as an oncogene across both solid and hematological malignancies (10). Expression of these same microRNAs decreases during the course of lung maturation. Investigators have demonstrated that the transgenic overexpression of the miR-17-92 cluster drives the proliferation of lung epithelial progenitor cells while inhibiting their differentiation (11). Other microRNAs, such as miR-127 and miR-351, are expressed primarily at the sacculo-alveolar stage of development, initially in the mesenchymal compartment and later in lung epithelial cells, suggesting their role in the cellular reorganization and differentiation processes during the mesenchymal-to-epithelial transition period (8). A study conducted by Ventura and colleagues demonstrated that miR-17-92 silencing led to defective lung development characterized by hypoplasia and ventral septal defects (12). These detrimental effects of both microRNA overexpression and silencing further support the importance of microRNAs as mediators of homeostasis during organ development. A separate study by Harris and colleagues demonstrated that targeted inactivation of the microRNA processing enzyme DICER significantly impaired lung epithelial development (13).
A study by Mujahid and colleagues further demonstrated the inherent importance of microRNA in the airway and vascular development of fetal lungs (14). Changes in airway morphology were associated with miR-221 inhibition and miR-130a overexpression. In particular, miR-221–treated lungs showed diminished airway branching, whereas miR-130a–treated lungs demonstrated increased branching into the central lung regions (14).
Sex hormones have similarly been implicated in the regulation of lung development. Androgens possess inhibitory effects on lung maturational events, owing to differences noted during male–female gestation. Prior analysis has identified 37 distinct microRNAs that changed in expression during fetal development. Of those, a group of 13 in particular changed significantly with sex and gestation (15).
Asthma
Asthma is defined by the hallmarks of airway inflammation, airway hyperresponsiveness, and reversible airway obstruction. A number of cellular-based changes occur throughout the course of disease progression, including airway remodeling (characterized by smooth muscle hyperplasia), subepithelial cell fibrosis, goblet cell hyperplasia, and neovascularization. MiRNAs appear to play pivotal roles in the overall pathogenesis of asthma though several mechanisms, including, for example, regulating an array of immune receptors and cytokines. The physiologic changes that occur are induced by Th2-driven inflammation, resulting in increased levels of proinflammatory cytokines (IL-4, IL-5, IL-9, and IL-13) and decreased antiinflammatory cytokines, such as IL-10 (16).
The inflammatory response is central to the progression and pathology of asthma and is manifested by a production of IgE and consistent recruitment of leukocytes, in particular eosinophils, together with Th2 cells and mast cells (17). MiR-21 is a microRNA that may be central to this process. Up-regulated in individuals with asthma, miR-21 targets IL-12, a potent cytokine responsible for Th1 cell activation. Ovalbumin is an established model allergen for the assessment of airway hyperresponsiveness. On ovalbumin administration, miR-21−/− mice demonstrated an augmented allergen response, which occurred as a direct result of the imbalance between Th1 and Th2. Consequently, these mice produced greater IFN and IL-12 and increased levels of IL-4, leading to an increased influx of eosinophils into the lung (18).
MiR-145 is another microRNA implicated in the Th2 response and subsequent eosinophil recruitment. In a study by Collison and colleagues, allergen-challenged mice treated with intranasal delivery of a miR-145 inhibitor demonstrated a reduction in levels of the proinflammatory cytokines IL-13 and IL-5 in conjunction with an alleviation of their symptoms (19). There were concomitant decreases in goblet cell hyperplasia, peribronchial eosinophils, and production of IL-5 and IL-13. The antiinflammatory effects of miR-145 inhibition were comparable to that of corticosteroid treatment. Similarly, the miR-221/222 family is transcriptionally induced on mast cell activation. Data generated by high-throughput human microRNA profiling by Tsitsiou and colleagues identified the selective down-regulation of miR-28-5p and miR-146 a/b in CD8+ and CD4+ cells from individuals with severe asthma (20). Based on these and other studies, it is apparent that a number of microRNAs work in concert by multiple mechanisms to either promote or abrogate disease progression in asthma. Furthermore, it is clear that patterns of microRNA expression may be both context- and cell-specific. Those up-regulated microRNAs in other studies include miR-126, -145a, -21, -221/222, -106a, and -155. Down-regulated expression is seen primarily with let-7, miR-20b, and miR-133a. Most of the current asthma treatment modalities are vastly hindered by inherent potential medication side effects. Targeting microRNAs may represent a novel potential therapeutic strategy in the treatment of asthma. By inhibiting expression of various microRNAs, such as miR-21, -106a, -126, -145, -155, and -221, abnormal cytokine expression and inflammation can potentially be mitigated. As is the case in developmental pharmacotherapy, the ideal mechanisms for delivery, exact efficacy, and specificity of specific miRs/anti-miRs remain unknown.
COPD
Few studies to date have examined the role for microRNAs in COPD. The majority of studies have focused on global patterns of microRNA expression. In two early studies, investigators evaluated the effects of cigarette smoke on microRNA expression in animal models. In the first study, rats were exposed to environmental cigarette smoke versus ambient air control over 4 weeks. Twenty-four microRNAs were significantly down-regulated between smoke-exposed and sham groups (21). In the second study, investigators evaluated the effects of smoke exposure on microRNA expression in mice. The majority of deregulated microRNAs in this study were also down-regulated (22).
Schembri and colleagues examined microRNA expression patterns in the bronchial epithelium from smokers and never smokers. They identified 28 differentially expressed microRNAs (23). In an independent study, investigators identified 70 deregulated microRNAs and more than 2,000 mRNAs within the lung tissue of smokers with normal lung function compared with those with varying degrees of COPD (24). Based on the comparison of microRNA and mRNA expression patterns and potential relationships, the authors identified enriched biological pathways for transforming growth factor (TGF)-β, MAP kinase, and cancer pathways in COPD tissue. Mizuno and colleagues also identified miR-199-5p and miR-34a as potential biomarkers in COPD lungs (25). Given the complex temporospatial contribution of individual cell types to lung disease, investigators have also focused on identifying functional cell-specific microRNAs. Christenson and colleagues conducted a comprehensive analysis for microRNAs and mRNAs within different regions of the lung (26). They profiled eight separate regions within the lungs of eight individuals (six with COPD and two control subjects). Using this strategy, the authors were able to not only identify select deregulated microRNAs but also build and enrich for specific biological pathways. They identified 63 microRNAs that were deregulated in regions of emphysema. Of note, three of the deregulated microRNAs (miR-638, miR-30c, and miR-181d) had an inverse relationship in expression to several of their predicted targets. A separate study by Savarimuthu and colleagues identified five microRNAs that were down-regulated in more advanced emphysema (miR-34b, miR-34c, miR-149, miR-133a, and miR-133b) (27). Two recent studies attempted to correlate microRNA expression with clinical phenotype. Sato and colleagues were able to correlate fibroblast-specific miR-146a expression with in vitro inflammatory response and clinical severity in COPD (28). Another study identified the presence of muscle-specific microRNAs (miR-1 and miR-499) in circulation and correlated the presence of these microRNAs with fat-free mass index and strength, respectively, in patients with COPD (29). MicroRNA investigation in COPD has now moved beyond profiling to identifying the role of microRNAs as direct drivers of disease pathogenesis. Perhaps one of the best such examples is a recent study by Hassan and colleagues, who identified miR-199a-5p as a key regulator of components of the unfolded protein response in MM and ZZ monocytes derived from individuals with COPD (30). In this study, the authors demonstrated that in asymptomatic individuals, ZZ monocytes harbor increased levels of miR-199a-5p, which in turn negatively regulates the endoplasmic reticulum stress response. Symptomatic individuals are susceptible to methylation of the miR-199a-5p promoter and microRNA silencing leading to an exaggerated unfolded protein response and inflammatory response. This study provides an additional potential actionable target in COPD.
Pulmonary Fibrosis
Pulmonary fibrosis carries a poor prognosis with limited therapeutic options. Given the numerous biological pathways implicated in pulmonary fibrosis, microRNAs are ideal biological targets. Several biological pathways have been identified in the molecular pathogenesis of fibrotic lung disease, including TGF-β/SMAD. Global profiling studies have been applied to demonstrate distinct patterns of microRNA expression in fibrotic lung disease (31) and identify select biological pathways as targets for microRNA including TGF-β signaling, focal adhesion, and Wnt (32). Among the profiling studies, several microRNAs have emerged as being of importance to fibrosis, including miR-155, miR-29, miR-200, miR-21, and miR-326 (33–35). Xiao and colleagues recently demonstrated in a mouse model that systemic delivery of miR-29 could abrogate the development of pulmonary fibrosis (36). A separate study demonstrated that decreased expression of let-7d contributed to the fibrotic process in a murine model of lung fibrosis (37). This seminal study was further supported by a more recent one showing reversal of mesenchymal phenotype after in cell culture followed by Let-7 administration (38). In addition, the intratracheal introduction of members of the miR-200 family, which have been implicated in epithelial–mesenchymal transition, attenuated the development of fibrosis in a murine model (39). MicroRNA patterns of expression have also been linked to clinical phenotype. For example, Oak and colleagues identified a distinct pattern of lung microRNAs (miR-302c, miR-423-5p, miR-210, miR-376c, miR-185) that were increased among patients with rapidly progressive disease as opposed to slowly progressive disease (40). Another study led by Dakhlallah demonstrated a correlation between expression of miR-17-92 and severity of pulmonary fibrosis as well as a link between patterns of miR-17-92 expression and epigenetic machinery (41).
Pulmonary Hypertension
PH is a deadly disease characterized by hyperproliferative and inflammatory vascular smooth muscle and endothelium. The contribution of microRNAs to the pathogenesis of PH remains largely unknown, with the majority of studies being limited to cell culture and murine models of disease (42). In the last few years, we have seen several novel and interesting studies aimed at better defining microRNAs in disease pathogenesis. Caruso and colleagues demonstrated a temporal deregulation of microRNAs in the lungs of rats exposed to two models (hypoxia and monocrotaline) of PH (43). In response to hypoxia, Let-7f, miR-30c, and miR-22 were down-regulated within 7 days of exposure, whereas miR-451 and miR-322 were up-regulated microRNAs. In response to monocrotaline, miR-30c, miR-22, and let-7f were significantly down-regulated, whereas miR-451 and miR-322 were up-regulated. Chen and colleagues demonstrated a functional association between polymorphisms in the 3′ UTR of the fibrinogen α gene, binding and regulation by the complementary microRNA miR-759, and susceptibility to chronic thromboembolic pulmonary hypertension (44). Circulating microRNAs may also function as potential biomarkers. Rhodes and colleagues recently examined independent cohorts of patients with PH for circulating microRNAs and identified circulating miR-150 as a biomarker linked to decreased survival (45).
Given the inherent complexity of PH biology and the contribution of various cell types to disease, investigators have taken to analyzing microRNA biology at the cell-specific level. Courboulin and colleagues (46) evaluated cell-specific microRNA expression in pulmonary artery smooth muscle cells (PASMCs) from patients with pulmonary arterial hypertension (PAH). They identified seven deregulated microRNAs from which they elected to investigate the only decreased microRNA, miR-204, in detail. They demonstrated in vitro that miR-204 gain of function could attenuate both proliferative capacity and antiapoptotic phenotype in PASMCs (46). These effects were mechanistically linked to the Src-STAT-NFAT axis. Last, nebulized delivery reduced pulmonary artery pressures and vascular remodeling in a rat model of monocrotaline-induced PH. Based on previous profiling, Pullamsetti and colleagues demonstrated that intravenous inhibition of miR-17 attenuated murine chronic hypoxia-induced PH (47). Recently, Wang and colleagues identified miR-124 as an additional actionable target in PH (48). The investigators determined that miR-124 down-regulation in PH fibroblasts contributed to a migratory and inflammatory phenotype (48). Restoration of miR-124 inhibited both proliferation and migration in fibroblasts. In a separate study, miR-124 was decreased in PASMCs after exposure to hypoxia (49). miR-124 directly targeted components of the nuclear factor of activated T cell (NFAT) pathway. In the endothelium, both miR-424 and miR-503 deregulation were shown to be critical links between apelin and fibroblast growth factor signaling, which have been implicated in pulmonary artery endothelial phenotype in PH (50)
Other investigators have linked microRNA biology to known molecular mutations within PAH. Among patients with familial forms of PAH, 50% harbor mutations in Bone Morphogenic Protein receptor 2 (BMPR2) (51). Among the potential mechanisms that exist for the observed down-regulation of BMPR2 signaling in PAH, we should now consider microRNA deregulation. Drake and colleagues showed that BMPR2 mutations led to a decrease in induction of specific antiproliferative microRNAs miR-21 and miR-27a and that this effect may be mediated by mutations in the downstream Smad-8 (52). Brock and colleagues determined that members of the miR-17-92 cluster (miR-17-5 and miR-20a) had direct regulatory effects on BMPR2 protein expression and that this regulation may be mediated through STAT3 signaling (53). They demonstrated that systemic delivery of antagomiR-20a restored BMPR2 signaling and attenuated hypoxia-induced pulmonary hypertension.
Lung Cancer
MicroRNAs have been well studied in lung cancer with a multitude of cell, animal, and human studies demonstrating deregulation in lung tumors compared with uninvolved lung tissues (54). The complexity of microRNA biology in lung cancer is evidenced by their context-dependent role as oncogenes, tumor suppressors, or, in some cases, both. Furthermore, it is clear that they have the capacity to regulate aspects of fundamental tumor biology, including proliferation, apoptosis, angiogenesis, and epithelial–mesenchymal transition. Global impairment in microRNA processing, epigenetic modifications, and regulatory effects of cigarette smoke have all been implicated as mechanisms for microRNA deregulation in lung cancer. In fact, early studies determined that global reduction in microRNA processing through targeting of key processing components such as Drosha, DGCR8, and Dicer could promote tumor development (55–57).
Up until recently, investigators have used patterns of microRNA expression and polymorphisms of both microRNA and target 3′ UTRs in lung cancers as both diagnostic and prognostic biomarkers; however, neither application has yet to reach the clinical arena. Yu and colleagues conducted one of the earliest profiling studies in a cohort of 122 patients and identified a five-microRNA signature of high-risk microRNAs (let-7a, miR-137, -182* [designating passenger strand], -221, and -372) within tumor tissue (58). Since that initial study, several global profiling studies have been conducted with variable results in reproducibility (59). Specific microRNAs, although not lung cancer–specific, including let-7, miR-21, miR-29, miR-126, miR-155, and miR-17-92, appear to be fundamental in tumor biology and thus ideal candidates as biomarkers in solid tumors (60–63). For example, a retrospective study determined that lung tumor levels of miR-21 correlated with disease progression and survival in stage I lung cancer (64). miR-21 has been linked to oncogenic KRAS activation with genetic manipulation altering the course of lung tumor development (65). Boeri and colleagues recently demonstrated that patterns of circulating microRNAs could be used clinically in lung cancer (66). By integrating patterns of tumor and circulating microRNA into an ongoing lung cancer computed tomography (CT) screening trial, the authors were able to correlate patterns of tumor and circulating microRNA expression with both clinical parameters and the development of lung cancer. As validation, Sozzi and colleagues conducted an independent study demonstrating that a distinct panel of circulating microRNAs could be applied as diagnostic and prognostic biomarkers in lung cancer (67).
MicroRNAs as Noninvasive Biomarkers
microRNAs have become attractive as potential noninvasive biomarkers of disease; however, circulating microRNA detection is fraught with challenges, including compartment dependence of microRNA expression (whole blood, serum, plasma, peripheral blood mononuclear cells), lack of a standardized method for detection, and, most importantly, determining the physiological relevance of circulating microRNAs; however, a major obstacle in translating microRNA biology to clinical practice has been achieving interstudy reproducibility to identify consistent signatures and elucidating the mechanisms that drive the presence of these molecules in body fluids and their relevant biology. Additionally, we are starting to realize that circulating microRNAs are highly compartment specific. Investigators have demonstrated that microRNAs may circulate both freely attached to select proteins (e.g., Ago) or lipids and packaged within extracellular vesicles including exosomes, apoptotic bodies, and microvesicles. The concept of “packaged” microRNAs is particularly intriguing as a mechanism for the intercellular transfer of genetic material (68). These paradigm-shifting discoveries have led to intense investigation for novel mechanisms by which circulating microRNAs may serve as mediators of intercellular communication. In order for circulating microRNAs to reach point-of-care application, assays will need to be both cost effective and easy to use. Despite these challenges, several such studies have been conducted in malignancies and cardiac, endocrine, rheumatological, and now lung disease. The majority of lung-related studies have been conducted in lung cancer. Recently, we have observed an increasing number of circulating microRNA studies in diseases such as pulmonary fibrosis (69), tuberculosis (70), sarcoidosis (71), COPD (72) and PH (73).
Investigators have also examined sputum microRNA as biomarkers, particularly in smoking-related lung disease. Two independent studies identified sputum microRNAs that distinguished patients with lung cancer from normal control patients. Xing and colleagues identified three microRNAs in sputum (miR-205, miR-210, and miR-708) that distinguished patients with squamous cell carcinoma of the lung from healthy control subjects with a reported sensitivity of 73% and specificity of 96% (74). A more recent investigation determined that when combined with CT scan, sputum-based microRNA analysis (miR-31 and miR-210) led to a higher specificity (91%) than either CT scan or microRNAs alone (75). Van Pottelberge and colleagues have also shown that sputum-based microRNA detection may be applied to COPD (76). The investigators identified reductions in both Let-7c and miR-125b in current smokers with COPD compared with never smokers, current smokers without COPD, and ex-smokers with COPD. Last, an intriguing study by Sinha and colleagues demonstrated the feasibility of microRNA detection in exhaled breath condensate as a biomarker for asthma (77).
Other ncRNAs
MicroRNAs present only one member of a family of ncRNAs, many of which were believed to have minimal biological function. We now know that other members of this group of ncRNAs are involved in cellular differentiation, proliferation, and imprinting. ncRNAs consist of short (< 200 nt) and lncRNAs (300 nt to 100 kb) forms (78). Within these groups, lncRNAs, transcribed ultraconserved regions (79), and small nucleolar RNAs are all the subject of investigation. The majority of studies to date investigating a functional role for other ncRNAs have been conducted in solid and hematological malignancies. For example, metastasis associated in lung adenocarcinoma transcript-1 (MALAT-1) is an intergenic transcript (7 kb) ncRNA whose expression correlates with metastases and prognosis in early-stage non-small cell lung cancer (80). Located on chromosome 11, MALAT-1 is a member of a subclass of lncRNA called nuclear-retained RNAs (nrRNAs) (81). Several other lncRNAs, including Highly up-regulated in liver cancer (HULC) and SPRY4-IT1, have also been described in malignancies (82, 83). Small nucleolar RNAs are smaller ncRNAs that have been shown to be deregulated in malignancies (84). With the increasing application of platforms such as RNA sequencing in lung disease, it is likely that other ncRNAs will be identified as potential actionable targets and biomarkers.
The Future of MicroRNAs in Lung Disease
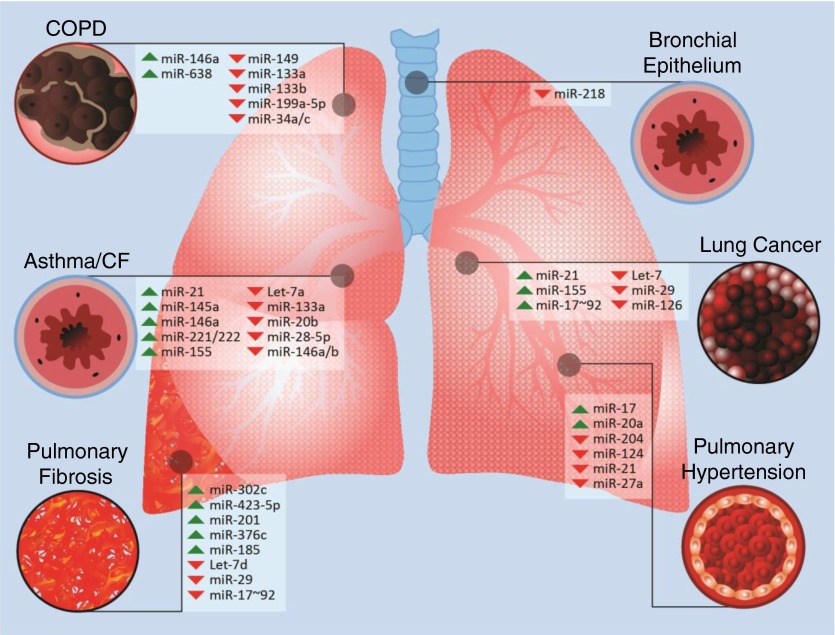
Since their initial discovery in the early 1990s, microRNAs have now become the focus of a multitude of avenues of investigation ranging from basic biology to translational applications in the clinic. In the case of disease of the lung, the majority of studies have targeted microRNA biology to better understand the fundamental biology that drives disease and microRNAs as potential biomarkers of disease (Figure 2). We have now transitioned from single miRNA and single-target relationships to better understanding the mechanisms by which a miRNAs can reprogram biological pathways. A key to these relationships will be the careful integration of miRNA expression with changes in the transcriptome and proteome. Despite these efforts, several obstacles remain to truly transitioning microRNA biology to clinical application in lung disease. These obstacles include the development of nontoxic targeted delivery to the lung, improved understanding of how microRNAs interact with other components of the genome, and standardization of microRNA detection. Although there have been several studies demonstrating in vivo targeting of microRNAs using various formulations, including nanoparticles as one vehicle, few studies have translated to human application. There are several obstacles to microRNAs as directed therapeutics in human lung disease, including selection of the proper vehicle for delivery, mode of delivery (systemic versus lung), potential toxicity, and off-organ effects. Two ongoing human clinical trials focused on anti–miR-122 therapy in hepatitis C and miR-34 in hepatocellular carcinoma represent encouraging signs that microRNAs as directed therapies in human disease are in the not-too-distant future (85–87). In addition, opportunities exist for applying patterns of microRNA expression as biomarkers that may inform the use of conventional therapies in lung disease. In the future, we are likely to see additional microRNA biomarker studies in the context of ongoing therapeutic trials in lung disease. In addition, the increasing importance of extracellular vesicle biology and its potential as both biomarkers and intercellular carriers of genetic material should continue to be investigated in lung disease. Although we have presented for the clinician examples of microRNA application in a small number of lung diseases, it is important to recognize that there are ongoing studies in many other conditions, including cystic fibrosis, sarcoidosis, acute lung injury, sepsis, and respiratory infections, each of which is currently at a different stage of maturity but could potentially lead to the development of novel therapies and, we hope, improved outcomes.
Figure 2.
Overview of microRNAs implicated in lung disease. CF = cystic fibrosis; COPD = chronic obstructive pulmonary disease; miR = microRNA.
Footnotes
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 3.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Gregory RI, Shiekhattar R. MicroRNA biogenesis and cancer. Cancer Res. 2005;65:3509–3512. doi: 10.1158/0008-5472.CAN-05-0298. [DOI] [PubMed] [Google Scholar]
- 7.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 8.Bhaskaran M, Wang Y, Zhang H, Weng T, Baviskar P, Guo Y, Gou D, Liu L. MicroRNA-127 modulates fetal lung development. Physiol Genomics. 2009;37:268–278. doi: 10.1152/physiolgenomics.90268.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams AE, Perry MM, Moschos SA, Lindsay MA. microRNA expression in the aging mouse lung. BMC Genomics. 2007;8:172. doi: 10.1186/1471-2164-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mendell JT. miRiad roles for the miR-17–92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu Y, Thomson JM, Wong HY, Hammond SM, Hogan BL. Transgenic over-expression of the microRNA miR-17–92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol. 2007;310:442–453. doi: 10.1016/j.ydbio.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris KS, Zhang Z, McManus MT, Harfe BD, Sun X. Dicer function is essential for lung epithelium morphogenesis. Proc Natl Acad Sci USA. 2006;103:2208–2213. doi: 10.1073/pnas.0510839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mujahid S, Nielsen HC, Volpe MV. MiR-221 and miR-130a regulate lung airway and vascular development. PLoS ONE. 2013;8:e55911. doi: 10.1371/journal.pone.0055911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mujahid S, Logvinenko T, Volpe MV, Nielsen HC. miRNA regulated pathways in late stage murine lung development. BMC Dev Biol. 2013;13:13. doi: 10.1186/1471-213X-13-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greene CM, Gaughan KP. microRNAs in asthma: potential therapeutic targets. Curr Opin Pulm Med. 2013;19:66–72. doi: 10.1097/MCP.0b013e32835a5bc8. [DOI] [PubMed] [Google Scholar]
- 17.Frieri M. Advances in the understanding of allergic asthma. Allergy Asthma Proc. 2007;28:614–619. doi: 10.2500/aap.2007.28.2952. [DOI] [PubMed] [Google Scholar]
- 18.Lu TX, Munitz A, Rothenberg ME. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J Immunol. 2009;182:4994–5002. doi: 10.4049/jimmunol.0803560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collison A, Mattes J, Plank M, Foster PS.Inhibition of house dust mite-induced allergic airways disease by antagonism of microRNA-145 is comparable to glucocorticoid treatment J Allergy Clin Immunol 2011128160–167.e4 [DOI] [PubMed] [Google Scholar]
- 20.Tsitsiou E, Williams AE, Moschos SA, Patel K, Rossios C, Jiang X, Adams OD, Macedo P, Booton R, Gibeon D, et al. Transcriptome analysis shows activation of circulating CD8+ T cells in patients with severe asthma. J Allergy Clin Immunol. 2012;129:95–103. doi: 10.1016/j.jaci.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 21.Izzotti A, Calin GA, Arrigo P, Steele VE, Croce CM, De Flora S. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB J. 2009;23:806–812. doi: 10.1096/fj.08-121384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Izzotti A, Calin GA, Steele VE, Croce CM, De Flora S. Relationships of microRNA expression in mouse lung with age and exposure to cigarette smoke and light. FASEB J. 2009;23:3243–3250. doi: 10.1096/fj.09-135251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schembri F, Sridhar S, Perdomo C, Gustafson AM, Zhang X, Ergun A, Lu J, Liu G, Bowers J, Vaziri C, et al. MicroRNAs as modulators of smoking-induced gene expression changes in human airway epithelium. Proc Natl Acad Sci USA. 2009;106:2319–2324. doi: 10.1073/pnas.0806383106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ezzie ME, Crawford M, Cho JH, Orellana R, Zhang S, Gelinas R, Batte K, Yu L, Nuovo G, Galas D, et al. Gene expression networks in COPD: microRNA and mRNA regulation. Thorax. 2012;67:122–131. doi: 10.1136/thoraxjnl-2011-200089. [DOI] [PubMed] [Google Scholar]
- 25.Mizuno S, Bogaard HJ, Gomez-Arroyo J, Alhussaini A, Kraskauskas D, Cool CD, Voelkel NF. MicroRNA-199a-5p is associated with hypoxia-inducible factor-1alpha expression in lungs from patients with COPD. Chest. 2012;142:663–672. doi: 10.1378/chest.11-2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christenson SA, Brandsma CA, Campbell JD, Knight DA, Pechkovsky DV, Hogg JC, Timens W, Postma DS, Lenburg M, Spira A. miR-638 regulates gene expression networks associated with emphysematous lung destruction. Genome Med. 2013;5:114. doi: 10.1186/gm519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Savarimuthu Francis SM, Davidson MR, Tan ME, Wright CM, Clarke BE, Duhig EE, Bowman RV, Hayward NK, Fong KM, Yang IA. MicroRNA-34c is associated with emphysema severity and modulates SERPINE1 expression. BMC Genomics. 2014;15:88. doi: 10.1186/1471-2164-15-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sato T, Liu X, Nelson A, Nakanishi M, Kanaji N, Wang X, Kim M, Li Y, Sun J, Michalski J, et al. Reduced miR-146a increases prostaglandin E(2)in chronic obstructive pulmonary disease fibroblasts. Am J Respir Crit Care Med. 2010;182:1020–1029. doi: 10.1164/rccm.201001-0055OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Donaldson A, Natanek SA, Lewis A, Man WD, Hopkinson NS, Polkey MI, Kemp PR. Increased skeletal muscle-specific microRNA in the blood of patients with COPD. Thorax. 2013;68:1140–1149. doi: 10.1136/thoraxjnl-2012-203129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hassan T, Carroll TP, Buckley PG, Cummins R, O'Neill SJ, McElvaney NG, Greene CM. miR-199a-5p silencing regulates the unfolded protein response in chronic obstructive pulmonary disease and alpha1-antitrypsin deficiency. Am J Respir Crit Care Med. 2014;189:263–273. doi: 10.1164/rccm.201306-1151OC. [DOI] [PubMed] [Google Scholar]
- 31.Xie T, Liang J, Guo R, Liu N, Noble PW, Jiang D. Comprehensive microRNA analysis in bleomycin-induced pulmonary fibrosis identifies multiple sites of molecular regulation. Physiol Genomics. 2011;43:479–487. doi: 10.1152/physiolgenomics.00222.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cho JH, Gelinas R, Wang K, Etheridge A, Piper MG, Batte K, Dakhallah D, Price J, Bornman D, Zhang S, et al. Systems biology of interstitial lung diseases: integration of mRNA and microRNA expression changes. BMC Med Genomics. 2011;4:8. doi: 10.1186/1755-8794-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 33.Cushing L, Kuang PP, Qian J, Shao F, Wu J, Little F, Thannickal VJ, Cardoso WV, Lu J. miR-29 is a major regulator of genes associated with pulmonary fibrosis. Am J Respir Cell Mol Biol. 2011;45:287–294. doi: 10.1165/rcmb.2010-0323OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu G, Friggeri A, Yang Y, Milosevic J, Ding Q, Thannickal VJ, Kaminski N, Abraham E. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med. 2010;207:1589–1597. doi: 10.1084/jem.20100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Das S, Kumar M, Negi V, Pattnaik B, Prakash YS, Agrawal A, Ghosh B. microRNA-326 regulates pro-fibrotic functions of transforming growth factor-β in pulmonary fibrosis. Am J Respir Cell Mol Biol. 2014;50:882–892. doi: 10.1165/rcmb.2013-0195OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiao J, Meng XM, Huang XR, Chung AC, Feng YL, Hui DS, Yu CM, Sung JJ, Lan HY. miR-29 inhibits bleomycin-induced pulmonary fibrosis in mice. Mol Ther. 2012;20:1251–1260. doi: 10.1038/mt.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pandit KV, Corcoran D, Yousef H, Yarlagadda M, Tzouvelekis A, Gibson KF, Konishi K, Yousem SA, Singh M, Handley D, et al. Inhibition and role of let-7d in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2010;182:220–229. doi: 10.1164/rccm.200911-1698OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huleihel L, Ben-Yehudah A, Milosevic J, Yu G, Pandit K, Sakamoto K, Yousef H, Lejeune M, Coon TA, Redinger CJ, et al. Let-7d microRNA affects mesenchymal phenotypic properties of lung fibroblasts. Am J Physiol Lung Cell Mol Physiol. 2014;306:L534–L542. doi: 10.1152/ajplung.00149.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang S, Banerjee S, de Freitas A, Sanders YY, Ding Q, Matalon S, Thannickal VJ, Abraham E, Liu G. Participation of miR-200 in pulmonary fibrosis. Am J Pathol. 2012;180:484–493. doi: 10.1016/j.ajpath.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oak SR, Murray L, Herath A, Sleeman M, Anderson I, Joshi AD, Coelho AL, Flaherty KR, Toews GB, Knight D, et al. A micro RNA processing defect in rapidly progressing idiopathic pulmonary fibrosis. PLoS ONE. 2011;6:e21253. doi: 10.1371/journal.pone.0021253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dakhlallah D, Batte K, Wang Y, Cantemir-Stone CZ, Yan P, Nuovo G, Mikhail A, Hitchcock CL, Wright VP, Nana-Sinkam SP, et al. Epigenetic regulation of miR-17∼92 contributes to the pathogenesis of pulmonary fibrosis. Am J Respir Crit Care Med. 2013;187:397–405. doi: 10.1164/rccm.201205-0888OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parikh VN, Jin RC, Rabello S, Gulbahce N, White K, Hale A, Cottrill KA, Shaik RS, Waxman AB, Zhang YY, et al. MicroRNA-21 integrates pathogenic signaling to control pulmonary hypertension: results of a network bioinformatics approach. Circulation. 2012;125:1520–1532. doi: 10.1161/CIRCULATIONAHA.111.060269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caruso P, MacLean MR, Khanin R, McClure J, Soon E, Southgate M, MacDonald RA, Greig JA, Robertson KE, Masson R, et al. Dynamic changes in lung microRNA profiles during the development of pulmonary hypertension due to chronic hypoxia and monocrotaline. Arterioscler Thromb Vasc Biol. 2010;30:716–723. doi: 10.1161/ATVBAHA.109.202028. [DOI] [PubMed] [Google Scholar]
- 44.Chen Z, Nakajima T, Tanabe N, Hinohara K, Sakao S, Kasahara Y, Tatsumi K, Inoue Y, Kimura A. Susceptibility to chronic thromboembolic pulmonary hypertension may be conferred by miR-759 via its targeted interaction with polymorphic fibrinogen alpha gene. Hum Genet. 2010;128:443–452. doi: 10.1007/s00439-010-0866-8. [DOI] [PubMed] [Google Scholar]
- 45.Rhodes CJ, Wharton J, Boon RA, Roexe T, Tsang H, Wojciak-Stothard B, Chakrabarti A, Howard LS, Gibbs JS, Lawrie A, et al. Reduced microRNA-150 is associated with poor survival in pulmonary arterial hypertension. Am J Respir Crit Care Med. 2013;187:294–302. doi: 10.1164/rccm.201205-0839OC. [DOI] [PubMed] [Google Scholar]
- 46.Courboulin A, Paulin R, Giguere NJ, Saksouk N, Perreault T, Meloche J, Paquet ER, Biardel S, Provencher S, Cote J, et al. Role for miR-204 in human pulmonary arterial hypertension. J Exp Med. 2011;208:535–548. doi: 10.1084/jem.20101812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pullamsetti SS, Doebele C, Fischer A, Savai R, Kojonazarov B, Dahal BK, Ghofrani HA, Weissmann N, Grimminger F, Bonauer A, et al. Inhibition of microRNA-17 improves lung and heart function in experimental pulmonary hypertension. Am J Respir Crit Care Med. 2012;185:409–419. doi: 10.1164/rccm.201106-1093OC. [DOI] [PubMed] [Google Scholar]
- 48.Wang D, Zhang H, Li M, Frid MG, Flockton AR, McKeon BA, Yeager ME, Fini MA, Morrell NW, Pullamsetti SS, et al. MicroRNA-124 controls the proliferative, migratory, and inflammatory phenotype of pulmonary vascular fibroblasts. Circ Res. 2014;114:67–78. doi: 10.1161/CIRCRESAHA.114.301633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kang K, Peng X, Zhang X, Wang Y, Zhang L, Gao L, Weng T, Zhang H, Ramchandran R, Raj JU, et al. MicroRNA-124 suppresses the transactivation of nuclear factor of activated T cells by targeting multiple genes and inhibits the proliferation of pulmonary artery smooth muscle cells. J Biol Chem. 2013;288:25414–25427. doi: 10.1074/jbc.M113.460287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim J, Kang Y, Kojima Y, Lighthouse JK, Hu X, Aldred MA, McLean DL, Park H, Comhair SA, Greif DM, et al. An endothelial apelin-FGF link mediated by miR-424 and miR-503 is disrupted in pulmonary arterial hypertension. Nat Med. 2013;19:74–82. doi: 10.1038/nm.3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lane KB, Machado RD, Pauciulo MW, Thomson JR, Phillips JA, III, Loyd JE, Nichols WC, Trembath RC. Heterozygous germline mutations in BMPR2, encoding a TGF-beta receptor, cause familial primary pulmonary hypertension. Nat Genet. 2000;26:81–84. doi: 10.1038/79226. [DOI] [PubMed] [Google Scholar]
- 52.Drake KM, Zygmunt D, Mavrakis L, Harbor P, Wang L, Comhair SA, Erzurum SC, Aldred MA. Altered MicroRNA processing in heritable pulmonary arterial hypertension: an important role for Smad-8. Am J Respir Crit Care Med. 2011;184:1400–1408. doi: 10.1164/rccm.201106-1130OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brock M, Samillan VJ, Trenkmann M, Schwarzwald C, Ulrich S, Gay RE, Gassmann M, Ostergaard L, Gay S, Speich R, et al. AntagomiR directed against miR-20a restores functional BMPR2 signalling and prevents vascular remodelling in hypoxia-induced pulmonary hypertension Eur Heart JIn press [DOI] [PubMed] [Google Scholar]
- 54.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 55.Karube Y, Tanaka H, Osada H, Tomida S, Tatematsu Y, Yanagisawa K, Yatabe Y, Takamizawa J, Miyoshi S, Mitsudomi T, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–115. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 57.Rotunno M, Zhao Y, Bergen AW, Koshiol J, Burdette L, Rubagotti M, Linnoila RI, Marincola FM, Bertazzi PA, Pesatori AC, et al. Inherited polymorphisms in the RNA-mediated interference machinery affect microRNA expression and lung cancer survival. Br J Cancer. 2010;103:1870–1874. doi: 10.1038/sj.bjc.6605976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu SL, Chen HY, Chang GC, Chen CY, Chen HW, Singh S, Cheng CL, Yu CJ, Lee YC, Chen HS, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13:48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 59.Voortman J, Goto A, Mendiboure J, Sohn JJ, Schetter AJ, Saito M, Dunant A, Pham TC, Petrini I, Lee A, et al. MicroRNA expression and clinical outcomes in patients treated with adjuvant chemotherapy after complete resection of non-small cell lung carcinoma. Cancer Res. 2010;70:8288–8298. doi: 10.1158/0008-5472.CAN-10-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 61.Seike M, Goto A, Okano T, Bowman ED, Schetter AJ, Horikawa I, Mathe EA, Jen J, Yang P, Sugimura H, et al. MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc Natl Acad Sci USA. 2009;106:12085–12090. doi: 10.1073/pnas.0905234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vosa U, Vooder T, Kolde R, Vilo J, Metspalu A, Annilo T. Meta-analysis of microRNA expression in lung cancer. Int J Cancer. 2013;132:2884–2893. doi: 10.1002/ijc.27981. [DOI] [PubMed] [Google Scholar]
- 64.Lu Y, Govindan R, Wang L, Liu PY, Goodgame B, Wen W, Sezhiyan A, Pfeifer J, Li YF, Hua X, et al. MicroRNA profiling and prediction of recurrence/relapse-free survival in stage I lung cancer. Carcinogenesis. 2012;33:1046–1054. doi: 10.1093/carcin/bgs100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, van Rooij E, Olson EN. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. 2010;18:282–293. doi: 10.1016/j.ccr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boeri M, Verri C, Conte D, Roz L, Modena P, Facchinetti F, Calabro E, Croce CM, Pastorino U, Sozzi G. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci USA. 2011;108:3713–3718. doi: 10.1073/pnas.1100048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sozzi G, Boeri M, Rossi M, Verri C, Suatoni P, Bravi F, Roz L, Conte D, Grassi M, Sverzellati N, et al. Clinical utility of a plasma-based miRNA signature classifier within computed tomography lung cancer screening: a correlative MILD trial study. J Clin Oncol. 2014;32:768–773. doi: 10.1200/JCO.2013.50.4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010;101:2087–2092. doi: 10.1111/j.1349-7006.2010.01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li P, Zhao GQ, Chen TF, Chang JX, Wang HQ, Chen SS, Zhang GJ. Serum miR-21 and miR-155 expression in idiopathic pulmonary fibrosis. J Asthma. 2013;50:960–964. doi: 10.3109/02770903.2013.822080. [DOI] [PubMed] [Google Scholar]
- 70.Zhang H, Sun Z, Wei W, Liu Z, Fleming J, Zhang S, Lin N, Wang M, Chen M, Xu Y, et al. Identification of serum microRNA biomarkers for tuberculosis using RNA-seq. PLoS ONE. 2014;9:e88909. doi: 10.1371/journal.pone.0088909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maertzdorf J, Weiner J, III, Mollenkopf HJ, Bauer T, Prasse A, Muller-Quernheim J, Kaufmann SH. Common patterns and disease-related signatures in tuberculosis and sarcoidosis. Proc Natl Acad Sci USA. 2012;109:7853–7858. doi: 10.1073/pnas.1121072109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lewis A, Riddoch-Contreras J, Natanek SA, Donaldson A, Man WD, Moxham J, Hopkinson NS, Polkey MI, Kemp PR. Downregulation of the serum response factor/miR-1 axis in the quadriceps of patients with COPD. Thorax. 2012;67:26–34. doi: 10.1136/thoraxjnl-2011-200309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schlosser K, White RJ, Stewart DJ. miR-26a linked to pulmonary hypertension by global assessment of circulating extracellular microRNAs. Am J Respir Crit Care Med. 2013;188:1472–1475. doi: 10.1164/rccm.201308-1403LE. [DOI] [PubMed] [Google Scholar]
- 74.Xing L, Todd NW, Yu L, Fang H, Jiang F. Early detection of squamous cell lung cancer in sputum by a panel of microRNA markers. Mod Pathol. 2010;23:1157–1164. doi: 10.1038/modpathol.2010.111. [DOI] [PubMed] [Google Scholar]
- 75.Shen J, Liao J, Guarnera MA, Fang H, Cai L, Stass SA, Jiang F. Analysis of MicroRNAs in sputum to improve computed tomography for lung cancer diagnosis. J Thorac Oncol. 2014;9:33–40. doi: 10.1097/JTO.0000000000000025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Van Pottelberge GR, Mestdagh P, Bracke KR, Thas O, van Durme YM, Joos GF, Vandesompele J, Brusselle GG. MicroRNA expression in induced sputum of smokers and patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2011;183:898–906. doi: 10.1164/rccm.201002-0304OC. [DOI] [PubMed] [Google Scholar]
- 77.Sinha A, Yadav AK, Chakraborty S, Kabra SK, Lodha R, Kumar M, Kulshreshtha A, Sethi T, Pandey R, Malik G, et al. Exosome-enclosed microRNAs in exhaled breath hold potential for biomarker discovery in patients with pulmonary diseases. J Allergy Clin Immunol. 2013;132:219–222. doi: 10.1016/j.jaci.2013.03.035. [DOI] [PubMed] [Google Scholar]
- 78.Gibb EA, Enfield KS, Stewart GL, Lonergan KM, Chari R, Ng RT, Zhang L, MacAulay CE, Rosin MP, Lam WL. Long non-coding RNAs are expressed in oral mucosa and altered in oral premalignant lesions. Oral Oncol. 2011;47:1055–1061. doi: 10.1016/j.oraloncology.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 79.Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 80.Schmidt LH, Spieker T, Koschmieder S, Schaffers S, Humberg J, Jungen D, Bulk E, Hascher A, Wittmer D, Marra A, et al. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol. 2011;6:1984–1992. doi: 10.1097/JTO.0b013e3182307eac. [DOI] [PubMed] [Google Scholar]
- 81.Zong X, Tripathi V, Prasanth KV. RNA splicing control: yet another gene regulatory role for long nuclear noncoding RNAs. RNA Biol. 2011;8:968–977. doi: 10.4161/rna.8.6.17606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y, Chen N, Sun F, Fan Q. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Khaitan D, Dinger ME, Mazar J, Crawford J, Smith MA, Mattick JS, Perera RJ. The melanoma-upregulated long noncoding RNA SPRY4–IT1 modulates apoptosis and invasion. Cancer Res. 2011;71:3852–3862. doi: 10.1158/0008-5472.CAN-10-4460. [DOI] [PubMed] [Google Scholar]
- 84.Gee HE, Buffa FM, Camps C, Ramachandran A, Leek R, Taylor M, Patil M, Sheldon H, Betts G, Homer J, et al. The small-nucleolar RNAs commonly used for microRNA normalisation correlate with tumour pathology and prognosis. Br J Cancer. 2011;104:1168–1177. doi: 10.1038/sj.bjc.6606076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Elmen J, Lindow M, Silahtaroglu A, Bak M, Christensen M, Lind-Thomsen A, Hedtjarn M, Hansen JB, Hansen HF, Straarup EM, et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36:1153–1162. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 87.Bader AG. miR-34 - a microRNA replacement therapy is headed to the clinic. Front Genet. 2012;3:120. doi: 10.3389/fgene.2012.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]