Abstract
The coordination of cell polarity within the plane of the tissue layer (planar polarity) is crucial for the development of diverse multicellular organisms. Small Rac/Rho-family GTPases and the actin cytoskeleton contribute to planar polarity formation at sites of polarity establishment in animals and plants. Yet, upstream pathways coordinating planar polarity differ strikingly between kingdoms. In the root of Arabidopsis thaliana, a concentration gradient of the phytohormone auxin coordinates polar recruitment of Rho-of-plant (ROP) to sites of polar epidermal hair initiation. However, little is known about cytoskeletal components and interactions that contribute to this planar polarity or about their relation to the patterning machinery. Here, we show that ACTIN7 (ACT7) represents a main actin isoform required for planar polarity of root hair positioning, interacting with the negative modulator ACTIN-INTERACTING PROTEIN1-2 (AIP1-2). ACT7, AIP1-2 and their genetic interaction are required for coordinated planar polarity of ROP downstream of ethylene signalling. Strikingly, AIP1-2 displays hair cell file-enriched expression, restricted by WEREWOLF (WER)-dependent patterning and modified by ethylene and auxin action. Hence, our findings reveal AIP1-2, expressed under control of the WER-dependent patterning machinery and the ethylene signalling pathway, as a modulator of actin-mediated planar polarity.
Keywords: AIP1, Arabidopsis, WEREWOLF, Actin, Patterning, Planar polarity
Summary: Planar polarity in Arabidopsis is shaped by ACTIN-INTERACTING PROTEIN1-2, which is under the control of WEREWOLF-dependent patterning and ethylene signalling.
INTRODUCTION
Development of multicellular organisms relies on formation and maintenance of structural asymmetries at the single-cell level as well as their coordination within the tissue context. Whereas cell polarity describes axially organised asymmetries in subcellular structures or intracellular molecule distributions (Yang, 2008), planar polarity defines the coordinated organisation of cells within a plane of a tissue layer (Nübler-Jung, 1987). The polar orientation of hairs in the Drosophila wing epithelium is widely used as a model system for planar polarity in animals (Adler, 2002). Similarly, the emergence of root hairs in the Arabidopsis root epidermis represents a model system to unveil mechanisms underlying planar polarity in plants (Nakamura et al., 2012). The root epidermis is composed of alternating files of hair-forming cells (trichoblasts) and non-hair-forming cells (atrichoblasts), the fate of which is specified by the WEREWOLF (WER) MYB transcription factor-dependent patterning system (Galway et al., 1994; Masucci et al., 1996; Lee and Schiefelbein, 1999). Root hair-forming cells develop root hairs as tubular protrusions from their outer membrane, where hairs are uniformly initiated towards, albeit not completely at, the root tip-oriented (basal) ends of cells (Masucci and Schiefelbein, 1994). In contrast to Drosophila, in which the long-range polarising cue instructing the Frizzled planar cell polarity pathway is provided by Wingless and its homologue dWnt4 (Wu et al., 2013), vectorial information for planar polarity in Arabidopsis is provided by a concentration gradient of the phytohormone auxin (Fischer et al., 2006; Ikeda et al., 2009). Formation of this gradient depends on local auxin biosynthesis in the root tip, where auxin concentration reaches its maximum, and on the basipetal (shootward) transport of auxin in the root epidermis (Ikeda et al., 2009). Local upregulation of auxin biosynthesis induced by mutations in the CONSTITUTIVE TRIPLE RESPONSE1 (CTR1) gene, a repressor of the auxin gradient, causes hyperpolar positioning of root hairs at basal-most ends of cells (Ikeda et al., 2009). In analogy to animal systems, in which Rac/Rho-family GTPases are downstream effectors of planar cell polarity (Goodrich and Strutt, 2011), auxin instructs polar accumulation of Rho-of-plant (ROP) GTPases at the outer plasma membrane (Fischer et al., 2006; Ikeda et al., 2009), thus marking the root hair initiation site (Molendijk et al., 2001; Jones et al., 2002). During planar morphogenesis in the leaf epidermis, ROP2 and ROP4 regulate actin organisation, whereas ROP6 modulates the microtubule cytoskeleton (Fu et al., 2009; Xu et al., 2010). In the root epidermis, ROP6 is thought to contribute to actin organisation (Lin et al., 2012). Among the eight expressed actin paralogues encoded in the Arabidopsis genome, ACT2, ACT7 and ACT8, and potentially also ACT11 (Cvrčková et al., 2010), contribute to root development (Kandasamy et al., 2009). Mutant alleles of ACT2 display weak defects in root hair positioning (Ringli et al., 2002), but mechanisms regulating the actin cytoskeleton during planar polarity formation in plants remain largely unknown. In Drosophila, the single actin-depolymerizing factor (ADF) Twinstar (Tsr) interacts with the single actin-interacting protein 1 (AIP1) Flare (Flr) during epithelial morphogenesis (Chu et al., 2012). Both negatively regulate actin filament organisation required for planar cell polarity downstream of the Frizzled pathway (Blair et al., 2006; Ren et al., 2007). AIP1 homologues from several systems, such as Aip1p from yeast and AIP1-1 from Arabidopsis, have been shown to enhance the F-actin depolymerizing activity of ADF (named Cofilin in yeast) in vitro (Rodal et al., 1999; Allwood et al., 2002). In Arabidopsis, simultaneous downregulation of the reproductive isoform AIP1-1 and the vegetative isoform AIP1-2 by RNA interference (RNAi), as well as ectopic overexpression of AIP1-1, affect actin organisation and root hair tip growth (Ketelaar et al., 2004, 2007). Yet, analyses of individual functions of the Arabidopsis AIP1 genes are lacking. Here, we report that AIP1-2 and ACT7 interact and are required for polar root hair positioning downstream of CTR1. We demonstrate that WEREWOLF-dependent patterning specifies AIP1-2 expression, suggesting that AIP1-2 function during auxin-mediated planar polarity becomes spatially restricted by cell fate patterning.
RESULTS
ACT2 and ACT7 are required for planar polarity downstream of CTR1
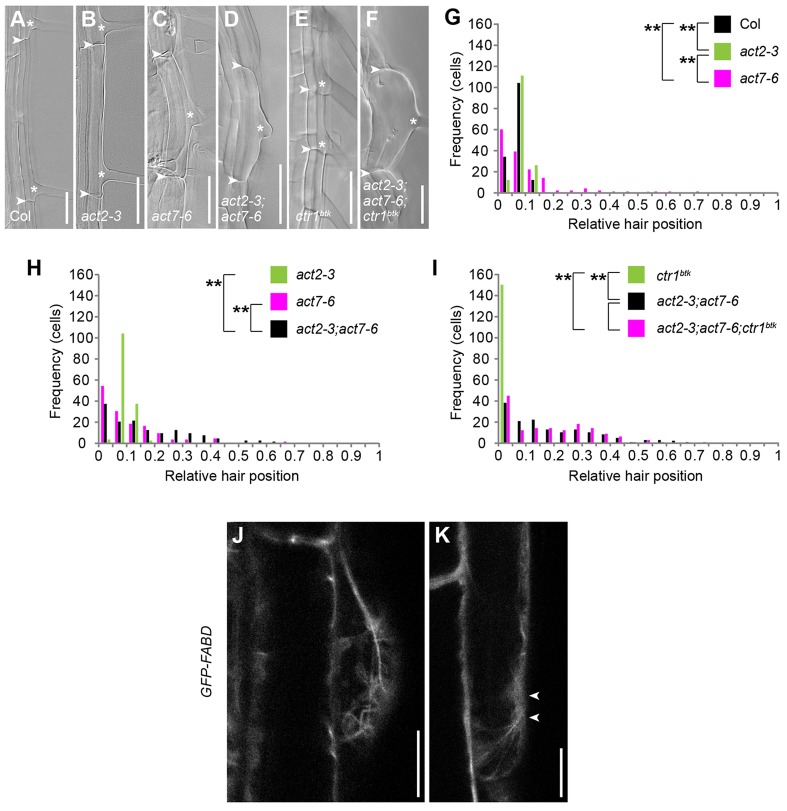
To further investigate the role of actin during planar polarity formation, we analysed root hair position phenotypes of act2, act7 and act8 loss-of-function mutants. We employed the act2-3 null allele (Nishimura et al., 2003) and an act7 T-DNA line with an insertion in the first exon (SALK_131610) that shows a twofold reduction of total actin levels (Guo et al., 2013), which we refer to as act7-6. In addition, we employed an act7 T-DNA line carrying a single insertion in the third exon of ACT7 (GK-498G06), which we named act7-7, as well as act8-2 (Kandasamy et al., 2009). We found that root hair position shifted slightly apically in act2-3 when compared with wild type (WT) (Fig. 1A,B,G). More strikingly, hair positions in act7-6 and act7-7 were widely distributed along the apical-basal axis of cells, revealing both an apical and a basal shift (Fig. 1A,C,G; supplementary material Fig. S1A,B). We established allelism between act7-6 and act7-7 by analysing the act7-6/act7-7 (act7-6/-7) trans-heterozygous mutant, which was indistinguishable from act7-6 and act7-7 homozygotes (supplementary material Fig. S1A-C). In comparison, the root hair position phenotype of act8-2 did not differ from WT (supplementary material Fig. S1D). Defects in polar hair positioning were significantly stronger in act7 compared with the act2 allele (Fig. 1B,C,G; supplementary material Fig. S1A,B) and much more pronounced in the act2-3;act7-6 double mutant when compared with the single mutants (Fig. 1B-D,H), suggesting that ACT7 contributes more strongly to planar polarity than ACT2, but that both synergistically act on polar root hair positioning.
Fig. 1.
ACT2 and ACT7 are required for planar polarity formation downstream of CTR1. (A-F) Root hair-forming cells of five-day-old (A) wild-type Columbia (Col), (B) act2-3, (C) act7-6, (D) act2-3;act7-6, (E) ctr1btk and (F) act2-3;act7-6;ctr1btk seedlings. Arrowheads mark apical and basal ends of cells; asterisks mark hair initiation sites. (G-I) Quantitative analysis of root hair-positioning phenotypes of lines depicted in A-F, displaying number of cells (frequency) with relative hair positions in classes from 0 (basal-most) to 1 (apical-most). n=150 cells from 30 roots per genotype. Significance P values were determined by non-parametric, two-sample Kolmogorov–Smirnov (KS) test. (G) **P=0.000 Col versus act2-3; **P=0.000 Col versus act7-6; **P=0.000 act2-3 versus act7-6. (H) **P=0.000 act2-3 versus act2-3;act7-6; **P=0.003 act7-6 versus act2-3;act7-6. (I) **P=0.000 ctr1btk versus act2-3;act7-6; **P=0.000 ctr1btk versus act2-3;act7-6;ctr1btk; P=0.706 act2-3;act7-6 versus act2-3;act7-6;ctr1btk. (J,K) Confocal laser scanning microscopy (CLSM) image projections of GFP-FABD-labelled actin filaments (J) in a forming root hair bulge and (K) at hair initiation site prior to bulging. Arrowheads in K indicate site of future hair emergence. Scale bars: 50 µm in A-F; 10 µm in J,K.
We next addressed the genetic relationship between ACT2, ACT7 and CTR1. Whereas act7-6;ctr1btk and act2-3;ctr1btk double mutants revealed partial suppression of the hyperpolar ctr1btk root hair-positioning phenotype (supplementary material Fig. S1E,F), the act2-3;act7-6;ctr1btk triple mutant displayed root hair placement indistinguishable from the act2-3;act7-6 double mutant (Fig. 1D,F,I), thus revealing full suppression of the ctr1btk effect on polar hair placement (Fig. 1D-F,I). This demonstrates the requirement of ACT2 and ACT7 for planar polarity downstream of CTR1.
To study the distribution of filamentous actin (F-actin) during polar hair positioning, we visualised actin during early stages of root hair formation using the actin-tubulin double-marker line GFP-FABD;mCherry-TUA5. We observed a dense actin network in forming root hair bulges (Fig. 1J), as reported previously (Baluska et al., 2000). In addition, we identified actin filaments in vicinity of the future hair initiation site prior to bulge formation (Fig. 1K), supporting a role for actin in polar site selection. Similar results were obtained using the GFP-ABD2-GFP and Lifeact-Venus markers or the F-actin-binding probe BODIPY FL phallacidin (supplementary material Fig. S1G-J). However, we did not observe a significant difference in actin cytoskeleton organisation in the basal region of trichoblasts when compared with the apical ends of the same cells (supplementary material Fig. S1K-M). Our findings reveal that planar polarity strongly depends on ACT7, which synergistically acts with ACT2 during selection of the polar hair initiation site downstream of CTR1. We also observed that F-actin is present during hair initiation, although not particularly enriched at the hair initiation site.
ACT7 interacts with AIP1-2 in yeast and in vitro
To identify interactors of actin, we conducted a yeast two-hybrid screen of a cDNA library prepared from Arabidopsis seedlings using ACT7 as bait, revealing AIP1-2 (At3g18060) as a single interactor. We confirmed the interaction in pairwise yeast two-hybrid assays by further including reproductive ACT1, displaying highest levels of gene expression in pollen (An et al., 1996), ACT2, ACT8 and AIP1-1. All actins strongly interacted with AIP1-1 and AIP1-2 (Fig. 2A; supplementary material Fig. S2A), but not a truncated form of AIP1-1 (ΔAIP1-1), lacking the first 137 amino acids of the protein when either used as bait or prey (supplementary material Fig. S2A-C). We independently evaluated interactions for the two actin isoforms that were of particular interest with respect to planar polarity, ACT2 and ACT7, by glutathione-S-transferase (GST) in vitro pull-down assays. Bacterially expressed GST-AIP1-1 or GST-AIP1-2, bound to glutathione sepharose beads, specifically precipitated actin from Arabidopsis protein extracts (Fig. 2B; supplementary material Fig. S2D). Furthermore, GST-AIP1-2 precipitated both 6×Histidine-tagged (6×His)-fusions with ACT2 and ACT7 from bacterial protein extracts (Fig. 2C; supplementary material Fig. S2E,F). These results identify AIP1-2 as an interactor of ACT7, ACT2 and other actin isoforms in yeast and in vitro.
Fig. 2.
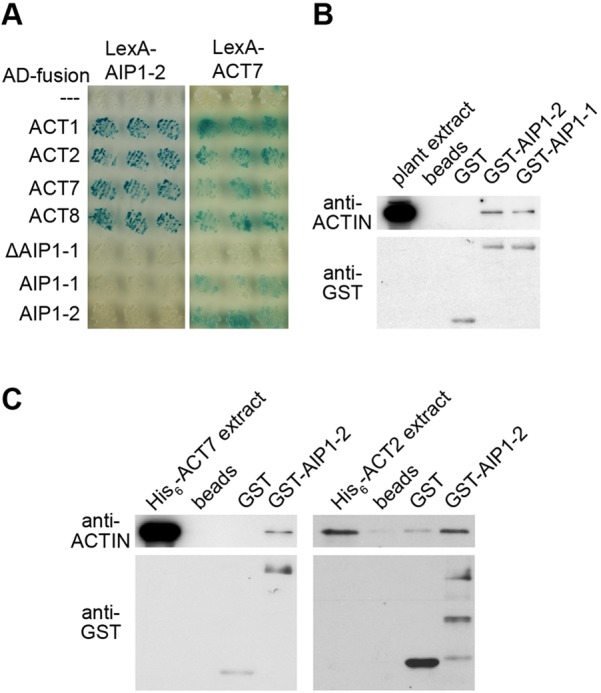
AIP1-2 interaction with Arabidopsis ACTINs in yeast and in vitro. (A) LexA-AIP1-2 interaction with B42 transcriptional activation domain (AD)-fusions of ACT1, ACT2, ACT7 and ACT8 (left panel), and LexA-ACT7 interaction with AD-fusions of ACT1, ACT2, ACT7, ACT8, AIP1-1 and AIP1-2 (right panel) in the yeast two-hybrid system. Yeasts were grown for 24 (left panel) and 48 h (right panel). Empty prey vector (---) and truncated AIP1-1 lacking amino acids 1-137 (ΔAIP1-1) are negative controls. (B,C) Empty glutathione sepharose (beads) or beads carrying GST only (GST) are negative controls. Loading of sepharose beads was tested using anti-GST antibody in western blots. (B) Glutathione-sepharose-bound GST-fusions of AIP1-2 (GST-AIP1-2) and AIP1-1 (GST-AIP1-1) precipitate actin from Arabidopsis protein extracts. (C) Glutathione-sepharose-bound GST-fusion of AIP1-2 (GST-AIP1-2) precipitates 6×His-fusions of ACT7 (His6-ACT7) and ACT2 (His6-ACT2) from bacterial protein extracts.
AIP1-2 genetically interacts with ACT7
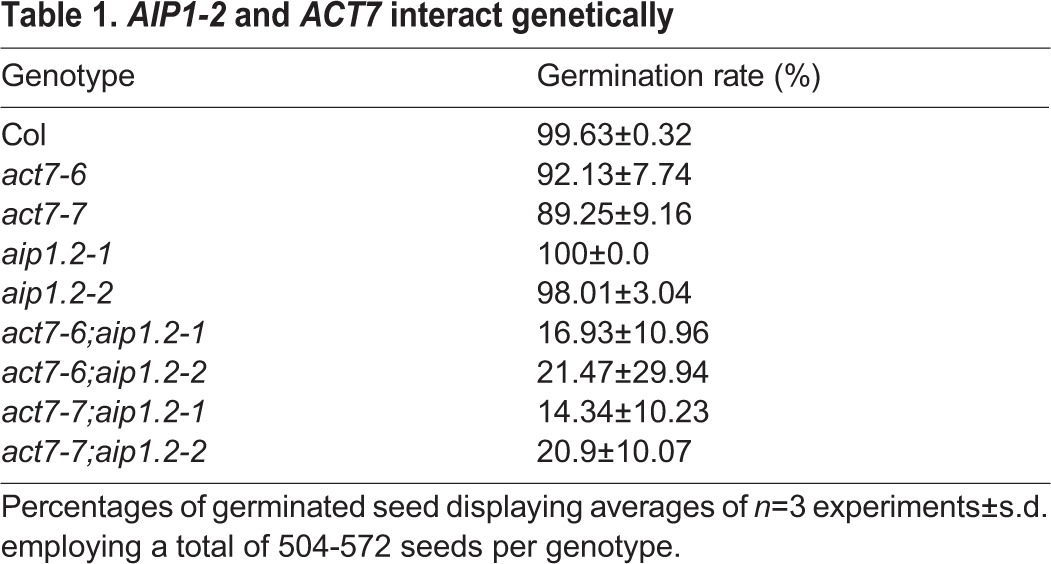
To address AIP1-2 function in planta, we characterised two lines carrying a T-DNA insertion in the AIP1-2 gene, GK-063F04 with an insertion in the fifth exon and SAIL_337_E05 with an insertion in the fifth intron, and named them aip1.2-1 and aip1.2-2, respectively (Fig. 3A). Semi-quantitative RT-PCR revealed that neither aip1.2-1 nor aip1.2-2 produced a full-length AIP1-2 transcript (Fig. 3B). To analyse potential genetic interactions between ACT7 and AIP1-2, we generated double mutants between aip1.2-1 or aip1.2-2 and act7-6 or act7-7. Whereas WT and single mutants germinated at rates between 89% and 100%, germination of act7;aip1-2 double mutants was reduced to 14-21% (Table 1), uncovering a synergistic effect of the aip1-2 and act7 mutations that resulted in synthetic lethality of most double mutants (Fig. 3C; supplementary material Fig. S3A). This synergistic effect might be due to strong interaction between ACT7 and AIP1-2 as well as to additional interactions between AIP1-2 and ACT7 with ACT2 or other actins. As the germination defect was not fully penetrant, some double-mutant seeds eventually germinated and could be used to analyse adult plants with this low-penetrance phenotype. These act7;aip1-2 double mutants displayed striking alterations of shoot architecture, such as twisted stems and siliques abnormally oriented along the shoot, when compared with the single mutants (Fig. 3D-G; supplementary material Fig. S3B-F). Our results reveal synergistic genetic interaction between AIP1-2 and ACT7, strongly suggesting that the AIP1-2 and ACT7 protein-protein interaction observed in yeast and in vitro is relevant during Arabidopsis development.
Fig. 3.
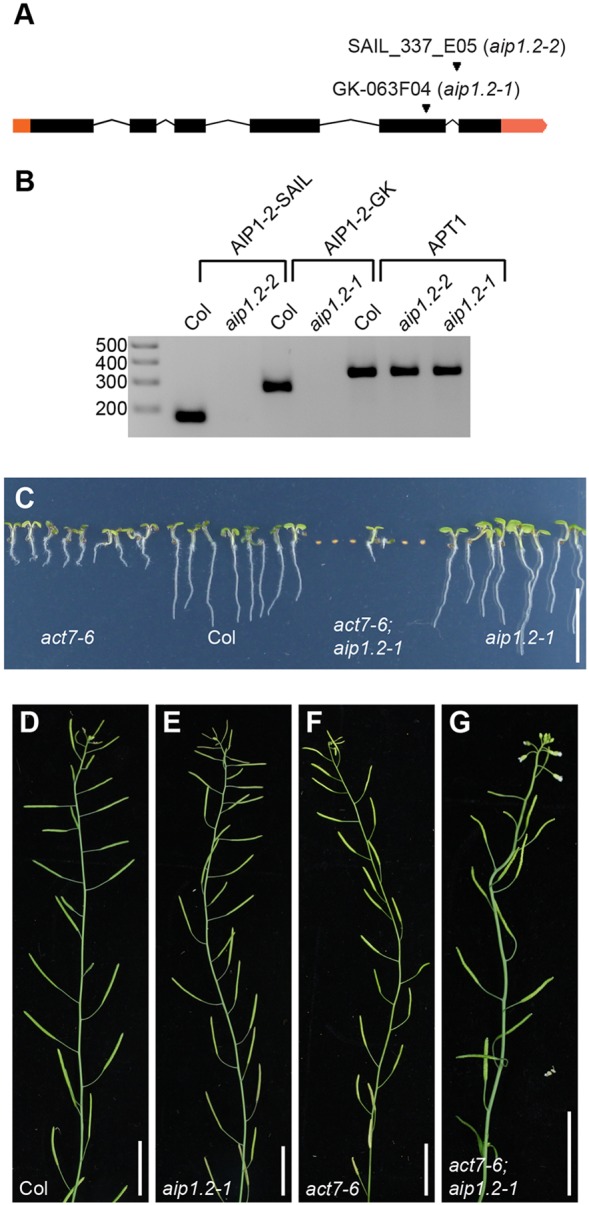
AIP1-2 and ACT7 genetically interact in Arabidopsis. (A) AIP1-2 locus map. Orange boxes, untranslated regions; black boxes, exons. Arrowheads, positions of T-DNA insertions. (B) Semi-quantitative RT-PCR analysis of Col wild-type and aip1-2 mutant seedlings. Regions surrounding the T-DNA insertion sites were amplified from Col but not from the respective aip1-2 mutant cDNA. APT1 was used as a reference gene. (C) Five-day-old act7-6, Col, act7-6;aip1.2-1 and aip1.2-1 seedlings. Germination of act7-6;aip1.2-1 seeds is strongly reduced compared with Col, act7-6 and aip1.2-1. (D-G) Main stems of three-week-old Col, aip1.2-1, act7-6 and act7-6;aip1.2-1 plants. Scale bars: 10 mm in C; 20 mm in D-G.
Table 1.
AIP1-2 and ACT7 interact genetically
AIP1-2 is predominantly expressed in hair cell files and contributes to planar polarity
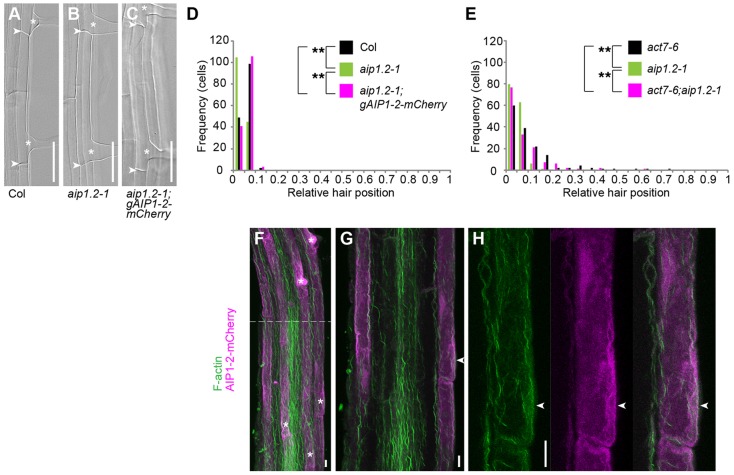
To address whether AIP1-2 contributes to planar polarity, we analysed root hair positioning in aip1-2 mutants. In comparison with the wild type (Fig. 4A), root hair placement was shifted basally in aip1.2-1 (Fig. 4B,D) and in aip1.2-2 (supplementary material Fig. S4A). These defects were caused by reduced AIP1-2 function, because when we reintroduced a C-terminally mCherry-tagged or a 3×Myc-tagged fusion of AIP1-2 expressed under control of AIP1-2 genomic sequences in the aip1.2-1 mutant background, aip1.2-1-mutant phenotypes were restored to wild-type levels (Fig. 4A-D; supplementary material Fig. S4B).
Fig. 4.
AIP1-2 modulates planar polarity together with ACT7 and co-localises with actin at root hair initiation sites. (A-C) Root-hair forming cells of five-day-old (A) wild-type Columbia (Col), (B) aip1.2-1 and (C) aip1.2-1;gAIP1-2-mCherry seedlings. Arrowheads, apical and basal ends of cells; asterisks, root hair initiation sites. (D,E) Quantitative analysis of relative root hair position of (D) aip1.2-1 and aip1.2-1;gAIP1-2-mCherry compared with Col, and (E) act7-6 and aip1.2-1 compared with act7-6;aip1.2-1. n=150 cells from 30 roots per genotype. Significance P values were determined by KS test. (D) **P=0.000 Col versus aip1.2-1; P=0.097 Col versus aip1.2-1;gAIP1-2-mCherry; **P=0.000 aip1.2-1 versus aip1.2-1;gAIP1-2-mCherry. (E) **P=0.000 act7-6 versus aip1.2-1; P=0.215 act7-6 versus act7-6;aip1.2-1; **P=0.000 aip1.2-1 versus act7-6;aip1.2-1. (F-H) CLSM images of early root differentiation zone of five-day-old aip1.2-1;gAIP1-2-mCherry seedlings probed for F-actin with BODIPY FL phallacidin. (F) Top view displaying AIP1-2-mCherry fluorescence preferentially in hair-forming cell files. Asterisks, root hair initiation sites. (G) Transversal section showing AIP1-2-mCherry and F-actin accumulation at sites of future hair emergence. (H) Magnification of root hair initiation site (arrowheads) shown in G. Scale bars: 50 µm in A-C; 10 µm in F-H.
To further investigate whether ACT7 interacts with AIP1-2 during planar polarity establishment, we analysed the phenotypes of act7;aip1-2 double mutants in which the genetic interaction was not fully penetrant. In these seedlings, root hair position was indistinguishable between act7-7 single and act7-7;aip1.2-1 double mutants as well as between four different allele combinations of act7;aip1-2 double mutants (Fig. 4E; supplementary material Fig. S4C,D), suggesting that act7 behaves epistatically over aip1-2 in these plants.
We next examined the localisation of AIP1-2 and actin during root hair initiation. Strikingly, AIP1-2-mCherry was most strongly expressed in the epidermis and specifically enriched in trichoblast cell files (Fig. 4F,G). At the subcellular level, AIP1-2-mCherry co-localised with the cytosolic marker WAVE 1Y except for the additional nuclear localisation of WAVE 1Y (supplementary material Fig. S4E,F). Together with actin, AIP1-2-mCherry was abundant in outgrowing root hair bulges (Fig. 4F) as well as at hair initiation sites (Fig. 4G,H). The abundance of actin at root hair initiation sites did not differ between the wild type and the aip1.2-1 mutant (supplementary material Fig. S4G,H). Similarly, quantitative evaluation of actin-filament bundling as a measure of actin filament organisation (see methods in the supplementary material) in trichoblasts did not reveal a difference in actin cytoskeleton organisation between aip1.2-1 and the wild type (supplementary material Fig. S4I). This is consistent with the only subtle defects in polar hair positioning observed in aip1-2 single mutants, which might be difficult to resolve at the level of actin organisation. Together, our results reveal a function for AIP1-2 in planar polarity, its genetic interaction with ACT7, hair cell file-specific expression of AIP1-2 and its co-localisation with F-actin at sites of root hair initiation. The basal polarity shift observed in aip1-2 mutants suggests AIP1-2 as a negative modulator of planar polarity.
AIP1-2 and ACT7 are required for polar ROP positioning
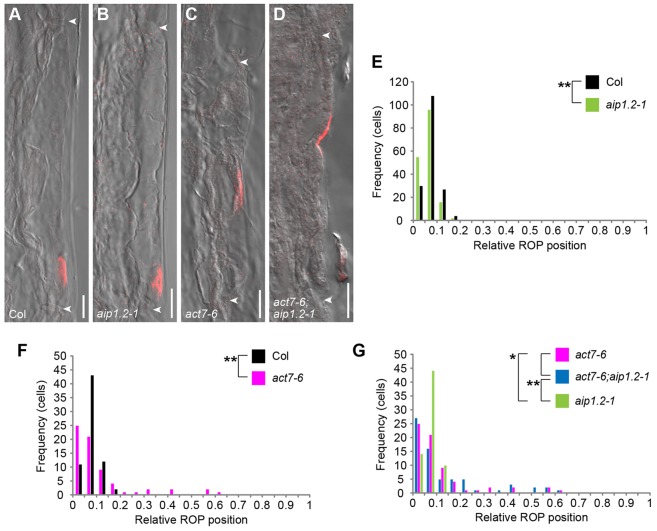
To address whether AIP1-2 and ACT7 affect polar positioning of ROP proteins at sites of hair initiation, we raised an antibody against an AtROP2-derived peptide. The affinity-purified antibody (see methods in the supplementary material) recognised a single band of the expected size at 21.5 kDa on western blots from Arabidopsis seedling protein extracts (see methods in the supplementary material), and we detected EYFP-ROP2 and GFP-ROP6 in lines expressing these proteins (supplementary material Fig. S5). Moreover, the purified serum marked the hair initiation site, where it co-labelled with EYFP-ROP2 (supplementary material Fig. S5). When compared with the wild type (Fig. 5A,E), polar ROP position was shifted basally in aip1.2-1 (Fig. 5B,E), whereas ROP patches were shifted basally and apically in act7-6 (Fig. 5C,F). Furthermore, act7-6 behaved epistatically over aip1.2-1, as the ROP position did not differ between the act7-6 single and the act7-6;aip1.2-1 double mutant (Fig. 5C,D,G). However, ROP patch distribution significantly differed between aip1.2-1 and act7-6 or act7-6;aip1.2-1 mutants (Fig. 5B-D,G). These results show that AIP1-2 and ACT7 are required for correct polar positioning of ROP proteins during establishment of planar polarity.
Fig. 5.
AIP1-2, ACT7 and their interaction mediate polar ROP positioning. (A-D) Anti-ROP (red) immunofluorescence in (A) Col, (B) aip1.2-1, (C) act7-6 and (D) act7-6;aip1.2-1, combined with differential interference contrast (DIC) microscopy image. Arrowheads, apical and basal ends of cells. Scale bars: 10 µm. (E-G) Quantitative analysis of relative ROP positioning of (E) aip1.2-1 and (F) act7-6 compared with Col, and of (G) aip1.2-1 and act7-6 compared with act7-6;aip1.2-1. Number of cells (frequency) with relative apical-basal ROP protein patch position in classes from 0 (basal-most) to 1 (apical-most). n=169 for Col and aip1.2-1, 68 for act7-6 and 78 for act7-6;aip1.2-1; significance P values were determined by KS test. (E) **P=0.006 Col versus aip1.2-1. (F) *P=0.014 Col versus act7-6. (G) P=0.707 act7-6 versus act7-6;aip1.2-1; **P=0.004 aip1.2-1 versus act7-6;aip1.2-1; *P=0.023 act7-6 versus aip1.2-1.
WEREWOLF and ethylene signalling mediate patterned AIP1-2 expression
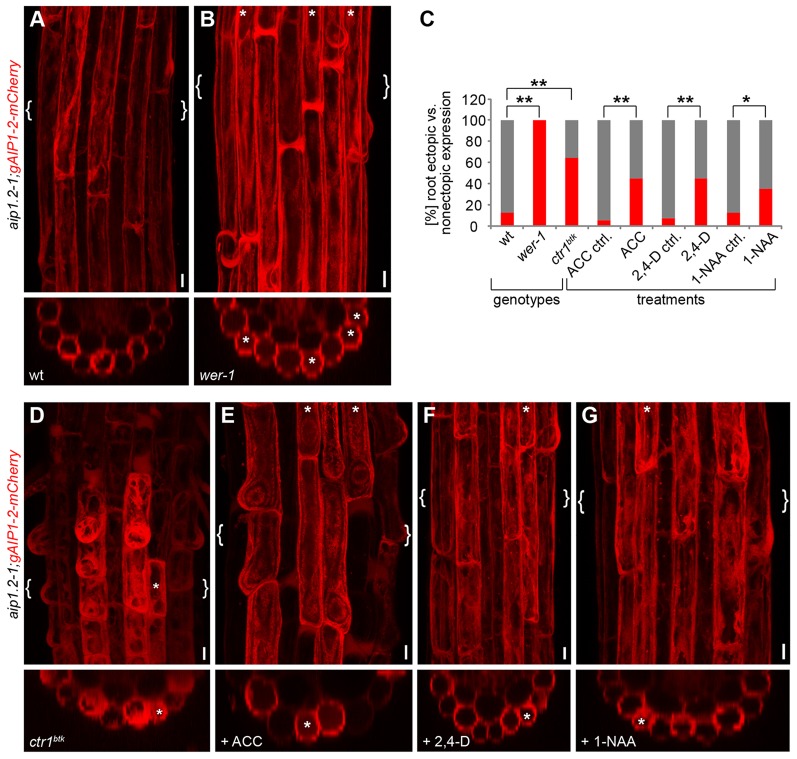
Our findings that AIP1-2 displays hair cell file-enriched expression prompted us to investigate whether its expression relies on WER, a key regulator of cell fate patterning in the root epidermis that controls specification of non-hair cell fate (Lee and Schiefelbein, 1999). Accordingly, most epidermal cells acquire hair cell fate in wer loss-of-function mutants, forming ectopic hairs in non-hair positions. Non-hair positions are those in which epidermal cells contact only one cell file of the underlying cortical tissue layer (Dolan et al., 1994). Strikingly, we observed strong AIP1-2-mCherry expression in all epidermal cell files in wer-1 mutants, (Fig. 6A,B), revealing that WER specifies AIP1-2 expression.
Fig. 6.
Hair cell file-specific AIP1-2 expression depends on WEREWOLF and ethylene signalling. (A,B) CLSM image projections displaying AIP1-2-mCherry fluorescence in root differentiation zone of five-day-old (A) aip1.2-1;gAIP1-2-mCherry (WT) and (B) wer-1;aip1.2-1;gAIP1-2-mCherry (wer-1) seedlings. Upper panels are top views and lower panels transversal sections of the regions indicated by curly brackets. Asterisks indicate ectopic AIP1-2-mCherry enrichment in non-hair cell files. (C) Percentage of five-day-old seedling roots displaying strictly root hair file-specific (grey) or additional ectopic (red) AIP1-2-mCherry expression in aip1.2-1;gAIP1-2-mCherry (wt; n=16 seedlings), wer-1;aip1.2-1;gAIP1-2-mCherry (wer-1; n=15 seedlings), ctr1btk;aip1.2-1;gAIP1-2-mCherry (ctr1btk; n=14 seedlings) seedlings and aip1.2-1;gAIP1-2-mCherry seedlings grown on media supplemented with 5 µM ACC, 20 nM 2,4-D and 100 nM 1-NAA (n=40 seedlings per treatment). Significances were determined by Fisher's exact test with significance levels *P<0.05 and **P<0.01. Supplementary material Table S1 shows exact P values. (D-G) CLSM projections displaying AIP1-2-mCherry fluorescence in root differentiation zone of five-day-old (D) ctr1btk;aip1.2-1;gAIP1-2-mCherry (ctr1btk) or aip1.2-1;gAIP1-2-mCherry seedlings grown on media supplemented with (E) 5 µM ACC, (F) 20 nM 2,4-D and (G) 100 nM 1-NAA. Upper panels are top views and lower panels transversal sections of the regions indicated by curly brackets. Asterisks, ectopic AIP1-2-mCherry enrichment in non-hair cell files. Scale bars: 10 µm.
Interestingly, both CTR1-dependent ethylene signalling and auxin signalling contribute to root hair cell differentiation downstream of cell fate regulation (Dolan et al., 1994; Tanimoto et al., 1995; Masucci and Schiefelbein, 1996). We therefore investigated AIP1-2-mCherry expression in the ctr1btk mutant or upon application of the ethylene precursor 1-Aminocyclopropane-1-carboxylic acid (ACC) and observed cells forming hairs in non-hair positions that expressed AIP1-2-mCherry in the ctr1btk;aip1.2-1 mutant background (Fig. 6C,D), as well as in aip1.2-1 seedling roots grown on ACC-containing medium (Fig. 6C,E). These findings revealed the dependence of patterned AIP1-2 expression on CTR1 function and balanced ethylene levels.
To test whether exogenous application of auxin had a similar effect on AIP1-2 expression, we applied the synthetic auxins 2,4-Dichlorophenoxyacetic acid (2,4-D) or 1-Naphthaleneacetic acid (1-NAA) to the growth medium of AIP1-2-mCherry-expressing seedlings. Strikingly, we observed an increase in ectopic AIP1-2-mCherry expression in non-hair positions in seedlings treated with 2,4-D (Fig. 6C,F) or 1-NAA (Fig. 6C,G). Together, these results demonstrate that cell file-specific AIP1-2 expression depends on WER function and ethylene signalling, and that it is sensitive to auxin concentration.
CTR1 is epistatic over AIP1-2 during root hair positioning
To further analyse the functional relationship between AIP1-2 and CTR1, we generated an aip1.2-1;ctr1btk double mutant that exceptionally displayed basal-most placement of root hairs along trichoblasts and was indistinguishable from ctr1btk (Fig. 7A,C,D), but significantly differed from aip1.2-1 seedlings (Fig. 7B-D). Moreover, average trichoblast length did not differ between aip1.2-1;ctr1btk and ctr1btk (Fig. 7E), although further reduction of trichoblast cell length is found in the strong ctr1-1 allele (Ikeda et al., 2009). Together, these findings suggest that CTR1 behaves epistatically over AIP1-2 during trichoblast elongation and root hair positioning.
Fig. 7.
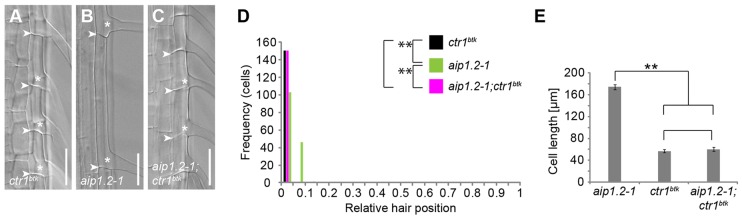
CTR1 is epistatic over AIP1-2. (A-C) Root-hair forming cells of five-day-old (A) ctr1btk, (B) aip1.2-1 and (C) aip1.2-1;ctr1btk seedlings. Arrowheads, apical and basal ends of cells; asterisks, root hair initiation sites. Scale bars: 50 µm. (D) Quantitative analysis of relative root hair-positioning phenotypes of ctr1btk and aip1.2-1 compared with aip1.2-1;ctr1btk. n=150 cells from 30 roots per genotype. Significance P values were determined by KS test. **P=0.000 ctr1btk versus aip1.2-1; P=0.882 ctr1btk versus aip1.2-1;ctr1btk; **P=0.000 aip1.2-1 versus aip1.2-1;ctr1btk. (E) Quantitative analysis of trichoblast cell length in aip1.2-1, ctr1btk and aip1.2-1;ctr1btk. Average cell length from n=150 cells from 30 roots. Data are means±s.d. from three independent experiments, each employing 50 cells from ten roots. Statistical differences determined by two-tailed, unpaired t-test with equal variance and significance level P<0.05 are indicated as **P<0.01; n=3.
DISCUSSION
In this study, we unravel a function for ACT2 and ACT7 downstream of CTR1 during planar polarity formation (a schematic model is found in supplementary material Fig. S6). We further discover AIP1-2 as an interactor of ACTINs and as a negative modulator of planar polarity, the expression of which depends on WER and ethylene signalling. Although a role for ACT2 during root hair positioning had been described (Ringli et al., 2002), our results reveal the strongest contribution of ACT7 to planar polarity among the three actins tested. The phenotypes of act2;act7 mutants further suggest synergistic interaction of the two genes during this process. Strikingly, analysis of double and triple mutants between ctr1btk, act2-3 and act7-6 placed the actin cytoskeleton downstream of ethylene signalling. Whereas ethylene signalling had been known to play a role in microtubule reorganisation (Steen and Chadwick, 1981; Lang et al., 1982; Roberts et al., 1985), our results reveal a genetic link with the actin cytoskeleton. Considering that upregulation of ethylene signalling promotes local auxin biosynthesis in the root tip (Ikeda et al., 2009), and that ACT7 can be induced by auxin (Kandasamy et al., 2001), it is conceivable that some of the observed effects are mediated by auxin.
When searching for ACT7 interactors in a yeast two-hybrid screen, we identified AIP1-2, the yeast homologue Aip1p of which was discovered in a screen for interactors of Act1p (Amberg et al., 1995). Interestingly, neither yeast two-hybrid nor in vitro pull-down assays revealed an obvious difference between the interactions of AIP1-1 or AIP1-2 with the tested ACTs. Therefore, spatio-temporal differences in expression of the investigated proteins or the presence of other actin-binding proteins that have evolved class-specific interactions (Kandasamy et al., 2007) might confer specificity to ACT-AIP1 interaction in vivo.
Our analysis of aip1-2 mutant alleles revealed a basal shift of root hair position, suggesting a negative role for AIP1-2 during planar polarity formation. In contrast to decreased tissue polarisation in act mutants, which might result from destabilisation of actin filaments such as the one observed in act2 single mutants (Kandasamy et al., 2009), the increase in tissue polarisation in aip1-2 mutants could be explained by stabilisation of actin filaments into bundles, as observed in Arabidopsis AIP1 RNAi lines (Ketelaar et al., 2004). However, we did not find an increased bundling of actin in aip1-2 mutants. This is most likely due to the subtle phenotypic defects that might not allow resolving differences in actin organisation when compared with the actin bundling in root epidermal cells observed in RNAi lines downregulated for both Arabidopsis AIP1-1 and AIP1-2 (Ketelaar et al., 2004). Increased actin bundling has also been observed in knockout mutants of the single AIP1 gene in the moss Physcomitrella patens (Augustine et al., 2011) and in hypomorphic flr mutant alleles in Drosophila (Ren et al., 2007). Interestingly, flr negatively affects F-actin accumulation and is required for establishment of wing hair polarity downstream of Frizzled signalling (Ren et al., 2007), suggesting evolutionary conservation of AIP1-mediated actin reorganisation during planar polarity formation in diverse organisms.
In contrast to single-celled yeast, the interaction between specific actin isoforms and AIP1-homologues has not been studied during development of multicellular organisms. To find out which actins might interact with AIP1-2 during planar polarity formation in roots, we studied double mutants between aip1-2 and mutant alleles of three ACTs, revealing strong genetic interaction between AIP1-2 and ACT7. Consistent with these findings, ACT2 and ACT8 function is mainly required during the later stage of root hair tip growth (Kandasamy et al., 2009). Accordingly, double mutants between aip1-2 and act2 or act8 only showed weak defects in root hair positioning (C.S.K. and M.G., unpublished). Interestingly, the synthetic lethality of act7;aip1-2 mutants resembles the situation in yeast, in which deletion of AIP1 is synthetically lethal only in combination with a hypomorphic act1 mutant allele (Rodal et al., 1999). Whereas analyses of single and double mutants suggest that AIP1-2 primarily interacts with ACT7 during planar polarity formation, the lethality of most act7;aip1-2 seeds provides the possibility for future analyses of this interaction during earlier stages of development, including embryogenesis.
The early action of AIP1-2 and ACT7 during planar polarity formation is further supported by the mis-positioning of ROP patches in these mutants, demonstrating a requirement for actin during polar ROP placement. As short-term treatments with F-actin-destabilising drugs did not affect ROP localisation during hair initiation (Molendijk et al., 2001), it is conceivable that the actin cytoskeleton affects polar ROP recruitment in an indirect manner. Nevertheless, whereas previous studies highlighted a role for actin downstream of ROP signalling (Fu et al., 2005, 2009; Nagawa et al., 2012; Lin et al., 2012), our findings reveal an additional requirement of AIP1-2 and ACT7 upstream of ROPs in planar polarity formation. In addition, patterned expression of AIP1-2 in hair files from the early elongation zone onwards supports an early role of actin reorganisation during planar polarity formation. This cell type-enriched AIP1-2 expression depends on WER function as well as on balanced levels of the phytohormones ethylene and auxin. The strong ectopic expression of AIP1-2 observed in wer-1 suggests that the WER pathway is at the core of specifying AIP1-2 expression, whereas the two hormones might modify events downstream of WER action, as previously suggested for their relationship towards other components of the cell fate machinery (Masucci and Schiefelbein, 1996). Consistent with this idea, auxin induces expression of RSL4, a downstream transcription factor of the WER pathway that positively regulates genes specifically expressed in hair cells with a function during hair differentiation (Yi et al., 2010). However, AIP1-2 was not identified among the genes regulated by RSL4, suggesting the existence of additional components or mechanisms that act to integrate hormone- and WER-controlled signals. Intriguingly, our observations that WER mediates patterned expression of AIP1-2 by restricting its expression to hair cell files is contrasted by the findings that expression of the AUX1 auxin influx carrier, which also contributes to planar polarity (Grebe et al., 2002), requires the function of WER and its closest homologue MYB23 for expression in non-hair cell files (Jones et al., 2009). These findings support the view that WER function negatively affects expression of factors involved in planar polarity in hair and non-hair files. Interestingly, whereas components of the Frizzled planar polarity pathway in Drosophila are uniformly expressed throughout the tissue layer, similar to downstream effectors of cytoskeletal organisation, such as Flr and Tsr (Goodrich and Strutt, 2011), our work reveals AIP1-2 as a component of the planar polarity pathway in Arabidopsis that is under control of a patterning system spatially restricting its expression. Our work thus opens the door for future studies on molecular and cellular mechanisms executing planar polarity, such as cytoskeletal reorganisation and its control by a plant-specific patterning system.
MATERIALS AND METHODS
Plant material and growth conditions
The following mutant lines of Arabidopsis thaliana accession Columbia-0 (Col-0) were obtained from the European Nottingham Arabidopsis Stock Centre (NASC): act2-3 (stock number N6959), act7-6 (N670149), act7-7 (N447790), act8-2 (N446015), Col-8 (N60000; genetically equivalent to Col-0), aip1.2-1 (N406016), aip1.2-2 (N873278) and wer-1 (N6349; Lee and Schiefelbein, 1999). Further mutant or marker lines were provided by the corresponding authors of the following articles: ctr1btk (Ikeda et al., 2009), GFP-FABD;mCherry-TUA5 (Sampathkumar et al., 2011; Ketelaar et al., 2004; Gutierrez et al., 2009), GFP-ABD2-GFP (Wang et al., 2008), Lifeact-Venus (Era et al., 2009), WAVE 1Y (Geldner et al., 2009). Primers employed for genotypic characterisation of the lines mentioned above are listed in supplementary material Table S2.
Arabidopsis plants were grown under a photoperiod of 16 h light (150 µmol/m2/s) at 22°C and 8 h dark at 18°C at 60% air humidity. Seeds were surface-sterilised and stratified as described (Fischer et al., 2006). Seedlings were vertically grown for five days on Murashige and Skoog (MS) medium (1× MS salt; Duchefa, Haarlem, The Netherlands), 10 g/l sucrose, 8 g/l plant agar (Duchefa), 0.5 g/l 2-(N-morpholino)ethanesulfonic acid hydrate (MES; Sigma-Aldrich, pH 5.7). Media containing 5 µM ACC (Sigma-Aldrich), 20 nM 2,4-D (Sigma-Aldrich) or 100 nM 1-NAA (Sigma-Aldrich) were prepared as described (Grebe et al., 2002; Fischer et al., 2006).
An Arabidopsis root-cell suspension culture (Col-0; Boutté et al., 2010) was grown in MS medium for Arabidopsis [MSAR; 1× MS basal salts (Sigma-Aldrich), 30 g/l sucrose, 0.24 mg/l 2,4-D, 14 µg/l kinetin (Sigma-Aldrich), 200 mg/l myo-inositol (Sigma-Aldrich), 1.0 mg/l nicotinic acid (Sigma-Aldrich), 1.0 mg/l pyridoxine (Sigma-Aldrich), 10.0 mg/l thiamine (Sigma-Aldrich), pH 5.7] under constant shaking. The cell suspension was sub-cultured weekly by diluting it 6.7-fold in fresh medium.
Quantitative analysis of root hair positioning
Quantitative analysis of relative root hair positions was performed as described previously (Fischer et al., 2006). One-hundred and fifty cells per genotype were analysed derived from three independent experiments performed on ten roots, 50 cells each. As root hair positioning does not follow a normal distribution, differences in distributions of relative hair position between genotypes were tested for significance using the non-parametric, two-sample Kolmogorov–Smirnov (KS)-test (http://www.physics.csbsju.edu/stats/KS-test.n.plot_form.html) as described (Pietra et al., 2013). The KS-test considers both location and shape of the distributions. Significance level was set at P<0.05.
Immunocytochemistry and quantitative analysis of ROP positioning
Whole-mount immunolabelling of roots was performed as described (Boutté and Grebe, 2014), in three independent experiments with two different secondary antibodies. Antibody dilutions were: affinity-purified rabbit anti-AtROP2 1:250 (generation of antibody is described in the supplementary material); DyLight 488 donkey anti-rabbit (Jackson ImmunoResearch, 711-485-152; 1:400); DyLight 549 donkey anti-rabbit (Jackson ImmunoResearch, 711-505-152; 1:400). Nuclei were counterstained with DAPI (1 µg/ml, Sigma-Aldrich). Immunolabelled roots were imaged by confocal laser scanning microscopy (CLSM) using a Zeiss LSM780 system. Images were processed and analysed using ImageJ (NIH) and Adobe Photoshop CS4 (Adobe), and ROP positioning in trichoblasts prior to root hair initiation was quantified as previously described (Fischer et al., 2006). As relative position of ROP patches along trichoblasts did not follow a normal distribution, significances of differences between distributions were analysed employing the KS-test with a significance level at P<0.05 as described (Pietra et al., 2013).
Bacterial protein expression and protein extraction
AIP1-1 and AIP1-2 cDNA sequences were cloned C-terminally of the GST-coding sequence, between the NcoI and the KpnI restriction sites of the pET-GST1b vector. This is a derivative of pET-GST1a (Bogomolovas et al., 2009; kindly provided by Dr Gunter Stier, Heidelberg University, Germany), in which the 6×Histidine (6×His) sequence is introduced C-terminally of the GST moiety instead of N-terminally as in pET-GST1a. A GST-only control vector was generated by introducing a STOP linker into the NcoI restriction site. Oligonucleotide sequences used for cloning are listed in supplementary material Table S2. Vectors encoding GST or GST-fusion proteins were transformed into E. coli Rosetta DE3 cells (Merck). According to an auto-induction protocol for protein expression in bacteria (Studier, 2005), single Rosetta DE3 colonies containing the GST-fusion expression vectors were transferred to 3 ml auto-inducing ZYM 5052 medium [1% N-Z-amine (Sigma-Aldrich), 0.5% yeast extract, 25 mM Na2HPO4, 25 mM KH2PO4, 50 mM NH4Cl, 5 mM Na2SO4, 2 mM MgSO4, 0.5% glycerol, 0.05% glucose, 0.2% lactose] supplemented with trace elements (50 µM FeCl3, 20 µM CaCl2, 10 µM MnCl2, 10 µM ZnSO4, 2 µM CoCl2, 2 µM CuCl2, 2 µM NiCl2, 2 µM Na2MoO4, 2 µM H3BO3), initially incubated for 3 h at 37°C under constant shaking before the temperature was reduced to 20°C for another 16 h of growth. The pelleted cultures were resuspended in 300 µl lysis buffer [140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, 1× protease inhibitor cocktail (Sigma-Aldrich), 1 mM phenylmethanesulfonyl fluoride (PMSF; Sigma-Aldrich), pH 7.3] and lysed using a standard sonication protocol (Ausubel et al., 1995). Triton X-100 was added to a relative concentration of 1% and the protein extracts were collected after 10 min centrifugation at 15,000 g.
6×His-fusions of ACT2 and ACT7 were expressed from the pColdI vector (Takara). ACT2 and ACT7 cDNAs were cloned C-terminally of the 6×His moiety between the BamHI and the EcoRI restriction sites of the vector. Primers used for cloning are listed in supplementary material Table S2. According to a modified isopropyl β-D-thiogalactoside (IPTG) induction protocol (Tamura et al., 2011), single E. coli Rosetta DE3 colonies containing the 6×His-fusion expression vectors were transferred to 5 ml LB medium (10 g/l bacto-tryptone, 5 g/l bacto yeast extract, 5 g/l NaCl) and incubated for 4 h at 37°C under constant shaking. The cells were then cooled to 15°C for 30 min, IPTG (Sigma-Aldrich) was added to a final concentration of 50 µM and the cultures were incubated for 20 h at 15°C under constant shaking. According to the protocol of Tamura et al. (2011), the pelleted cultures were cooled down to −80°C for 2 h, resuspended in 1 ml buffer A [1 mM ethylenediamine tetraacetic acid (EDTA), 5 mM DL-dithiothreitol (DTT), 1 mM adenosine 5′-diphosphate (ADP), 0.1 mM PMSF, 2 mM diisopropyl fluorophosphate (DIFP), 10 mM tris(hydroxymethyl)aminomethane (Tris), pH 8.0], lysed using a standard sonication protocol (Ausubel et al., 1995) and protein extracts were collected after 10 min centrifugation at 15,000 g.
Extraction of plant proteins
Extraction of plant proteins from a Col-0 root-cell suspension culture (Boutté et al., 2010) was performed as described previously (Grebe et al., 2000). Four days after subculturing, a 50 ml suspension culture was filtered through grade-3 qualitative filter paper (Munktell Filter, Falun, Sweden). Cells were frozen in liquid nitrogen, homogenised and suspended in 6 ml bead-binding buffer (50 mM K3PO4, 150 mM KCl, 1 mM MgCl2, pH 7.5), containing 1× protease inhibitor cocktail for use with plant cell extracts (Sigma-Aldrich) and 1 mM PMSF. Cells were disrupted by sonication using a standard protocol as applied to bacterial protein extracts (Ausubel et al., 1995), and 6 ml bead-binding buffer, containing 10% glycerol, 2% Triton X-100 and 1 mM DTT, was added. After initial centrifugation for 10 min at 10,000 g, the supernatant was transferred to a fresh vial, centrifuged for 10 min at 30,000 g and the supernatant containing plant proteins was used for western blotting and in vitro binding assays.
In vitro binding assays
We prepared a bead volume of 40 µl glutathione sepharose (Amersham) according to the manufacturer's protocol and applied 300 µl bacterial extracts containing GST, GST-AIP1-1 or GST-AIP1-2. Beads pre-absorbed with GST constructs or beads just after preparation according to the manufacturer's protocol were incubated with 300 µl plant extract obtained from root-cell suspension culture or 300 µl bacterial extracts containing 6×His-ACT2 or 6×His-ACT7. Following steps of the in vitro binding assay were conducted as previously described (Grebe et al., 2000).
Western blot analyses
Dilutions and distributors of antibodies employed for western blot detection are listed in supplementary material Table S3. Enhanced chemiluminescence (ECL) prime western blotting detection reagents were obtained from Amersham and used according to the manufacturer's instruction. AGFA Cronex 5 medical X-ray films (Agfa-Gevaert) were developed using a Curix 60 processor (Agfa-Gevaert) or images of the blots were acquired using a LAS-3000 imaging system (Fujifilm).
Yeast two-hybrid analyses
Initial screening for interactors of the full-length Arabidopsis ACT7 bait was performed at Hybrigenics (Paris, France) using ULTImate Y2H technology to screen a one-week-old Arabidopsis seedling cDNA expression library. Several full-length AIP1-2 (At3g18060) cDNA clones revealed it as the only interactor. Different ACTIN and AIP1 cDNA sequences from Arabidopsis were re-cloned in original plasmid vectors of the LexA-based interaction trap version of the two-hybrid system (Gyuris et al., 1993; Ausubel et al., 1995), employing the pJG4-5 derivative pMG5 and the pEG202 derivative pMG8 (Grebe et al., 2000) as prey and bait vectors, respectively. Coding sequences were cloned in frame between the NotI and XbaI restriction sites, except for ACT1 cDNA, which was cloned solely employing the NotI restriction site. Primers used for cloning of the analysed constructs and accession numbers for cDNA sequences are listed in supplementary material Table S2.
Reverse transcription (RT)-PCR
Isolation and purification of mRNA from roots of five-day-old Arabidopsis seedlings and subsequent cDNA synthesis were performed as described (Pietra et al., 2013). For semi-quantitative PCR, primer pairs were employed that flanked the T-DNA insertions in AIP1-2 and, furthermore, spanned introns in order to detect potential genomic DNA contaminations (see supplementary material Table S2 for primer sequences). PCR products were analysed after 28 cycles.
Complementation tests
The genomic AIP1-2 sequence (gAIP1-2) was cloned into the pGreenII0229 vector (John Innes Centre, Norwich, UK; Hellens et al., 2000) between the XbaI and ApaI restriction sites. The 2162 bp region upstream of the start codon of AIP1-2 was employed as a promoter and cloned between the NotI and the XbaI restriction sites. The 465 bp region downstream of the stop codon of AIP1-2 was employed as terminator and cloned into the KpnI restriction site. The 3×Myc coding sequence provided in supplementary material Table S2 was synthesised by GenScript, based on the 3×Myc sequence from pNIGEL 07 (Geldner et al., 2009) and provided in the vector pUC57. The coding sequence for mCherry obtained in the plasmid pmCherry (Clontech) was amplified by PCR, and 3×Myc was restricted from pUC57-3×Myc using ApaI and KpnI. It was then fused in-frame to the 3′-end of the gAIP1-2 coding sequence using the ApaI and the KpnI restriction sites. Primers used for cloning are listed in supplementary material Table S2. Integrity of all constructs was confirmed by nucleic acid sequencing. Arabidopsis plants ecotype Col-0 were transformed by the floral dip method (Clough and Bent, 1998) employing the Agrobacterium tumefaciens strain GV3101 (Koncz and Schell, 1986).
Confocal laser scanning microscopy (CLSM)
All analyses using CLSM were conducted with a Zeiss LSM780 laser scanning microscope (Zeiss). Samples mounted in aqueous solutions were imaged using a water-corrected C-Apochromat 40× objective (Zeiss) with a numerical aperture (NA) of 1.2. Samples mounted in glycerol-based solutions were imaged using an oil-corrected Plan-Apochromat 63× objective with NA 1.4 (Zeiss). Detailed CLSM settings are listed in supplementary material Table S4.
Visualisation of F-actin networks and AIP1-2-mCherry expression
F-actin networks at root hair initiation sites were visualised in seedlings expressing GFP-FABD, GFP-ABD2-GFP, Lifeact-Venus or in seedlings probed with BODIPY FL phallacidin (Nishimura et al., 2003). For display, images of early trichoblast cells acquired on longitudinal cross-sections at distances of 0.52 µm (GFP-FABD or Lifeact-Venus), 0.91 µm (GFP-ABD2-GFP) or 0.45 µm (BODIPY FL phallacidin) from each other, respectively, were processed by maximum intensity projection, employing ZEN lite 2012 analysis software (Zeiss).
To visualise cell file-specific AIP1-2-mCherry expression, longitudinal sections at a distance of 3.54 µm from a top view of the analysed seedling root were processed by maximum-intensity projection, employing ZEN lite 2012 analysis software. Cross-sectional overviews were generated by transversally projecting selected areas of 15.1 µm in length into one image.
Image processing
Image analysis software programs used for acquiring and processing graphical data were: AxioVs40 V 4.8.2.0 (Zeiss), ZEN lite 2012 analysis software, ImageJ (NIH), Adobe Photoshop CS3 and CS4 and Adobe Illustrator CS4 (Adobe).
Supplementary Material
Acknowledgements
We thank E. Blancaflor, N. Geldner, T. Ketelaar, S. Persson, B. Scheres, T. Ueda and Z. Yang for sharing published materials, the European Nottingham Arabidopsis Stock Centre for distributing seed stocks, G. Stier for advice on expressing recombinant actin, Y. Boutté for help with RT-PCR, T. Vain for help with image analysis and D. Gendre for commenting on the manuscript.
Footnotes
Competing interests
The authors declare no competing financial interests.
Author contributions
M.G., C.S.K. and Y.I. designed the experiments. C.S.K. carried out the majority of the experiments. C.S.K., A.R.C., J.-C.N., S.P., Y.I. and M.G. carried out experiments, analysed and interpreted the data. T.S. and A.H. performed experiments to characterise the anti-ROP2 serum. C.S.K. and M.G. wrote the manuscript. All authors read and edited the manuscript prior to submission.
Funding
This work was supported by the Swedish Research Council (‘Vetenskapsrådet’) grant [2003-2744 to M.G.], by a Marie Curie Incoming International Fellowship [022215-epol to Y.I.] and by an Umeå University Young Researcher Award (‘karriärbidrag’) to M.G. Deposited in PMC for immediate release.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.111013/-/DC1
References
- Adler P. N. (2002). Planar signaling and morphogenesis in Drosophila. Dev. Cell 2, 525-535 10.1016/S1534-5807(02)00176-4 [DOI] [PubMed] [Google Scholar]
- Allwood E. G., Anthony R. G., Smertenko A. P., Reichelt S., Drobak B. K., Doonan J. H., Weeds A. G. and Hussey P. J. (2002). Regulation of the pollen-specific actin-depolymerizing factor LlADF1. Plant Cell 14, 2915-2927 10.1105/tpc.005363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg D. C., Basart E. and Botstein D. (1995). Defining protein interactions with yeast actin in vivo. Nat. Struct. Biol. 2, 28-35 10.1038/nsb0195-28 [DOI] [PubMed] [Google Scholar]
- An Y. Q., Huang S., McDowell J. M., McKinney E. C. and Meagher R. B. (1996). Conserved expression of the Arabidopsis ACT1 and ACT3 actin subclass in organ primordia and mature pollen. Plant Cell 8, 15-30 10.1105/tpc.8.1.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustine R. C., Pattavina K. A., Tuzel E., Vidali L. and Bezanilla M. (2011). Actin interacting protein1 and actin depolymerizing factor drive rapid actin dynamics in Physcomitrella patens. Plant Cell 23, 3696-3710 10.1105/tpc.111.090753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A. and Struhl K. (1995). Current Protocols in Molecular Biology. New York: Greene Publishing Associates and Wiley-Interscience. [Google Scholar]
- Baluška F., Salaj J., Mathur J., Braun M., Jasper F., Šamaj J., Chua N.-H., Barlow P. W. and Volkmann D. (2000). Root hair formation: F-actin-dependent tip growth is initiated by local assembly of profilin-supported F-actin meshworks accumulated within expansin-enriched bulges. Dev. Biol. 227, 618-632 10.1006/dbio.2000.9908 [DOI] [PubMed] [Google Scholar]
- Blair A., Tomlinson A., Pham H., Gunsalus K. C., Goldberg M. L. and Laski F. A. (2006). Twinstar, the Drosophila homolog of cofilin/ADF, is required for planar cell polarity patterning. Development 133, 1789-1797 10.1242/dev.02320 [DOI] [PubMed] [Google Scholar]
- Bogomolovas J., Simon B., Sattler M. and Stier G. (2009). Screening of fusion partners for high yield expression and purification of bioactive viscotoxins. Protein Expr. Purif. 64, 16-23 10.1016/j.pep.2008.10.003 [DOI] [PubMed] [Google Scholar]
- Boutté Y. and Grebe M. (2014). Immunocytochemical fluorescent in situ visualization of proteins in Arabidopsis. Methods Mol. Biol. 1062, 453-472 10.1007/978-1-62703-580-4_24 [DOI] [PubMed] [Google Scholar]
- Boutté Y., Frescatada-Rosa M., Men S., Chow C.-M., Ebine K., Gustavsson A., Johansson L., Ueda T., Moore I., Jürgens G. et al. (2010). Endocytosis restricts Arabidopsis KNOLLE syntaxin to the cell division plane during late cytokinesis. EMBO J. 29, 546-558 10.1038/emboj.2009.363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D., Pan H., Wan P., Wu J., Luo J., Zhu H. and Chen J. (2012). AIP1 acts with cofilin to control actin dynamics during epithelial morphogenesis. Development 139, 3561-3571 10.1242/dev.079491 [DOI] [PubMed] [Google Scholar]
- Clough S. J. and Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735-743 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Cvrčková F., Bezvoda R. and Zárský V. (2010). Computational identification of root hair-specific genes in Arabidopsis. Plant Signal. Behav. 5, 1407-1418 10.4161/psb.5.11.13358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan L., Duckett C. M., Grierson C., Linstead P., Schneider K., Lawson E., Dean C., Poethig S. and Roberts K. (1994). Clonal relationships and cell patterning in the root epidermis of Arabidopsis. Development 120, 2465-2474. [Google Scholar]
- Era A., Tominaga M., Ebine K., Awai C., Saito C., Ishizaki K., Yamato K. T., Kohchi T., Nakano A. and Ueda T. (2009). Application of Lifeact reveals F-actin dynamics in Arabidopsis thaliana and the liverwort, Marchantia polymorpha. Plant Cell Physiol. 50, 1041-1048 10.1093/pcp/pcp055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U., Ikeda Y., Ljung K., Serralbo O., Singh M., Heidstra R., Palme K., Scheres B. and Grebe M. (2006). Vectorial information for arabidopsis planar polarity is mediated by combined AUX1, EIN2, and GNOM activity. Curr. Biol. 16, 2143-2149 10.1016/j.cub.2006.08.091 [DOI] [PubMed] [Google Scholar]
- Fu Y., Gu Y., Zheng Z., Wasteneys G. and Yang Z. (2005). Arabidopsis interdigitating cell growth requires two antagonistic pathways with opposing action on cell morphogenesis. Cell 120, 687-700 10.1016/j.cell.2004.12.026 [DOI] [PubMed] [Google Scholar]
- Fu Y., Xu T., Zhu L., Wen M. and Yang Z. (2009). A ROP GTPase signaling pathway controls cortical microtubule ordering and cell expansion in Arabidopsis. Curr. Biol 19, 1827-1832 10.1016/j.cub.2009.08.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galway M. E., Masucci J. D., Lloyd A. M., Walbot V., Davis R. W. and Schiefelbein J. W. (1994). The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev. Biol. 166, 740-754 10.1006/dbio.1994.1352 [DOI] [PubMed] [Google Scholar]
- Geldner N., Denervaud-Tendon V., Hyman D. L., Mayer U., Stierhof Y. D. and Chory J. (2009). Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J. 59, 169-178 10.1111/j.1365-313X.2009.03851.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich L. V. and Strutt D. (2011). Principles of planar polarity in animal development. Development 138, 1877-1892 10.1242/dev.054080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebe M., Gadea J., Steinmann T., Kientz M., Rahfeld J.-U., Salchert K., Koncz C. and Jürgens G. (2000). A conserved domain of the Arabidopsis GNOM protein mediates subunit interaction and cyclophilin 5 binding. Plant Cell 12, 343-356 10.1105/tpc.12.3.343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebe M., Friml J., Swarup R., Ljung K., Sandberg G., Terlou M., Palme K., Bennett M. J. and Scheres B. (2002). Cell polarity signaling in Arabidopsis involves a BFA-sensitive auxin influx pathway. Curr. Biol. 12, 329-334 10.1016/S0960-9822(02)00654-1 [DOI] [PubMed] [Google Scholar]
- Guo B., Chen Y., Zhang G., Xing J., Hu Z., Feng W., Yao Y., Peng H., Du J., Zhang Y. et al. (2013). Comparative proteomic analysis of embryos between a maize hybrid and its parental lines during early stages of seed germination. PLoS ONE 8, e65867 10.1371/journal.pone.0065867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez R., Lindeboom J. J., Paredez A. R., Emons A. M. C. and Ehrhardt D. W. (2009). Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat. Cell Biol. 11, 797-806 10.1038/ncb1886 [DOI] [PubMed] [Google Scholar]
- Gyuris J., Golemis E., Chertkov H. and Brent R. (1993). Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75, 791-803 10.1016/0092-8674(93)90498-F [DOI] [PubMed] [Google Scholar]
- Hellens R. P., Edwards E. A., Leyland N. R., Bean S. and Mullineaux P. M. (2000). pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42, 819-832 10.1023/A:1006496308160 [DOI] [PubMed] [Google Scholar]
- Ikeda Y., Men S., Fischer U., Stepanova A. N., Alonso J. M., Ljung K. and Grebe M. (2009). Local auxin biosynthesis modulates gradient-directed planar polarity in Arabidopsis. Nat. Cell Biol. 11, 731-738 10.1038/ncb1879 [DOI] [PubMed] [Google Scholar]
- Jones M. A., Shen J.-J., Fu Y., Li H., Yang Z. and Grierson C. S. (2002). The Arabidopsis Rop2 GTPase is a positive regulator of both root hair initiation and tip growth. Plant Cell 14, 763-776 10.1105/tpc.010359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A. R., Kramer E. M., Knox K., Swarup R., Bennett M. J., Lazarus C. M., Leyser H. M. O. and Grierson C. S. (2009). Auxin transport through non-hair cells sustains root-hair development. Nat. Cell Biol. 11, 78-84 10.1038/ncb1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy M. K., Gilliland L. U., McKinney E. C. and Meagher R. B. (2001). One plant actin isovariant, ACT7, is induced by auxin and required for normal callus formation. Plant Cell 13, 1541-1554 10.1105/tpc.13.7.1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy M. K., Burgos-Rivera B. McKinney E. C. Ruzicka D. R. and Meagher R. B. (2007). Class-specific interaction of profilin and ADF isovariants with actin in the regulation of plant development. Plant Cell 19, 3111-3126 10.1105/tpc.107.052621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy M. K., McKinney E. C. and Meagher R. B. (2009). A single vegetative actin isovariant overexpressed under the control of multiple regulatory sequences is sufficient for normal Arabidopsis development. Plant Cell 21, 701-718 10.1105/tpc.108.061960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketelaar T., Allwood E. G., Anthony R., Voigt B., Menzel D. and Hussey P. J. (2004). The actin-interacting protein AIP1 is essential for actin organization and plant development. Curr. Biol. 14, 145-149 10.1016/j.cub.2004.01.004 [DOI] [PubMed] [Google Scholar]
- Ketelaar T., Allwood E. G. and Hussey P. J. (2007). Actin organization and root hair development are disrupted by ethanol-induced overexpression of Arabidopsis actin interacting protein 1 (AIP1). New Phytol. 174, 57-62 10.1111/j.1469-8137.2007.01979.x [DOI] [PubMed] [Google Scholar]
- Koncz C. and Schell J. (1986). The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. MGG 204, 383-396 10.1007/BF00331014 [DOI] [Google Scholar]
- Lang J. M., Eisinger W. R. and Green P. B. (1982). Effects of ethylene on the orientation of microtubules and cellulose microfibrils of pea epicotyl cells with polylamellate cell walls. Protoplasma 110, 5-14 10.1007/BF01314675 [DOI] [Google Scholar]
- Lee M. M. and Schiefelbein J. (1999). WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99, 473-483 10.1016/S0092-8674(00)81536-6 [DOI] [PubMed] [Google Scholar]
- Lin D., Nagawa S., Chen J., Cao L., Chen X., Xu T., Li H., Dhonukshe P., Yamamuro C., Friml J. et al. (2012). A ROP GTPase-dependent auxin signaling pathway regulates the subcellular distribution of PIN2 in Arabidopsis roots. Curr. Biol. 22, 1319-1325 10.1016/j.cub.2012.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci J. D. and Schiefelbein J. W. (1994). The rhd6 mutation of Arabidopsis thaliana alters root-hair initiation through an auxin- and ethylene-associated process. Plant Physiol. 106, 1335-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci J. D. and Schiefelbein J. W. (1996). Hormones act downstream of TTG and GL2 to promote root hair outgrowth during epidermis development in the Arabidopsis root. Plant Cell 8, 1505-1517 10.1105/tpc.8.9.1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci J. D., Rerie W. G., Foreman D. R., Zhang M., Galway M. E., Marks M. D. and Schiefelbein J. W. (1996). The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122, 1253-1260. [DOI] [PubMed] [Google Scholar]
- Molendijk A. J., Bischoff F., Rajendrakumar C. S. V., Friml J., Braun M., Gilroy S. and Palme K. (2001). Arabidopsis thaliana Rop GTPases are localized to tips of root hairs and control polar growth. EMBO J. 20, 2779-2788 10.1093/emboj/20.11.2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagawa S., Xu T., Lin D., Dhonukshe P., Zhang X., Friml J., Scheres B., Fu Y. and Yang Z. (2012). ROP GTPase-dependent actin microfilaments promote PIN1 polarization by localized inhibition of clathrin-dependent endocytosis. PLoS Biol. 10, e1001299 10.1371/journal.pbio.1001299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M., Kiefer C. S. and Grebe M. (2012). Planar polarity, tissue polarity and planar morphogenesis in plants. Curr. Opin. Plant Biol. 15, 593-600 10.1016/j.pbi.2012.07.006 [DOI] [PubMed] [Google Scholar]
- Nishimura T., Yokota E., Wada T., Shimmen T. and Okada K. (2003). An Arabidopsis ACT2 dominant-negative mutation, which disturbs F-actin polymerization, reveals its distinctive function in root development. Plant Cell Physiol. 44, 1131-1140 10.1093/pcp/pcg158 [DOI] [PubMed] [Google Scholar]
- Nübler-Jung K., Bonitz R. and Sonnenschein M. (1987). Cell polarity during wound healing in an insect epidermis. Development 100, 163-170. [DOI] [PubMed] [Google Scholar]
- Pietra S., Gustavsson A., Kiefer C., Kalmbach L., Hörstedt P., Ikeda Y., Stepanova A. N., Alonso J. M. and Grebe M. (2013). Arabidopsis SABRE and CLASP interact to stabilize cell division plane orientation and planar polarity. Nat. Commun. 4, 2779 10.1038/ncomms3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren N., Charlton J. and Adler P. N. (2007). The flare gene, which encodes the AIP1 protein of Drosophila, functions to regulate F-actin disassembly in pupal epidermal cells. Genetics 176, 2223-2234 10.1534/genetics.107.072959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringli C., Baumberger N., Diet A., Frey B. and Keller B. (2002). ACTIN2 is essential for bulge site selection and tip growth during root hair development of Arabidopsis. Plant Physiol. 129, 1464-1472 10.1104/pp.005777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts I. N., Lloyd C. W. and Roberts K. (1985). Ethylene-induced microtubule reorientations: mediation by helical arrays. Planta 164, 439-447 10.1007/BF00395959 [DOI] [PubMed] [Google Scholar]
- Rodal A. A., Tetreault J. W., Lappalainen P., Drubin D. G. and Amberg D. C. (1999). Aip1p interacts with cofilin to disassemble actin filaments. J. Cell Biol. 145, 1251-1264 10.1083/jcb.145.6.1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampathkumar A., Lindeboom J. J., Debolt S., Gutierrez R., Ehrhardt D. W., Ketelaar T. and Persson S. (2011). Live cell imaging reveals structural associations between the actin and microtubule cytoskeleton in Arabidopsis. Plant Cell 23, 2302-2313 10.1105/tpc.111.087940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen D. A. and Chadwick A. V. (1981). Ethylene effects in pea stem tissue: EVIDENCE OF MICROTUBULE MEDIATION. Plant Physiol. 67, 460-466 10.1104/pp.67.3.460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W. (2005). Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 41, 207-234 10.1016/j.pep.2005.01.016 [DOI] [PubMed] [Google Scholar]
- Tamura M., Ito K., Kunihiro S., Yamasaki C. and Haragauchi M. (2011). Production of human beta-actin and a mutant using a bacterial expression system with a cold shock vector. Protein Expr. Purif. 78, 1-5 10.1016/j.pep.2010.09.007 [DOI] [PubMed] [Google Scholar]
- Tanimoto M., Roberts K. and Dolan L. (1995). Ethylene is a positive regulator of root hair development in Arabidopsis thaliana. Plant J. 8, 943-948 10.1046/j.1365-313X.1995.8060943.x [DOI] [PubMed] [Google Scholar]
- Wang Y. S., Yoo C. M. and Blancaflor E. B. (2008). Improved imaging of actin filaments in transgenic Arabidopsis plants expressing a green fluorescent protein fusion to the C- and N-termini of the fimbrin actin-binding domain 2. New Phytol. 177, 525-536. [DOI] [PubMed] [Google Scholar]
- Wu J., Roman A.-C., Carvajal-Gonzalez J. M. and Mlodzik M. (2013). Wg and Wnt4 provide long-range directional input to planar cell polarity orientation in Drosophila. Nat. Cell Biol. 15, 1045-1055 10.1038/ncb2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Wen M., Nagawa S., Fu Y., Chen J.-G., Wu M.-J., Perrot-Rechenmann C., Friml J., Jones A. M. and Yang Z. (2010). Cell surface- and Rho GTPase-based auxin signaling controls cellular interdigitation in Arabidopsis. Cell 143, 99-110 10.1016/j.cell.2010.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (2008). Cell polarity signaling in Arabidopsis. Annu. Rev. Cell Dev. Biol. 24, 551-575 10.1146/annurev.cellbio.23.090506.123233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi K., Menand B., Bell E. and Dolan L. (2010). A basic helix-loop-helix transcription factor controls cell growth and size in root hairs. Nat. Genet. 42, 264-267 10.1038/ng.529 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.