Abstract
An important pool of cardiovascular progenitor cells arises from the epicardium, a single layer of mesothelium lining the heart. Epicardium-derived progenitor cell (EPDC) formation requires epithelial-to-mesenchymal transition (EMT) and the subsequent migration of these cells into the sub-epicardial space. Although some of the physiological signals that promote EMT are understood, the functional mediators of EPDC motility and differentiation are not known. Here, we identify a novel regulatory mechanism of EPDC mobilization. Myocardin-related transcription factor (MRTF)-A and MRTF-B (MKL1 and MKL2, respectively) are enriched in the perinuclear space of epicardial cells during development. Transforming growth factor (TGF)-β signaling and disassembly of cell contacts leads to nuclear accumulation of MRTFs and the activation of the motile gene expression program. Conditional ablation of Mrtfa and Mrtfb specifically in the epicardium disrupts cell migration and leads to sub-epicardial hemorrhage, partially stemming from the depletion of coronary pericytes. Using lineage-tracing analyses, we demonstrate that sub-epicardial pericytes arise from EPDCs in a process that requires the MRTF-dependent motile gene expression program. These findings provide novel mechanisms linking EPDC motility and differentiation, shed light on the transcriptional control of coronary microvascular maturation and suggest novel therapeutic strategies to manipulate epicardium-derived progenitor cells for cardiac repair.
Keywords: Heart, Epicardium, Migration, Progenitor, Pericyte, Transcription
Highlighted article: Myocardin-related transcription factors respond to TGFβ signalling and control both the motility of mouse epicardium-derived progenitors and the maturation of coronary vessels.
INTRODUCTION
The formation of a functioning heart depends upon the coordinated migration and differentiation of a progenitor cell population from the epicardium, a single cell layer of mesothelium lining the heart. Epicardium-derived progenitor cells (EPDC) arise following epithelial-to-mesenchymal transition (EMT) (Martinez-Estrada et al., 2010; von Gise et al., 2011), and impact heart development by secretion of paracrine factors or by directly contributing to various cardiac lineages, including vascular endothelial cells (ECs), vascular smooth muscle cells (SMCs) and fibroblasts (Cai et al., 2008; Dettman et al., 1998; Li et al., 2011; Martinez-Estrada et al., 2010; Mikawa and Gourdie, 1996; Tian et al., 2013; Wilm et al., 2005; Zhou et al., 2008). Physical disruption or ablation of the epicardium leads to aberrant vascular plexus formation and myocardial hypoperfusion, highlighting the role of EPDCs in coronary vessel formation (Eralp et al., 2005; Gittenberger-de Groot et al., 2000; Manner et al., 2005). Epicardial cells can also become migratory in the adult, providing a potential source of resident progenitor cells that have the capacity to repopulate damaged cardiac tissue (Lepilina et al., 2006; Limana et al., 2007; Smart et al., 2011; Zhou et al., 2011).
Epicardial EMT is regulated by a complex combination of extracellular molecules, including Wnts, platelet-derived growth factor (PDGF), transforming growth factor (TGF)-β, fibroblast growth factor, and retinoic acid (Austin et al., 2008; Kim et al., 2010; Mellgren et al., 2008; Merki et al., 2005; Pennisi and Mikawa, 2009). These upstream signals are thought to impact upon EMT partially through the induction of Snai1 and Snai2 expression (Martinez-Estrada et al., 2010). However, recent evidence suggests that canonical pathways might not be essential for epicardial EMT (Casanova et al., 2013). More importantly, the functional mediators of EPDC mobilization and cell motility remain poorly defined.
Serum response factor (SRF) controls gene expression by binding to and activating a DNA element called a CArG box (CCA/T6GG) (Miano, 2003). Although SRF is expressed in multiple tissues, including the epicardium (Lu et al., 2001; Nelson et al., 2004), SRF-dependent gene expression largely depends upon the availability of tissue-specific or signal-responsive cofactors (Olson and Nordheim, 2010; Wang et al., 2001, 2004). Cell motility and contractility is driven by interactions between SRF and myocardin-related transcription factor-A (MRTF-A, also known as MKL1, MAL and BSAC) and MRTF-B (MKL1) (Medjkane et al., 2009; Olson and Nordheim, 2010; Wang et al., 2002). In vitro, MRTFs are sequestered in the cytoplasm through interactions with G-actin; however, upon mechanical tension, TGF-β stimulation or Rho-kinase (ROCK) activation, changes in actin dynamics allow for MRTF nuclear accumulation (Busche et al., 2010; Fan et al., 2007; Guettler et al., 2008; Miralles et al., 2003; Olson and Nordheim, 2010; Sandbo et al., 2011; Small et al., 2010). Genetic deletion of Mrtfa in mice disrupts the differentiation of mammary myoepithelial cells (Li et al., 2006), attenuates myofibroblast differentiation and scar formation in various injury models (Small et al., 2010; Zhou et al., 2013), and blocks metastatic cancer cell migration (Medjkane et al., 2009). In contrast, Mrtfb deletion results in embryonic lethality at midgestation, largely attributed to reduced vascular SMC differentiation and aortic arch malformation (Li et al., 2005; Oh et al., 2005). MRTF-A and -B play redundant roles in certain contexts, such as neuronal migration and retinal vessel growth (Mokalled et al., 2010; Weinl et al., 2013). Taken together, MRTFs link extracellular signals and cytoskeletal dynamics to a gene expression program that drives cell contractility, motility and differentiation.
Here, we show that MRTF-A and -B play a crucial role in coronary vessel maturation and integrity by stimulating EPDC motility and mobilizing cardiovascular progenitors. We demonstrate that MRTFs are enriched in the epicardium prior to EMT and are required and sufficient for EPDC migration in vitro and in vivo. Mice that lack both Mrtfa and Mrtfb in the epicardium display disorganized coronary plexus formation, EC dysfunction and sub-epicardial hemorrhage, stemming in part from the depletion of epicardium-derived coronary pericytes. These findings provide novel insight into the developmental mechanisms driving coronary vessel formation and might lead to strategies for cardiac repair.
RESULTS
SRF and MRTFs are expressed in the epicardium
To test whether members of the SRF-MRTF signaling axis are expressed in a manner consistent with potentiating epicardial EMT and EPDC differentiation, we isolated epicardial cells from embryonic day (E)11.5 embryos by outgrowth from cardiac explants (Austin et al., 2008). Expression analysis by quantitative real-time RT-PCR (qPCR) revealed robust expression of various epicardial marker genes, including Wt1, Tcf21 and Krt14, and low expression of cardiomyocyte genes (Myh7 and Nkx2-5), confirming the purity of our culture system (Fig. 1A). Srf is expressed in both EPDCs and whole heart fractions (Fig. 1A). Somewhat unexpectedly, although myocardin (Myocd) displayed cardiomyocyte-specific expression, both Mrtfa and Mrtfb showed significant enrichment in EPDCs (Fig. 1A).
Fig. 1.
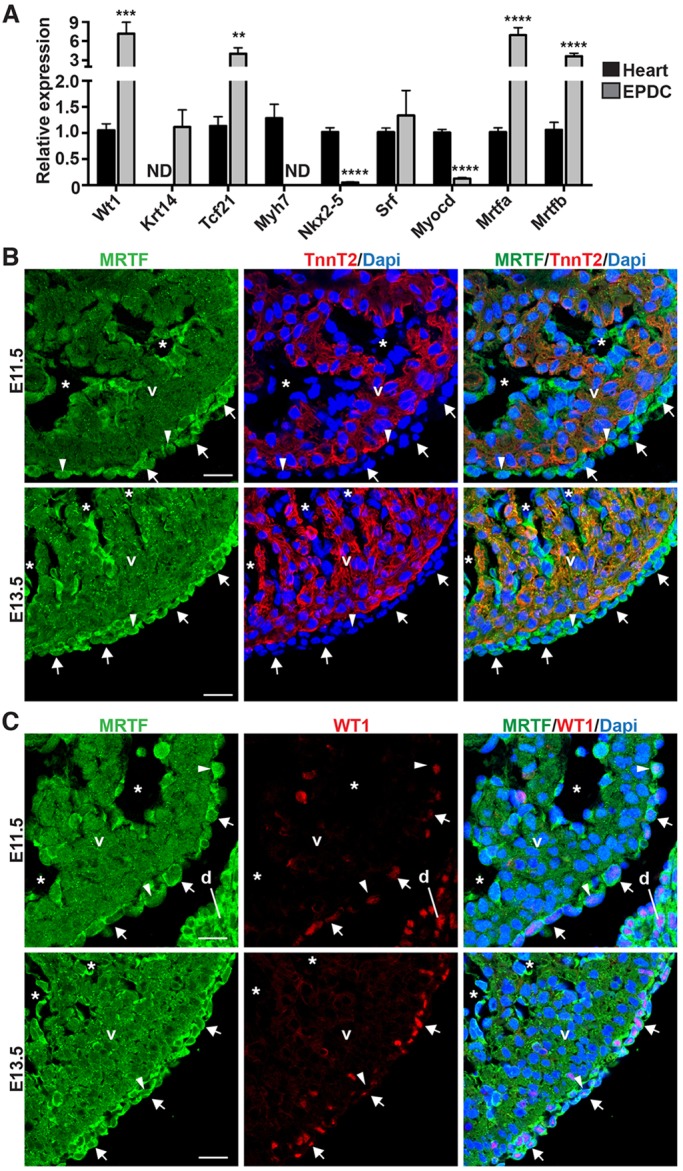
MRTFs are expressed in the epicardium. (A) qPCR analysis of primary EPDC cultures and corresponding EPDC-depleted E11.5 hearts. ND, not detected. Data represent the mean±s.e.m. [n=7 (heart) or n=4 (EPDC)]; **P<0.01; ***P<0.001; ****P<0.0001. (B,C) Co-immunofluorescent staining of E11.5 and E13.5 ventricles using antibodies directed against MRTF (green) and TNNT2 (red, B) or WT1 (red, C). MRTF proteins are enriched in the epicardium (arrows) and endocardium (asterisks). Nuclear accumulation of MRTF is observed in a subset of epicardial cells (arrowheads). d, diaphragm; v, ventricle. Scale bars: 20 µm.
Staining with an antibody that recognizes both MRTF-A and MRTF-B (supplementary material Fig. S1A-D) confirmed the enrichment of MRTF proteins in the epicardium (Fig. 1B,C; supplementary material Fig. S1E,F). Robust epicardial and endocardial expression was observed at E11.5 and E13.5, and MRTFs were found to colocalize with WT1-positive epicardial cells in an expression domain distinct from cardiac troponin T2 (TNNT2)-positive myocardium (Fig. 1B,C). Beginning at ∼E15.5, MRTFs are enriched in cells within the myocardial wall, including interstitial and vascular cells, indicative of an epicardial origin (supplementary material Fig. S1E). By E17.5, MRTFs are expressed in the epicardium, sub-epicardial mesenchymal cells, interstitial cells and vascular cells (supplementary material Fig. S1F). Although MRTFs are primarily localized to the perinuclear space, we occasionally observed nuclear accumulation in the epicardium and sub-epicardial mesenchymal cells. Taken together, these data show that SRF and MRTFs are expressed in a spatial and temporal manner consistent with a role in epicardial EMT and EPDC migration.
SRF and MRTFs are required for the epicardial motile gene program
To examine the potential role of SRF and MRTFs in epicardial EMT, we first asked whether MRTF-A shuttled to the nucleus of epicardial cells in a manner consistent with stimulating their motility. Epicardial-mesothelial cells (EMCs) (Wada et al., 2003) were transduced with an epitope-tagged MRTF-A adenovirus and stimulated to undergo EMT by treatment with TGF-β1. TGF-β1 treatment led to a striking accumulation of MRTF-A within the nucleus, primarily in cells migrating out of the periphery of the culture (Fig. 2A,B).
Fig. 2.
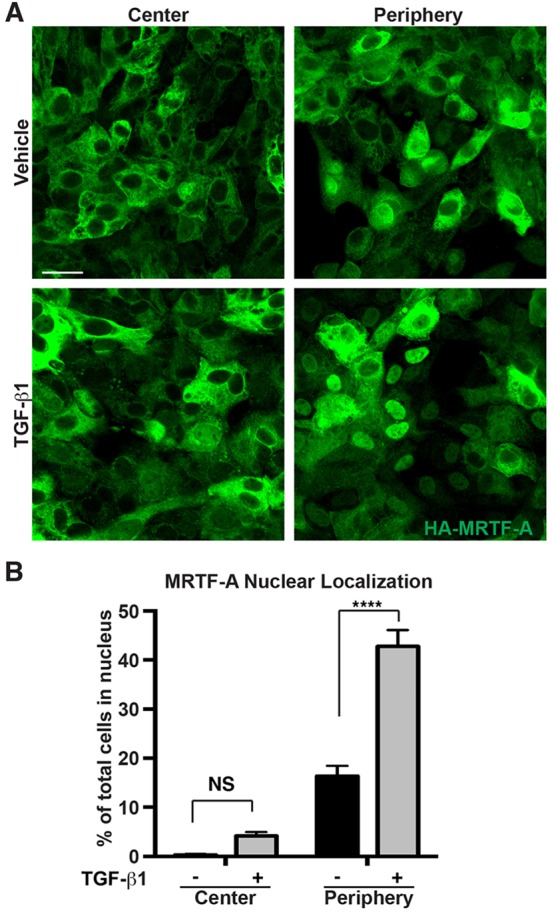
MRTF nuclear accumulation is regulated by TGF-β1 and cell contact. (A) Serum-starved EMCs transduced with HA-tagged MRTF-A adenovirus and treated with TGF-β1 (10 ng/ml) or vehicle. HA immunostaining identifies nuclear MRTF-A at the center or periphery of EMC monolayers. Scale bar: 25 µm. (B) Quantification of the data presented in A. Nuclear accumulation of MRTF-A was significantly enhanced by both TGF-β1 and low cell confluence as determined by a two-way ANOVA. Data show the mean±s.e.m. (≥1000 cells scored per treatment group within three independent experiments). ****P<0.0001; NS, not significant (Tukey post-hoc test).
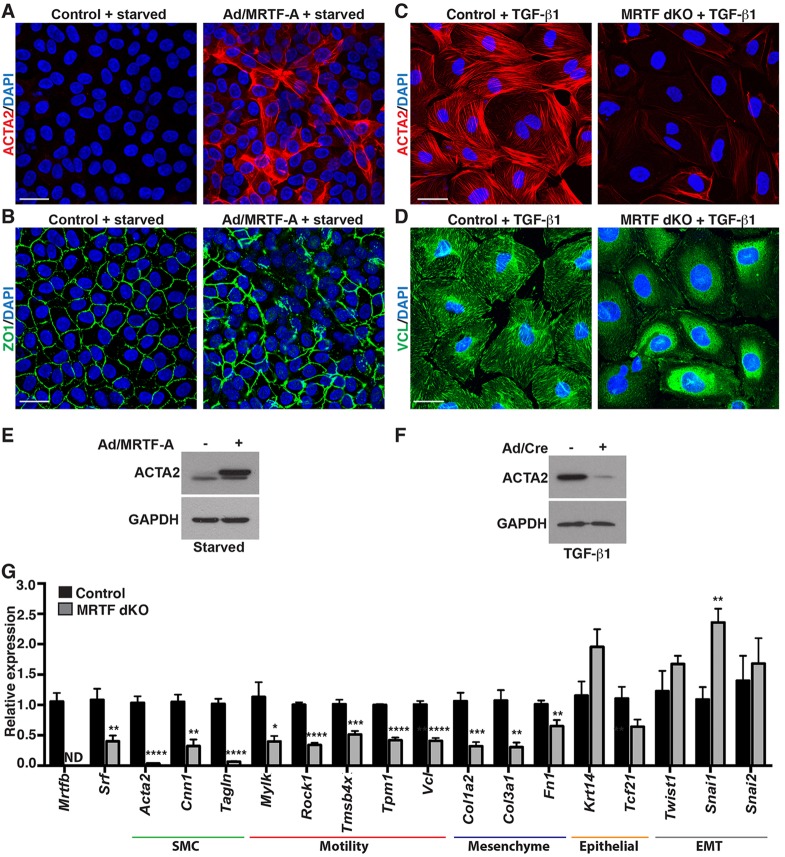
Next, we manipulated SRF or MRTF levels and examined markers of EMT and cell motility. Compared with control serum-starved EMC cultures, which displayed minimal ACTA2 (also known as SMA, smooth muscle α actin) staining, ∼12-fold overexpression of MRTF-A driven from an adenoviral vector (supplementary material Fig. S2A,B) resulted in robust staining that was localized to stress fibers (Fig. 3A). Zona occludens protein 1 (ZO1; also known as TJP1 – Mouse Genome Informatics), which marks epithelial tight junctions and is disrupted upon mesenchymal transformation (Kim et al., 2010), was lost in MRTF-A-transduced cells that also expressed high levels of ACTA2 (Fig. 3B). These cells exhibited an elongated morphology indicative of mesenchymal cells, in contrast to the ‘cobblestone-like’ epithelial arrangement in control cultures.
Fig. 3.
MRTFs are required for the epicardial motile gene program. (A) Adenovirus expressing MRTF-A (Ad/MRTF-A) in serum-starved EMCs induced robust ACTA2 protein expression compared with βgal (control)-transduced cultures. (B) ZO1 staining corresponding to A. (C,D) EPDCs isolated from Mrtfa−/−; Mrtfbfl/fl embryos, cultured in 1% serum and transduced with adenovirus expressing βgal (control) or Cre (MRTF dKO) were treated with TGF-β1 and immunostained for ACTA2 (C) or VCL (D). Scale bars: 25 µm. (E,F) Western blot analysis of serum-starved EMCs transduced with control or Ad/MRTF-A (E) or TGF-β1-treated EPDCs isolated from Mrtfa−/−; Mrtfbfl/fl embryos and transduced with adenovirus expressing βgal or Cre (Ad/Cre) (F). (G) qPCR analysis of TGF-β1-treated Mrtfa−/−; Mrtfbfl/fl EPDCs transduced with control or Cre adenovirus. ND, not detectable. Data are presented as the mean±s.e.m. (n=6-7 per group; data represent at least two independent experiments); *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001.
Similarly, TGF-β1-stimulated primary epicardial cells isolated from embryos harboring a homozygous null allele of Mrtfa and floxed alleles of Mrtfb displayed robust ACTA2-positive stress fibers associated with well-defined vinculin (VCL)-positive focal adhesions (Fig. 3C,D). In contrast, transduction with a Cre-expressing adenoviral vector resulted in the efficient deletion of the Mrtfb allele (supplementary material Fig. S2C; called MRTF dKO), a near complete elimination of ACTA2 staining (Fig. 3C) and focal adhesion disassembly (Fig. 3D). The alteration of ACTA2 protein levels upon MRTF-A overexpression and knockout was substantiated by western blotting (Fig. 3E,F).
qPCR analysis confirmed that MRTF dKO EPDCs displayed a significant reduction in the expression of the SMC and cell motility markers Acta2, Cnn1, Tagln (SM22), myosin light chain kinase (Mylk), Rho-associated protein kinase 1 (Rock1), thymosin b4 (Tmsb4x), tropomyosin 1 (Tpm1) and Vcl, and the extracellular matrix (ECM)/mesenchyme markers Col1a2, Col3a1 and Fn1 (Fig. 3G). In contrast, MRTF dKO EPDCs retained the expression of epithelium/epicardium markers (Krt14 and Tcf21). The attenuation of ACTA2 staining and cell motility gene expression was also confirmed upon deletion of Srf in primary epicardial explant cultures (supplementary material Fig. S3A,B). Surprisingly, the reduced motile gene program in MRTF dKO EPDCs was not associated with a reduced gene expression of Twist1, Snai1 or Snai2, which are important transcriptional regulators of the EMT program (Fig. 3G).
SRF and MRTFs are required for epicardial cell migration
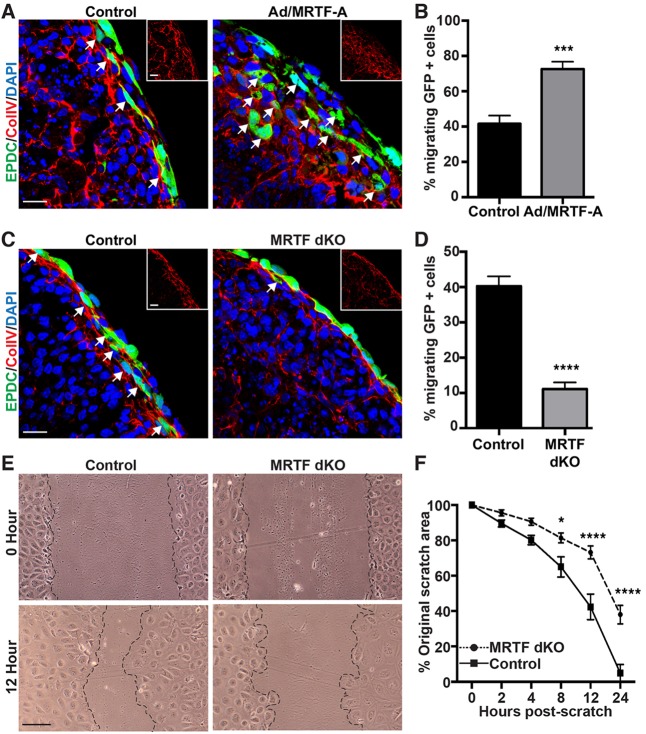
Although the motile gene expression program is altered in SRF-null and MRTF dKO epicardial cells, these cells might still take on mesenchymal characteristics and migrate into the sub-epicardial space of the intact heart. Therefore, we undertook an ex vivo embryonic heart culture assay to track EPDC migration following transduction with a GFP-expressing adenovirus, previously reported to label only the epicardium (Mellgren et al., 2008). Hearts of E12.5 embryos co-transduced with an MRTF-A-expressing adenovirus and cultured until the equivalent of E14.5 displayed exaggerated and disorganized migration of GFP-positive EPDCs into the sub-epicardial space and beyond, which was associated with a disruption of the type IV collagen (ColIV)-positive epicardial basement membrane (Fig. 4A,B). In contrast, deletion of either Mrtfaand Mrtfb (Fig. 4C) or Srf (supplementary material Fig. S3C) with a Cre-expressing adenovirus significantly attenuated EPDC migration across the ColIV-positive basement membrane. Quantification of the movement of EPDCs revealed a 75% reduction in migration in MRTF-depleted hearts (Fig. 4D) and a 25% reduction in SRF-depleted hearts (supplementary material Fig. S3D).
Fig. 4.
MRTFs drive epicardial cell migration. (A-D) E12.5 hearts from C57BL/6 (A,B) or Mrtfa−/−; Mrtfbfl/fl (C,D) embryos were explanted and transduced with adenovirus expressing GFP to mark the epicardium. (A,C) Immunostaining for GFP (green) and ColIV (red, inset) reveals migration of EPDCs across the basement membrane. Migrating cells are marked by arrows. (A,B) Forced expression of MRTF-A promotes EPDC migration. (C,D) Deletion of MRTF-A and -B with adenovirus expressing Cre attenuates EPDC migration. (E) EPDCs isolated from Mrtfa−/−; Mrtfbfl/fl embryos were transduced with adenovirus expressing βgal (control) or Cre (MRTF dKO), scratched with a pipette tip and imaged at the indicated time points. Scale bars: 20 µm (A,C) or 100 µm (E). (F) Migration in E is presented as percentage of the original scratch area. Significant differences in F were determined using a two-way ANOVA and Tukey post-hoc test. Data are presented as the mean±s.e.m. (n=4 for each condition, and data represents the results of at least two independent experiments); *P<0.05; ***P<0.001; ****P<0.0001.
We also examined the role of MRTFs in the migration of primary EPDCs using an in vitro scratch wound assay. Deletion of MRTFs with a Cre adenovirus significantly abrogated wound closure (Fig. 4E,F). We observed a similar reduction in the migration of SRF-knockout EPDCs (supplementary material Fig. S3E,F). Altered proliferation of EPDCs is unlikely to account for the differences in wound closure, as we observed equivalent numbers of cells positive for Ki67 in the MRTF dKO cultures (supplementary material Fig. S3G,H). These data suggest the SRF-MRTF gene regulatory axis is necessary and sufficient for epicardial cell motility.
MRTFs are required for epicardial integrity
To ascertain the functional role of SRF-MRTF-dependent transcriptional regulation of EPDC motility during embryonic development, we utilized the tamoxifen-inducible Wt1CreERT2 mouse line (Rudat and Kispert, 2012; Zhou and Pu, 2012) to delete the conditional Mrtfb allele in mice harboring a global Mrtfa deletion. Tamoxifen administration at E9.5 and E10.5 led to consistent and specific recombination in the epicardium as demonstrated by β-galactosidase (βgal) expression from the Cre reporter locus (RosalacZ) (supplementary material Fig. S4A,B). Isolation of EPDCs from mice lacking both Mrtfa and Mrtfb in the epicardium from E9.5 onward (called MRTFepiDKO) revealed ∼50-70% reduction of Mrtfb in EPDCs, but no change of expression within the myocardium (supplementary material Fig. S4C).
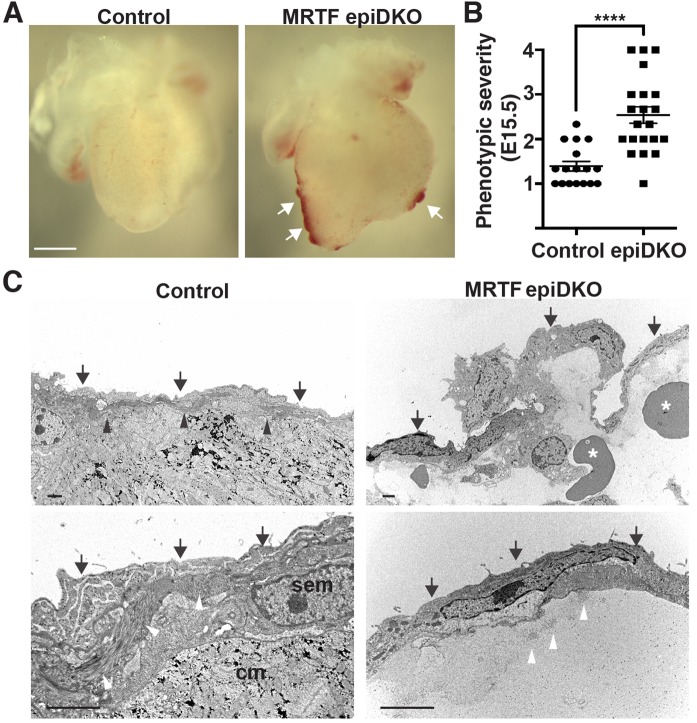
Gross examination of MRTFepiDKO embryos revealed sub-epicardial hemorrhage at E15.5 and E17.5 (Fig. 5A; supplementary material Fig. S5A,B). Quantification by three blinded reviewers based upon a phenotypic severity scale (supplementary material Fig. S5A) confirmed extreme sub-epicardial hemorrhage in ∼50% of MRTFepiDKO embryos (Fig. 5B; supplementary material Fig. S5C). Sub-epicardial hemorrhage was also observed in embryos devoid of Srf in EPDCs (supplementary material Fig. S5D,E). Hematoxylin and Eosin (HE)-stained hearts from MRTFepiDKO embryos displayed a non-adherent epicardium (supplementary material Fig. S6A). Proliferation of epicardial cells was subtly but not significantly reduced in MRTFepiDKO embryos, as demonstrated by co-staining for Ki67 and WT1 (supplementary material Fig. S6B,C).
Fig. 5.
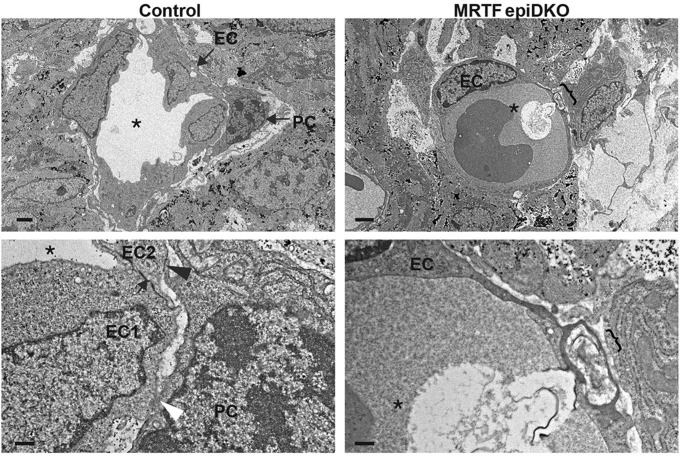
MRTFs are required for epicardial integrity. (A) Gross morphology of hearts isolated from control or MRTFepiDKO embryos at E15.5. Sub-epicardial hemorrhage is marked by arrows. Scale bar: 500 µm. (B) Hemorrhage severity scored at E15.5. n=17 (control), n=21 (MRTFepiDKO). Data are presented as the mean±s.e.m. (combined results of five independent litters); ****P<0.0001 (Mann–Whitney test). (C) TEM of E15.5 control hearts reveals a highly organized epicardial layer (arrows) that is closely associated with the basal lamina (black arrowheads). MRTFepiDKO hearts display a disorganized epicardium, which is often dissociated from the underlying myocardium and filled with red blood cells (asterisks) and exudate. Higher magnification images (lower panels) highlight the collagen-rich basal lamina in control hearts (white arrowheads) and tightly associated sub-epicardial mesenchymal cells (sem) and cardiomyocytes (cm). Individual collagen bundles (white arrowheads) are observed within the lesion of MRTFepiDKO hearts. Scale bars: 2 µm (upper panels) and 1 µm (lower panels). n=2 (control) and 3 (MRTFepiDKO).
We next examined the ultrastructure of E15.5 hearts by transmission electron microscopy (TEM). Hearts isolated from control embryos displayed a highly organized epicardium in close apposition with underlying sub-epicardial mesenchymal cells and cardiomyocytes (Fig. 5C; supplementary material Fig. S7). In contrast, MRTFepiDKO hearts displayed a profoundly disorganized epicardial layer that was often dissociated from the underlying myocardium. Individual collagen bundles were displaced from the basal lamina within the sub-epicardial lesion, which was often filled with blood cells and protein-rich exudate (Fig. 5C; supplementary material Fig. S7). Occasionally, MRTFepiDKO embryos displayed aberrant ECM organization within the sub-epicardial space (supplementary material Fig. S7).
MRTFepiDKO embryos were observed at a normal Mendelian ratio at E11.5, E13.5 and E15.5. Although we observed fewer MRTFepiDKO embryos than expected at E17.5, this failed to reach significance (supplementary material Table S1). Given that we were unable to evaluate postnatal viability owing to non-specific tamoxifen-associated lethality, we next utilized a Wt1CreBAC transgenic mouse line to delete Mrtfb within the epicardium of Mrtfa-null embryos (called MRTFBACdKO) (Wessels et al., 2012). MRTFBACdKO embryos phenocopied the deficits in the mesenchymal gene program observed upon tamoxifen-mediated deletion (M.A.T. and E.M.S., data not shown). Importantly, 5 out of 16 MRTFBACdKO offspring died within 48 h of birth, resulting in ∼50% of the expected number of this genotype (P=0.0113; supplementary material Table S2). These data suggest that loss of MRTF expression in the epicardium causes a ∼50% penetrant sub-epicardial hemorrhage phenotype invariably resulting in embryonic or neonatal lethality.
Disruption of coronary angiogenesis in MRTFepiDKO embryos
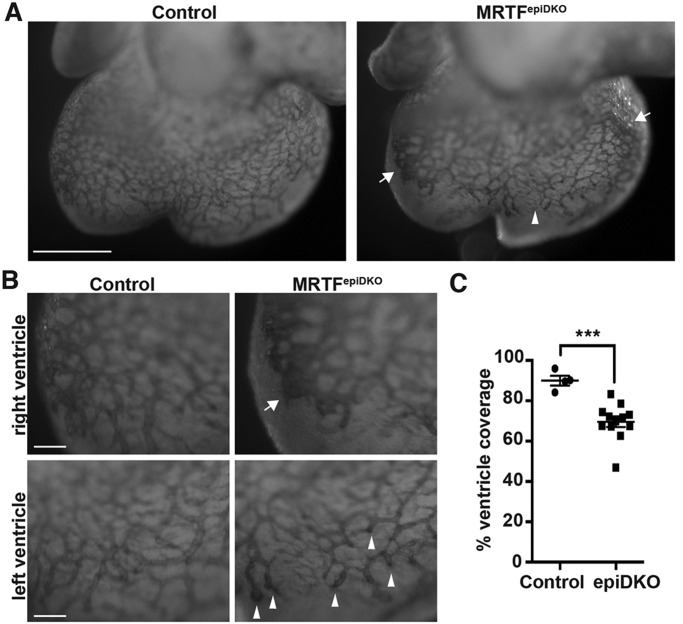
To gain insight into the structural basis of sub-epicardial hemorrhage in MRTFepiDKO embryos, we visualized the growing EC plexus by whole-mount PECAM1 (also known as CD31) staining. Control embryos display angiogenic sprouting from the sinus venosus and sub-epicardial growth of the primary endothelial plexus, which reaches the apex of the heart by ∼E14.5 (Red-Horse et al., 2010). E13.5 MRTFepiDKO hearts presented with aberrant vessels terminating prematurely at endothelial bundles and large sheets of PECAM1-positive cells resembling blood islands (Fig. 6A,B). Aberrant vessel formation led to a consistent reduction in ventricular coverage by the coronary plexus (Fig. 6C).
Fig. 6.
Epicardial MRTFs are required for normal coronary plexus formation. (A-B) Whole-mount PECAM1 staining of hearts isolated from E13.5 embryos. Arrows mark EC sheets and arrowheads mark EC foci observed in MRTFepiDKO hearts. Scale bars: 500 µm (A) and 50 µm (B). (C) Quantification of the area of vessel coverage expressed as a percentage of the area of the ventricles, as depicted in A. Data are presented as the mean±s.e.m. [n=4 (control) or n=12 (MRTFepiDKO); combined results of two independent litters]; ***P<0.001.
Ultrastructural analysis of sub-epicardial vessels in control E15.5 hearts revealed tight junctions linking neighboring ECs, which were often associated with an overlying pericyte (Fig. 7; supplementary material Fig. S8). Pericytes displayed cytoplasmic processes extending around the endothelial tube, with characteristic peg and socket connections and adhesion plaques between the pericyte and EC, based upon electron-dense foci. In contrast, reduced vessel integrity was observed in MRTFepiDKO embryos, which often contained dysfunctional ECs displaying nuclear and cytoplasmic granulation. Thin-walled microvessel ECs occasionally exhibited convoluted junctions, macrovesicular uptake and disrupted continuity (Fig. 7; supplementary material Fig. S8). Although MRTFepiDKO microvessels often contained ultrastructurally normal perictyes, qualitatively, these seemed to be sparse compared with those of controls. These results are consistent with the hemorrhage observed in MRTFepiDKO embryos; however, the cellular cause of EC disruption was not apparent. In fact, our lineage tracing analyses using mice harboring both Wt1CreERT2 and RosamTmG alleles, in which EPDCs are irreversibly labeled with membrane-bound GFP, confirmed recent reports suggesting that most sub-epicardial ECs do not originate from Wt1Cre-positive cells (supplementary material Fig. S9A,B) (Katz et al., 2012; Tian et al., 2013; Wilm et al., 2005; Zhou et al., 2008). Thus, we reasoned that EC disruption upon conditional deletion of MRTFs within WT1CreERT2-positive epicardial cells might occur largely through a non-cell-autonomous mechanism.
Fig. 7.
Ultrastructural analysis of sub-epicardial vessels. TEM of sub-epicardial microvessels in E15.5 hearts reveals EC tight junctions (arrows), peg and socket interactions (white arrowhead), adhesion plaques (black arrowhead) and vessel lumen (asterisk) in a control embryo. The bracket marks a disrupted EC layer in the MRTFepiDKO embryo. Scale bars:2 µm (upper) and 0.5 µm (lower). PC, pericyte; EC, endothelial cell. n=2 (control) and n=3 (MRTFepiDKO).
The differentiation of epicardium-derived pericytes requires MRTF-dependent cell motility
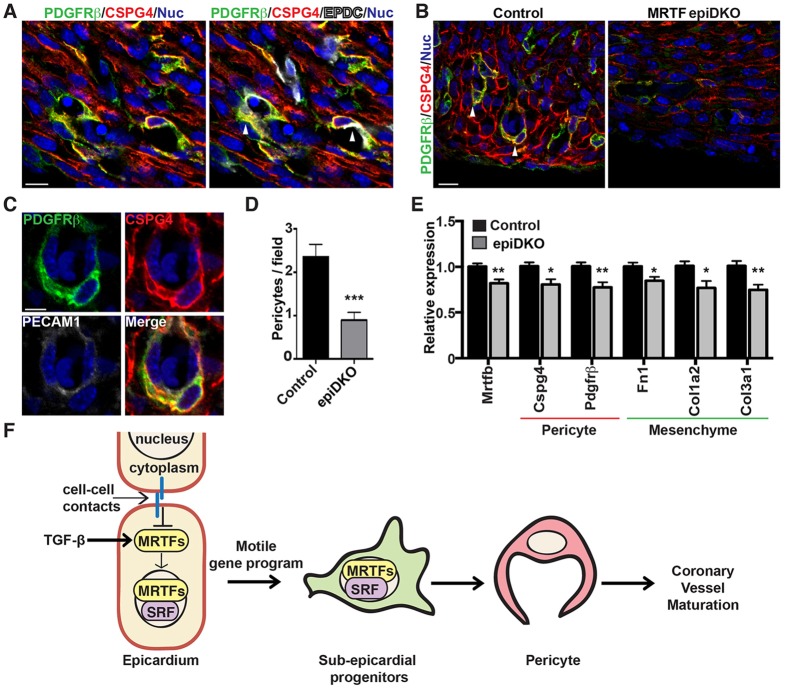
Maturation and homeostasis of the vascular endothelial plexus in the embryo and adult depends upon appropriate investment of perivascular cells. Although perivascular cells, including vascular SMCs, fibroblasts and pericytes, are generally thought to originate from the mesothelium in coelomic organs (Armulik et al., 2011; Mikawa and Gourdie, 1996), the source of pericytes within the developing heart has not been unambiguously defined. We defined pericytes as CSPG4+ PDGFRβ+ cells that displayed cytoplasmic projections ensheathing a microvessel wall. Triple staining with antibodies against GFP, CSPG4 and PDGFRβ in Wt1CreERT2;RosamTmG embryos thus allowed for identification of epicardium-derived pericytes (Fig. 8A). Although our analysis was limited by the efficiency of Cre-mediated recombination, quantification revealed that ∼25% of sub-epicardial coronary pericytes are derived from a Wt1CreERT2-positive epicardial source.
Fig. 8.
MRTFs are essential for pericyte differentiation from EPDCs. (A) Wt1CreERT2; RosamTmG embryos were isolated at E15.5 from dams gavaged with tamoxifen at E12.5. Immunofluorescence analysis reveals colocalization of GFP-labeled EPDCs (white) with pericyte markers, PDGFRβ (green) and CSPG4 (red). Nuclei are labeled with TOPRO (blue). (B,C) E15.5 control and MRTFepiDKO embryos stained with antibodies against PDGFRβ (green), CSPG4 (red) and PECAM1 (white) to identify sub-epicardial pericytes (arrowheads). Scale bars: 10 µm (A,B) and 5 µm (C). (D) Quantification of images from B revealed significantly fewer pericytes lining sub-epicardial microvessels in MRTFepiDKO embryos. n=5 per group. (E) qPCR analysis of pericyte and mesenchyme markers in whole ventricles isolated from control and MRTFepiDKO embryos at E15.5. n=9 (control) and n=14 (epiDKO). Data represent the combined results of three independent litters. All data are presented as the mean±s.e.m.; *P<0.05; **P<0.01; ***P<0.001. (F) Physiological cues such as TGF-β1 signaling and/or cell junction disassembly induce the nuclear accumulation of MRTFs and expression of target genes that drive the migration of EPDCs into the sub-epicardial space. MRTF-directed EPDC motility is essential for pericyte differentiation and coronary vessel maturation.
We speculated that the loss of coronary vessel integrity in MRTFepiDKO embryos might be due to defective pericyte investment from the epicardium. As expected, E15.5 control embryos exhibited well-formed, CSPG4+ PDGFRβ+ pericytes lining sub-epicardial PECAM1-positive vessels (Fig. 8B,C). In contrast, CSPG4 and PDGFRβ expression was attenuated in MRTFepiDKO embryos, correlating with a ∼50% reduction in the number of sub-epicardial pericytes (Fig. 8B,D). Although the expression of various pericyte, EC and cell motility genes was reduced in E15.5 MRTFepiDKO ventricles, we did not observe appreciable attenuation of the fibroblast or SMC programs at this stage by qPCR (Fig. 8E; supplementary material Fig. S10). Taken together, our data implicate the SRF-MRTF gene regulatory axis in coronary vessel maturation and homeostasis, at least in part through the control of EPDC motility and the subsequent differentiation of sub-epicardial pericytes.
DISCUSSION
Despite more than a century of research since the pericyte was first described (Rouget, 1873), the origin and function of these cells within the heart remains obscure. Pericytes in the neurovascular niche have proven to be essential in diverse processes such as angiogenesis, vessel maturation, homeostasis and permeability, and vascular tone (Aguilera and Brekken, 2014; Armulik et al., 2011). Cardiotoxicity in sunitinib-treated patients has recently been attributed to the depletion of coronary pericytes and subsequent microvascular dysfunction (Chintalgattu et al., 2013), suggesting an equally important role for pericytes in the heart. However, there remains a significant lag in the understanding of coronary pericyte biology, which is partially due to the difficulty in unambiguously identifying and isolating pericytes in the heart. In this study, we present evidence that coronary pericytes originate in part from the epicardium in a process that is dependent upon activation of the motile gene program by SRF-MRTFs (Fig. 8F).
MRTFs accumulate within the nucleus of a subset of epicardial and sub-epicardial cells, and are essential for their migration. Although various signaling pathways that induce MRTF nuclear shuttling are active in the epicardium, including TGF-β and Rho-ROCK, it is not clear why all MRTF-expressing cells are not capable of responding. The initial stages of EMT involve the dissolution of epithelial cell junctions, which promote changes in cell polarity and cause reorganization of the actin cytoskeleton (Lamouille et al., 2014). In addition, loss of cell-cell contacts has also been shown to potentiate MRTF-A activity in vitro (Busche et al., 2010; Fan et al., 2007), and we observe nuclear MRTF-A primarily within migrating epicardial cells at the periphery of a monolayer. Although other physiological cues might be at work, our results suggest epicardial cells with weaker contacts might be particularly susceptible to MRTF-mediated motility.
The primary phenotype in MRTFepiDKO embryos is sub-epicardial hemorrhage due to loss of EC integrity, which we hypothesize is secondary to pericyte deficiency. Given that the epicardium is made up of a heterogeneous population of progenitor cells with the capacity to give rise to multiple cardiac cell types, it is conceivable that MRTFs also control EC differentiation and migration directly, as reported in the postnatal retinal vasculature and adult neoangiogenesis (Hinkel et al., 2014; Weinl et al., 2013). However, our current study confirms previous findings that coronary ECs originating from the sub-epicardium are primarily descendants of a Wt1Cre-negative lineage (Katz et al., 2012; Tian et al., 2013; Wilm et al., 2005; Zhou et al., 2008). These results imply that the loss of EC integrity upon WT1CreERT2-specific deletion of MRTFs within the epicardium arises at least partially non-cell autonomously. However, further studies are required to rule out a role for MRTFs in ECs derived from other epicardial cell populations or a potential role for epicardial MRTFs in cardiac fibroblast differentiation in the embryo or after injury.
Given the considerable promiscuity of currently available pericyte markers, we have taken great care to identify and quantify this cell type unambiguously. Our stringent methods proved essential, as 80% of CSPG4+ PDGFRβ+ cells did not exhibit clear pericyte morphology. Of the cells that we define as pericytes, 25% originate from the epicardium. Upon deletion of MRTF-A and -B in the epicardium, we observed an ∼50% reduction in pericyte number. It is possible that, in addition to the cell autonomous loss of pericytes due to deficient EPDC migration, inefficient recruitment of pericytes by ECs might contribute to the overall phenotype.
Dormant throughout postnatal life, the epicardium is activated in response to cardiac injury or ischemic events, contributing to fibroblast and nascent vessel formation in experimental models and human patient samples (Braitsch et al., 2013; Limana et al., 2007; Smart et al., 2011; Zhou et al., 2011). Harnessing the natural regenerative capacity of epicardium-derived cardiovascular progenitor cells by modulating MRTF activity presents an exciting therapeutic strategy. Unlike most transcription factors, which are difficult to target pharmacologically, affecting MRTF localization and activity has proven feasible (Evelyn et al., 2007; Haak et al., 2014; Velasquez et al., 2013). It is interesting to speculate that epicardium-derived progenitor cells might be manipulated to promote neoangiogenesis and cardiac repair at the expense of fibroblast accumulation and scar formation.
MATERIALS AND METHODS
Mice
All experiments using animals were approved by the University Committee on Animal Resources at the University of Rochester. Mrtfa−/− and Mrtfbflox mice were as described previously (Li et al., 2006; Oh et al., 2005) and were kind gifts from Dr Eric Olson (UT Southwestern, Dallas, TX, USA). The Srfflox mouse was as described previously (Miano et al., 2004) and was a kind gift from Dr Joseph Miano (University of Rochester, Rochester, NY, USA). Wt1CreERT2, RosamTmG and RosalacZ mice were as described previously (Rudat and Kispert, 2012; Zhou and Pu, 2012) and are available from Jackson Laboratories (stock numbers 010912, 007576 and 003309), as were the C57BL/6 mice. The Wt1CreBAC line was as reported previously (Wessels et al., 2012) and was a kind gift from Dr Sylvia Evans (UC San Diego, La Jolla, CA, USA). Rosa mice were kept on a mixed genetic background and all other mice were kept on a C57BL/6 background. The breeding strategy for lineage tracing was as follows: Wt1CreERT2/+ sire crossed to RosalacZ/+ or mTmG/+ dam. The breeding strategy for MRTFepiDKO was as follows: Wt1CreERT2/+; Mrtfa−/−; Mrtfbflox/flox sire crossed to Mrtfa−/−; Mrtfbflox/flox dam. The breeding strategy for SRFepiKO was: Wt1CreBAC; Srfflox/+ sire crossed to Srfflox/flox dam. For inducible deletion, 4-hydroxytamoxifen (Sigma) was suspended in sunflower seed oil at 10 mg/ml by sonication and administered to pregnant dams (75 mg/kg) by gavage at the time points noted in the text.
Epicardial explant culture
Epicardial explant cultures were carried out as reported previously (Austin et al., 2008). Briefly, E11.5 hearts were placed dorsal side down on collagen-coated slides (BD Biosciences). After culture overnight in M199 (HyClone) with 5% FBS at 37°C and 5% CO2, the hearts were collected, revealing an EPDC monolayer attached to the collagen surface. EPDCs isolated from Srffl/fl or Mrtfa−/−; Mrtfbfl/fl embryos were then transduced with adenovirus expressing Cre or βgal at a multiplicity of infection (MOI) of ∼10 and cultured for 24 h in M199 with 1% FBS, followed by an additional incubation for 24 h in medium supplemented with TGF-β1 (R&D Systems, 10 ng/ml) to stimulate EMT. EPDCs isolated from Wt1CreERT2/+ or +/+; Mrtfa−/−; Mrtfbfl/fl embryos were cultured for 48 h in medium supplemented with TGF-β1 (10 ng/ml) or TGF-β2 (2 ng/ml, R&D Systems) and PDGF-BB (20 ng/ml, R&D Systems). After 72 h of culture, EPDCs were processed for immunostaining or RNA or protein isolation. For scratch wound migration assays, Srffl/fl or Mrtfa−/−; Mrtfbfl/fl explants were transduced with adenovirus expressing Cre or βgal at a multiplicity of infection (MOI) of ∼10 and cultured for 24 h in M199 with 1% FBS. Explants were then scratched with a 10 μl pipette tip and treated with TGF-β1 (10 ng/ml) to stimulate EMT. Images were taken at the indicated time points over ∼24 h.
Ex vivo embryonic heart culture
Srffl/fl, Mrtfa−/−; Mrtfbfl/fl or C57BL/6 embryos were collected at E12.5 and dissected into M199 (HyClone) with 10% FBS. All ex vivo hearts were immediately transduced with adenovirus expressing GFP (2.8×106 pfu/ml) in the presence of adenovirus expressing Cre (9.6×104 pfu/ml), MRTF-A (4.2×105 pfu/ml) or βgal (1.3×107 pfu/ml). Hearts were then cultured for 48 h at 37°C and 5% CO2 in the presence of TGF-β2 (2 ng/ml) and PDGF-BB (20 ng/ml) (R&D Systems). Samples were then cryopreserved for immunostaining.
Histology, immunohistochemistry and immunocytochemistry
Cells were fixed in 4% paraformaldehyde (PFA) at 4°C (Ki67) or methanol at −20°C for 10 min. Tissues were fixed in 4% PFA or 10% acetic acid/60% methanol in distilled H2O overnight at 4°C and processed for paraffin sectioning (5 μM), or cryopreserved in tissue freezing medium (Triangle Biomedical Science) for subsequent cryosectioning (10 μM). Details regarding the primary antibodies and immunostaining conditions are listed in supplementary materials and methods and Table S3.
Whole-mount X-gal staining
E12.5 embryos were fixed and permeabilized for 1 h at 4°C (4% PFA, 0.01% deoxycholic acid, 0.02% NP40 in PBS), stained overnight at room temperature in X-gal solution [0.01% deoxycholic acid, 0.02% NP40, 2 mM MgCl2, 1 mg/ml X-gal (Roche), 5 mM K4Fe(CN)6, 5 mM K3Fe(CN)6 in PBS], and then post fixed overnight at 4°C (4% PFA, 0.2% glutaraldehyde in PBS).
Whole-mount PECAM1 immunostaining
Hearts dissected from E13.5 embryos were fixed overnight at 4°C in 4% PFA. Following dehydration, endogenous peroxidase was quenched with 5% H2O2 in methanol for 2 h at 4°C, and the samples were then rehydrated. Hearts were blocked for 2 h at room temperature in PBSMT (3% milk, 0.5% Triton X-100 in PBS), incubated with rat anti-mouse PECAM1 (BD Pharmingen 553370, 10 μg/ml in PBSMT) at 4°C overnight, followed by 4°C overnight with horseradish peroxidase (HRP)-conjugated goat anti-rat-IgG (Kirkgaad and Perry Laboratories, 1:100 in PBSMT). Following colorimetric detection of PECAM1 with diaminobenzidine peroxidase substrate, hearts were post-fixed in 4% PFA at 4°C overnight and imaged.
Immunoblot
Whole cell extracts were collected from EMCs (Wada et al., 2003) (courtesy of Dr David Bader, Vanderbilt) or COS-7 cells, or pooled from two epicardial explant cultures per treatment. Lysates were subjected to SDS-PAGE and immunoblotted onto PVDF membranes (Millipore) before incubating with the following antibodies at 4°C overnight: mouse anti-ACTA2 (clone 1A4, Sigma A2547, 1:1000), rabbit anti-MRTF (a kind gift from Guido Posern, Martin Luther University Halle-Wittenberg, Germany, 1:500), mouse anti-FLAG (Sigma F1804, 1:5000) or mouse anti-GAPDH (Millipore MAB374 1:30,000). Membranes were incubated with secondary HRP-conjugated goat anti-mouse-IgG or anti-rabbit-IgG antibody (Bio-Rad) for 1 h at room temperature and developed with luminol reagent (Santa Cruz Biotechnology).
Cell culture
COS-7 cells and EMCs were cultured in DMEM at 37°C and 5% CO2. Additional details can be found in supplementary materials and methods.
RNA isolation and analysis
Total RNA was isolated from cell cultures and tissue samples using Trizol reagent (Life Technologies) and cDNA was generated using iScript cDNA Synthesis Kit (Bio-Rad) following the manufacturer's protocol. RT-PCR conditions and primer sequences are listed in supplementary materials and methods and Table S4.
Transmission electron microscopy
Hearts isolated from E15.5 embryos were fixed in 2.5% glutaraldehyde buffered with 0.1 M sodium cacodylate, postfixed in 1% osmium tetroxide, dehydrated in a graded series of ethanol and transitioned into propylene oxide, then into Epon/Araldite resin. The tissue was embedded into molds containing fresh resin and polymerized for 2 days at 65°C. Sections of 1 μm thickness were stained with Toluidine Blue to determine the area to be thin sectioned (at 70 nm) with a diamond knife. Thin sections were placed onto carbon-coated Formvar slot grids and stained with aqueous uranyl acetate and lead citrate. The grids were examined using a Gatan 11 megapixel Erlansheng digital camera and Digital micrograph software.
Imaging and image quantification
Whole-mount samples were imaged with a Leica MZ125 microscope. Immunofluorescence images were captured using an Olympus IX81 confocal microscope. All image analyses were conducted using NIH ImageJ software. For ex vivo migration assays, sections were stained with anti-ColIV antibody to label the epicardial basement membrane as described previously (Mellgren et al., 2008). Migration was quantified as the percentage of GFP-positive cells migrating in or beyond the epicardial basement membrane relative to the total number of GFP-positive cells in a given 40× field of view (FOV). Cells were counted in five non-consecutive sections of a given heart for each condition. For in vitro scratch wound assays, the total area of the scratch was measured using NIH ImageJ at the indicated time points and presented as a percentage of the original scratch area. Cell proliferation was determined as the percentage of Ki67-positive nuclei at the leading edge of migration or within the entire explant. For in vivo cell proliferation analysis, the total number of Ki67-positive nuclei was quantified by a blinded observer from four non-consecutive sections per heart at a 40× FOV. WT1 co-staining marked epicardial cells and ColIV co-staining marked the epicardial basement membrane. For coronary plexus formation experiments, the area of PECAM1-positive vessel coverage was measured and presented as a percentage of the total area of the ventricles. For pericyte quantification, CSPG4+ PDGFRβ+ cells were counted in seven areas along the sub-epicardial space (50-60 μm from the epicardial layer) in three non-consecutive sections of a given heart sample. Four hearts were examined per genotype. Pericytes were identified as double positive cells with pericyte morphology that were in close association with a microvessel having a 10-25 μm lumen. Images were captured in 5-10 μM z-stacks at 40× FOV. For lineage tracing, epicardium-derived pericytes were identified as cells with CSPG4+ PDGFRβ+ GFP+ staining and pericyte morphology along a vessel.
Statistical analysis
Statistical differences were determined by a Student's two-tailed t-test with unequal variance, unless otherwise noted in the text. Significance was considered as P<0.05.
Supplementary Material
Acknowledgements
We thank C. Lowenstein and J. Miano for insightful comments, T. Carroll (UT Southwestern Medical Center at Dallas) and R. Bell for critical reading of the manuscript, and C. Hoffman for technical support.
Footnotes
Competing interests
The authors declare no competing financial interests.
Author contributions
M.A.T. and L.S.V. designed and performed experiments, interpreted data, maintained and genotyped mouse lines and prepared the manuscript. K.L.deM.B. generated and interpreted TEM images. E.M.S. designed and performed experiments, interpreted data and wrote and edited the manuscript.
Funding
Work in the laboratory of E.M.S. was supported by grants from the National Institutes of Health (NIH) [grant number R01HL120919]; the American Heart Association [grant number 10SDG4350046]; University of Rochester CTSA award from the NIH [grant number UL1 TR000042]; and startup funds from the Aab CVRI. M.A.T. was supported in part by a grant to the University of Rochester School of Medicine and Dentistry from the Howard Hughes Medical Institute Med-Into-Grad Initiative; and an Institutional Ruth L. Kirschstein National Research Service Award from the NIH [grant number GM068411]. Deposited in PMC for release after 12 months.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.116418/-/DC1
References
- Aguilera K. Y. and Brekken R. A. (2014). Recruitment and retention: factors that affect pericyte migration. Cell. Mol. Life Sci. 71, 299-309 10.1007/s00018-013-1432-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armulik A., Genové G. and Betsholtz C. (2011). Pericytes: developmental, physiological, and pathological perspectives, problems, and promises. Dev. Cell 21, 193-215 10.1016/j.devcel.2011.07.001 [DOI] [PubMed] [Google Scholar]
- Austin A. F., Compton L. A., Love J. D., Brown C. B. and Barnett J. V. (2008). Primary and immortalized mouse epicardial cells undergo differentiation in response to TGFbeta. Dev. Dyn. 237, 366-376 10.1002/dvdy.21421 [DOI] [PubMed] [Google Scholar]
- Braitsch C. M., Kanisicak O., van Berlo J. H., Molkentin J. D. and Yutzey K. E. (2013). Differential expression of embryonic epicardial progenitor markers and localization of cardiac fibrosis in adult ischemic injury and hypertensive heart disease. J. Mol. Cell. Cardiol. 65, 108-119 10.1016/j.yjmcc.2013.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busche S., Kremmer E. and Posern G. (2010). E-cadherin regulates MAL-SRF-mediated transcription in epithelial cells. J. Cell Sci. 123, 2803-2809 10.1242/jcs.061887 [DOI] [PubMed] [Google Scholar]
- Cai C.-L., Martin J. C., Sun Y., Cui L., Wang L., Ouyang K., Yang L., Bu L., Liang X., Zhang X. et al. (2008). A myocardial lineage derives from Tbx18 epicardial cells. Nature 454, 104-108 10.1038/nature06969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova J. C., Travisano S. and de la Pompa J. L. (2013). Epithelial-to-mesenchymal transition in epicardium is independent of Snail1. Genesis 51, 32-40 10.1002/dvg.22353 [DOI] [PubMed] [Google Scholar]
- Chintalgattu V., Rees M. L., Culver J. C., Goel A., Jiffar T., Zhang J., Dunner K. Jr, Pati S., Bankson J. A., Pasqualini R. et al. (2013). Coronary microvascular pericytes are the cellular target of sunitinib malate-induced cardiotoxicity. Sci. Transl. Med. 5, 187ra169 10.1126/scitranslmed.3005066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettman R. W., Denetclaw W. Jr, Ordahl C. P. and Bristow J. (1998). Common epicardial origin of coronary vascular smooth muscle, perivascular fibroblasts, and intermyocardial fibroblasts in the avian heart. Dev. Biol. 193, 169-181 10.1006/dbio.1997.8801 [DOI] [PubMed] [Google Scholar]
- Eralp I., Lie-Venema H., DeRuiter M. C., van den Akker N. M. S., Bogers A. J. J. C., Mentink M. M. T., Poelmann R. E. and Gittenberger-de Groot A. C. (2005). Coronary artery and orifice development is associated with proper timing of epicardial outgrowth and correlated Fas ligand associated apoptosis patterns. Circ. Res. 96, 526-534 10.1161/01.RES.0000158965.34647.4e [DOI] [PubMed] [Google Scholar]
- Evelyn C. R., Wade S. M., Wang Q., Wu M., Iniguez-Lluhi J. A., Merajver S. D. and Neubig R. R. (2007). CCG-1423: a small-molecule inhibitor of RhoA transcriptional signaling. Mol. Cancer Ther. 6, 2249-2260 10.1158/1535-7163.MCT-06-0782 [DOI] [PubMed] [Google Scholar]
- Fan L., Sebe A., Peterfi Z., Masszi A., Thirone A. C. P., Rotstein O. D., Nakano H., McCulloch C. A., Szaszi K., Mucsi I. et al. (2007). Cell contact-dependent regulation of epithelial-myofibroblast transition via the rho-rho kinase-phospho-myosin pathway. Mol. Biol. Cell 18, 1083-1097 10.1091/mbc.E06-07-0602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittenberger-de Groot A. C., Vrancken Peeters M.-P. F. M., Bergwerff M., Mentink M. M. T. and Poelmann R. E. (2000). Epicardial outgrowth inhibition leads to compensatory mesothelial outflow tract collar and abnormal cardiac septation and coronary formation. Circ. Res. 87, 969-971 10.1161/01.RES.87.11.969 [DOI] [PubMed] [Google Scholar]
- Guettler S., Vartiainen M. K., Miralles F., Larijani B. and Treisman R. (2008). RPEL motifs link the serum response factor cofactor MAL but not myocardin to Rho signaling via actin binding. Mol. Cell. Biol. 28, 732-742 10.1128/MCB.01623-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haak A. J., Tsou P.-S., Amin M. A., Ruth J. H., Campbell P., Fox D. A., Khanna D., Larsen S. D. and Neubig R. R. (2014). Targeting the myofibroblast genetic switch: inhibitors of myocardin-related transcription factor/serum response factor-regulated gene transcription prevent fibrosis in a murine model of skin injury. J. Pharmacol. Exp. Ther. 349, 480-486 10.1124/jpet.114.213520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinkel R., Trenkwalder T., Petersen B., Husada W., Gesenhues F., Lee S., Hannappel E., Bock-Marquette I., Theisen D., Leitner L. et al. (2014). MRTF-A controls vessel growth and maturation by increasing the expression of CCN1 and CCN2. Nat. Commun. 5, 3970 10.1038/ncomms4970 [DOI] [PubMed] [Google Scholar]
- Katz T. C., Singh M. K., Degenhardt K., Rivera-Feliciano J., Johnson R. L., Epstein J. A. and Tabin C. J. (2012). Distinct compartments of the proepicardial organ give rise to coronary vascular endothelial cells. Dev. Cell 22, 639-650 10.1016/j.devcel.2012.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Wu Q., Zhang Y., Wiens K. M., Huang Y., Rubin N., Shimada H., Handin R. I., Chao M. Y., Tuan T.-L. et al. (2010). PDGF signaling is required for epicardial function and blood vessel formation in regenerating zebrafish hearts. Proc. Natl. Acad. Sci. USA 107, 17206-17210 10.1073/pnas.0915016107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamouille S., Xu J. and Derynck R. (2014). Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15, 178-196 10.1038/nrm3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepilina A., Coon A. N., Kikuchi K., Holdway J. E., Roberts R. W., Burns C. G. and Poss K. D. (2006). A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell 127, 607-619 10.1016/j.cell.2006.08.052 [DOI] [PubMed] [Google Scholar]
- Li J., Zhu X., Chen M., Cheng L., Zhou D., Lu M. M., Du K., Epstein J. A. and Parmacek M. S. (2005). Myocardin-related transcription factor B is required in cardiac neural crest for smooth muscle differentiation and cardiovascular development. Proc. Natl. Acad. Sci. USA 102, 8916-8921 10.1073/pnas.0503741102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Chang S., Qi X., Richardson J. A. and Olson E. N. (2006). Requirement of a myocardin-related transcription factor for development of mammary myoepithelial cells. Mol. Cell. Biol. 26, 5797-5808 10.1128/MCB.00211-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Cavallero S., Gu Y., Chen T. H. P., Hughes J., Hassan A. B., Bruning J. C., Pashmforoush M. and Sucov H. M. (2011). IGF signaling directs ventricular cardiomyocyte proliferation during embryonic heart development. Development 138, 1795-1805 10.1242/dev.054338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limana F., Zacheo A., Mocini D., Mangoni A., Borsellino G., Diamantini A., De Mori R., Battistini L., Vigna E., Santini M. et al. (2007). Identification of myocardial and vascular precursor cells in human and mouse epicardium. Circ. Res. 101, 1255-1265 10.1161/CIRCRESAHA.107.150755 [DOI] [PubMed] [Google Scholar]
- Lu J., Landerholm T. E., Wei J. S., Dong X.-R., Wu S.-P., Liu X., Nagata K.-i., Inagaki M. and Majesky M. W. (2001). Coronary smooth muscle differentiation from proepicardial cells requires rhoA-mediated actin reorganization and p160 rho-kinase activity. Dev. Biol. 240, 404-418 10.1006/dbio.2001.0403 [DOI] [PubMed] [Google Scholar]
- Männer J., Schlueter J. and Brand T. (2005). Experimental analyses of the function of the proepicardium using a new microsurgical procedure to induce loss-of-proepicardial-function in chick embryos. Dev. Dyn. 233, 1454-1463 10.1002/dvdy.20487 [DOI] [PubMed] [Google Scholar]
- Martínez-Estrada O. M., Lettice L. A., Essafi A., Guadix J. A., Slight J., Velecela V., Hall E., Reichmann J., Devenney P. S., Hohenstein P. et al. (2010). Wt1 is required for cardiovascular progenitor cell formation through transcriptional control of Snail and E-cadherin. Nat. Genet. 42, 89-93 10.1038/ng.494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medjkane S., Perez-Sanchez C., Gaggioli C., Sahai E. and Treisman R. (2009). Myocardin-related transcription factors and SRF are required for cytoskeletal dynamics and experimental metastasis. Nat. Cell Biol. 11, 257-268 10.1038/ncb1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellgren A. M., Smith C. L., Olsen G. S., Eskiocak B., Zhou B., Kazi M. N., Ruiz F. R., Pu W. T. and Tallquist M. D. (2008). Platelet-derived growth factor receptor beta signaling is required for efficient epicardial cell migration and development of two distinct coronary vascular smooth muscle cell populations. Circ. Res. 103, 1393-1401 10.1161/CIRCRESAHA.108.176768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merki E., Zamora M., Raya A., Kawakami Y., Wang J., Zhang X., Burch J., Kubalak S. W., Kaliman P., Izpisua Belmonte J. C. et al. (2005). Epicardial retinoid X receptor alpha is required for myocardial growth and coronary artery formation. Proc. Natl. Acad. Sci. USA 102, 18455-18460 10.1073/pnas.0504343102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miano J. M. (2003). Serum response factor: toggling between disparate programs of gene expression. J. Mol. Cell. Cardiol. 35, 577-593 10.1016/S0022-2828(03)00110-X [DOI] [PubMed] [Google Scholar]
- Miano J. M., Ramanan N., Georger M. A., de Mesy Bentley K. L., Emerson R. L., Balza R. O. Jr, Xiao Q., Weiler H., Ginty D. D. and Misra R. P. (2004). Restricted inactivation of serum response factor to the cardiovascular system. Proc. Natl. Acad. Sci. USA 101, 17132-17137 10.1073/pnas.0406041101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikawa T. and Gourdie R. G. (1996). Pericardial mesoderm generates a population of coronary smooth muscle cells migrating into the heart along with ingrowth of the epicardial organ. Dev. Biol. 174, 221-232 10.1006/dbio.1996.0068 [DOI] [PubMed] [Google Scholar]
- Miralles F., Posern G., Zaromytidou A.-I. and Treisman R. (2003). Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell 113, 329-342 10.1016/S0092-8674(03)00278-2 [DOI] [PubMed] [Google Scholar]
- Mokalled M. H., Johnson A., Kim Y., Oh J. and Olson E. N. (2010). Myocardin-related transcription factors regulate the Cdk5/Pctaire1 kinase cascade to control neurite outgrowth, neuronal migration and brain development. Development 137, 2365-2374 10.1242/dev.047605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson T. J., Duncan S. A. and Misra R. P. (2004). Conserved enhancer in the serum response factor promoter controls expression during early coronary vasculogenesis. Circ. Res. 94, 1059-1066 10.1161/01.RES.0000125296.14014.17 [DOI] [PubMed] [Google Scholar]
- Oh J., Richardson J. A. and Olson E. N. (2005). Requirement of myocardin-related transcription factor-B for remodeling of branchial arch arteries and smooth muscle differentiation. Proc. Natl. Acad. Sci. USA 102, 15122-15127 10.1073/pnas.0507346102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson E. N. and Nordheim A. (2010). Linking actin dynamics and gene transcription to drive cellular motile functions. Nat. Rev. Mol. Cell Biol. 11, 353-365 10.1038/nrm2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi D. J. and Mikawa T. (2009). FGFR-1 is required by epicardium-derived cells for myocardial invasion and correct coronary vascular lineage differentiation. Dev. Biol. 328, 148-159 10.1016/j.ydbio.2009.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Red-Horse K., Ueno H., Weissman I. L. and Krasnow M. A. (2010). Coronary arteries form by developmental reprogramming of venous cells. Nature 464, 549-553 10.1038/nature08873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouget C. (1873). Memoire sur le developpement, la structures et les proprietes des capillaires sanguins et lymphatiques. Archs. Physiol. Norm. Pathol. 5, 603-633. [Google Scholar]
- Rudat C. and Kispert A. (2012). Wt1 and epicardial fate mapping. Circ. Res. 111, 165-169 10.1161/CIRCRESAHA.112.273946 [DOI] [PubMed] [Google Scholar]
- Sandbo N., Lau A., Kach J., Ngam C., Yau D. and Dulin N. O. (2011). Delayed stress fiber formation mediates pulmonary myofibroblast differentiation in response to TGF-beta. Am. J. Physiol. Lung Cell Mol. Physiol. 301, L656-L666 10.1152/ajplung.00166.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small E. M., Thatcher J. E., Sutherland L. B., Kinoshita H., Gerard R. D., Richardson J. A., DiMaio J. M., Sadek H., Kuwahara K. and Olson E. N. (2010). Myocardin-related transcription factor-a controls myofibroblast activation and fibrosis in response to myocardial infarction. Circ. Res. 107, 294-304 10.1161/CIRCRESAHA.110.223172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart N., Bollini S., Dubé K. N., Vieira J. M., Zhou B., Davidson S., Yellon D., Riegler J., Price A. N., Lythgoe M. F. et al. (2011). De novo cardiomyocytes from within the activated adult heart after injury. Nature 474, 640-644 10.1038/nature10188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X., Hu T., Zhang H., He L., Huang X., Liu Q., Yu W., He L., Yang Z., Zhang Z. et al. (2013). Subepicardial endothelial cells invade the embryonic ventricle wall to form coronary arteries. Cell Res. 23, 1075-1090 10.1038/cr.2013.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasquez L. S., Sutherland L. B., Liu Z., Grinnell F., Kamm K. E., Schneider J. W., Olson E. N. and Small E. M. (2013). Activation of MRTF-A-dependent gene expression with a small molecule promotes myofibroblast differentiation and wound healing. Proc. Natl. Acad. Sci. USA 110, 16850-16855 10.1073/pnas.1316764110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gise A., Zhou B., Honor L. B., Ma Q., Petryk A. and Pu W. T. (2011). WT1 regulates epicardial epithelial to mesenchymal transition through beta-catenin and retinoic acid signaling pathways. Dev. Biol. 356, 421-431 10.1016/j.ydbio.2011.05.668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada A. M., Smith T. K., Osler M. E., Reese D. E. and Bader D. M. (2003). Epicardial/Mesothelial cell line retains vasculogenic potential of embryonic epicardium. Circ. Res. 92, 525-531 10.1161/01.RES.0000060484.11032.0B [DOI] [PubMed] [Google Scholar]
- Wang D.-Z., Chang P. S., Wang Z., Sutherland L., Richardson J. A., Small E., Krieg P. A. and Olson E. N. (2001). Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell 105, 851-862 10.1016/S0092-8674(01)00404-4 [DOI] [PubMed] [Google Scholar]
- Wang D.-Z., Li S., Hockemeyer D., Sutherland L., Wang Z., Schratt G., Richardson J. A., Nordheim A. and Olson E. N. (2002). Potentiation of serum response factor activity by a family of myocardin-related transcription factors. Proc. Natl. Acad. Sci. USA 99, 14855-14860 10.1073/pnas.222561499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Wang D.-Z., Hockemeyer D., McAnally J., Nordheim A. and Olson E. N. (2004). Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature 428, 185-189 10.1038/nature02382 [DOI] [PubMed] [Google Scholar]
- Weinl C., Riehle H., Park D., Stritt C., Beck S., Huber G., Wolburg H., Olson E. N., Seeliger M. W., Adams R. H. et al. (2013). Endothelial SRF/MRTF ablation causes vascular disease phenotypes in murine retinae. J. Clin. Invest. 123, 2193-2206 10.1172/JCI64201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels A., van den Hoff M. J. B., Adamo R. F., Phelps A. L., Lockhart M. M., Sauls K., Briggs L. E., Norris R. A., van Wijk B., Perez-Pomares J. M. et al. (2012). Epicardially derived fibroblasts preferentially contribute to the parietal leaflets of the atrioventricular valves in the murine heart. Dev. Biol. 366, 111-124 10.1016/j.ydbio.2012.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilm B., Ipenberg A., Hastie N. D., Burch J. B. E. and Bader D. M. (2005). The serosal mesothelium is a major source of smooth muscle cells of the gut vasculature. Development 132, 5317-5328 10.1242/dev.02141 [DOI] [PubMed] [Google Scholar]
- Zhou B. and Pu W. T. (2012). Genetic Cre-loxP assessment of epicardial cell fate using Wt1-driven Cre alleles. Circ. Res. 111, e276-e280 10.1161/CIRCRESAHA.112.275784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Ma Q., Rajagopal S., Wu S. M., Domian I., Rivera-Feliciano J., Jiang D., von Gise A., Ikeda S., Chien K. R. et al. (2008). Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature 454, 109-113 10.1038/nature07060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Honor L. B., He H., Ma Q., Oh J.-H., Butterfield C., Lin R.-Z., Melero-Martin J. M., Dolmatova E., Duffy H. S. et al. (2011). Adult mouse epicardium modulates myocardial injury by secreting paracrine factors. J. Clin. Invest. 121, 1894-1904 10.1172/JCI45529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Huang X., Hecker L., Kurundkar D., Kurundkar A., Liu H., Jin T.-H., Desai L., Bernard K. and Thannickal V. J. (2013). Inhibition of mechanosensitive signaling in myofibroblasts ameliorates experimental pulmonary fibrosis. J. Clin. Invest. 123, 1096-1108 10.1172/JCI66700 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.