Abstract
In many invertebrates, the nuclearization of β-catenin at one pole of the embryo initiates endomesoderm specification. An intriguing possibility is that a gradient of nuclear β-catenin (nβ-catenin), similar to that operating in vertebrate neural tube patterning, functions to distinguish cell fates in invertebrates. To test this hypothesis, we determined the function of nβ-catenin during the early development of the sea star, which undergoes a basal deuterostomal mode of embryogenesis. We show that low levels of nβ-catenin activity initiate bra, which is expressed in the future posterior endoderm-fated territory; intermediate levels are required for expression of foxa and gata4/5/6, which are later restricted to the endoderm; and activation of ets1 and erg in the mesoderm-fated territory requires the highest nβ-catenin activity. Transcription factors acting downstream of high nβ-catenin segregate the endoderm/mesoderm boundary, which is further reinforced by Delta/Notch signaling. Significantly, therefore, in sea stars, endomesoderm segregation arises through transcriptional responses to levels of nβ-catenin activity. Here, we describe the first empirical evidence of a dose-dependent response to a dynamic spatiotemporal nβ-catenin activity that patterns cell fates along the primary axis in an invertebrate.
Keywords: Echinoderm, Endomesoderm, Gene regulatory network, Nuclear β-catenin, Patiria
Summary: A spatiotemporal gradient of Wnt signaling, which is known to pattern early vertebrate embryos, regulates the segregation of cell fate in the invertebrate sea star embryo.
INTRODUCTION
During development, animals construct a complex organism comprising multiple distinct cell types, beginning with only a limited number of maternal anisotropies. The specification of these cell types is controlled by interacting networks of regulatory genes that transduce early asymmetries into distinct combinations of genes expressed in different cell types. Global, systems-level approaches that compile these interactions into gene regulatory networks (GRNs) have been key to understand how early maternal processes establish zygotic regulatory hierarchies that direct cell specification (see e.g. Davidson, 2006; Levine and Davidson, 2005). The specification of endomesoderm, a transient bipotential territory that gives rise to both endoderm and mesoderm, is a crucial early step in animal development. Detailed GRNs explaining the genesis of this territory have been described for the invertebrates Nematostella vectensis, Caenorhabditis elegans and Strongylocentrotus purpuratus, and for vertebrates, such as Xenopus laevis and Danio rerio (Chan et al., 2009; Loose and Patient, 2004; Maduro, 2009; Peter and Davidson, 2011; Röttinger et al., 2012). In these and other, less well-studied organisms (Darras et al., 2011; Henry et al., 2008; Imai et al., 2000), nuclear β-catenin (nβ-catenin) is required for endomesoderm specification, or, in vertebrates, for the specification of anterior endomesoderm (Logan and Nusse, 2004). Therefore, the polar nuclearization of β-catenin is an ancestral mechanism among the metazoa used to establish endomesoderm.
Several well-characterized model systems, most notably vertebrate species and Drosophila, make extensive use of molecular gradients during early development (see e.g. Green and Smith, 1991; Ip et al., 1992). These gradients are thought to be formed via diffusion of a factor from a localized source, and function to activate gene expression in a dose-dependent manner to organize territories of discrete cell types along embryonic axes. In vertebrate embryos, a Wnt-driven, posterior-to-anterior gradient of nβ-catenin functions to specify positional cell fates in the neural tube during axial patterning (see e.g. Kiecker and Niehrs, 2001). It has been suggested that endoderm and mesoderm are patterned by this gradient (In der Rieden et al., 2010; Kiecker and Niehrs, 2001; McLin et al., 2007; Schneider and Mercola, 2001). Early Drosophila development is also characterized by the use of gradients to pattern the embryo, although there is no known requirement for a gradient of nβ-catenin during axis specification (see e.g. Jaeger, 2011). However, as numerous studies have shown that β-catenin is nuclearized in one pole of invertebrate embryos, it has been proposed that a gradient of nβ-catenin might also act to specify fates along the primary axis during early development in many taxa, and that it might be an ancestral feature of metazoan development (see e.g. Niehrs, 2010). As yet, there is no evidence in any invertebrate for a long-range nβ-catenin gradient being used to establish different territories, although differential accumulation has been observed within restricted domains (Darras et al., 2011; Henry et al., 2008; Imai et al., 2000; Logan et al., 1999; Miyawaki et al., 2003; Röttinger et al., 2012).
In several taxa, nβ-catenin is selectively restricted to one pole of the embryo during early cleavage (Henry et al., 2008, 2010; Miyawaki et al., 2003; Wikramanayake et al., 1998, 2003). In sea urchins and the cnidarian Nematostella vectensis, nβ-catenin is stabilized in the vegetal pole by cortically localized Dishevelled (Weitzel et al., 2004; Wikramanayake et al., 2003). This differential stability in sea urchins establishes a gradient of nβ-catenin, such that its levels are higher in the vegetal-most cells (micromeres) and lower in the blastomeres fated to endomesoderm (Lhomond et al., 2012; Logan et al., 1999; Weitzel et al., 2004). Therefore, a mechanism to localize and generate gradients of nβ-catenin activity along one pole of invertebrate embryos might be a widespread phenomenon. However, to date there is no indication that this gradient functions to direct different cell fates within the endomesoderm. How such a gradient might regulate development, or whether different levels of nβ-catenin themselves elicit distinct transcriptional responses, is currently not known.
Once endomesoderm is specified, it is segregated into spatially and molecularly distinct endoderm and mesoderm territories. In several organisms, this asymmetry is also controlled by a change in nβ-catenin localization. In C. elegans and ascidians, an asymmetric cell division rapidly clears nβ-catenin from the presumptive mesoderm (Hudson et al., 2013; Thorpe et al., 1997). In sea urchins, Delta/Notch signaling from the micromeres induces mesoderm in the overlying cells, in part by inhibiting nβ-catenin function (Croce and McClay, 2010; Materna and Davidson, 2012; Ransick and Davidson, 2006; Röttinger et al., 2006; Sethi et al., 2012; Sherwood and McClay, 1999; Sweet et al., 2002; Ben-Tabou de-Leon and Davidson, 2010). Therefore, restriction of nβ-catenin localization might be a key step in the formation of separate endoderm and mesoderm territories commonly found among animals.
The central role of nβ-catenin in early endomesoderm development is a highly conserved mechanism among many taxa, and might thus constitute a common connection between early, maternal anisotropies and the later, taxon-specific networks that drive subsequent endoderm and mesoderm development. It remains to be determined whether gradients of nβ-catenin could operate in a dose-dependent manner to segregate distinct territories along the primary axis in invertebrates. If this is the case, the use of nβ-catenin dose dependence in territory segregation in both invertebrates and vertebrates could be a unifying feature of their development.
To address these issues, we analyzed the early specification and segregation of endoderm and mesoderm in the sea star Patiria miniata. Sea stars form hollow blastulae of a few thousand cells by the onset of gastrulation. Mesoderm forms at the central-most vegetal pole, with a torus of endoderm-fated cells situated above (Kuraishi and Osanai, 1992). In a closely related sea star species, β-catenin has been shown to accumulate in the nuclei of endomesoderm-fated vegetal blastomeres, and the vegetal-most cells appear to have higher levels of nβ-catenin than more peripheral vegetal blastomeres (Kuraishi and Osanai, 1992; Miyawaki et al., 2003). Given the basal mode of sea star development, and the fact that nβ-catenin is known to accumulate in endomesoderm-fated cells, the sea star is an ideal invertebrate model for assessing the function of nβ-catenin in endomesoderm segregation.
In P. miniata blastulae, three concentric territories are established in the vegetal pole in response to a changing profile of nβ-catenin transcriptional activity. Low levels of nβ-catenin activity drive bra expression, which is later restricted to the veg1, endoderm/ectoderm boundary territory; intermediate levels drive foxa and gata4/5/6 expression, which later are restricted to the endoderm-fated cells; and high levels of nβ-catenin are necessary for the expression of ets1 and erg in the vegetal-most territory fated for mesoderm. The changing spatial distribution of nβ-catenin activity through time, coupled to a zygotic GRN, establishes the boundaries of endoderm and mesoderm. We also show that the expression of factors that ultimately define endoderm is cleared from the central mesoderm territory through the activation of repressive network circuitry established by high levels of nβ-catenin activity. The specification of endoderm is further reinforced by Delta/Notch induction and a positive-feedback circuitry between hex and gata4/5/6 that is coincident with the loss of nβ-catenin from this territory. Thus, in this system, segregation of endoderm and mesoderm arises as the result of distinct transcriptional responses to levels of nβ-catenin activity. This dosage response to the dynamic spatiotemporal activity of nβ-catenin provides the first empirical demonstration of its use in invertebrate territory segregation, and establishes this response as a potentially widespread mechanism among metazoans.
RESULTS
Spatiotemporal regulatory states of the sea star vegetal pole
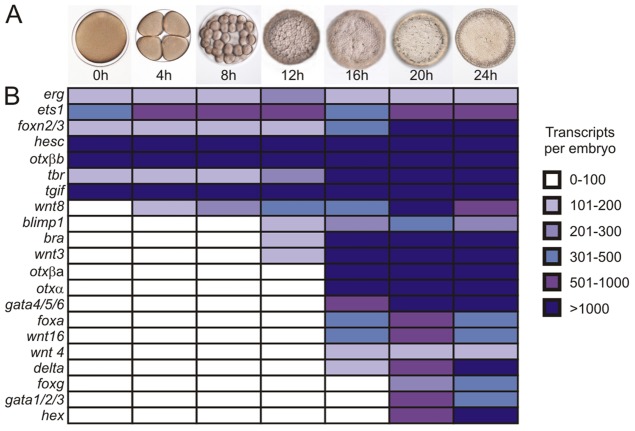
To understand the dynamics of endomesoderm territory specification, we determined the regulatory state of the sea star vegetal pole through the late blastula stage. Previous gene expression profiling identified three concentric domains, defined by distinct patterns of gene expression within the endomesoderm of hatched blastulae (Hinman and Davidson, 2003a,b, 2007; Hinman et al., 2003a; McCauley et al., 2010, 2013). However, little is known about gene expression prior to the formation of these domains, although this information is essential for understanding how these territories are established. Quantitative RT-PCR (qPCR) was used to assess the temporal profile of expression of 21 genes, namely erg, ets1, foxn2/3, hesc, otxβb, tbr, tgif, wnt8, blimp1, bra, wnt3, otxβa, otxα, gata4/5/6, foxa, wnt16, wnt4, delta, foxg, gatac and hex, which have been implicated in the studies mentioned above to play a role in sea star endomesoderm development. The results are summarized in Fig. 1. Two trends are evident: (1) seven transcripts (erg, ets1, foxn2/3, hesc, otxβb, tbr and tgif) are present maternally; and (2) an increase in gene expression is observed between 12 and 16 h post fertilization (h).
Fig. 1.
Temporal gene expression in early sea star embryos. (A) Representative embryo morphology at the indicated time points. (B) Quantitation of expression levels of indicated transcripts at the listed time points. White signifies that a gene is not expressed; intensity of blue scales with increasing gene transcript abundance. Transcript levels were assessed by qPCR (to calculate transcripts per embryo). h, hours post fertilization.
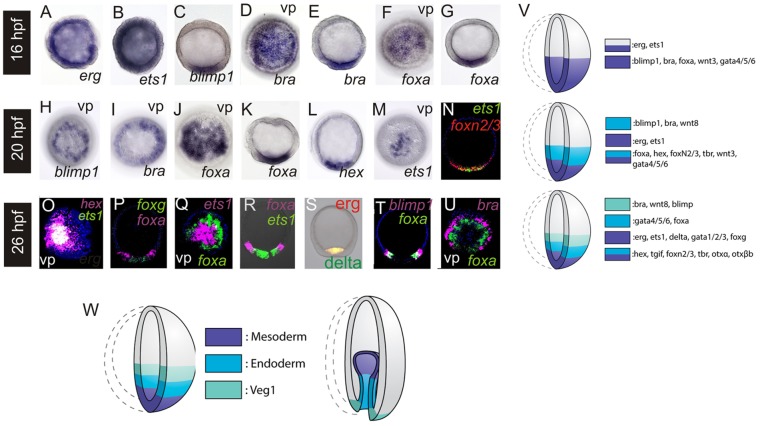
As expression of most zygotic genes is initiated by the 16 h early blastula stage, we next assessed spatial patterns of gene expression in the vegetal pole, starting at this time point. Maternal factors, e.g. erg and ets1, are expressed ubiquitously (Fig. 2A,B). Conversely, genes that are first expressed after cleavage, e.g. blimp1, bra, foxa, (Fig. 2C-G), wnt3 (supplementary material Fig. S1) and gata4/5/6 (McCauley et al., 2012), are restricted to one embryonic pole. Therefore, at this stage, there is a single vegetal territory that localizes to the overlap of ubiquitously expressed maternal transcripts and the polar expression of zygotically activated regulatory genes (summarized in Fig. 2V). It is noteworthy that bra and blimp1, which are among the first genes expressed zygotically, are localized to a somewhat broader domain than that of foxa and gata4/5/6, although we cannot confirm the precise expression boundaries at this stage.
Fig. 2.
Spatial gene expression in early sea star embryos. Whole-mount in situ hybridization (WMISH) was used to determine localization of gene expression in embryos at 16-26 h. Probe used is indicated on right. (A-G) 16-h-embryos (early blastulae); (H-N) 20-h-embryos (mid blastulae); (O-U) 26-h-embryos (late blastulae). Embryos are shown in the lateral view with vegetal towards the bottom unless indicated by vp, which is a view from the vegetal pole. N-U show two-color FISH. (V,W) Schematic depicting dynamics of gene expression in the sea star vegetal pole during blastula (V), and corresponding territories at blastula and gastrula (W) stages.
By the mid-blastula-stage (20 h), the earliest zygotically expressed factors in our study, blimp1, bra (Fig. 2H,I) and wnt8 (supplementary material Fig. S1), are localized to a torus surrounding the central vegetal pole. Other genes, including foxa, hex, foxn2/3 (Fig. 2J-L,N; supplementary material Fig. S1), tbr, wnt3 (supplementary material Fig. S1) and gata4/5/6 (McCauley et al., 2012), remain expressed throughout the vegetal pole. By contrast, ets1 (Fig. 2M,N) and erg (supplementary material Fig. S1) expression is observed in a domain more closely restricted to the vegetal pole. Thus, at this stage, the initial restriction of gene expression to the veg1 territory is observed, which will contribute to the posterior endoderm and blastopore region of the gastrula [see Kuraishi and Osanai (1992); and schematic in Fig. 2W]. We also note the first restriction of gene expression solely to the mesoderm-fated territory. This territory invaginates first and contributes to the coeloms and mesenchyme of the gastrula [Kuraishi and Osanai (1992); and schematic in Fig. 2W]. Another suite of genes, namely foxa, hex, foxn2/3, tbr, wnt3 and gata4/5/6, is expressed within an endomesodermal domain that encompasses both presumptive mesoderm and endoderm.
By the late blastula stage (26 h), foxa and gata4/5/6 expression is now cleared from the central domain and is thus, for the first time, specifically restricted to the torus of endoderm-fated territory. There are three distinct territories, as defined by localized patterns of regulatory gene expression: ets1, erg, foxg, delta (Fig. 2O-S; supplementary material Fig. S1) and gata1/2/3 (Hinman and Davidson, 2007) are expressed within the central-most domain; expression of foxa and gata4/5/6 is localized to a torus surrounding this domain [Fig. 2P-R; see Hinman and Davidson (2003b)]; and bra, blimp1 (Fig. 2T,U) and wnt8 (McCauley et al., 2013) are restricted to a torus above the endoderm-fated cells (summarized in Fig. 2V). These are the mesoderm, endoderm and veg1 endoderm territories, respectively (Fig. 2W). Some factors in the vegetal pole domain remain expressed throughout both endoderm and mesoderm, namely hex (Fig. 2O), tgif, foxn/3, tbr, otxα and otxβa (Hinman et al., 2003a; McCauley et al., 2010).
These data show that a largely uniform, initial vegetal endomesoderm region is segregated into three, molecularly distinct territories. Expression of genes that will be restricted to a veg1 territory occurs first, whereas the restricted zygotic expression of genes localized to the mesoderm occurs last. The early, ubiquitous phase of expression of erg and ets1 can be attributed to maternally deposited transcripts. The GRN developed here, therefore, aims to explain: (1) the earliest activation of genes later expressed in the veg1 territory; (2) the initial activation of gene expression in a broad territory within the vegetal pole of the embryo; (3) the later expression of factors in the mesoderm-restricted territory; and (4) the clearing of gene expression from the central territory of the vegetal plate to form discrete mesoderm and endoderm. At this time, the GRN model does not attempt to describe the mechanism that leads to the early segregation of the bra, blimp1 and wnt8 expression domain from the remaining endomesoderm.
nβ-catenin is progressively restricted to the vegetal pole as its transcriptional activity increases
Given the dynamic changes in the regulatory states of the vegetal pole starting at the early blastula stage, we characterized the localization of nβ-catenin during this time window and assayed its ability to activate gene expression. A β-catenin:GFP fusion construct (Logan et al., 1999) was used to visualize the localization of this protein during early development. The domain of nβ-catenin becomes more restricted toward the vegetal pole between 16 h and 20 h (Fig. 3A,B; compare with Fig. 3C,D and schematic in Fig. 3E). We compared the number of β-catenin:GFP positive nuclei with the number of ets1- or foxa-positive cells as determined by WMISH at the same stage, and found that the number of β-catenin:GFP-positive nuclei at 16 h and 20 h is not significantly different from the number of foxa- or ets1-positive cells, respectively, at these same stages (Fig. 3F; two-tailed t-test, P>0.05). This indicates that, at 16 h, β-catenin is nuclearized within the foxa-expressing endomesoderm domain, whereas at 20 h, nβ-catenin is restricted to the ets1-expressing, mesoderm-fated territory (schematic in Fig. 3E).
Fig. 3.
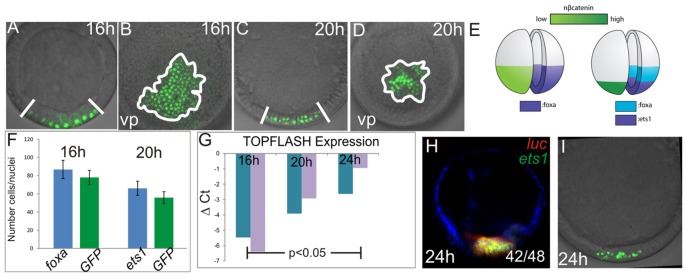
Spatiotemporal gradient of nβ-catenin in the vegetal pole. (A-D) Subcellular localization of β-catenin in 16-h- and 20-h-embryos assessed by confocal microscopy of GFP derived from the β-catenin:GFP fusion construct. 16-h-embryos are shown in A,B and 20-h-embryos in C,D. (A,C) Lateral views and (B,D) vegetal pole views. (E) Schematic of β-catenin nuclearization at 16 h and 20 h compared with expression of foxa and ets1 at these stages. (F) Comparison of number of GFP-positive nuclei and foxa- or ets1-positive cells at time points shown. Error bars are from one s.d. of ten individuals. (G) qPCR analysis of TOPFlash reporter expression in 16-h-, 20-h- and 24-h-embryos. Increased ΔCT (cycle threshold) values indicate increased reporter transcript abundance normalized to lamin2β receptor. (H) FISH showing co-expression of luciferase driven by the TOPFlash reporter and the mesoderm marker ets1 at 24 h. Co-expression of ets1 and luciferase was detected in 42 of 48 embryos (expression in the presumptive endoderm was detected in five embryos). (I) Subcellular localization of the β-catenin:GFP fusion construct in 24-h-embryos for comparison with (H) at the same stage.
nβ-catenin-driven gene expression was then quantified using the exogenous TOPFlash reporter. This construct contains six TCF-binding sites linked to a luciferase reporter gene (Veeman et al., 2003), expression of which is activated by nβ-catenin. As assayed by qPCR, normalized luciferase expression increases significantly (ANOVA, P=0.0405) as development proceeds from 16 h to 24 h (Fig. 3G). Thus, nβ-catenin-driven transcription increases during the time that the endomesoderm territories are being specified. We also used two-color fluorescent in situ hybridization (FISH) to show that, at 24 h, luciferase expression from the TOPFlash reporter co-localizes with ets1 gene expression in 42/48 embryos, and is thus restricted to the mesoderm (Fig. 3H). This corresponds visually with the restriction of β-catenin:GFP-positive nuclei to the central-most vegetal pole at this stage (Fig. 3I). At this point, we cannot examine the activity of nβ-catenin in individual nuclei. Although there might be differential activity within each territory, our measurements for now are an average of the nβ-catenin activity at each time point.
Taken together, these data (Fig. 3) show that, although gene expression driven by nβ-catenin activity increases as development proceeds, the domain in which β-catenin is nuclearized becomes more restricted. At 16 h, when foxa is first expressed, the transcriptional activity of nβ-catenin is lower than when ets1 is first expressed within the presumptive mesoderm. An increase in nβ-catenin activity over time has also been reported during early development in sea urchins (Lhomond et al., 2012; Logan et al., 1999), although whether this has a consequence for subsequent development has not been explored. As protein levels are not directly measured, we do not know whether there is an increase in β-catenin protein accumulation in the vegetal nuclei of sea star embryos or whether the increased transcriptional activity is due to its prolonged accumulation in the mesoderm-fated central-most territory compared with the endoderm-fated cells.
A dose-dependent response to the spatiotemporal profile of nβ-catenin establishes distinct regulatory territories in early endomesoderm development
The changing spatiotemporal profile of nβ-catenin transcriptional activity (Fig. 3), along with the description of the regulatory state of the vegetal territories through time (Figs 1 and 2), led us to hypothesize that differential nβ-catenin activity is required to specify distinct cell fates within the sea star vegetal pole. Specifically, we propose that the low transcriptional activity of nβ-catenin present throughout the vegetal pole prior to 16 h is sufficient to initiate expression of the earliest-expressed factors, e.g. bra and blimp; that increasing levels of nβ-catenin activity initiate expression of endoderm-restricted genes, e.g. foxa and gata4/5/6; and, finally, that the highest activity, which is only achieved when nβ-catenin is restricted to the central-most mesoderm-fated territory, drives ets1 and erg expression. To test this, we manipulated the levels of nβ-catenin by either sequestering β-catenin to the plasma membrane, using the Δ-cadherin construct (Logan et al., 1999) or driving it into nuclei with LiCl (Poustka et al., 2007; Stambolic et al., 1996). The appropriate amount of Δ-cadherin transcripts to use in these experiments was empirically determined by examining the phenotype of gastrula larvae expressing varying levels of the construct. When Δ-cadherin is injected at low levels (250 ng/µl), there is no or a greatly reduced mesodermal bulb and the archenteron is somewhat reduced (supplementary material Fig. S2). At medium levels (500 ng/µl) of introduced Δ-cadherin, there is no mesoderm, and a greatly reduced endoderm (supplementary material Fig. S2). At the highest levels of Δ-cadherin expression (750 ng/µl), we observe no gastrulation and no endomesoderm formation (Yankura et al., 2013).
Next, we assessed the effects of low and medium levels of Δ-cadherin on gene expression in 24-h-blastulae. Strikingly, when a low dose of Δ-cadherin is injected, bra expression is contracted towards the vegetal pole, although expression remains in a torus (Fig. 4F,F′; compare with Fig. 4A,A′). foxa and gata4/5/6 remain expressed but are now localized to a domain that encompasses the central vegetal pole (Fig. 4G-H′; compare with Fig. 4B-C′). Expression of the mesodermal markers ets1 and erg is entirely lost (Fig. 4I,J; compare with Fig. 4D,E). Thus, at a low dose of Δ-cadherin, there is a loss of mesodermal markers, and a vegetal retraction of the endoderm and veg1 territory markers (summarized in Fig. 4Q).
Fig. 4.
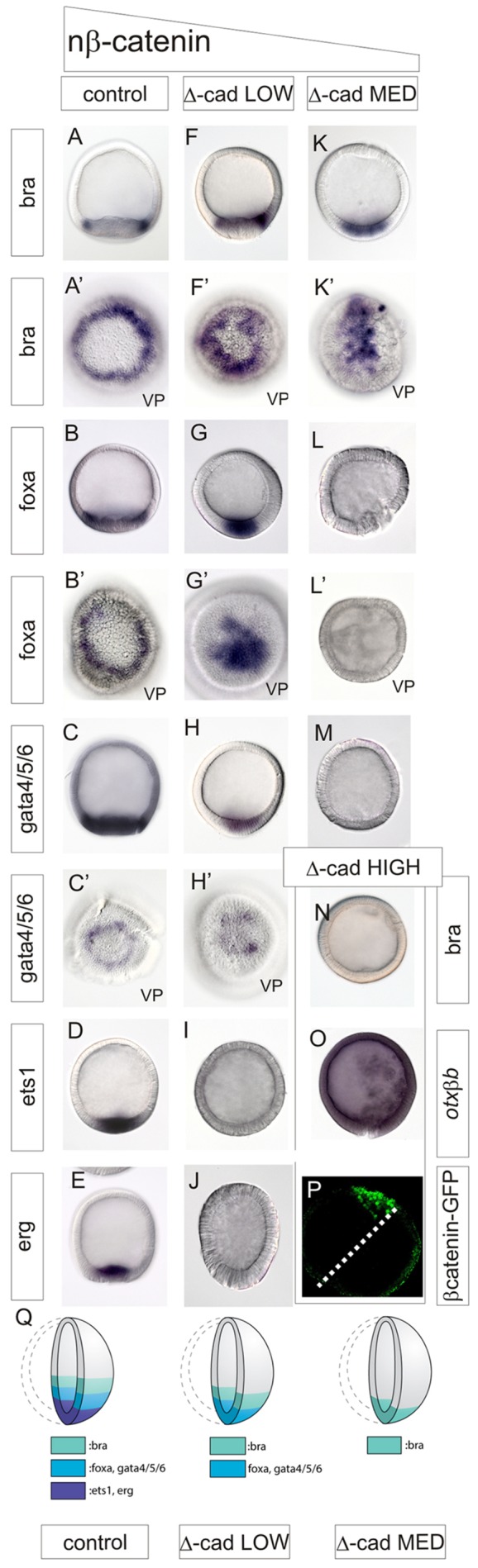
Progressive loss of mesoderm, then of endoderm, with decreasing levels of nβ-catenin. Localization of transcripts shown boxed on right or left as visualized by WMISH in 24 h-blastulae that were injected at single cell-stage, with GFP as a control (A-E), low levels of Δ-cadherin (F-J), medium levels of Δ-cadherin (K-M) or high levels of Δ-cadherin (N,O). (P) A 24 h-embryo that was injected at the one-cell stage with β-catenin:GFP and additionally in a single cell at the two-blastomere stage with high levels of Δ-cadherin. GFP is visualized at 24 h by confocal microcopy and shows vegetal nβ-catenin in one half of the embryo and complete loss of nβ-catenin in the half of the embryo that developed from the Δ-cadherin-injected blastomere. Embryos are depicted in a lateral view, unless indicated by vp for vegetal pole view. (Q) Schematic showing the effects of different levels of Δ-cadherin on vegetal territory specification.
When Δ-cadherin is injected at medium levels, the expression of the veg1 marker bra is localized within the vegetal pole region of the embryo (Fig. 4K,K′; compare with Fig. 4A,A′), and expression of foxa and gata4/5/6 is lost entirely (Fig. 4L-M; compare with Fig. 4B-C; summarized in Fig. 4Q). At the highest levels of Δ-cadherin injected, expression of bra is also lost (Fig. 4N; compare with Fig. 4A) and the ectodermal markers onecut and otxβb are now expressed throughout the resultant blastula (Hinman et al., 2003a; Yankura et al., 2013) (Fig. 4O). We visually inspected nβ-catenin:GFP in embryos overexpressing Δ-cadherin to confirm that the domain of nuclearization was also reduced (Fig. 4P). These patterns of gene expression changes are largely maintained through to the gastrula stage, reflecting the continued loss of these expression domains (supplementary material Fig. S3).
As blocking the nuclearization of β-catenin inhibited endomesoderm development in a dose-dependent manner, we wanted to determine whether increased nβ-catenin also has a graded effect on sea star endomesoderm development. Embryos were bathed in high (50 mM) or medium (30-40 mM) concentrations of LiCl, and the expression of ets1, foxa and bra was assessed at 24 h. Levels of LiCl used were also determined empirically by observing morphological effects on gastrulae (supplementary material Figs S4 and S5). At medium levels of LiCl, bra expression extends throughout the animal ectoderm, whereas foxa is expressed in a torus abutting this (Fig. 5C; compare with Fig. 5A). Both bra and foxa are expressed in concentric domains that extend further from the vegetal pole than in controls. The ets1 expression domain remains associated with the vegetal pole but is expanded further toward the animal pole than in untreated embryos (Fig. 5D; compare with Fig. 5B). When embryos were treated with high levels of LiCl, the domains of bra and foxa expression were expanded even further toward the animal pole and, notably, are observed to overlap in the animal-most region of the embryo, although expression of foxa extends further toward the vegetal pole than that of bra (Fig. 5E). ets1 expression remains within the vegetal pole, but this domain is even broader than at medium LiCl treatments (Fig. 5F; compare with Fig. 5D). We further confirmed that these changes in gene expression are maintained during gastrulation (supplementary material Fig. S5). In each of these perturbations, the ets1- and foxa-expressing territories remain distinct, which suggests that a robust mechanism functions to segregate mesoderm and endoderm cell fates. This is consistent with our later observation that an Ets1-driven GRN sub-circuit establishes the mesoderm/endoderm boundary (see below). The results are summarized in Fig. 5G. We also confirmed that the domain of nβ-catenin, as visualized using the β-catenin:GFP fusion construct, is greatly expanded in LiCl-treated embryos (Fig. 5H). Taken together, the results summarized in Fig 4Q and Fig. 5G suggest that the increase in nβ-catenin transcriptional activity through time leads first to the expression of at least some genes that will later be restricted to the presumptive veg1 and endoderm territories, and last to those expressed in the presumptive mesoderm.
Fig. 5.
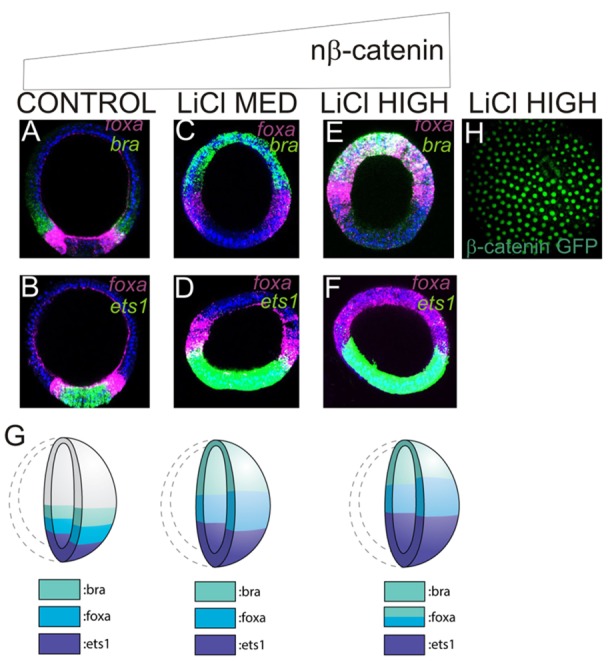
LiCl dose-dependent expansion of endoderm and mesoderm territories. FISH of pairs of transcripts shown in top right in NaCl (control)-treated (A,B), and in embryos treated with medium (C,D) or high (E,F) doses of LiCl. (G) Gene expression patterns in late blastulae treated with increasing doses of LiCl. (H) Confocal image of β-catenin:GFP in high LiCl-treated late blastula showing extensive expansion of nuclear β-catenin. All embryos are shown at the late blastula stage in the lateral view.
FoxA repressive circuitry distinguishes mesoderm and endoderm territories
So far, our model explains the establishment of overlapping foxa-expressing endoderm and ets1-expressing mesoderm territories through the 20-h mid-blastula stage. However, foxa and gata4/5/6 are subsequently cleared from the central vegetal pole territory. Previous work has shown that FoxA represses its own expression within the mesoderm, as foxa transcripts are not cleared from the mesoderm-fated territory in FoxA morphants (Hinman et al., 2003b). We are now able to extend this model to explain why this auto-repression only occurs in the mesoderm. At least two mesodermally restricted factors, gata1/2/3 and foxg, both of which are first expressed between 16 h and 20 h, require normal FoxA function for their expression (Fig. 6A,D; compare with Fig. 6C,F). Ets1 function is additionally required for the expression of both these genes (Fig. 6A,D; compare with Fig. 6B,E), so that the combinatorial regulation drives their expression solely in the mesoderm-fated territory. That is, foxg and gata1/2/3 require an input from FoxA at a time when it is expressed broadly through the vegetal pole, but are restricted to the mesoderm due to an additional requirement for an input from Ets1. We envision that Gata1/2/3 or FoxG then function to repress foxa and, possibly, gata4/5/6 expression specifically from the presumptive mesoderm. This clears the endodermal factors from the mesoderm-fated territory. According to this model, the expansion of foxa into the central vegetal pole territory in FoxA morphants is due to the loss of gata1/2/3 or foxg, as the product of one of these genes is what represses foxa expression. As these scenarios have not been explicitly tested, the gene that represses foxa expression in the mesoderm is referred to as geneX in our model, and it might also be another factor that is regulated similarly to gata1/2/3 and foxg. These data are summarized in the GRN circuit shown in Fig. 6G.
Fig. 6.
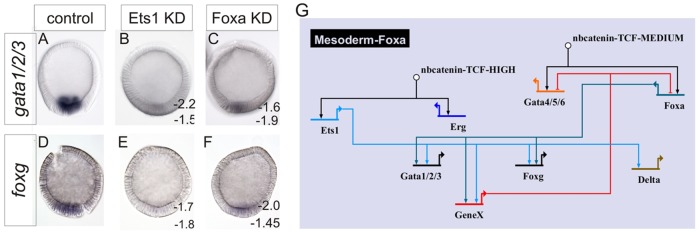
Mesoderm and endoderm segregation is achieved downstream of FoxA dual-positive/repressive circuitry. WMISH visualization of gata1/2/3 (A-C) or foxg (D-F) expression in 24-h-blastula in control MASO- (A,D), Ets1 MASO- (B,E) or FoxA MASO-injected embryos (C,F). (G) BioTapestry (Longabaugh, 2012) schematic detailing the GRN representation of these data. Numbers are ΔCT measurements from two independent qPCR analyses of control minus perturbation, as indicated in the figure. A twofold difference, i.e. ΔCT>±1.6, is considered significant.
Delta/Notch signaling maintains endoderm specification in late blastulae
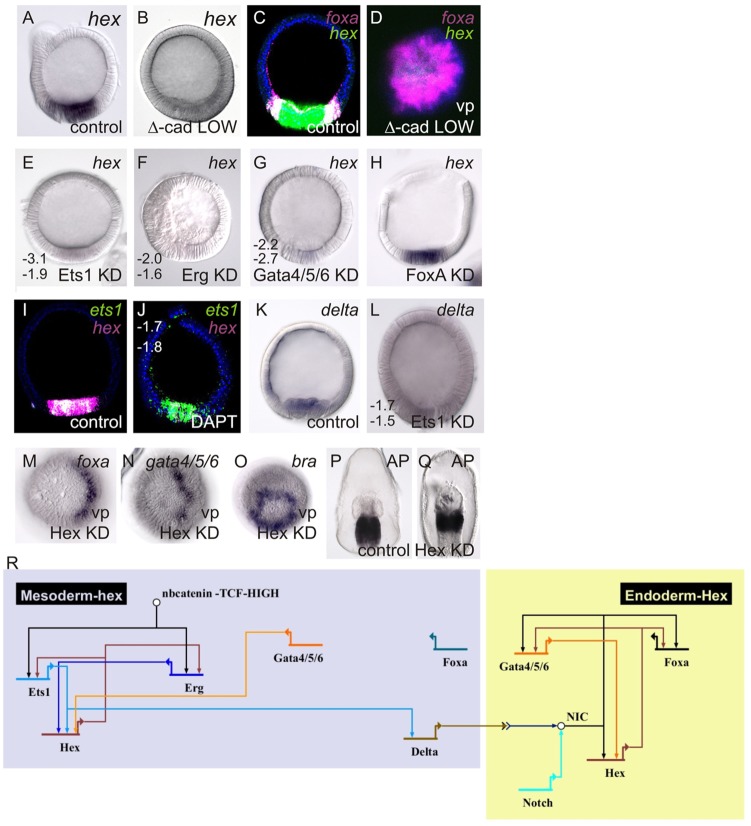
The expression of ets1 and erg in the mesoderm, and of foxa and gata4/5/6 in the endoderm, are a consequence of the spatiotemporal dynamics of β-catenin nuclearization and its activity level-dependent gene regulation. A central tenet of this model is that ets1 and erg expression are confined to only the central-most vegetal pole territory. This is due to the spatial restriction of nβ-catenin to only this region of the embryo by the time sufficient nβ-catenin activity is achieved to drive expression of these genes. The endoderm-fated territory consequently becomes an nβ-catenin-free domain shortly after its specification. This suggests that, at around the 20-h blastula stage, there is a transition from maternally initiated nβ-catenin-mediated regulation to a zygotically driven regulatory program in the endoderm. Expression of three genes in this study – foxg, gata1/2/3 and hex – is first detected at 20 h (Fig. 1); of these genes, the first two are expressed in the presumptive mesoderm, whereas hex is expressed throughout the endomesoderm (Figs 2 and 7; Hinman and Davidson, 2007). We therefore questioned how hex, which is initially expressed at 20 h, could be expressed in both the endoderm and mesoderm at a time when nβ-catenin is only present in the mesoderm-fated territory.
To determine how hex expression might be regulated by nβ-catenin, we first assessed how hex expression responds to Δ-cadherin perturbations. Strikingly, hex expression is lost throughout the endomesoderm territory in embryos expressing low levels of Δ-cadherin (Fig. 7A-D). Thus, hex does not respond to Δ-cadherin in the manner that we describe for foxa and gata4/5/6 (Fig. 4), as expression of these factors is first lost in medium levels of Δ-cadherin expression. An explanation for the dependence of hex on high nβ-catenin activity is that its expression requires the correct formation of the ets1- and erg-expressing mesoderm domain. We therefore examined more carefully the regulation and function of hex to understand which other regulatory events might be directing the formation of endoderm and mesoderm in 20-h embryos.
Fig. 7.
Delta/Notch regulation of hex is required to maintain endoderm specification. (A-O) WMISH localization of transcripts shown in the top right; C, D, I and J show FISH. Perturbation type or control is shown in bottom right. Numbers are ΔCT measurements from two independent qPCR analyses of control minus perturbation, as indicated in the figure. A twofold difference, i.e. ΔCT>±1.6, is considered significant. M and N show half-morphant blastulae with the morphant half to the left. (P,Q) Staining of endogenous alkaline phosphatase, which is a marker of endoderm specification (Hinman et al., 2003a). All embryos are late blastulae, shown in a lateral view unless indicated by vp for vegetal pole view. KD, antisense morpholino oligonucleotide-based knockdown. (R) Results are summarized in a BioTapestry GRN wiring diagram.
hex expression is lost throughout the endomesoderm in Ets1, Erg and Gata4/5/6 morphants, but not in FoxA morphants (Fig. 7E-H). At the time of hex activation (20 h), ets1 and erg expression is already restricted to the mesoderm, whereas gata4/5/6 remains expressed throughout the endomesoderm (Fig. 2; supplementary material Fig. S1); (McCauley et al., 2012). The requirement for Gata4/5/6 in particular, which is first expressed between 12 h and 16 h, can explain the later activation of hex and its expression throughout the endomesoderm. The loss of hex expression throughout the endomesoderm in Ets1 and Erg morphants, however, further requires that our model incorporates a mesoderm-derived signal for correct hex expression. Previous work has shown that a Delta/Notch signal is needed for the expression of gata4/5/6 and, to a lesser extent, foxa in the endoderm (Hinman and Davidson, 2007). Therefore, we tested whether hex expression in the endoderm requires a Delta/Notch input. Embryos bathed in the γ-secretase inhibitor DAPT, which prevents activation of Notch, showed a significant reduction in hex expression in the endoderm and mesoderm (Fig. 7I,J). delta expression, in turn, is significantly reduced in Ets1 morphants (Fig. 7K,L). Together, these data explain the loss of hex expression in Ets1morphants and low Δ-cadherin-expressing embryos. A functional ets1-expressing mesoderm is required for the expression of delta, and Delta/Notch signaling is in turn required for the expression of hex in the endoderm. As Hex is required for mesoderm development (McCauley et al., 2010), these data also suggest that Delta signaling has a role in mesoderm formation through the regulation of hex.
To better understand the role of Delta/Notch signaling in endoderm development, we next examined the function of Hex in this process. Although hex is the last gene activated in the endomesoderm that was analyzed in this study, Hex is required for expression of ets1 and erg in the mesoderm (McCauley et al., 2010) and, as we show now, for the expression of gata4/5/6 and foxa in the endoderm (Fig. 7M,N). Hex, however, does not regulate expression of the veg1-restricted bra (Fig. 7O). This is consistent with the phenotype of Hex morphant gastrulae, which show impaired endoderm differentiation (as assayed by alkaline phosphatase staining) and a reduction in mesoderm, but maintenance of posterior endoderm (Fig. 7P,Q). Therefore, Hex is engaged in positive feedback between ets1 and erg in the mesoderm (McCauley et al., 2010) and gata4/5/6 in the endoderm; that is, Ets1, Erg and Gata4/5/6 regulate hex expression and, in turn, are regulated by Hex. In this model, Delta/Notch, signaling through the activation of hex, does not function to induce gata4/5/6 in the endoderm, but instead to maintain its expression during the period in which nβ-catenin is being lost from the presumptive endoderm.
DISCUSSION
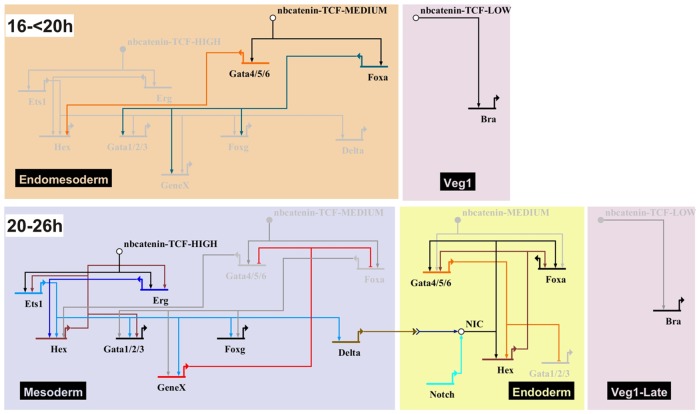
The data presented here are compiled into a GRN (Fig. 8). Although incomplete, this model explains the dynamic regulatory states that we have described for endomesoderm development in the sea star blastula. A dose-dependent response to the changing spatiotemporal transcriptional activity of nβ-catenin functions to segregate territories within the endomesoderm. Low levels of nβ-catenin activity initiate the expression of bra; intermediate levels that of gata4/5/6 and foxa; whereas the highest levels, which are only achieved when nβ-catenin is restricted to the central-most vegetal plate, drive zygotic expression of ets1 and erg. These data provide a mechanistic explanation for the embryonic manipulations which showed that a loss of endomesoderm occurs when the vegetal pole of sea star eggs is depleted. Furthermore, defects in mesoderm development occurred at smaller depletion volumes than are required to abrogate endomesoderm formation (Kuraishi and Osanai, 1994). We hypothesize that a vegetally localized maternal determinant is required for the nuclearization of β-catenin. Depletion of a small proportion of the vegetal pole reduces the amount of this determinant, such that only low levels of nβ-catenin accumulate, resulting in defects in mesoderm development. However, when most of the determinant is removed, along with a larger volume of the vegetal pole, β-catenin is not nuclearized at sufficiently high levels to initiate endoderm development. In sea urchins and cnidarians, a cortical restriction of Dishevelled mediates the stabilization of nβ-catenin in the vegetal pole during early cleavage (Weitzel et al., 2004; Wikramanayake et al., 2003). Our data are also consistent with this model, and we hypothesize that Dishevelled or another component of the canonical Wnt signaling pathway act as this maternal determinant in sea stars, and might represent a conserved mechanism among metazoans.
Fig. 8.
The GRN for endomesoderm specification in the sea star P. miniata. A GRN model developed in BioTapestry, depicting regulation of gene expression in the endomesoderm. Arrowheads represent positive regulation; bars represent repression. Genes are depicted on colored background according to their expression domains. Gray genes and lines are not active at the time shown. Gene names are as described in text. NIC, Notch intracellular domain.
This work presents the first empirical demonstration of a changing spatiotemporal profile of nβ-catenin transcriptional activity that functions in a dose-dependent manner to segregate territories along the primary axis of an invertebrate. It has been proposed that a gradient of nβ-catenin is the ancestral mechanism used to pattern the primary axis (Niehrs, 2010). However, no evidence for a long-range gradient of nβ-catenin has been found in any invertebrate (Darras et al., 2011; Röttinger et al., 2012). Although we demonstrate a dose-dependent response to nβ-catenin activity in territory specification in sea star embryos, this does not arise through a response to a long-range spatial gradient. Therefore, the formation of a long range nβ-catenin gradient in axial patterning might be unique to vertebrate embryos. Given the widespread occurrence of nβ-catenin localization to one pole in early invertebrate embryogenesis (Darras et al., 2011; Henry et al., 2008; Hudson et al., 2013; Logan et al., 1999; Röttinger et al., 2012; Thorpe et al., 1997), and especially in light of the increased transcriptional activity of nβ-catenin over time as seen in sea stars (Fig. 3) and sea urchins (Lhomond et al., 2012), we suggest that a dose-response to changing nβ-catenin activity is a broad phenomenon among invertebrates.
GRN subcircuits established by nβ-catenin segregate endoderm and mesoderm
In early sea star embryos, low levels of nβ-catenin transcriptional activity establish a vegetal pole domain in which the complement of genes that will later be exclusively expressed in endoderm or veg1 are expressed. foxa, which is initially expressed in this broad domain, and ets1, which is zygotically expressed specifically in the mesoderm, coordinately activate mesodermally restricted factors. We hypothesize that these, in turn, repress foxa expression in the mesoderm. Thus, a positive-/negative-feedback circuit operates to clear foxa, and, probably, gata4/5/6, from the mesoderm. Our model also predicts that hex expression is regulated by Delta/Notch signaling. In addition to its previously characterized role in mesoderm maintenance, hex is engaged in a positive-feedback loop in the endoderm, i.e. hex regulates and is regulated by gata4/5/6. Positive feedback in developmental GRNs has been proposed to function as a mechanism to ‘lock down’ or stabilize regulatory states (see e.g. Davidson and Erwin, 2006; Hinman and Cheatle Jarvela, 2014; Hinman et al., 2009). These types of regulatory interactions are therefore predicted to be especially prevalent downstream of transient signaling events (Davidson and Erwin, 2006). The time frame of this function of Hex coincides with the loss of nβ-catenin from the endoderm-fated territory. We therefore propose that, as predicted by these theories of GRN control, positive feedback between hex and gata4/5/6 in the endoderm functions to maintain gata4/5/6 and foxa expression, as the regulation of this territory changes from maternally driven nβ-catenin-dependent to a zygotic GRN.
A change in nβ-catenin localization or activity is implicated in endomesoderm segregation in several taxa. In sea urchins, a Delta signal from the micromeres promotes nuclear export of TCF, the binding partner of β-catenin, in adjacent cells that will be fated to mesoderm (Röttinger et al., 2006). Loss of TCF extinguishes foxa expression and thereby suppresses the activation of the endoderm GRN in this territory (Oliveri et al., 2006; Sethi et al., 2012; Ben-Tabou de-Leon and Davidson, 2010). In ascidians, nβ-catenin is present in blastomeres fated to endomesoderm at the 16-cell stage, but are restricted to the presumptive endoderm one cell division later. The loss of nβ-catenin from the presumptive mesoderm is required for the development of this territory (Hudson et al., 2013). This has been proposed to function as an ‘ON/OFF’ type mechanism of nβ-catenin activity, whereby endoderm segregates from mesoderm due to this binary change nβ-catenin. Platynereis dumerilii also has a reiterative binary ‘ON/OFF’ pattern in nβ-catenin accumulation in vegetal blastomere cell fate choices, although this is not related to endomesoderm segregation (Schneider and Bowerman, 2007). The lophotrochozoan nemertean worm Cerebratulus lacteus also shows a pattern in which nβ-catenin is present in the endomesoderm-fated blastomeres, but is retained only in the endoderm progenitors one cell division later (Henry et al., 2008). Common to ascidians, sea urchins and nemerteans, therefore, is that loss of nβ-catenin occurs in the mesoderm lineage, whereas continued β-catenin nuclearization is a feature of the presumptive endoderm. This is distinctly different from what we report for the sea star, P. miniata, in which nβ-catenin is clearly lost from the presumptive endoderm and maintained in the mesoderm-fated cells. β-catenin is shown to be localized to the nuclei of mesoderm progenitors in blastula and even maintained during gastrulation in a closely related sea star, Asterina pectinifera (Miyawaki et al., 2003). Our data are consistent with a model of ‘ON/OFF’ nβ-catenin function operating in the endoderm, whereas an ‘ON/ON’ mechanism operates in the mesoderm. In this scenario, we hypothesize that the maintained ‘ON’ state in the mesoderm causes the increase in transcriptional activity measured by our luciferase assays, therefore generating a spatiotemporal gradient of activity that then functions to segregate territories.
Thus, changing the localization of nβ-catenin is a common mechanism for segregation of endoderm and mesoderm in at least nemerteans, echinoderms and ascidians, although the territory from which it is lost in each organism is different. This implies that nβ-catenin might have a common role in fate choices that distinguish these two territories, although whether the presence or absence of nβ-catenin drives endoderm or mesoderm is evolutionarily labile. Our results extend the use of nβ-catenin dose responses to establish territories along the primary axis from vertebrates to invertebrates, although the means of forming this gradient are not necessarily conserved and many of its other features are also different.
MATERIALS AND METHODS
Embryo culture
Embryos were cultured and injected following the protocol by Cheatle Jarvela and Hinman (2014).
Time-course quantitative RT-PCR (qPCR) analysis
RNA was extracted, cDNA synthesized and qPCR performed as previously described (Hinman et al., 2003b), except that the number of transcripts was normalized to lamin2β receptor (GenBank ID: 1728045; McCauley et al., 2013). See supplementary material for primer sequences. The transcript number was calculated as described (Wang et al., 1995) and genes were considered as expressed if >100 transcripts were present per embryo.
Whole-mount in situ hybridization (WMISH) and two-color fluorescent in situ hybridization (FISH)
Embryos were fixed and WMISH was performed as previously described (McCauley et al., 2010). Two-color FISH was performed as described (Yankura et al., 2010).
Preparation of mRNA constructs
mRNA was synthesized with the mMessage mMachine kit (Life Technologies) using the pBluescript.Δ-cadherin plasmid (Logan et al., 1999) and the pCS2+β-catenin:GFP fusion construct (Weitzel et al., 2004) as templates.
Perturbation of gene expression
Zygotes were injected with mRNA or morpholino antisense oligonucleotides (MASOs; GeneTools) following the protocol by Cheatle Jarvela and Hinman (2014). In some cases, a single blastomere was injected following the first embryonic cleavage. As the first cleavage plane maps to the plane of bilateral symmetry, this will generate half-normal/half-perturbed embryos. See supplementary material for MASO sequences (Hinman and Davidson, 2007; Hinman et al., 2003b; McCauley et al., 2010). For all MASOs, the GeneTools standard control MASO was injected into sibling embryos. For Δ-cadherin mRNA injections, sibling embryos were injected with eGFP mRNA as a control. Li+ perturbations were achieved by bathing embryos in 20-50 mM LiCl or 20-50 mM NaCl as a control from the two-cell stage. Notch perturbations were achieved by bathing embryos in 64 µM DAPT (Materna and Davidson, 2012) or DMSO as a control from the two-cell stage. WMISH was performed on at least three independent sets of perturbed embryos. At least ten embryos were assessed in each replicate and phenotypes were highly consistent, with >90% showing the reported phenotype. Quantitative measures of perturbation were achieved by performing qPCR on perturbed compared with control siblings. Each assay was performed on at least two qPCR replicates in each of two biological replicates.
Analysis of β-catenin nuclearization
β-catenin:GFP mRNA was injected into embryos and subcellular localization was assayed by confocal microscopy. The TOPFlash β-catenin-inducible luciferase reporter (Addgene plasmid 12456) (Veeman et al., 2003) was linearized and injected into embryos. luciferase abundance was determined using qPCR and normalized to endogenous expression levels of the lamin2β receptor. Whereas significant luciferase expression was detected, expression was always lower than that of highly abundant lamin2β receptor and is therefore reported as a negative number. Increasing ΔCT values indicate higher luciferase expression. TOPFlash-injected embryos were co-injected with eGFP to control for batch variation in injection size bolus, and it was confirmed that GFP levels were equivalent across batches using qPCR on these samples.
Supplementary Material
Acknowledgements
The authors thank Drs Cheatle Jarvela and Cary for helpful discussion and comments. Dr McClay provided the Δ-cadherin plasmid and Dr Ettensohn the β-catenin:GFP plasmid. Sara Cary provided schematic illustrations for figures. We also thank Peter Halmay, Patrick Leahy and Marinus Inc. for animal collection.
Footnotes
Competing interests
The authors declare no competing financial interests.
Author contributions
B.S.M. and V.F.H. designed research; B.S.M., V.F.H., E.A. and H.R.S. performed research; and B.S.M. and V.F.H. analyzed data and wrote the manuscript.
Funding
This work was funded by the National Science Foundation (NSF) [IOS-0844948]. Additional funding was received through the Howard Hughes Medical Institute (HHMI) [52006917 to E.A.] and the NSF [REU 0851735 to H.R.S.]. Deposited in PMC for release after 6 months.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.113043/-/DC1
References
- Ben-Tabou de-Leon S. and Davidson E. H. (2010). Information processing at the foxa node of the sea urchin endomesoderm specification network. Proc. Natl. Acad. Sci. USA 107, 10103-10108 10.1073/pnas.1004824107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan T.-M., Longabaugh W., Bolouri H., Chen H.-L., Tseng W.-F., Chao C.-H., Jang T.-H., Lin Y.-I., Hung S.-C., Wang H.-D. et al. (2009). Developmental gene regulatory networks in the zebrafish embryo. Biochim. Biophys. Acta 1789, 279-298 10.1016/j.bbagrm.2008.09.005 [DOI] [PubMed] [Google Scholar]
- Cheatle Jarvela A. M. and Hinman V. (2014). A method for microinjection of Patiria miniata zygotes. J. Vis. Exp. 1, e51913 10.3791/51913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce J. C. and McClay D. R. (2010). Dynamics of Delta/Notch signaling on endomesoderm segregation in the sea urchin embryo. Development 137, 83-91 10.1242/dev.044149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darras S., Gerhart J., Terasaki M., Kirschner M. and Lowe C. J. (2011). β-Catenin specifies the endomesoderm and defines the posterior organizer of the hemichordate Saccoglossus kowalevskii. Development 138, 959-970 10.1242/dev.059493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson E. H. (2006). The Regulatory Genome: Gene Regulatory Networks in Development and Evolution. Academic Press. [Google Scholar]
- Davidson E. H. and Erwin D. H. (2006). Gene regulatory networks and the evolution of animal body plans. Science 311, 796-800 10.1126/science.1113832 [DOI] [PubMed] [Google Scholar]
- Green J. B. A. and Smith J. C. (1991). Growth factors as morphogens: do gradients and thresholds establish body plan? Trends Genet. 7, 245-250 10.1016/0168-9525(91)90323-I [DOI] [PubMed] [Google Scholar]
- Henry J. Q., Perry K. J., Wever J., Seaver E. and Martindale M. Q. (2008). β-Catenin is required for the establishment of vegetal embryonic fates in the nemertean, Cerebratulus lacteus. Dev. Biol. 317, 368-379 10.1016/j.ydbio.2008.02.042 [DOI] [PubMed] [Google Scholar]
- Henry J. Q., Perry K. J. and Martindale M. Q. (2010). β-catenin and early development in the gastropod, Crepidula fornicata. Integr. Comp. Biol. 50, 707-719 10.1093/icb/icq076 [DOI] [PubMed] [Google Scholar]
- Hinman V. F. and Cheatle Jarvela A. M. (2014). Developmental gene regulatory network evolution: Insights from comparative studies in echinoderms. Genesis 52, 193-207 10.1002/dvg.22757 [DOI] [PubMed] [Google Scholar]
- Hinman V. F. and Davidson E. H. (2003a). Expression of AmKrox, a starfish ortholog of a sea urchin transcription factor essential for endomesodermal specification. Gene Expr. Patterns 3, 423-426 10.1016/S1567-133X(03)00083-8 [DOI] [PubMed] [Google Scholar]
- Hinman V. F. and Davidson E. H. (2003b). Expression of a gene encoding a Gata transcription factor during embryogenesis of the starfish Asterina miniata. Gene Expr. Patterns 3, 419-422 10.1016/S1567-133X(03)00082-6 [DOI] [PubMed] [Google Scholar]
- Hinman V. F. and Davidson E. H. (2007). Evolutionary plasticity of developmental gene regulatory network architecture. Proc. Natl. Acad. Sci. USA 104, 19404-19409 10.1073/pnas.0709994104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinman V. F., Nguyen A. T. and Davidson E. H. (2003a). Expression and function of a starfish Otx ortholog, AmOtx: a conserved role for Otx proteins in endoderm development that predates divergence of the eleutherozoa. Mech. Dev. 120, 1165-1176 10.1016/j.mod.2003.08.002 [DOI] [PubMed] [Google Scholar]
- Hinman V. F., Nguyen A. T., Cameron R. A. and Davidson E. H (2003b). Developmental gene regulatory network architecture across 500 million years of echinoderm evolution. Proc. Natl. Acad. Sci. USA 100, 13356-13361 10.1073/pnas.2235868100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinman V. F., Yankura K. A. and McCauley B. S. (2009). Evolution of gene regulatory network architectures: examples of subcircuit conservation and plasticity between classes of echinoderms. Biochim. Biophys. Acta 1789, 326-332 10.1016/j.bbagrm.2009.01.004 [DOI] [PubMed] [Google Scholar]
- Hudson C., Kawai N., Negishi T. and Yasuo H. (2013). β-catenin-driven binary fate specification segregates germ layers in ascidian embryos. Curr. Biol. 23, 491-495 10.1016/j.cub.2013.02.005 [DOI] [PubMed] [Google Scholar]
- Imai K., Takada N., Satoh N. and Satou Y. (2000). (beta)-catenin mediates the specification of endoderm cells in ascidian embryos. Development 127, 3009-3020. [DOI] [PubMed] [Google Scholar]
- In der Rieden P. M. J., Vilaspasa F. L. and Durston A. J. (2010). Xwnt8 directly initiates expression of labial Hox genes. Dev. Dyn. 239, 126-139. [DOI] [PubMed] [Google Scholar]
- Ip Y. T., Levine M. and Small S. J. (1992). The bicoid and dorsal morphogens use a similar strategy to make stripes in the Drosophila embryo. J. Cell Sci. 1992Suppl. 16, 33-38 10.1242/jcs.1992.Supplement_16.5 [DOI] [PubMed] [Google Scholar]
- Jaeger J. (2011). The gap gene network. Cell. Mol. Life Sci. 68, 243-274 10.1007/s00018-010-0536-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiecker C. and Niehrs C. (2001). A morphogen gradient of Wnt/β-catenin signalling regulates anteroposterior neural patterning in Xenopus. Development 128, 4189-4201. [DOI] [PubMed] [Google Scholar]
- Kuraishi R. and Osanai K. (1992). Cell movements during gastrulation of starfish larvae. Biol. Bull. 183, 258-268 10.2307/1542213 [DOI] [PubMed] [Google Scholar]
- Kuraishi R. and Osanai K. (1994). Contribution of maternal factors and cellular interaction to determination of archenteron in the starfish embryo. Development 120, 2619-2628. [Google Scholar]
- Levine M. and Davidson E. H (2005). Gene regulatory networks for development. Proc. Natl. Acad. Sci. USA 102, 4936-4942 10.1073/pnas.0408031102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lhomond G., McClay D. R., Gache C. and Croce J. C. (2012). Frizzled1/2/7 signaling directs β-catenin nuclearisation and initiates endoderm specification in macromeres during sea urchin embryogenesis. Development 139, 816-825 10.1242/dev.072215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan C. Y. and Nusse R. (2004). The wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 20, 781-810 10.1146/annurev.cellbio.20.010403.113126 [DOI] [PubMed] [Google Scholar]
- Logan C. Y., Miller J. R., Ferkowicz M. J. and McClay D. R. (1999). Nuclear β-catenin is required to specify vegetal cell fates in the sea urchin embryo. Development 126, 345-357. [DOI] [PubMed] [Google Scholar]
- Longabaugh W. J. R. (2012). BioTapestry: a tool to visualize the dynamic properties of gene regulatory networks. Methods Mol. Biol. 786, 359-394 10.1007/978-1-61779-292-2_21 [DOI] [PubMed] [Google Scholar]
- Loose M. and Patient R. (2004). A genetic regulatory network for Xenopus mesendoderm formation. Dev. Biol. 271, 467-478 10.1016/j.ydbio.2004.04.014 [DOI] [PubMed] [Google Scholar]
- Maduro M. F. (2009). Structure and evolution of the C. elegans embryonic endomesoderm network. Biochim. Biophys. Acta 1789, 250-260 10.1016/j.bbagrm.2008.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Materna S. C. and Davidson E. H. (2012). A comprehensive analysis of Delta signaling in pre-gastrular sea urchin embryos. Dev. Biol. 364, 77-87 10.1016/j.ydbio.2012.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauley B. S., Weideman E. P. and Hinman V. F. (2010). A conserved gene regulatory network subcircuit drives different developmental fates in the vegetal pole of highly divergent echinoderm embryos. Dev. Biol. 340, 200-208 10.1016/j.ydbio.2009.11.020 [DOI] [PubMed] [Google Scholar]
- McCauley B. S., Wright E. P., Exner C., Kitazawa C. and Hinman V. F. (2012). Development of an embryonic skeletogenic mesenchyme lineage in a sea cucumber reveals the trajectory of change for the evolution of novel structures in echinoderms. EvoDevo 3, 17 10.1186/2041-9139-3-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauley B. S., Akyar E., Filliger L. and Hinman V. F. (2013). Expression of wnt and frizzled genes during early sea star development. Gene Expr. Patterns 13, 437-444 10.1016/j.gep.2013.07.007 [DOI] [PubMed] [Google Scholar]
- McLin V. A., Rankin S. A. and Zorn A. M. (2007). Repression of Wnt/beta-catenin signaling in the anterior endoderm is essential for liver and pancreas development. Development 134, 2207-2217 10.1242/dev.001230 [DOI] [PubMed] [Google Scholar]
- Miyawaki K., Yamamoto M., Saito K., Saito S., Kobayashi N. and Matsuda S. (2003). Nuclear localization of beta-catenin in vegetal pole cells during early embryogenesis of the starfish Asterina pectinifera. Dev. Growth Differ. 45, 121-128 10.1034/j.1600-0854.2004.00681.x [DOI] [PubMed] [Google Scholar]
- Niehrs C. (2010). On growth and form: a Cartesian coordinate system of Wnt and BMP signaling specifies bilaterian body axes. Development 137, 845-857 10.1242/dev.039651 [DOI] [PubMed] [Google Scholar]
- Oliveri P., Walton K. D., Davidson E. H. and McClay D. R. (2006). Repression of mesodermal fate by foxa, a key endoderm regulator of the sea urchin embryo. Development 133, 4173-4181 10.1242/dev.02577 [DOI] [PubMed] [Google Scholar]
- Peter I. S. and Davidson E. H. (2011). A gene regulatory network controlling the embryonic specification of endoderm. Nature 474, 635-639 10.1038/nature10100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poustka A. J., Kühn A., Groth D., Weise V., Yaguchi S., Burke R. D., Herwig R., Lehrach H. and Panopoulou G. (2007). A global view of gene expression in lithium and zinc treated sea urchin embryos: new components of gene regulatory networks. Genome Biol. 8, pR85. 10.1186/gb-2007-8-5-r85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransick A. and Davidson E. H. (2006). cis-regulatory processing of Notch signaling input to the sea urchin glial cells missing gene during mesoderm specification. Dev. Biol. 297, 587-602 10.1016/j.ydbio.2006.05.037 [DOI] [PubMed] [Google Scholar]
- Röttinger E., Croce J., Lhomond G., Besnardeau L., Gache C. and Lepage T. (2006). Nemo-like kinase (NLK) acts downstream of Notch/Delta signalling to downregulate TCF during mesoderm induction in the sea urchin embryo. Development 133, 4341-4353 10.1242/dev.02603 [DOI] [PubMed] [Google Scholar]
- Röttinger E., Dahlin P. and Martindale M. Q. (2012). A framework for the establishment of a cnidarian gene regulatory network for “Endomesoderm” specification: the inputs of β-catenin/TCF signaling. PLoS Genet. 8, e1003164. 10.1371/journal.pgen.1003164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider S. Q. and Bowerman B. (2007). β-catenin asymmetries after all animal/vegetal- oriented cell divisions in Platynereis dumerilii embryos mediate binary cell-fate specification. Dev. Cell 13, 73-86 10.1016/j.devcel.2007.05.002 [DOI] [PubMed] [Google Scholar]
- Schneider V. A. and Mercola M. (2001). Wnt antagonism initiates cardiogenesis in Xenopus laevis. Genes Dev. 15, 304-315 10.1101/gad.855601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi A. J., Wikramanayake R. M., Angerer R. C., Range R. C. and Angerer L. M. (2012). Sequential signaling crosstalk regulates endomesoderm segregation in sea urchin embryos. Science 335, 590-593 10.1126/science.1212867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood D. R. and McClay D. R. (1999). LvNotch signaling mediates secondary mesenchyme specification in the sea urchin embryo. Development 125, 1703-1713. [DOI] [PubMed] [Google Scholar]
- Stambolic V., Ruel L. and Woodgett J. R. (1996). Lithium inhibits glycogen synthase kinase-3 activity and mimics wingless signalling in intact cells. Curr. Biol. 6, 1664-1669 10.1016/S0960-9822(02)70790-2 [DOI] [PubMed] [Google Scholar]
- Sweet H. C., Gehring M. and Ettensohn C. A. (2002). LvDelta is a mesoderm-inducing signal in the sea urchin embryo and can endow blastomeres with organizer-like properties. Development 129, 1945-1955. [DOI] [PubMed] [Google Scholar]
- Thorpe C. J., Schlesinger A., Carter J. C. and Bowerman B. (1997). Wnt signaling polarizes an early C. elegans blastomere to distinguish endoderm from mesoderm. Cell 90, 695-705 10.1016/S0092-8674(00)80530-9 [DOI] [PubMed] [Google Scholar]
- Veeman M. T., Slusarski D. C., Kaykas A., Louie S. H. and Moon R. T. (2003). Zebrafish prickle, a modulator of noncanonical wnt/fz signaling, regulates gastrulation movements. Curr. Biol. 13, 680-685 10.1016/S0960-9822(03)00240-9 [DOI] [PubMed] [Google Scholar]
- Wang D. G., Britten R. J. and Davidson E. H. (1995). Maternal and embryonic provenance of a sea urchin embryo transcription factor, SpZ12-1. Mol. Marine Biol. Biotechnol. 4, 148-153. [PubMed] [Google Scholar]
- Weitzel H. E., Illies M. R., Byrum C. A., Xu R., Wikramanayake A. H. and Ettensohn C. A. (2004). Differential stability of beta-catenin along the animal-vegetal axis of the sea urchin embryo mediated by dishevelled. Development 131, 2947-2956 10.1242/dev.01152 [DOI] [PubMed] [Google Scholar]
- Wikramanayake A. H., Huang L. and Klein W. H (1998). beta-Catenin is essential for patterning the maternally specified animal-vegetal axis in the sea urchin embryo. Proc. Natl. Acad. Sci. USA 95, 9343-9348 10.1073/pnas.95.16.9343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikramanayake A. H., Hong M., Lee P. N., Pang K., Byrum C. A., Bince J. M., Xu R. and Martindale M. Q. (2003). An ancient role for nuclear β-catenin in the evolution of axial polarity and germ layer segregation. Nature 426, 446-450 10.1038/nature02113 [DOI] [PubMed] [Google Scholar]
- Yankura K. A., Martik M. L., Jennings C. K. and Hinman V. F. (2010). Uncoupling of complex regulatory patterning during evolution of larval development in echinoderms. BMC Biol. 8, 143 10.1186/1741-7007-8-143 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.