Significance
To date, inadequate analytical methods have severely limited our ability to understand virulence strategies used by intracellular bacterial pathogens to evade reactive oxygen species (ROS) during infection. We have developed a system that is based on redox-sensitive GFP for real-time and nondisruptive measurement of redox stress experienced by Salmonella inside phagocytic cells. Using this system, we directly report intrabacterial redox dynamics during Salmonella infection of macrophages. This biosensor, which can easily be applied to other bacteria, enables a detailed study of ROS/reactive nitrogen species (RNS) evasion strategies, even detecting differences between individual bacteria. Additionally, this technique can be used for directly measuring redox responses in different environmental conditions for many bacteria.
Keywords: oxidative stress, reactive oxygen species, Salmonella, bacterial pathogenesis, redox-sensitive GFP
Abstract
Many significant bacterial pathogens have evolved virulence mechanisms to evade degradation and exposure to reactive oxygen (ROS) and reactive nitrogen species (RNS), allowing them to survive and replicate inside their hosts. Due to the highly reactive and short-lived nature of ROS and RNS, combined with limitations of conventional detection agents, the mechanisms underlying these evasion strategies remain poorly understood. In this study, we describe a system that uses redox-sensitive GFP to nondisruptively measure real-time fluctuations in the intrabacterial redox environment. Using this system coupled with high-throughput microscopy, we report the intrabacterial redox dynamics of Salmonella enterica Typhimurium (S. Typhimurium) residing inside macrophages. We found that the bacterial SPI-2 type III secretion system is required for ROS evasion strategies and this evasion relies on an intact Salmonella-containing vacuole (SCV) within which the bacteria reside during infection. Additionally, we found that cytosolic bacteria that escape the SCV experience increased redox stress in human and murine macrophages. These results highlight the existence of specialized evasion strategies used by intracellular pathogens that either reside inside a vacuole or “escape” into the cytosol. Taken together, the use of redox-sensitive GFP inside Salmonella significantly advances our understanding of ROS and RNS evasion strategies during infection. This technology can also be applied to measuring bacterial oxidative and nitrosative stress dynamics under different conditions in a wide variety of bacteria.
A central mechanism of the innate immune response to defend against pathogens is the production of reactive oxygen species (ROS) and reactive nitrogen species (RNS) by specialized phagocytic immune cells (1). Macrophages and neutrophils generate ROS after detection of pathogen-associated molecular patterns (PAMPs) through the NADPH oxidase complex, whereas RNS is produced by inducible nitric oxide synthase (iNOS), which generates NO• through the conversion of l-arginine and oxygen (1–3).
To survive, replicate, and disseminate throughout the body, bacterial pathogens—especially those that reside in intracellular niches—must overcome the antimicrobial oxidative and nitrosative burst (1). General ROS and RNS enzymes such as catalases, peroxidases, superoxide dismutases, and DNA repair enzymes are used by most bacteria to survive exposure to ROS and RNS (4). In addition to these general defenses, many intracellular pathogens have evolved specific evasion strategies that allow them to live inside their host cells. For example, the intracellular pathogens Shigella flexneri and Listeria monocytogenes, which cause shigellosis and listeriosis, respectively, “escape” the phagolysosome to proliferate within the cytosol of macrophages. In contrast, the Gram-negative pathogen Salmonella enterica subsp. Typhimurium (S. Typhimurium), which is a major cause of gastroenteritis and some systemic diseases, remains inside a specific Salmonella-containing vacuole (SCV), where it injects bacterial effector proteins directly into the host cell through a type III secretion system (T3SS). A specific set of type III effectors associated with a Salmonella pathogenicity island (SPI), known as SPI-2 effectors, have been implicated in ROS and RNS evasion strategies (5, 6); however, the relationship between SPI-2 and oxidative stress evasion is a contentious topic after a recent study concluded that the contribution of SPI-2 to Salmonella pathogenesis is unrelated to its interaction with oxidative stress (7). Inadequate analytic tools for directly measuring ROS/RNS and rapid fluctuations due to the short-lived nature of ROS/RNS have also contributed to poor reproducibility in the studies of ROS and RNS evasion strategies (1, 8, 9).
Herein, we describe a previously unidentified use of a redox-sensitive GFP (roGFP2) that has been engineered to form a reversible disulfide bond upon oxidation or S-nitrosylation of specific cysteines. Formation of the disulfide bond leads to a slight shift in its conformation (Fig. S1A) resulting in two isoforms (roGFP2ox and roGFP2red) of roGFP2. These isoforms have distinct excitation spectra with specific fluorescence signals after excitation at 405 and 480 nm, respectively (10). Consequently, the 405/480-nm ratio can be used as a measure for the roGFP2ox/roGFP2red ratio, which corresponds with the intrabacterial redox potential (11). Because roGFP2 reports the redox potential by ratiometric analysis, this system excludes variations due to differences in roGFP2 concentrations. Until now, redox-sensitive GFP has almost exclusively been used in eukaryotes (12). In this study, we used roGFP2 to measure the intrabacterial redox potential in bacteria (Salmonella) after challenges with exogenous oxidative agents and during infection of HeLa cells (epithelial), THP-1 cells (monocytic), and bone marrow-derived macrophages (BMDMs) from mice. Finally, with the use of high-throughput microscopy, we demonstrated the involvement of the SPI-2 T3SS in ROS evasion strategies, as well as the requirement for an intact SCV to evade ROS and RNS inside macrophages.
Results
Characterization of roGFP2 and Real-Time Measurement of Intrabacterial Redox Potential.
We first determined the excitation wavelength spectra of roGFP2ox and roGFP2red. Purified HIS-tagged roGFP2 was exposed to 100 mM hydrogen peroxide (H2O2) and 10 mM DTT, respectively, and fluorescent signal was measured in a fluorescence plate reader (Fig. S1B). Consistent with previous studies, we found specific peaks in fluorescence signals after excitation at 405 and 480 nm (10, 11). For the duration of this study, these excitation wavelengths were used to measure the intrabacterial redox potential. Next, the stability of the roGFP2 signal under different pH conditions was tested. As S. Typhimurium encounters acidified environments during infection of macrophages, it is crucial that the roGFP2 ratio signal is unaffected by pH. None of the physiologically relevant pH values tested (pH 5–8) affected the maximum or minimum ratiometric signals of roGFP2-expressing S. Typhimurium (Fig. S1C). Throughout this study, each ratiometric signal of roGFP2 was normalized to a scale of 0.1 (most reduced) and 1.0 (most oxidized). Internal reduced and oxidized controls are therefore obtained within each experiment.
To test whether the system was able to monitor real-time fluctuations in the intrabacterial redox potential, S. Typhimurium was challenged with 2.5 mM H2O2 and subsequently with 10 mM DTT (Fig. S1D). After these challenges, the roGFP2 405/480-nm ratio directly reported real-time changes to the redox potential. The signal was entirely reversible and allowed for measurement of oxidation as well as reduction of the intrabacterial environment. The fast response time of roGFP2 in S. Typhimurium benefits real-time measurements; however, certain experiments require analysis at later time points. To “freeze” the ratio of roGFP2, addition of the alkylating compound N-ethyl maleimide (NEM) and subsequent fixation by paraformaldehyde (PFA) has been used previously (13). NEM covalently interacts with thiol groups and blocks formation of disulfide bonds. Addition of 20 mM NEM 5 min before H2O2 challenge effectively blocked oxidation of roGFP2 (Fig. S1D).
The response time of roGFP2 inside Salmonella was >50-fold faster than that of purified roGFP2 protein alone (Fig. S1 E and F). This is most likely due to bacterial enzymes catalyzing the formation and disruption of the reversible disulfide bond. Thus, H2O2 stress triggers formation of a disulfide bond in roGFP2, but the mechanism is uncertain and apparently indirect. Nitric oxide reacts with thiols to form S-nitrosylated cysteines, and oxidation following S-nitrosylation leads to subsequent disulfide bond formation (14). Previously, other groups have created fusions between roGFP2 and catalyzing enzymes to speed up the response time and increase specificity (15, 16). Because the intrabacterial roGFP2 response was immediate and to ensure sensitivity to different sources of redox stress, we used unaltered roGFP2 for our analyses.
Using the Nernst equation, the 405/480-nm ratiometric signal can be converted into a quantitative value for the redox potential (EroGFP2) inside S. Typhimurium (SI Materials and Methods). The EroGFP2 for S. Typhimurium can be calculated for each of the different conditions. Many in vitro measurements in this study were done in low-phosphate medium (LPM) at pH 5.8. This medium mimics intracellular conditions and induces expression of the SPI-2 T3SS and secretion of SPI-2 effectors by S. Typhimurium. Under these conditions, the resting EroGFP2 for S. Typhimurium was calculated to be −311 ± 4 mV, whereas the EroGFP2 in Escherichia coli (DH10B) under similar conditions was slightly higher at −302 ± 4 mV.
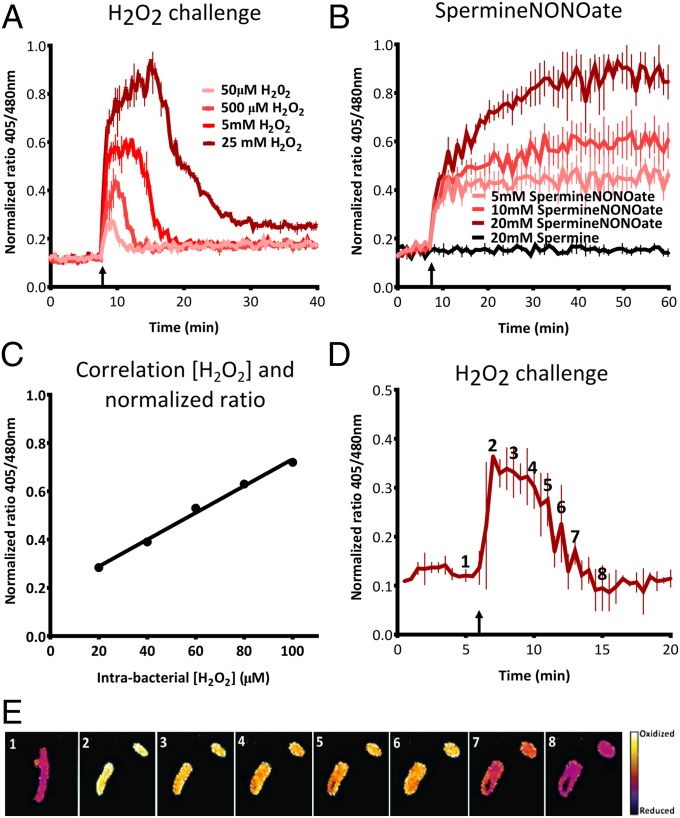
To further analyze the sensitivity and responsiveness of roGFP2 in S. Typhimurium after challenges with different oxidative agents, the intrabacterial redox potential was measured in real time with a fluorescence plate reader. Following the addition of different concentrations of H2O2 (Fig. 1A) or SpermineNONOate (Fig. 1B), a dose-dependent increase in the 405/480-nm ratio was observed. A one-time addition of H2O2 led to a sudden increase in the redox potential after which the bacteria detoxified H2O2 and restored a more reduced redox environment (Fig. 1A). After addition of SpermineNONOate (decay into NO•), a continuous generation of RNS resulted in a sustained increase in the redox potential (Fig. 1B).
Fig. 1.
Real-time measurement of roGFP2 in S. Typhimurium. (A and B) Dose-dependent response of intrabacterial redox potential after a challenge with H2O2 (A) or SpermineNONOate (B), respectively. The upward arrow indicates injection of the oxidative compound. These experiments are done in LPM at pH 5.8. (C) Correlation between the intrabacterial H2O2 concentration and the normalized 405/480 ratio in HpxF(−) Salmonella. (D and E) Microscopic analysis of real-time response to challenge with 2.5 mM H2O2 in PBS. The false-colored ratio images are obtained after analysis by ImageJ. The numbers in the images in E correspond to the time points indicated in D.
Models predict that ROS concentrations inside macrophages are in the micromolar range (8, 17, 18). To test whether intrabacterial roGFP2 was able to measure low concentrations of H2O2, a Salmonella mutant without three catalases (KatE, KatG, and KatN) and two alkyl hydroperoxide reductases (AhpC and TsaA) was used to calibrate the system (19). This HpxF(−) strain was almost deprived of detoxifying power, and using roGFP2 we were able to show micromolar sensitivity to H2O2. This demonstrates that roGFP2 is able to detect biologically relevant concentrations of ROS inside macrophages (Fig. 1C).
Intrabacterial redox dynamics can also be analyzed by fluorescent microscopy. After addition of 2.5 mM H2O2, images were taken by fluorescence microscopy and the ratio values of these images were obtained (Fig. 1D). Individual images were analyzed by ImageJ to create pseudocolored images for better visualization (Fig. 1E). A real-time fluorescence movie of an equivalent challenge showing the changes is included (Movie S1). These images and movie confirm our results from the fluorescence plate reader and allow for visualization of redox differences between individual bacteria.
Intrabacterial Redox Potential During S. Typhimurium Infection of Cultured Human Cells.
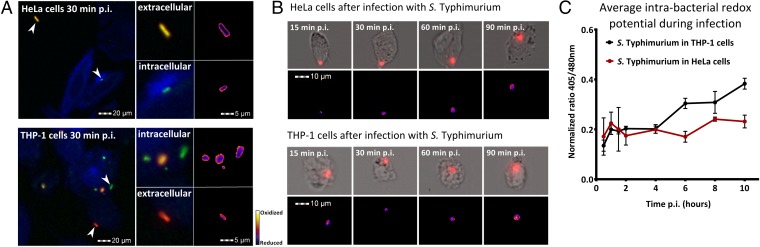
Following the verification of roGFP2 in S. Typhimurium, the system was used to analyze redox dynamics during infection of cultured cells. Both epithelium-like HeLa cells and macrophage-like THP-1 cells were infected with S. Typhimurium. HeLa cells are permissive for intracellular S. Typhimurium, in contrast to THP-1 cells, which kill the majority of S. Typhimurium (20). Images of infected cells were taken at 30 min postinfection (p.i.) with fluorescence microscopy. Extracellular bacteria were labeled with a red fluorescent antibody to distinguish them from intracellular bacteria (Fig. 2A).
Fig. 2.
Intrabacterial redox potential after infection of HeLa and THP-1 cells. (A) Infected HeLa cells and THP-1 cells at 30 min postinfection (p.i.) with fluorescence microscopy. All S. Typhimurium bacteria are shown in green, extracellular bacteria are shown in red, and actin is shown in blue. The upward arrows indicate extracellular bacteria, and the downward arrows indicate intracellular bacteria. Images were analyzed by ImageJ, and pseudocolored ratio images of individual bacteria are shown in the right panels. (B) For each time point, an image from the AMNIS ImageStream was selected and analyzed by ImageJ. In the upper panels, a bright-field image of individual infected cells is shown. Intracellular S. Typhimurium is shown in red. In the lower panels, pseudocolored ratio images of the corresponding bright-field images are shown. The pseudocolored legend is similar to the legend for A. (C) “Redox stress” timeline showing the average normalized 405/480 ratio value of intracellular S. Typhimurium in HeLa and THP-1 cells for the first 10 h p.i. Each value represents analysis of more than 15,000 pictures of individual cells over three separate experiments.
In HeLa cells, no significant overall H2O2 production was measured by the Amplex red assay at 2 h p.i. (data not shown), and no oxidative stress was observed in intracellular bacteria compared with extracellular bacteria. For THP-1 cells, we measured a significant efflux of H2O2 at 2 h p.i., but analysis of roGFP2 did not show increased oxidative stress inside intracellular S. Typhimurium by fluorescence microscopy.
To increase sampling size and ensure an unbiased approach, consecutive measurements of the redox potential in intracellular S. Typhimurium were done using an AMNIS ImageStream system. This system integrates flow cytometry and fluorescence microscopy, enabling very high-throughput fluorescence microscopy by acquiring at least 15,000 images of individual cells per time point and condition tested. A minimum of three individual experiments were done. Infected cells were fixed with NEM and PFA at various time points p.i., after which the cells were passed through a flow cytometry system. The ImageStream recorded each individual cell and all images were processed automatically. The results of the AMNIS ImageStream confirmed our previous findings with conventional fluorescence microscopy and enabled the visualization of infected cells during the first 90 min of infection (Fig. 2B). The average 405/480 ratio was obtained for each experiment, and together these results created a “redox stress” timeline for intracellular S. Typhimurium in HeLa cells and THP-1 cells (Fig. 2C). In HeLa cells, intracellular bacteria did not experience significant redox stress, whereas inside THP-1 cells S. Typhimurium experienced gradually increasing redox stress. Collectively, these results generate, for the first time (to our knowledge), an accurate portrayal of the redox stress dynamics experienced by WT Salmonella inside host cells during the first 10 h of infection.
Salmonella SPI-2 Evasion of the Oxidative Burst.
After determining redox stress dynamics during regular Salmonella infection, we used the roGFP2 system to investigate specific Salmonella ROS and RNS evasion mechanisms. From within the SCV, S. Typhimurium injects SPI-2 effectors into the cell via a T3SS. The SPI-2 system has been linked to ROS and RNS evasion strategies by interfering with proper colocalization of NADPH-oxidase and iNOS with the SCV (5, 6, 21). However, a recent study was unable to find a correlation between SPI-2 involvement and oxidative stress evasion (7), which has contributed to the ongoing controversy regarding SPI-2 involvement in ROS evasion (8, 9). These inconsistencies emphasize the need for better analytical tools to analyze redox stress of pathogens.
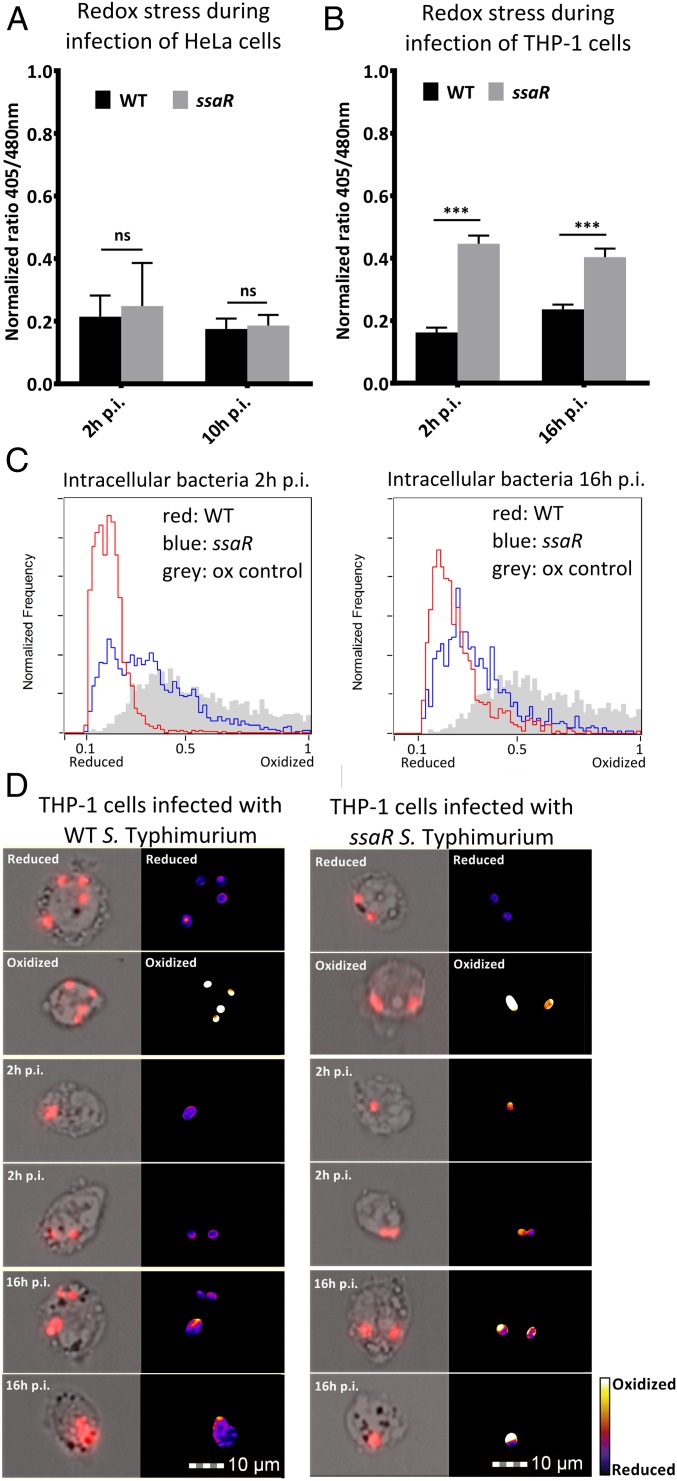
To investigate the involvement of the SPI-2 system in ROS and RNS evasion, redox dynamics in the ssaR mutant, which is deficient in the assembly of a functional SPI-2 T3SS (22), were compared with infection with WT S. Typhimurium. By plating colony-forming units (CFUs), it was confirmed that ssaR is attenuated for survival in THP-1 cells but not for survival in HeLa cells (Fig. S2A). Next, the redox potential of intracellular WT or ssaR S. Typhimurium during infection of HeLa and THP-1 cells was obtained with the AMNIS ImageStream. Infected THP-1 cells were harvested at 2 and 16 h p.i. and redox stress was measured. HeLa cells were harvested at 2 and 10 h p.i. because many HeLa cells did not survive past 10 h p.i. No significant redox stress was observed after infection of HeLa cells with the ssaR mutant (Fig. 3A). In THP-1 cells, however, a significant increase in redox stress at 2 and 16 h p.i. was observed in ssaR S. Typhimurium (Fig. 3B). To demonstrate the distribution of redox stress per cell, ratio values for all infected THP-1 cells were plotted in histograms (Fig. 3C). A random selection of images from each time point was processed by ImageJ to create pseudocolored ratio images (Fig. 3D).
Fig. 3.
Involvement of SPI-2 system in ROS and RNS evasion. (A) Average intrabacterial redox potential of WT and ssaR S. Typhimurium in HeLa cells at 2 and 10 h p.i. (B) Average intrabacterial redox potential of WT and ssaR S. Typhimurium in THP-1 cells at 2 and 16 h p.i. over three separate experiments. Error bars indicate the SD. Each value represents analysis of at least 15,000 pictures. Significance was analyzed by a Student t test. (C) Representative pictures of infected THP-1 cells at 2 and 16 h p.i. Images on the Right are pseudocolored ratio images after analysis with ImageJ. (D) The 405/480-nm ratio histograms of intrabacterial redox potential after infection of THP-1 cells with WT and ssaR at 2 and 16 h p.i. The gray histogram represents the oxidized control, the red line represents WT ratio values, and the blue line represents ssaR ratio values.
To determine the source of bacterial redox stress, we infected THP-1 cells with inhibitors of NADPH oxidase [diphenylene iodonium chloride (DPI)] or iNOS [NG-monomethyl l-arginine (l-NMMA)] (Fig. S2 B and C). These measurements revealed that, under the conditions that we used, increased redox stress in ssaR S. Typhimurium resulted from phagosomal ROS.
Because inhibitors can also alter other cellular processes, we next wanted to confirm our results in macrophages lacking phagosomal ROS and RNS production. In infected BMDMs from C57BL/6J mice, no redox stress was observed at 2 h p.i., indicating a difference in timing of the oxidative burst. At 16 h p.i., however, we measured increased redox stress in ssaR similar to that in THP-1 cells (Fig. S2D). Infection of gp91phox−/− and iNOS−/− BMDMs (which are unable to generate phagosomal ROS or RNS, respectively) also showed that, under our conditions, the source of redox stress is ROS rather than RNS (Fig. S2E).
To further demonstrate that increased bacterial redox stress in ssaR S. Typhimurium was not caused by an increase in overall H2O2 and NO• generation, the H2O2 and NO• production of infected THP-1 cells was measured using the Amplex red assay and the Griess assay (Fig. S2 F and G). THP-1 cells were infected with WT and ssaR S. Typhimurium, and the SPI-2 system was not found to inhibit overall H2O2 or NO• production. These results are consistent with previous studies that show SPI-2–mediated exclusion of NADPH-oxidase from the SCV, but no inhibition of overall H2O2 or NO• production in the host cells (6, 23). Interestingly, when examining individual infected THP-1 cells that contained many intracellular bacteria, the majority of bacteria in the cell experienced similar levels of oxidative stress (2 h p.i.); however, we observed heterogeneity in redox stress between intracellular bacteria at 16 h p.i. (Fig. S3).
An Intact Salmonella-Containing Vacuole Is Essential for ROS or RNS Evasion in Macrophages.
The above experiments demonstrate that S. Typhimurium uses the SPI-2 system to evade ROS and that these evasion strategies are most likely the result of SPI-2–mediated prevention of effective ROS colocalization with the SCV. We hypothesized that, for Salmonella to control ROS colocalization, it is essential to reside within the SCV. It is known that from inside the SCV, S. Typhimurium regulates fusion with vesicles through the actions of SPI-2 effectors. For example, SPI-2 effector SifA directs trafficking of mannose-6-phospate receptors and thereby inhibits lysosome function (24). Additionally, SifA is implicated in maintaining the integrity of the SCV, and it is well documented that infection with the sifA mutant leads to loss of SCV integrity (25). In contrast to S. flexneri and L. monocytogenes, S. Typhimurium is unable to survive in the cytosol of macrophages, which results in severe attenuation of the sifA mutant (22).
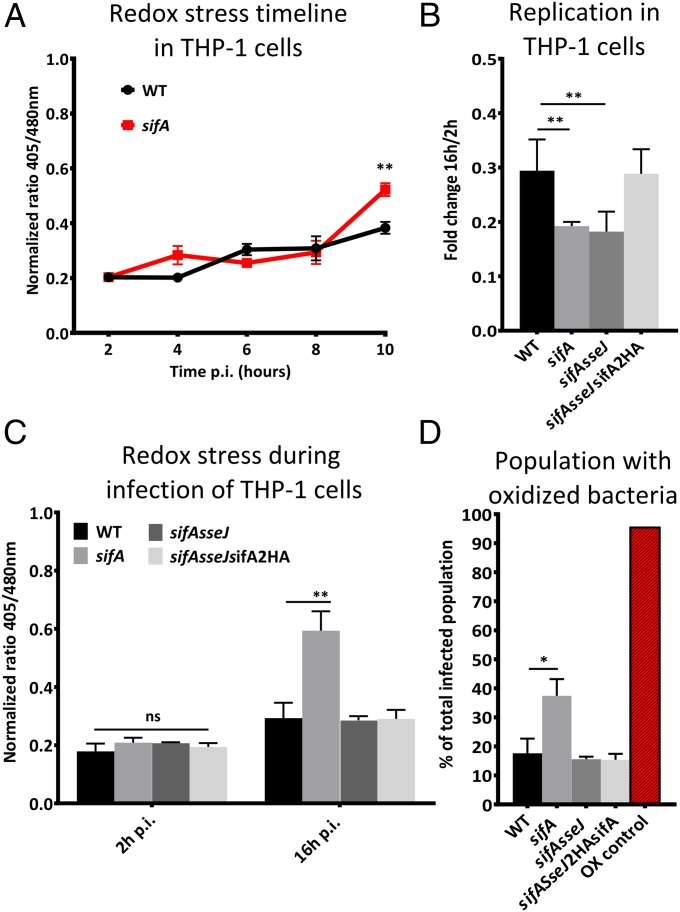
To investigate whether the SCV is important for ROS or RNS evasion, THP-1 cells were infected with WT or sifA S. Typhimurium. A redox stress timeline from 2 h p.i. until 10 h p.i. was created that showed increased redox stress at 10 h p.i. for sifA S. Typhimurium (Fig. 4A). To determine whether this redox stress was caused by vacuole disruption or inhibition of lysosome fusion, a sifAsseJ double mutant was constructed. This mutant maintains SCV integrity but is unable to inhibit lysosome fusion (24, 25). To determine the contribution of SPI-2 effector SseJ to redox stress in sifAsseJ, the double mutant was complemented by integration of sifA2HA in the chromosome, effectively creating a sseJ single-deletion mutant. THP-1 cells were infected by all four strains and both the sifA mutant as well as the sifAsseJ double mutant were similarly attenuated for intracellular survival in THP-1 cells (Fig. 4B). Complementation with sifA2HA reversed replication to levels of WT infection, indicating that deletion of sifA is the major reason for attenuated virulence.
Fig. 4.
SCV is crucial for RNS evasion. (A) “Redox stress” timeline showing the average normalized 405/480 ratio value of intracellular WT and sifA S. Typhimurium in THP-1 cells for the first 10 h p.i. (B) Intracellular survival of intracellular WT, sifA, sifAsseJ, and sifAsseJ2HAsifA S. Typhimurium in THP-1 cells determined by CFU counts fold change between 2 and 16 h p.i. Each experiment was repeated at least 12 times. (C) The average intrabacterial redox potential in THP-1 cells at 2 and 16 h p.i. Each value represents analysis of more than 15,000 individual cells over three separate experiments. (D) Subset of infected THP-1 cells that contain highly oxidized bacteria. Error bars indicate the SD. Significance was analyzed by a Student t test.
Analysis of WT and sifA-infected THP-1 cells at 2 and 16 h p.i. using the AMNIS ImageStream demonstrated that only the sifA single mutant experienced significantly more redox stress than WT S. Typhimurium (Fig. 4C). Both the sifA single mutant and the sifAsseJ double mutant are deficient in the inhibition of lysosome fusion, but only the sifA single mutant “escapes” the SCV and resides in the cytosol (24). Therefore, these results clearly suggest that cytosolic bacteria experience more redox stress than bacteria that reside inside the SVC.
To confirm that the increase in redox stress observed in sifA S. Typhimurium was not caused by increased generation of H2O2 or NO•, the total production was measured with the Amplex red and Griess assay (Fig. S4 A and B). No significant increase in H2O2 or NO• production was observed. Further in-depth analysis of redox stress in sifA S. Typhimurium in infected THP-1 cells revealed a significantly larger subset of cells containing “stressed” bacteria than WT infected cells (Fig. 4D). Furthermore, for THP-1 cells that contained higher numbers of intracellular bacteria were examined, more heterogeneity was observed in sifA-infected cells than for any of the other mutants (Fig. S5).
To test whether the redox stress of cytosolic bacteria was caused by ROS or RNS, we infected THP-1 cells with WT or sifA and added DPI and l-NMMA. In human THP-1 cells, we found increased redox stress in sifA resulting from RNS (Fig. S6A). Infection of murine BMDMs with sifA did also result in increased redox stress (Fig. S6B), but infection of gp91phox−/− and iNOS−/− BMDMs revealed that the source was phagosomal ROS rather than RNS (Fig. S6C).
Collectively, these results highlight that Salmonella uses specific oxidative stress evasion strategies that rely on SPI-2–mediated processes to prevent colocalization of ROS with the SCV. S. Typhimurium that escape the SCV into the cytosol experience more redox stress. The source and intensity of redox stress may be dependent on the host donor and the stimulation of macrophages by different cytokines and stimuli.
Discussion
Despite the successful use of roGFP2 in eukaryotic systems, this biosensor had yet to be extensively applied in bacterial systems. Here, we showed that roGFP2 can be used as a robust and direct redox biosensor in S. Typhimurium. Moreover, this provides a nondisruptive measure of real-time fluctuations in the intrabacterial redox potential. All of these properties make roGFP2 a superior biosensor for the study of redox dynamics in bacteria compared with other systems that are currently being used.
With this approach, we studied Salmonella redox dynamics during infection of human cultured cells and BMDMs. By measuring roGFP2 fluorescence, “redox stress” was measured inside S. Typhimurium from within the infected cells. Significantly more redox stress was observed inside antimicrobial macrophages than in permissive HeLa cells. For the study of infected cells, we used the AMNIS ImageStream to ensure an unbiased and highly sampled approach. High-throughput acquisition of images by this machine made it possible to detect subpopulations of THP-1 cells, whereas the processing of individual images allowed for analysis of individual bacteria. Because of the large sampling size and high image resolution, we observed heterogeneity between cells and even between bacteria within one cell. Recently, several studies showed that not all S. Typhimurium bacteria within one cell replicate at similar rates and not all bacteria are equally affected by antimicrobial immune responses (18, 26, 27). Our observations clearly show variance in redox stress among bacteria within one cell. What determines the level of redox stress that intracellular bacteria undergo is still unclear; however, we speculate that diversity of expression profiles and variation in effector secretion play important roles. It is crucial to better understand heterogeneity because this is thought to be the core of bacterial persistence and resistance to current antimicrobial therapies (27).
The role of the SPI-2 system in evasion of oxidative stress by S. Typhimurium has been discussed for many years and continues to be a hotly debated topic. Earlier studies found that abrogation of the oxidative burst rescued attenuation of SPI-2–deficient S. Typhimurium and concluded involvement of SPI-2 effectors in ROS evasion (6, 23). Additionally, the authors showed ineffective SPI-2–mediated colocalization of NADPH-oxidase with the SCV (6, 23). Although neither of these studies was able to directly measure oxidative stress inside S. Typhimurium during infection, these results implicate SPI-2 involvement in ROS evasion.
A more recent attempt to indirectly measure oxidative stress inside S. Typhimurium used induction of an oxidation sensitive ahpCp-GFP transcriptional fusion (7). Unable to measure differences in oxidative stress between WT and SPI-2–deficient S. Typhimurium during infection of macrophages and mice, the authors concluded that Salmonella detoxifying enzymes are sufficient to cope with the host oxidative burst and there is no role for the SPI-2 system in evasion strategies (7). These seemingly conflicting results created some confusion about the role of SPI-2 in ROS evasion strategies (8, 9). Recently, researchers using a similar approach had to substitute the ahpC promotor for the katGp promoter because of lower baseline activity and a larger dynamic range (18). It is therefore possible that the original ahpC promotor did not offer enough sensitivity to measure subtle but important differences in oxidative stress. With the roGFP2 biosensor, we clearly demonstrate the involvement of the SPI-2 system in ROS evasion strategies by directly measuring redox stress inside S. Typhimurium in human and murine macrophages. Furthermore, our results show that SPI-2–mediated evasion relies on discordant localization of ROS with the SCV.
In human and murine macrophages, we found that cytosolic S. Typhimurium experience increased redox stress compared with vacuolar bacteria. Interestingly, we found that the source of redox stress varied between human and murine macrophages. Macrophages are highly heterogeneous cells that can quickly alter their function in response to local signals and stimuli (28). It is most likely that both ROS and RNS can play a role in the antimicrobial response, but the intensity and nature of redox stress is dependent on the host and how the macrophages are stimulated. Specifically, the differences in the source of redox stress for cytosolic bacteria in human macrophages compared with murine macrophages may be explained by differences in cytosolic PAMP recognition. In murine macrophages, for example, it was found that caspase-11 protects against cytosolic bacteria (29), whereas human macrophages do not express caspase-11 but instead express caspase-4. Although caspase-4 and caspase-11 share some similarities, there may be differences with regard to the immune response toward cytosolic bacteria. Another difference is the expression of TLR-11 in murine macrophages, which recognizes Salmonella flagellin. Human macrophages do not express TLR-11, which has already be shown to have a dramatic impact on Salmonella infection (30).
From these results, one can speculate that intracellular pathogens that can survive and replicate in the macrophage cytosol, e.g., S. flexneri and L. monocytogenes, most likely have evolved specific virulence strategies to evade redox stress within the cytosol. Remarkably, both S. flexneri and L. monocytogenes use actin polymerization to facilitate continuous movement inside the cytosol (31), which may play a role in evasion of ROS and RNS.
In this study, we describe the use of roGFP2 as a biosensor to study redox dynamics in S. Typhimurium; however, this biosensor could easily be used in a multitude of bacteria. During the writing of this manuscript, a study showed that roGFP2 had been engineered for specific use in the intracellular pathogen Mycobacterium tuberculosis to investigate intracellular oxidative stress after infection of macrophages (32). ROS and RNS evasion strategies of many intracellular pathogens remain elusive, and we propose that roGFP2 will have broad utility as biosensor to study redox-related virulence mechanisms and will assist in further characterization of intracellular pathogenesis. Furthermore, roGFP2 can be used to study bacterial redox dynamics after exposure to antimicrobial compounds or during biofilm formation and thereby may contribute to potential clinical applications.
Materials and Methods
roGFP2 was cloned into the pfpv25 vector for constitutive expression of roGFP2. Deletions in S. Typhimurium SL1344 were made by allele exchange with vector pRE112 as described previously (33).
Detailed materials and methods can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We sincerely thank Dr. James Remington from the University of Oregon for providing the original pRSETB roGFP2 construct and Dr. Laurent Aussel from the Institut de Microbiologie de la Méditerranée for providing the HpxF(−) Salmonella strain. We also thank W. Deng for invaluable advice; S. Vogt, E. Brown, and R. Scholz for help with the manuscript; and other members of our group for insightful discussions. This work was supported by operating grants from the Canadian Institutes of Health Research. B.B.F. is the University of British Columbia Peter Wall Distinguished Professor.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1414569112/-/DCSupplemental.
References
- 1.Fang FC. Antimicrobial reactive oxygen and nitrogen species: Concepts and controversies. Nat Rev Microbiol. 2004;2(10):820–832. doi: 10.1038/nrmicro1004. [DOI] [PubMed] [Google Scholar]
- 2.Azenabor AA, Kennedy P, York J. Free intracellular Ca2+ regulates bacterial lipopolysaccharide induction of iNOS in human macrophages. Immunobiology. 2009;214(2):143–152. doi: 10.1016/j.imbio.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Berger SB, et al. SLAM is a microbial sensor that regulates bacterial phagosome functions in macrophages. Nat Immunol. 2010;11(10):920–927. doi: 10.1038/ni.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Imlay JA. The molecular mechanisms and physiological consequences of oxidative stress: Lessons from a model bacterium. Nat Rev Microbiol. 2013;11(7):443–454. doi: 10.1038/nrmicro3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chakravortty D, Hansen-Wester I, Hensel M. Salmonella pathogenicity island 2 mediates protection of intracellular Salmonella from reactive nitrogen intermediates. J Exp Med. 2002;195(9):1155–1166. doi: 10.1084/jem.20011547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vazquez-Torres A, et al. Salmonella pathogenicity island 2-dependent evasion of the phagocyte NADPH oxidase. Science. 2000;287(5458):1655–1658. doi: 10.1126/science.287.5458.1655. [DOI] [PubMed] [Google Scholar]
- 7.Aussel L, et al. Salmonella detoxifying enzymes are sufficient to cope with the host oxidative burst. Mol Microbiol. 2011;80(3):628–640. doi: 10.1111/j.1365-2958.2011.07611.x. [DOI] [PubMed] [Google Scholar]
- 8.Slauch JM. How does the oxidative burst of macrophages kill bacteria? Still an open question. Mol Microbiol. 2011;80(3):580–583. doi: 10.1111/j.1365-2958.2011.07612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang FC. Antimicrobial actions of reactive oxygen species. MBio. 2011;2(5):pii:e00141-11. doi: 10.1128/mBio.00141-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morgan B, Sobotta MC, Dick TP. Measuring E(GSH) and H2O2 with roGFP2-based redox probes. Free Radic Biol Med. 2011;51(11):1943–1951. doi: 10.1016/j.freeradbiomed.2011.08.035. [DOI] [PubMed] [Google Scholar]
- 11.Hanson GT, et al. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J Biol Chem. 2004;279(13):13044–13053. doi: 10.1074/jbc.M312846200. [DOI] [PubMed] [Google Scholar]
- 12.Meyer AJ, Dick TP. Fluorescent protein-based redox probes. Antioxid Redox Signal. 2010;13(5):621–650. doi: 10.1089/ars.2009.2948. [DOI] [PubMed] [Google Scholar]
- 13.Albrecht SC, Barata AG, Grosshans J, Teleman AA, Dick TP. In vivo mapping of hydrogen peroxide and oxidized glutathione reveals chemical and regional specificity of redox homeostasis. Cell Metab. 2011;14(6):819–829. doi: 10.1016/j.cmet.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 14.Lipton SA, et al. Cysteine regulation of protein function—as exemplified by NMDA-receptor modulation. Trends Neurosci. 2002;25(9):474–480. doi: 10.1016/s0166-2236(02)02245-2. [DOI] [PubMed] [Google Scholar]
- 15.Gutscher M, et al. Real-time imaging of the intracellular glutathione redox potential. Nat Methods. 2008;5(6):553–559. doi: 10.1038/nmeth.1212. [DOI] [PubMed] [Google Scholar]
- 16.Gutscher M, et al. Proximity-based protein thiol oxidation by H2O2-scavenging peroxidases. J Biol Chem. 2009;284(46):31532–31540. doi: 10.1074/jbc.M109.059246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winterbourn CC, Hampton MB, Livesey JH, Kettle AJ. Modeling the reactions of superoxide and myeloperoxidase in the neutrophil phagosome: Implications for microbial killing. J Biol Chem. 2006;281(52):39860–39869. doi: 10.1074/jbc.M605898200. [DOI] [PubMed] [Google Scholar]
- 18.Burton NA, et al. Disparate impact of oxidative host defenses determines the fate of Salmonella during systemic infection in mice. Cell Host Microbe. 2014;15(1):72–83. doi: 10.1016/j.chom.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 19.Hébrard M, Viala JP, Méresse S, Barras F, Aussel L. Redundant hydrogen peroxide scavengers contribute to Salmonella virulence and oxidative stress resistance. J Bacteriol. 2009;191(14):4605–4614. doi: 10.1128/JB.00144-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forest CG, Ferraro E, Sabbagh SC, Daigle F. Intracellular survival of Salmonella enterica serovar Typhi in human macrophages is independent of Salmonella pathogenicity island (SPI)-2. Microbiology. 2010;156(Pt 12):3689–3698. doi: 10.1099/mic.0.041624-0. [DOI] [PubMed] [Google Scholar]
- 21.Suvarnapunya AE, Stein MA. DNA base excision repair potentiates the protective effect of Salmonella Pathogenicity Island 2 within macrophages. Microbiology. 2005;151(Pt 2):557–567. doi: 10.1099/mic.0.27555-0. [DOI] [PubMed] [Google Scholar]
- 22.Brumell JH, Rosenberger CM, Gotto GT, Marcus SL, Finlay BB. SifA permits survival and replication of Salmonella typhimurium in murine macrophages. Cell Microbiol. 2001;3(2):75–84. doi: 10.1046/j.1462-5822.2001.00087.x. [DOI] [PubMed] [Google Scholar]
- 23.Gallois A, Klein JR, Allen LA, Jones BD, Nauseef WM. Salmonella pathogenicity island 2-encoded type III secretion system mediates exclusion of NADPH oxidase assembly from the phagosomal membrane. J Immunol. 2001;166(9):5741–5748. doi: 10.4049/jimmunol.166.9.5741. [DOI] [PubMed] [Google Scholar]
- 24.McGourty K, et al. Salmonella inhibits retrograde trafficking of mannose-6-phosphate receptors and lysosome function. Science. 2012;338(6109):963–967. doi: 10.1126/science.1227037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruiz-Albert J, et al. Complementary activities of SseJ and SifA regulate dynamics of the Salmonella typhimurium vacuolar membrane. Mol Microbiol. 2002;44(3):645–661. doi: 10.1046/j.1365-2958.2002.02912.x. [DOI] [PubMed] [Google Scholar]
- 26.Helaine S, et al. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science. 2014;343(6167):204–208. doi: 10.1126/science.1244705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helaine S, Holden DW. Heterogeneity of intracellular replication of bacterial pathogens. Curr Opin Microbiol. 2013;16(2):184–191. doi: 10.1016/j.mib.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 28.Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011;11(11):723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aachoui Y, et al. Caspase-11 protects against bacteria that escape the vacuole. Science. 2013;339(6122):975–978. doi: 10.1126/science.1230751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mathur R, et al. A mouse model of Salmonella typhi infection. Cell. 2012;151(3):590–602. doi: 10.1016/j.cell.2012.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gouin E, et al. A comparative study of the actin-based motilities of the pathogenic bacteria Listeria monocytogenes, Shigella flexneri and Rickettsia conorii. J Cell Sci. 1999;112(Pt 11):1697–1708. doi: 10.1242/jcs.112.11.1697. [DOI] [PubMed] [Google Scholar]
- 32.Bhaskar A, et al. Reengineering redox sensitive GFP to measure mycothiol redox potential of Mycobacterium tuberculosis during infection. PLoS Pathog. 2014;10(1):e1003902. doi: 10.1371/journal.ppat.1003902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edwards RA, Keller LH, Schifferli DM. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene. 1998;207(2):149–157. doi: 10.1016/s0378-1119(97)00619-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.