Significance
It is important to understand antiviral mechanisms in potential new arbovirus vectors, such as Anopheles mosquitoes, in order to assess risks associated with arbovirus spread. Using an arbovirus naturally transmitted by Anopheles, we find that important immune mechanisms involved in the first bottleneck to Anopheles infection, the midgut, have distinct effects on arbovirus or malaria. This result is, to our knowledge, the first concrete evidence of protection tradeoffs for different human pathogens in a human disease vector, and it suggests that design of genetically immune-modified mosquitoes could result in unexpected outcomes. These results also indicate that different mosquito tissues display distinct antiviral protection that probably imposes divergent selection pressures upon viral replication during different stages of the infection.
Keywords: arbovirus, insect immunity, host–pathogen interactions, innate immunity, malaria
Abstract
Arboviruses are transmitted by mosquitoes and other arthropods to humans and animals. The risk associated with these viruses is increasing worldwide, including new emergence in Europe and the Americas. Anopheline mosquitoes are vectors of human malaria but are believed to transmit one known arbovirus, o’nyong-nyong virus, whereas Aedes mosquitoes transmit many. Anopheles interactions with viruses have been little studied, and the initial antiviral response in the midgut has not been examined. Here, we determine the antiviral immune pathways of the Anopheles gambiae midgut, the initial site of viral infection after an infective blood meal. We compare them with the responses of the post-midgut systemic compartment, which is the site of the subsequent disseminated viral infection. Normal viral infection of the midgut requires bacterial flora and is inhibited by the activities of immune deficiency (Imd), JAK/STAT, and Leu-rich repeat immune factors. We show that the exogenous siRNA pathway, thought of as the canonical mosquito antiviral pathway, plays no detectable role in antiviral defense in the midgut but only protects later in the systemic compartment. These results alter the prevailing antiviral paradigm by describing distinct protective mechanisms in different body compartments and infection stages. Importantly, the presence of the midgut bacterial flora is required for full viral infectivity to Anopheles, in contrast to malaria infection, where the presence of the midgut bacterial flora is required for protection against infection. Thus, the enteric flora controls a reciprocal protection tradeoff in the vector for resistance to different human pathogens.
Arthropod-borne viruses (arboviruses) are a growing burden to human and animal health (1). These viruses are characterized by their alternating cycle of transmission between a mammalian host and an arthropod vector. The most important arboviruses are grouped into the Flaviviridae, Bunyaviridae, Togaviridae, and Reoviridae RNA virus families. Anopheles and Aedes mosquito species are two major vectors of human pathogens. Although their distributions are often sympatric, the range of pathogens transmitted by anopheline and aedine species is thought to be highly divergent. Aedes mosquitoes are the main vector of arboviruses, such as dengue virus (DENV; genus Flavivirus, family Flaviviridae), yellow fever virus (genus Flavivirus, family Flaviviridae), chikungunya (CHIKV, genus Alphavirus, family Togaviridae), and others, but not human malaria. In contrast, Anopheles mosquitoes are the only vectors of human malaria parasites (Plasmodium falciparum, Plasmodium vivax, Plasmodium ovale, and Plasmodium malariae). However, Anopheles and Aedes mosquitoes are each constantly exposed to the alternate pathogens in human blood meals, and the genomic and cellular factors that govern these pathogen restrictions to a particular mosquito genus are not yet understood, including the possibility of vector shifts by a pathogen.
Only one arbovirus is known to be consistently transmitted by Anopheles mosquitoes, the alphavirus o’nyong-nyong (ONNV, genus Alphavirus, family Togaviridae), which is closely related to CHIKV (2). However, the perceived division of labor between Aedes and Anopheles mosquitoes for transmission of arboviruses and Plasmodium, respectively, has led to a relative lack of study about anopheline mosquitoes and arbovirus transmission. In fact, other arboviruses, such as Rift Valley fever virus (RVFV), have been found in different anopheline species when they were examined during recent epidemics (3, 4). RVFV was present in females, males, and larvae, indicating vertical, presumably transovarial transmission, clearly suggesting that Anopheles could contribute to the transmission and maintenance of arboviruses other than ONNV. These observations pose the question of what it would require for other arboviruses to adapt to anopheline mosquitoes as transmission vectors and highlight the importance of studying Anopheles antiviral immunity. Throughout this article, the term Anopheles antiviral response refers to the response to ONNV, which is the only arbovirus that has been studied in Anopheles.
The small RNA-based RNA interference (siRNA) pathway, which has been extensively studied in Drosophila melanogaster (5), is generally thought to be the main cellular antiviral pathway in insects. The first description of siRNA as a mosquito antiviral mechanism was in Anopheles gambiae infected with ONNV (6). The main components of siRNA pathways are conserved between Drosophila and mosquitoes (7), and include the key factors of the siRNA pathway, Dicer-2 (Dcr2) and Argonaute-2 (Ago2), which are crucial in mediating antiviral activities. The classical trigger of this small RNA response in insects is long double-stranded RNA (dsRNA), which is recognized by the RNaseIII-like enzyme Dcr2 and processed into small RNA duplexes 21 nt in length. Two arms of the siRNA pathway are defined based on whether the source of the processed dsRNA is exogenous to the organism (exo-siRNA) or endogenous (endo-siRNA). Viral dsRNA is processed by the exo-siRNA pathway, yielding virus-derived siRNA (viRNA). These viRNAs are loaded into the RNA-induced silencing complex (RISC), containing the “slicer” enzyme Ago2, to produce the antiviral effector complex. The loaded RISC scans for an RNA sequence complementary to the loaded viRNA, and the Ago2 subunit cleaves the recognized viral mRNA.
Functional studies using Aedes aegypti mosquitoes and aedine cell lines have elucidated the role of the siRNA pathway in controlling flavivirus and alphavirus infection of mosquitoes (7–12), and studies with transgenic Aedes mosquitoes have implicated the siRNA pathway as an antiviral mechanism in the midgut against an alphavirus [Sindbis virus (SINV)] and a flavivirus (DENV) (13, 14). However, these latter studies examined the midgut and carcass of mosquitoes only at 7 or 14 d post-blood meal (PBM), which is well after virus dissemination from the midgut, thus analyzing the response in the systemic (i.e., post-midgut) compartment rather than the initial midgut response. Therefore, these results did not address the function of the siRNA pathway in the midgut early following infection but, rather, the ability of this pathway to control viral load once persistent infection was established.
Other immune signaling pathways have also been implicated in the response of Aedes to arboviruses. Both Toll and JAK/STAT pathways participate in the control of DENV infection in A. aegypti (15, 16). When positive regulators of Toll or JAK/STAT were silenced, DENV titers in the mosquito increased, and silencing of negative regulators of the pathways decreased viral load (15, 16). Both DENV and Semliki forest virus (SFV, genus Alphavirus, family Togaviridae) are able to inhibit signaling of these antiviral immune pathways (17, 18).
Studies of immune signaling in An. gambiae have mainly focused on interactions with malaria parasites, and the important pathways and factors have been reviewed recently (19–21). It is well documented that the presence of live midgut bacterial flora strongly inhibits Plasmodium infection (22–27). However, little is known about virus interactions with Anopheles. In previous analyses of the An. gambiae immune response to arbovirus infection, ONNV was delivered by intrathoracic injection (6, 28) or by feeding mosquitoes on an infectious blood meal (29). Blood feeding is the natural route of virus entry, but the latter study analyzed the mosquito response only at 14 d PBM, which consequently, as mentioned above, analyzed the systemic response to a disseminated infection and not the initial midgut response. The midgut environment is the initial mosquito tissue exposed to arbovirus infection and is an important key to establishment of subsequent immune responses. To our knowledge, the early events of antiviral immunity in the mosquito midgut after viral infection by natural blood feeding, as distinct from the later events occurring after viral dissemination, have not been previously examined.
Here, we have elucidated the immune pathways involved in the early control of arbovirus in the Anopheles midgut compartment and distinguished them from the pathways controlling viral load in the systemic compartment during the later stages of infection after dissemination from the midgut. Interplay between the immune deficiency (Imd) and JAK/STAT pathways, soluble effectors, and midgut flora controls the capacity of the virus to infect and replicate in the midgut. We show that the exo-siRNA pathway, generally considered as the canonical mosquito antiviral mechanism, has no detectable role in the initial midgut infection. However, we confirm that exo-siRNA does indeed control viral load once the virus has escaped from the midgut into the systemic compartment. Our results also suggest a Toll-mediated antiviral activity in the systemic compartment, which might have been previously masked by a potential Toll-inhibitory mechanism of ONNV. These results shed light on the immune pathways of the mosquito midgut after a blood meal, which is the initial critical infection bottleneck for multiple pathogens.
Results
ONNV Is Restricted to the Mosquito Midgut at Day 3 PBM.
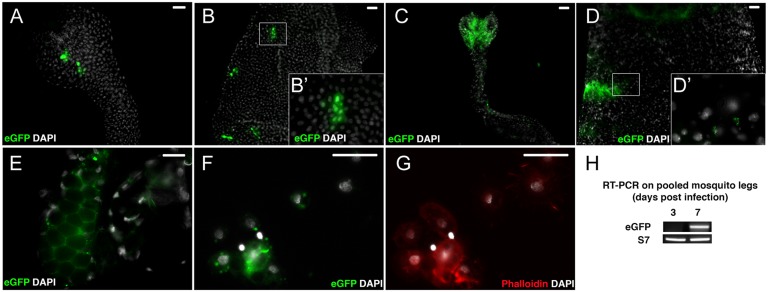
We first determined the optimal time point PBM for measurement of midgut ONNV infection, when the virus is still limited to the digestive tube. We fed mosquitoes an infective blood meal containing ONNV expressing the enhanced green fluorescent protein, EGFP (ONN-EGFP) (30), and used EGFP fluorescence to assess the presence of replicating virus in different tissues at 1, 3, and 7 d PBM (Fig. 1). Throughout, we used the lowest passage of the stable 5′ double-subgenomic ONNV-EGFP (30, 31) to minimize potential loss of EGFP fluorescence that could occur from using virus with a 3′ insertion. Tissues from control mosquitoes after a noninfective normal blood meal displayed only low nonspecific background. At 1 d PBM (Fig. 1 A and B) and 3 d PBM (Fig. 1 C and D), EGFP signal was restricted to the midgut. By 7 d PBM, we observed virus in salivary glands (Fig. 1E) and in circulating cells perfused along with hemolymph, presumably hemocytes (Fig. 1 F and G). We did not detect fluorescent signal in the fat body of the insect, which is a secretory and immune organ, suggesting that the virus is not able to disseminate to the fat body after infection by a blood meal, as previously observed in ONNV-EGFP blood-feeding experiments (30). To confirm systemic viral dissemination, we extracted RNA from the legs of 10 mosquitoes at 3 and 7 d, and tested it for the presence of the viral genome by RT-PCR. A viral signal was not present in legs at 3 d but was detected at 7 d (Fig. 1H), consistent with the microscopic results. We controlled for the possibility that viruses that lost the EGFP cassette could disseminate more rapidly but be undetectable by fluorescence by repeating the experiment using wild-type (WT) virus (Fig. S1), which confirmed the same absence of viral dissemination from the midgut at 3 d as observed by microscopy. Thus, at 1 and 3 d PBM, the ONNV infection of An. gambiae is detectable exclusively in the mosquito digestive tube, although infection is disseminated beyond the midgut into the systemic compartment by 7 d PBM. The 3-d time point was chosen for subsequent experiments because unbound virus particles are still present in blood and debris in the midgut at 1–2 d PBM, before digestion and defecation eliminate most of them.
Fig. 1.
ONNV replication is restricted to midgut cells in An. gambiae 3 d after an infective blood meal. Labeled ONN-EGFP virus was fed to mosquitoes in a blood meal, and tissues were dissected and examined for EGFP fluorescence to detect cellular sites of viral replication. Anterior midgut (A) and posterior midgut (B) 1 d PBM are shown (Scale bar: 50 μm.) Anterior midgut (C) and posterior midgut (D) 3 d PBM are shown. (Scale bar: 50 μm. B′ and D′ enlarge boxed regions.) (E) Salivary glands 7 d PBM. (Scale bar: 20 μm.) (F and G) Circulating hemocyte cells perfused from mosquitoes 7 d PBM. (Scale bar: 20 μm.) (H) RT-PCR detection of viral RNA extracted from mosquito legs shows an absence of detectable virus in systemic circulation 3 d PBM and positive detection of circulating virus 7 d PBM. eGFP, RT-PCR detection of EGFP target in ONN-EGFP viral genome; S7, transcripts of mosquito ribosomal protein S7 as control.
siRNA Is Not the First Line of Natural Antiviral Defense in the Vector.
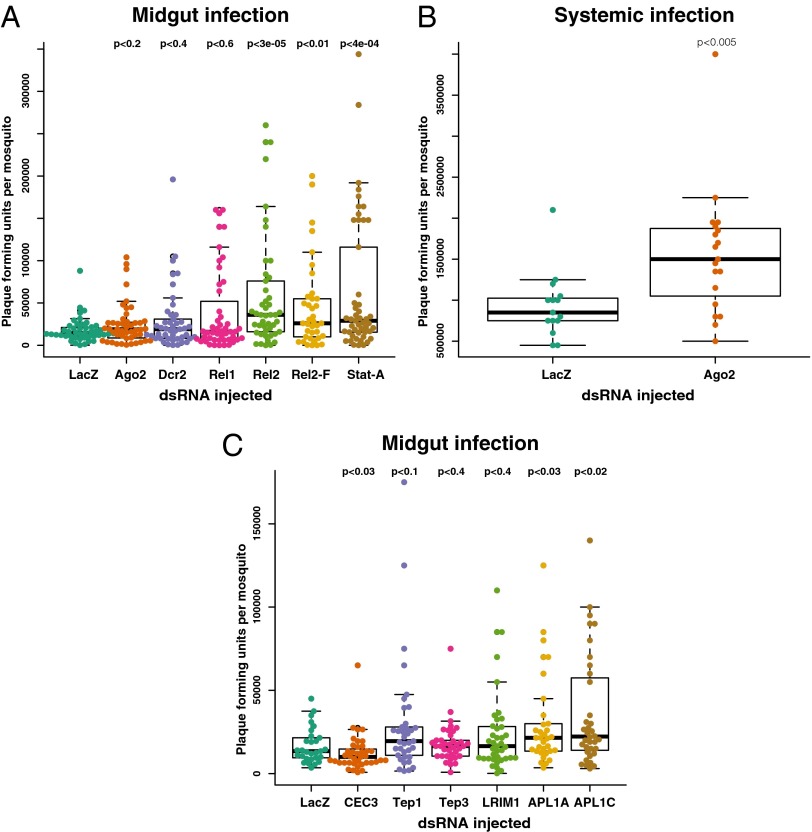
Previous studies reported that the siRNA pathway of An. gambiae, particularly the exo-siRNA pathway, controls systemic immunity against ONNV following virus injection into the hemocoel (6, 28). Direct hemocoel injection bypasses the midgut barrier and the blood-meal milieu, which is the natural route of mosquito exposure to arboviruses. We sought to characterize the natural protective antiviral mechanisms of the Anopheles midgut after an infective blood meal. Thus, we first assessed the contribution of the exo-siRNA pathway to midgut antiviral immunity by silencing two critical pathway components, Ago2 and Dcr2, followed by challenge with WT ONNV and measurement of viral loads in all mosquitoes that took a blood meal (Fig. 2A). We confirmed that dsRNA injection efficiently silenced the target genes, both in the whole mosquito as well as in midguts dissected from dsRNA-treated mosquitoes (Fig. S2 A and E). In complete contrast to the results after virus and dsRNA coinjection into the hemocoel (6, 28) (confirmed in Fig. 2B), dsRNA directed against Ago2 and Dcr2 injected 2 d before infection had no effect on viral titers in the midgut after blood-meal infection (Fig. 2A). To control for the possibility that measuring infectious viral particles might not detect an effect of pathway silencing upon viral replication, we also measured viral mRNA levels in the same samples (Fig. S3), which yielded the same result as viral titers.
Fig. 2.
Imd and JAK/STAT pathways are required for midgut protection against ONNV infection of An. gambiae after an infective blood meal. Panels of genes involved in immune function were assayed for influence upon ONNV infection (all mosquitoes that took blood are represented in the figures, with an infection prevalence of 100%). Genes were silenced by treatment with the dsRNA indicated on the x axis. Mosquitoes were challenged by blood-feeding with ONNV (A and C) or by injection with ONNV (B). Three days after infection, mosquitoes were individually assessed for viral titers by plaque assay. (A) Silencing of key intracellular immune signaling factors shows strong antiviral protection in the midgut mediated by the Imd pathway (Rel2) and JAK/STAT pathway (Stat-A). No detectable midgut protection is mediated by the siRNA pathway (Ago2, Dcr2) or the Toll pathway (Rel1). (B) As a positive control, the same reagents detect antiviral protection by the siRNA pathway (Ago2) against injected ONNV in the systemic hemocoel compartment. (C) Silencing of extracellular immune factors reveals an antiviral function for the LRR proteins APL1A and APL1C, and an absence of antiviral activity for Tep1, Tep3, and LRIM1, whereas loss of CEC3 function depressed the viral titer. LacZ, irrelevant control dsRNA targeting the E. coli lacZ gene encoding β-galactosidase.
Additional technical and biological controls strengthened the conclusion that the exo-siRNA pathway is not an antiviral protective mechanism in Anopheles after natural infection by means of a blood meal. First, we tested our dsRNA reagents in an in vitro cell infection model, and as previously reported, silencing of Ago2 increased viral replication (Fig. S4). Next, coinjection of dsAgo2 along with virus into the hemocoel, bypassing the midgut, caused elevated ONNV titers (Fig. 2B), which is also consistent with previously reported findings of others (6, 28). These results indicate that the exo-siRNA pathway plays a role in controlling disseminated viral infection in the systemic hemocoel compartment but not in the midgut during the initial period of infection.
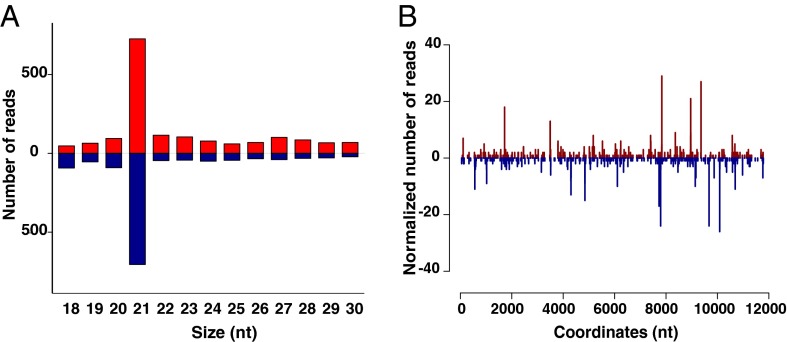
Finally, we tested for functional activity of the exo-siRNA pathway in the midgut after ONNV infection by means of a blood meal. We sequenced small RNAs in ONNV-infected mosquitoes at 3 d PBM and detected the classical 21-nt viRNA peak, a hallmark of siRNA induction (Fig. 3A). When aligned to the ONNV genome, the 20- to 22-nt viRNAs produced a dicing pattern in both sense and antisense orientations (Fig. 3B). Reads from uninfected samples were aligned using the same methods and, as expected, showed <10 reads aligning to the viral genome. These results indicate that the exo-siRNA pathway is active and functional in the midgut, but without protective antiviral function (as determined by virus titers and RNA levels; Fig. 2A and Fig. S3) at a time point when the infection is confined to the midgut. Thus, the exo-siRNA pathway does not mount a detectable natural barrier to ONNV infection in the midgut, although it is implicated in antiviral protection in the systemic compartment after dissemination from the midgut. Our results leave open the possibility that the reaction products of midgut exo-siRNA activity might generate an early surveillance signal that influences later responses in the systemic compartment.
Fig. 3.
Exogenous siRNA pathway is functionally active but not antiviral in the An. gambiae midgut. (A) Size distribution of viRNAs in An. gambiae mosquitoes 3 d after an ONNV-infective blood meal, when viral replication is restricted to the midgut. Positive numbers on the y axis indicate the frequency of viRNAs mapping to the ONNV genome (red bars), and negative numbers on the y axis indicate the frequency of viRNAs mapping to the ONNV antigenome (blue bars). (B) Frequency distribution of loci in the ONNV genome generating 21-nt viRNA (5′ to 3′; viRNAs starting with the 5′ terminal nucleotide) or antigenome (3′ to 5′; viRNAs starting with the 3′ terminal nucleotide), starting at nucleotide 1 on the x axis. Numbers on the y axis correspond to the frequencies of viRNAs mapping to the ONNV genome (positive) or antigenome (negative). Blue peaks indicate loci of viRNAs mapping to the ONNV genome, and red peaks indicate loci of viRNAs mapping to the ONNV antigenome. Results shown are representative of two replicates.
Immune Signaling and Effector Activity Create a Midgut Antiviral Defense Barrier.
We then tested involvement of innate immune pathways during midgut infection by silencing key intracellular immune signaling nodes, followed by virus challenge (Fig. 2A). The JAK/STAT pathway transcription factor Stat-A positively regulates the Stat-B transcription factor, thus creating a positive amplification loop upon activation of the pathway. Silencing of Stat-A is sufficient to inhibit both Stat-A and Stat-B (16, 32). Inhibition of the JAK/STAT pathway by silencing Stat-A caused an increased viral titer (Fig. 2A). The Imd pathway signals to two isoforms of the NF-κB transcription factor Rel2, the long isoform Rel2-F with a C-terminal ankyrin domain and the short Rel2-S isoform, which is identical but lacking the ankyrin domain (33). Silencing of both isoforms by targeting a shared region [double-stranded Rel2 (dsRel2)] or specifically silencing only the long isoform (dsRel2-F) both yielded increased susceptibility to viral infection (Fig. 2A). Thus, the JAK/STAT and Imd pathways are protective against ONNV infection of the midgut, and Imd protection requires at least the long Rel2-F isoform. Conversely, ablating Toll activity by silencing the NF-κB transcription factor, Rel1, had no effect on ONNV midgut infection (Fig. 2A).
We next queried a panel of known extracellular immune factors for their influence on midgut protection after challenge with an ONNV-infective blood meal (Fig. 2C): the antimicrobial peptide Cecropin 3 (CEC3); the complement analogs Tep1 and Tep3; and their immune complex partner subunits LRIM1, APL1A, and APL1C (34–36). Silencing of CEC3 transcript by treatment with dsCEC3 caused an inhibition of ONNV titers following blood-meal infection. Silencing of the genes for Leu-rich repeat (LRR) proteins APL1A and APL1C allowed significantly higher viral titers, indicating that these factors are protective against ONNV (Fig. 2C). Surprisingly, silencing the known APL1C binding partners that play a role in Plasmodium protection (LRIM1, Tep1, or Tep3) did not show any effect on ONNV titers in mosquitoes (Fig. 2C). This result indicates that the anti-Plasmodium ternary immune complex composed of APL1C, LRIM1, and TEP1 (34, 35) is not functional against virus infection and suggests that APL1C interacts in distinct immune complexes with other unknown subunit proteins to protect against viruses.
Viral Infection Suppresses Immune Activation of An. gambiae Cells in Vitro.
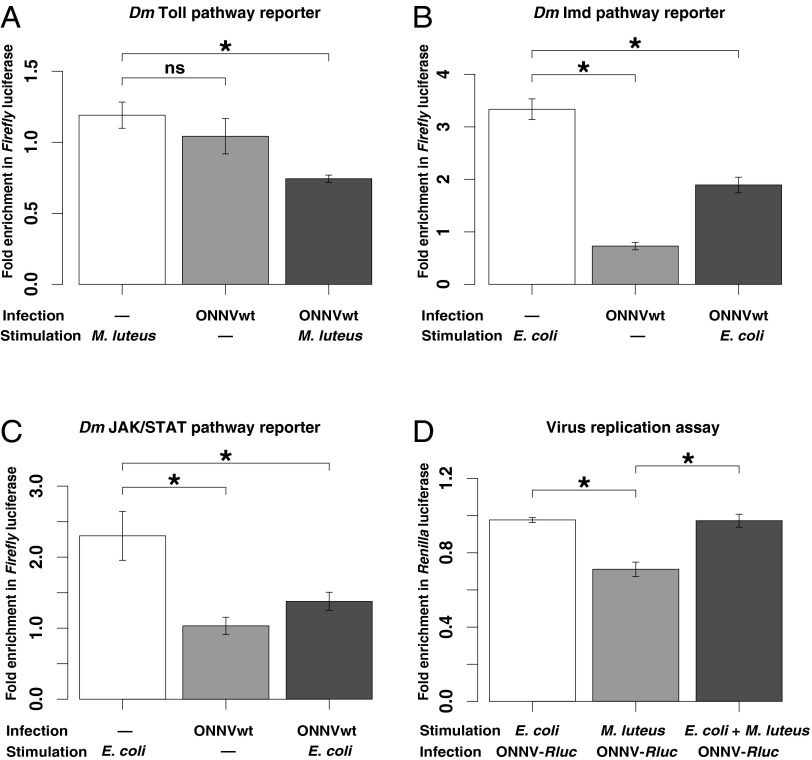
We queried viral activation of systemic immunity, and the effect of systemic immunity on viral replication, using an in vitro model of the systemic hemocoel compartment. The hemocoel is the open body cavity of insects, and it is the site of systemic immunity due, in large part, to the presence of immune hemocytes (37, 38). For the systemic hemocoel model, we used the An. gambiae hemocyte-like cell line 4A3A (39). We transfected cells with reporter plasmids coding for the Firefly luciferase reporter under control of Drosophila promoters specific for the Toll, Imd, and JAK/STAT pathways (18), and then infected the cells with ONNV. Reporter assays with truncated or multimerized promoter elements have limits, and there are differences between Drosophila and a number of mosquito species in activation of immune responses. Nonetheless, there are also strong similarities in Toll, Imd, and JAK/STAT pathways between Drosophila and Anopheles (32, 40, 41). As with any in vitro model, the results form hypotheses to be verified in vivo. Activation of the pathways was measured as the fold induction of reporter signal compared with unstimulated control cells. The activity of all three pathways was significantly inhibited by virus infection (Fig. 4 A–C, right histogram bar in each panel) compared with the activity after pathway elicitor alone (Fig. 4 A–C, left histogram bars). Of these pathways, ONNV displayed the strongest inhibitory activity against Toll, because elicitor-stimulated cells challenged with virus expressed even less Toll activity than the virus-infected control cells (Fig. 4A, compare middle and right histogram bars). Cells activated for Imd or JAK/STAT and challenged with virus displayed equivalent or higher pathway activity than only virus-infected controls (Fig. 4A, middle vs. right histogram bar), although pathway activity was still significantly inhibited by virus compared with elicitor alone (Fig. 4A, left vs. right histogram bar).
Fig. 4.
ONNV inhibits activation of multiple immune pathways in an in vitro model of the An. gambiae systemic compartment, but only Toll inhibits systemic viral replication. (A–C) Suppression of immune pathway activation by ONNV infection was tested using a Firefly luciferase reporter driven by D. melanogaster (Dm) immune pathway-specific promoters transfected into An. gambiae 4A3A hemocyte-like cells. A reporter plasmid responding to activation of a Drosophila Toll pathway promoter (A), activation of a Drosophila Imd pathway promoter (B), or activation of a Drosophila JAK/STAT pathway promoter (C) was cotransfected with an internal control constitutively expressing RLuc, and immediately infected with ONNV [multiplicity of infection (MOI) of 10] or mock-infected. Immune pathways were then stimulated by adding, as indicated, heat-inactivated E. coli (JAK/STAT and Imd stimulation) or M. luteus (Toll stimulation) immediately postinfection. Luciferase activity was determined at 18 h postinjection. Each bar represents the mean fold induction of Firefly luciferase signal (normalized against the RLuc internal control), compared with a mock-infected and mock-stimulated control (defined as 1.0), and error bars indicate the SD. (D) Efficiency of viral replication was measured in a cellular environment specifically activated for immune pathway Imd and JAK/STAT (bar 1, E. coli); Toll (bar 2, M. luteus); or Toll, Imd, and JAK/STAT (bar 3, E. coli and M. luteus), and then infected (MOI of 1) with recombinant ONN-RLuc. Rluc activity was measured at 12 h postinfection. Each bar represents the level of viral replication relative to mock-stimulated control (defined as 1.0), and error bars indicate the SD. Only Toll-activated cells suppressed viral replication relative to the control. *P < 0.05; ns, not significant.
The generalized immune inhibition of activation potential by ONNV in the in vitro hemocoel model is consistent with previous observations that the same pathways display little effect upon viral load in mosquitoes infected with ONNV by hemocoel injection (6, 28), because virus inhibition of pathway activation might subvert otherwise protective pathways. To test directly whether these pathways can, in fact, control viral replication, we prestimulated cells with elicitor before ONNV infection and then measured the level of virus replication in the immune-activated cellular environment (Fig. 4D). To facilitate measurement of virus replication, we constructed an ONN-Renilla luciferase (RLuc) infectious clone from the ONN-EGFP infectious clone, in which EGFP is replaced by RLuc. Luciferase activity thus reflects viral genome replication and translation in the same way as EGFP in the parent clone; however, unlike EGFP, RLuc activity allows simple quantitative measurement. Of the three immune pathways, only activated Toll inhibited viral replication. Mechanistically, the functional antiviral activity of Toll may explain the adaptive significance of the strong virus inhibition of Toll activation observed above. Simultaneous activation of all three pathways (Imd, JAK/STAT, and Toll) abolished the protective effect produced by stimulation of Toll alone. This result suggests that the antiviral effect of Toll is context-dependent and may depend on the correct temporally staged activation of different pathways, such that concurrent activation generates noise rather than protection.
Live Midgut Flora Is Required for Full Viral Infectivity to An. gambiae.
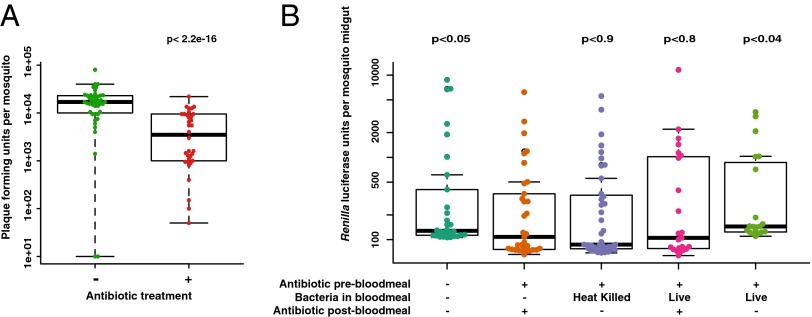
Because Imd and JAK/STAT activity may be important to manage balance of the midgut flora in the midgut of Drosophila (42) and Anopheles (22–27), and because altering the level of the antimicrobial effector CEC3 influenced viral loads, we tested whether the mosquito midgut flora might influence ONNV infection. Mosquitoes were treated with antibiotics to reduce the general midgut flora population before an infectious blood meal with ONNV. Antibiotic treatment was either continued or discontinued after the blood meal, as specified in Fig. 5. Antibiotic feeding reduced bacterial abundance in the midgut by at least fivefold at the time of the blood meal, detected by comparing colony-forming units in treated and untreated midguts.
Fig. 5.
Presence of live midgut bacterial flora is required for full viral infectivity of ONNV to An. gambiae. All mosquitoes that took a blood meal are represented in the figures. (A) Mosquitoes were orally treated from adult emergence with (+) or without (−) antibiotic for the duration of the experiment, and were challenged with virus by feeding an ONNV-infective blood meal. ONNV titers were assessed by plaque assay 3 d after the blood meal. Treatment with antibiotic reduced levels of bacterial flora greater than fivefold and strongly inhibited viral infectivity to mosquitoes. (B) Parameters of bacterial flora influence upon viral infection were dissected by the following treatment grid. Mosquitoes were orally treated (+) or not treated (−) with antibiotics before a blood meal (as indicated in the figure), and were infected orally by a blood meal containing ONN-RLuc. At 3 d postinfection, viral load was measured by RLuc quantitation in individual midguts. Bars 1 and 2 reproduce the nontreated and antibiotic-treated samples in A, but are shown here with viral quantitation by measurement of luciferase activity. For bar 3, the viral blood meal was supplemented with heat-killed culture of midgut bacteria, not followed by (−) antibiotic PBM. For bar 4, the viral blood meal was supplemented with live culture of midgut bacteria, but was followed by (+) antibiotic PBM. None of the treatments that eliminate live bacteria from the midgut restore virus infectivity to the mosquito. Only bar 5, which supplements the viral blood meal with live culture of midgut bacteria not followed by (−) antibiotic, complements the loss of viral infectivity caused by the initial antibiotic treatment. All P values were calculated using the second bar as the reference.
Reduction of the midgut flora led to a significant concomitant reduction of ONNV infection in mosquitoes, indicating a positive cooperative role of the bacterial flora in promoting viral infection of the midgut (Fig. 5A). Reduction of midgut flora also leads to a significant increase in Anopheles infection prevalence with P. falciparum, as has been extensively documented (22–27), and we confirmed the same phenomenon in the specific An. gambiae mosquito colony used in the current report (Fig. S5). As expected, treatment with antibiotics does not have a direct antiviral effect in mosquito cells (Fig. S6). To distinguish between the requirement for bacterial molecules or live bacteria, we provisioned antibiotic-treated mosquitoes with midgut bacteria followed by challenge with infectious ONNV. The bacteria, cultured from mosquito midguts, were either heat-inactivated or live, and they were used to supplement the ONN-RLuc–containing blood meal. The provision of heat-killed bacteria with the ONN-RLuc infective blood meal did not restore the viral infectivity lost due to antibiotic treatment, nor did the provision of live bacteria if the blood meal was also followed by antibiotic treatment (Fig. 5B). Only provision of live bacteria without subsequent antibiotic treatment produced significant restoration of ONN-RLuc infectivity (Fig. 5B). Thus, the full infectivity of ONN-RLuc for the mosquito by natural blood-meal exposure requires the presence of live bacteria in the vector midgut.
Discussion
Arboviruses cross multiple physical, physiological, and immune barriers in mosquitoes to traverse from the blood meal to the salivary glands. The midgut environment, including the cellular epithelium, is the first obstacle. Midgut infection is a determinative factor in vectorial capacity because an efficient midgut blockade produces sterile immunity, thereby preventing subsequent virus dissemination and transmission. The goal of the current study was to elucidate the initial immune mechanisms controlling arbovirus infection in An. gambiae and, in particular, the contribution of the midgut compartment to limiting and controlling infection by the natural blood-meal route of entry. We compared midgut antiviral protection with responses taking place only in the post-midgut systemic compartment and found an array of defenses unique to each compartment.
The antiviral activity of the mosquito midgut early after the blood meal, when the infection is still restricted to the midgut, has not been previously examined to our knowledge. The digestive tube is the site of the first encounter between the virus and vector, and it is a key bottleneck for transmission. We find that the exo-siRNA pathway does not influence the success of midgut infection during this initial period before virus dissemination from the midgut. Thus, siRNA activity is not a component of first-line antiviral defense, at least in Anopheles. The question has not been examined in Aedes. Other work, discussed below, has reported on the antiviral response to disseminated infections in the systemic compartment and has indicated that the exo-siRNA pathway is the main antiviral pathway. Our results agree with the importance of siRNA-based protection after viral dissemination from the midgut but highlight the crucial distinction with protective mechanisms before dissemination from the midgut.
In many previous studies of antiviral immunity, mosquitoes were infected by intrathoracic injection of virus, which bypasses the normal midgut route of virus entry (6, 8, 28, 43). Alternately, SINV expressing the B2 protein of Flock House virus, which inhibits the activity of small RNA pathways, was injected into Aedes mosquitoes (44), or Aedes mosquitoes were modified to express B2 or an inverse repeat against Dcr2, followed by SINV challenge (13, 14). Viral levels in the midgut and carcass were compared at 7 or 14 d postinfection. These siRNA-suppressed mosquitoes displayed higher SINV infection levels, although the earliest time point tested, 1 wk PBM, was well after virus dissemination from the midgut. Thus, these experiments uniformly indicated that siRNA pathways control viral load in later stages of infection, but none of the studies queried early immune events in the midgut compartment.
In our experimental conditions, all tested mosquitoes were positive for virus at 3 d PBM, and this fraction decreased at later infection stages. Thus, the initial infection rate of mosquitoes before dissemination from the midgut was higher than after dissemination. We do not know the relative contributions of the predissemination midgut protective mechanisms that we describe, or the postdissemination siRNA pathway, to the decrease of infection prevalence. The viRNAs that are produced in the midgut compartment could potentiate or influence systemic antiviral immunity at later stages of infection by distant signaling (45, 46). Therefore, the balance between the largely distinct and nonoverlapping suites of pre- and postdissemination protective mechanisms in controlling the establishment or resolution of infection remains to be addressed.
We detected ONNV-induced exo-siRNA activity in the midgut, yielding cleaved viRNAs from the ONNV genome, but this activity nevertheless does not reduce viral load or viral RNA levels (Fig. 2A and Fig. S3). Because the exo-siRNA pathway dices viral RNA, it has been assumed that it must be an effector mechanism for virus elimination, but effector function of exo-siRNA has not been proved. Our data indicate that exo-siRNA pathway direct activity upon viral RNA in the midgut does not detectably reduce virus levels. These results may suggest a different function for exo-siRNA. Mammalian antigen-presenting cells also destroy pathogens, but their main function is pathogen sensing and immune signaling, not quantitative pathogen reduction. By analogy, the main function of viral dicing by exo-siRNA, at least in the midgut, may be surveillance, virus genome “antigen” detection, and immune signaling rather than direct pathogen reduction.
We identified a previously unrecognized role for the Imd pathway in mosquito antiviral immunity. A previous study of An. gambiae needle-infected with ONNV found no role for the Imd pathway in the systemic hemocoel compartment (28). In contrast, our current results show a strong midgut protective effect of the Imd pathway against ONNV after blood-meal infection, mediated, at least in part, by the Rel2-F long isoform of the transcription factor Rel2. Activity of the short Rel2-S isoform is required for protection of An. gambiae against P. falciparum infection (36, 47). Thus, Imd-mediated antiviral activity in An. gambiae is functionally different from the Imd anti-P. falciparum response, where the Rel2-F isoform has no effect (36), suggesting that Imd signaling is diversified for response to distinct classes of pathogens (Fig. 6). Studies in Drosophila (48, 49) or mosquito cells (18) have suggested a role for Imd in controlling alphavirus infection. We similarly identified a previously unrecognized role for the JAK/STAT pathway in midgut antiviral immunity. Stat-A has no effect on virus after needle infection in the systemic compartment of the mosquito (28), whereas we find an important role for this pathway in controlling viral infection in midgut tissues after blood-meal infection.
Fig. 6.
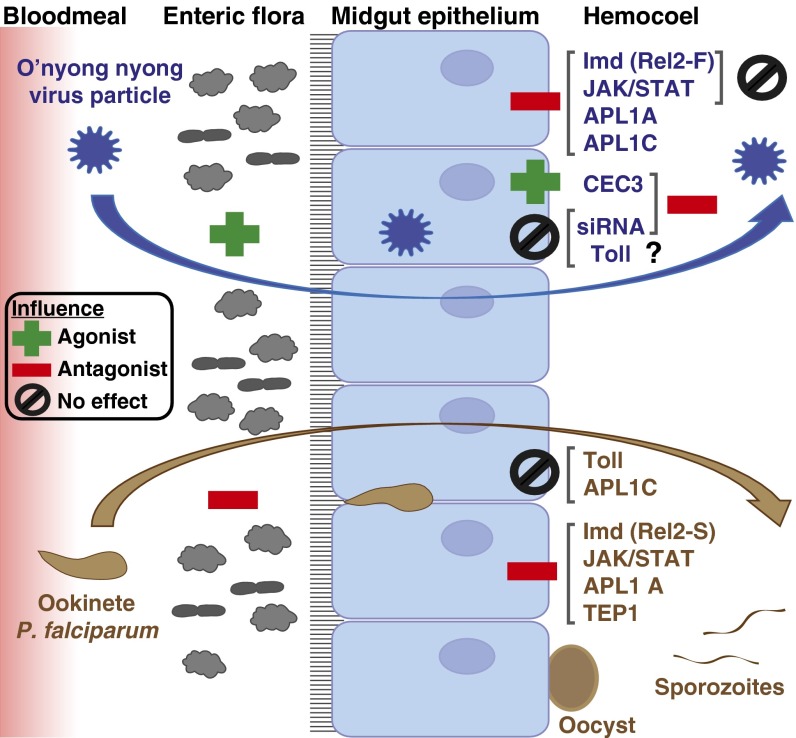
Influence of An. gambiae host-defense mechanisms upon blood-meal infection by ONNV and the human malaria parasite P. falciparum. Largely distinct suites of protective mechanisms operate within the midgut and hemocoel tissue compartments. (Left) Midgut entry of ONNV (blue) and P. falciparum (brown) within the blood meal is depicted. The enteric flora displays an opposite effect on infection by the two pathogens, facilitating infection by ONNV (acting as an agonist) and diminishing infection by P. falciparum (acting as an antagonist). (Middle) In the midgut epithelium, the JAK/STAT and Imd pathways limit infection by both pathogens, although the protective Imd response to ONNV or P. falciparum is mediated by distinct isoforms of the transcription factor Rel2 (Results). (Right) However, JAK/STAT and Imd have no detectable influence on viral load in the systemic hemocoel compartment when the midgut is bypassed by virus injection (28). The siRNA pathway displays the inverse pattern, controlling viral infection in the hemocoel but with no influence over virus in the midgut. Little is known of the systemic hemocoel response to P. falciparum sporozoites. The antimicrobial effector CEC3 cooperates with viral infectivity in the midgut after blood exposure but was shown to protect against virus in the hemocoel after needle infection (28). Two LRR protein immune factors, APL1A and APL1C, control ONNV infection in the midgut, whereas only APL1A protects against P. falciparum and APL1C has no effect on this parasite (36). The mosquito complement protein TEP1, a subunit partner with APL1C in a functional immune complex, protects against P. falciparum (65) but has no effect on ONNV, which points to the likely existence of other unknown immune subunits interacting with APL1 proteins in distinct functional complexes that exhibit antiviral activity. The Toll pathway is not involved in either anti-ONNV or anti-P. falciparum midgut immunity, although Toll is involved in midgut protection against the rodent malaria parasite anti-Plasmodium berghei (36) and displays anti-ONNV activity in Anopheles cell culture (Results).
Our results show that the presence of live flora is necessary for full midgut infectivity of ONNV, in contrast to Plasmodium, where the presence of midgut flora is required for normal immune protection against infection of the Anopheles midgut (22–27) (Fig. S5). The only other arbovirus tested for influence of mosquito midgut flora was the flavivirus DENV in A. aegypti, where clearance of the midgut flora resulted in twofold higher viral titers (15). The mechanism by which bacteria exert an agonist effect for ONNV infection in Anopheles is not clear. If the effect is due to basal immune homeostasis maintained by the midgut flora, then the same state of protective readiness that blocks Plasmodium midgut infection subverts antiviral protection (Fig. 6). Alternatively, bacterially derived molecules can serve as a molecular adapter for virus binding and invasion of host cells, or for evasion of host immunity (50, 51). However, a molecular adapter mechanism may be less likely in our system, because newly dead bacteria (i.e., heat-killed or live bacteria in the blood meal followed by postfeeding antibiotics) did not complement the loss of ONNV infectivity. Only the presence of currently live bacteria (i.e., live bacteria not followed by antibiotics), cultivable in aerobic LB conditions, restored viral infectivity. These findings underline the complexity of the mosquito midgut biotic environment and its potentially conflicting influence upon different blood-borne pathogens.
We find that Toll activity does not influence ONNV midgut infection in An. gambiae exposed to virus by means of a blood meal (Fig. 2), and in previous work, Toll was not involved in protection from ONNV in the systemic compartment (28). However, surprisingly, in cell culture experiments, we find that the strongest inhibitory effect upon host immune signaling by the virus is exerted upon the Toll pathway and that stimulation of the Toll pathway by Micrococcus luteus is the only inducer capable of inhibiting viral replication (Fig. 4). These results suggest that the Toll pathway possesses antiviral activity and that ONNV might possess a mechanism allowing specific Toll pathway inhibition. In A. aegypti, Toll and JAK/STAT are both key anti-DENV immune pathways (15, 16, 52). In the case of the related alphaviruses SINV (family Togaviridae) and SFV, in Drosophila or A. aegypti, Toll was not involved in antiviral protection (18, 48). Thus, comparative studies indicate that there are diverse strategies of infection and host defense, although deeper examination could reveal shared themes, perhaps including an Alphavirus Toll-inhibition mechanism.
We show that the LRR proteins APL1A and APL1C play a role in midgut antiviral defense against virus infection. The role of these molecules as protective factors against Plasmodium infection is well documented (34, 36, 53–55). APL1C is a subunit, along with LRIM1 and Tep1, in a ternary immune complex with activity against rodent malaria parasites (34, 35, 56), whereas its paralog APL1A, but not APL1C, protects against P. falciparum infection (36). Surprisingly, we find that the LRIM1 and Tep1 subunits of the known ternary immune complex are not involved in antiviral activity, suggesting that APL1A and APL1C must interact in other functional immune complexes that are relatively specific to virus infection. Further work will be required to determine the recognized viral motif and whether the site of interaction is on the virus particle or within infected cells.
The current study expands our understanding of the functions and diversity of Anopheles antiviral responses by focusing on the role of the midgut barrier in the initial infection phase, which has not been previously examined. We highlight the role of pathways that we newly describe as antiviral and reevaluate the importance and localization of previously described main antiviral pathways, such as RNAi. This understanding of antiviral midgut immunity, and the interplay between the different pathways and different pathogens in the African malaria vector, is important in assessment of potential risks linked with the development of immune-modified mosquitoes for control of pathogen transmission. The apparent protection tradeoff in the vector between ONNV and malaria infection raises the possibility of creating a better vector for arboviruses at the same time as decreasing vectorial capacity for malaria, and the potential for human-provoked outbreaks, if multiple pathogen interactions with mosquito immunity are not carefully considered.
Experimental Procedures
Mosquitoes.
The An. gambiae Ngousso strain was originally initiated with mosquitoes collected in Cameroon in January 2006 and belongs to the M molecular and Forest chromosomal forms (57). Mosquitoes were reared under standard conditions, as previously described (36), in insectaries of the Center for Production and Infection of Anopheles core facility of the Institut Pasteur.
Viruses and Mosquito Infection.
WT ONNV strain SG650 and an ONN-EGFP infectious clone (6) were kindly provided by Brian Foy (Colorado State University, Fort Collins, CO). ONNV SG650 was first isolated from human serum in Uganda in 1996. Aliquots of virus received had been passed seven times in BHK-21 cells and then once in Aedes albopictus C6/36 cells. Stock virus was then amplified once more in C6/36 cells to produce stocks for mosquito infections or in An. gambiae 4A3A cells before in vitro experiments. ONN-RLuc was constructed on the ONN-EGFP backbone by replacing EGFP with the RLuc sequence between the AscI and PacI restriction sites. Infectious clone cDNA was linearized by NotI, and viral RNA was transcribed using T7 RNA polymerase (New England Biolabs), m7(5′)ppp(5′)A-Cap Analog (New England Biolabs), and rNTPs (Promega). Transcribed RNA was electroporated into BHK-21 cells. Virus was recovered after 72 h and passaged once on 4A3A cells to increase titers.
Mosquitoes were infected by feeding on an infectious blood meal containing viral titers at the high end of natural exposure, as found at the peak of clinical viremia (58). These conditions ensure that all mosquitoes are positive for virus infection at 3 d PBM, which augments experimental and statistical power. Viral challenge with ONNV was done 2 d after dsRNA treatment. Female mosquitoes deprived of sugar for 12 h were allowed to feed for 15 min through a Hemotek membrane (Hemotek) covering a glass feeder containing the blood/virus mixture maintained at 37 °C. The infectious blood meal was composed of a virus suspension diluted (1:3) in washed rabbit erythrocytes from arterial blood and resuspended at 50% (vol/vol) in dialyzed rabbit serum (R4505; Sigma). ATP was added to a final concentration of 5 μM. Fully engorged females were transferred to small cardboard containers and maintained with 10% (wt/vol) sucrose at 28 ± 1 °C until needed. The final blood-meal titer fed to mosquitoes was between 1–3 × 107 pfu/mL.
For Fig. 2B, mosquitoes were infected with ONNV by intrathoracic injection, as described (6, 28), without a blood meal, thus bypassing the midgut entirely and directly infecting the systemic hemocoel compartment. Mosquitoes were injected with ≈8,750 plaque-forming units of WT ONNV per mosquito in a volume of 100 nL. Injection titer was assessed by plaque assay on five individual mosquitoes harvested immediately after injection.
Detection of ONN-EGFP in Perfused Cells and Tissues.
Circulating cells from adult female mosquitoes were collected by perfusion. Briefly, mosquitoes were cold-anesthetized, and the last abdominal segment was removed. With a microinjection needle, 1× PBS was injected in the thorax, and the first five drops exiting from the abdomen were collected on a glass slide (10 perfused mosquitoes per well). After 45 min of cell attachment at 27 °C, cells were fixed in 4% (vol/vol) paraformaldehyde (PFA) diluted in PBS for 15 min. After three washes in PBS, cells were incubated overnight at 4 °C in PBS with 1 μg/mL DAPI (Sigma) and 0.33 μM phalloidin 647 nm (New England Biolabs). Cells were then washed three times in PBS and mounted in BrightMount Plus (Abcam). Midguts, salivary glands, and abdomens were dissected from females and fixed overnight at 4 °C in 4% (vol/vol) PFA diluted in PBS-Tween 0.05%. Tissues were then washed three times for 15 min in PBS. Samples were incubated overnight at 4 °C in PBS with 1 μg/mL DAPI, washed three times for 15 min in PBS, and mounted on a glass slide. All steps were carried out in the dark. All images were taken using an Axiovert II fluorescence microscope (Zeiss).
Gene Silencing in Vivo.
dsRNAs were synthesized from PCR amplicons using the T7 Megascript Kit (Ambion) (primer sequences are listed in Table S1). The dsRNA sequences were either previously published or designed using the program E-RNAi (www.dkfz.de/signaling/e-rnai3/evaluation.php). LacZ throughout indicates a control treatment targeting the irrelevant lacZ gene of E. coli, which encodes β-galactosidase. Primers for synthesis of the dsRNA used in the LacZ treatments are indicated in Table S1 as T7-βGal-F and T7-βGal-R. For each targeted gene, 500 ng of dsRNA in a final volume of 70 nL maximum was injected into the thorax of cold-anesthetized 1- to 2-d-old An. gambiae females using a nanoinjector (Nanoject II; Drummond Scientific) and glass capillary needle as previously described (36). For gene silencing in mosquitoes infected by ONNV injection (used in Fig. 2B), mosquitoes were coinjected with 500 ng of dsRNA and ≈8,750 plaque-forming units of WT ONNV per mosquito in a volume of 100 nL.
Gene silencing efficiency was monitored 2–3 d after dsRNA injection with RNA from a pool of five whole mosquitoes collected at the time of the blood meal by RT-quantitative PCR (qPCR) or RT-PCR (Fig. S2). RNA was extracted using TriReagent (Sigma) or TRIzol Reagent (Ambion), followed by Turbo DNase treatment (Ambion). One step RT-qPCR (Power SYBR Green RNA-to-Ct 1 Step Kit; Applied Biosystems) was then performed following the manufacturer’s instructions. Alternatively, RT-PCR was performed with RT enzyme (Moloney murine leukemia virus; Invitrogen) followed by PCR (DreamTaq; Thermo Fischer). Analysis of the expression of transcript relative to ribosomal protein S7 as a reference gene was performed according to the 2−ΔΔCt (cycle threshold) method (59). Primer sequences are indicated in Table S2.
Gene Silencing in Vitro.
For gene silencing in cultured cells (used in Fig. S4), experiments were performed in 96-well plates. A total of 8 × 104 An. gambiae 4A3A cells were seeded and left to adhere overnight. Cells were incubated for 30 min on a rocker with 2.5 μg of dsRNA (prepared as described above) in 50 μL of OptiMEM (Gibco). Fifty microliters of Schneider’s medium with 20% (vol/vol) FBS was then added, and cells were incubated for 12 h. Cells were infected with ONN-RLuc at a multiplicity of infection of 1 for 1 h, followed by a further 12-h incubation in 100 μL of Schneider’s medium complemented with 10% (vol/vol) FBS, 100 units/mL penicillin, and 100 μg/mL streptomycin, containing 2.5 μg of dsRNA. Cells were lysed in 1× passive lysis buffer (Promega), and virus replication was assessed by luciferase assay.
Measurement of Virus Infection.
Virus titers or virus replication in mosquitoes infected with ONNV SG650 or luciferase-expressing virus, respectively, was determined by plaque assay or luciferase activity. All live mosquitoes that took a blood meal were assayed and are represented in the figures (infection prevalence at 3 d was 100%). Briefly, individual mosquitoes were collected 3 d after an infectious blood meal, crushed in 250 μL of DMEM (Gibco), and centrifuged for 5 min at 660 × g before storage at −80 °C until standard plaque assays were performed. Ten-fold serial dilutions of each sample were added in duplicate to confluent monolayers of Vero cells in six-well plates and immobilized using a 1% agarose DMEM containing 2% FBS and 1× Antibiotic-Antimycotic (Gibco). Cells were stained after incubation at 37 °C for 3 d with 4% formaldehyde crystal violet solution. Alternatively, mosquitoes infected with ONN-RLuc were dissected, and individual midguts were collected and lysed in passive lysis buffer before luciferase assay.
Small RNA Sequencing.
Twelve mosquitoes per pool, in each of two replicate pools, were collected 3 d after infection with ONN and homogenized in TriReagent (Sigma). Small RNA fractions (<200 nt) were extracted using Nucleospin miRNA (Machery Nagel) utilizing the TRIzol protocol and submitted to Bioanalyser for quality assessment. Briefly, the 10- to 30-nt RNA fragments were purified from small RNA fractions on 15% urea PAGE (BioRad). Small RNA libraries were then prepared using TruSeq Small RNA Library reagents (Illumina). Libraries were sequenced using an Illumina Hiseq 2000 sequencing system in a multiplexed 51 + 7 bases single read using TruSeq SR cluster kit v3 cBot HS and TruSeq SBS kit v3 HS 50 cycles (both from Illumina). Primary analysis of the sequences was performed with Casava software (v1.7; Illumina). Library preparation and sequencing were performed at the Transcriptomics and Epigenomics core facility of the Institut Pasteur. Analysis was performed on a Galaxy Project (galaxyproject.org) server instance hosted by the Drosophila Genetics and Epigenetics Laboratory (Université Pierre et Marie Curie VI). Data are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession no. E-MTAB-2595.
Cell Lines and Immune Pathway Signaling.
Vero and BHK-21 cells were maintained on DMEM complemented with 10% FBS and 1× penicillin/streptomycin (Gibco) at 37 °C in 5% CO2. A. albopictus C6/36 and An. gambiae 4A3A (39) insect cells were maintained in Schneider’s medium complemented with 10% FBS (Gibco) at 28 °C. For cell signaling experiments (used in Fig. 4 A–C), plasmids p6 × 2DRAF-Luc [multimerized Drosophila STAT-responsive element with a Firefly luciferase reporter (60)], pJL169 (Imd-response Drosophila Attacin A promoter driving a Firefly luciferase reporter), pJM648 [Toll-responsive Drosphila Drosomycin promoter driving a Firefly luciferase reporter (61)], and pAct-Renilla (an RLuc reporter driven by the constitutive Drosophila actin 5C promoter as a positive control) were used as previously described (18). Briefly, 4 × 105 4A3A cells were plated in 24-well plates. Twelve hours later, plasmids were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. Stimulation with heat-inactivated (10 min at 80 °C in PBS) overnight culture of Escherichia coli or M. luteus and infection with ONNVwt were performed in the order stated in the legend for Fig. 4. In Fig. 4D, cells were infected with RLuc-labeled virus, ONN-RLuc, to measure virus replication in the presence of stimulated pathways (the Firefly luciferase pathway reporters were not used). For all panels, E. coli was used to stimulate Imd and JAK/STAT pathways and M. luteus was used to stimulate the Toll pathway. Reporter activity was assessed by the Dual-Luciferase Reporter Assay System (Promega).
Midgut Bacterial Flora.
When indicated, mosquitoes were treated with antibiotics by a standard procedure (62), as follows. Immediately following emergence from pupae, adult mosquitoes were maintained on a 10% sucrose solution complemented with 1× penicillin/streptomycin and 15 μg/mL gentamicin. To measure the influence of antibiotic treatment on the abundance of midgut flora, midguts dissected at the time of the blood meal were homogenized in pools of 10 in LB and plated on LB-agar, and colonies were counted. The antibiotic effect was determined as the ratio of colony counts between treated and untreated midguts. To cultivate midgut bacteria for supplementation in the blood meal, 10 midguts from unfed mosquitoes were homogenized in 5 mL of LB and cultured overnight. The pelleted overnight culture was resuspended in 200 μL of PBS. Half of the culture was heat-inactivated, and half was left untreated and used to complement the ONN-RLuc–containing blood meal at 1:100 vol/vol.
Luciferase Assay.
Luciferase activities were determined on a Glomax-Multi+ Microplate Multimode reader (Promega) and a Dual-Luciferase Reporter Assay System for in vitro studies using RLuca and Firefly luciferase, or on a 1420 Victor 2 System (EG&G Wallac) and Renilla-Glo luciferase assay kit (Promega) for the in vivo studies.
Statistical Analysis.
Box plots were created using R (63) and the beeswarm package (64). Statistical significance was determined using the Wilcoxon–Mann–Whitney rank-sum test, and P values were assessed with a null distribution of the test statistic approximated using Monte-Carlo resampling with 1,000,000 permutations. Bar plots were created using R. Statistical significance was established using the exact Wilcoxon–Mann–Whitney rank-sum test.
Supplementary Material
Acknowledgments
We thank B. D. Foy (Colorado State University) for ONNV and infectious cDNA clones used in this study and N. Puchot for assistance with hemocoel perfusion experiments. We thank the Center for Production and Infection of Anopheles platform at the Institut Pasteur for rearing mosquitoes. This work received financial support from the European Commission FP7 Infrastructures for Vector Biology (InfraVec) program, the European Research Council, and the UK Medical Research Council.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Small RNA sequence data are available in the ArrayExpress database, www.ebi.ac.uk/arrayexpress (accession no. E-MTAB-2595).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1412984112/-/DCSupplemental.
References
- 1.Weaver SC, Reisen WK. Present and future arboviral threats. Antiviral Res. 2010;85(2):328–345. doi: 10.1016/j.antiviral.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Powers AM, et al. Evolutionary relationships and systematics of the alphaviruses. J Virol. 2001;75(21):10118–10131. doi: 10.1128/JVI.75.21.10118-10131.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ratovonjato J, et al. Detection, isolation, and genetic characterization of Rift Valley fever virus from Anopheles (Anopheles) coustani, Anopheles (Anopheles) squamosus, and Culex (Culex) antennatus of the Haute Matsiatra region, Madagascar. Vector Borne Zoonotic Dis. 2011;11(6):753–759. doi: 10.1089/vbz.2010.0031. [DOI] [PubMed] [Google Scholar]
- 4.Seufi AM, Galal FH. Role of Culex and Anopheles mosquito species as potential vectors of rift valley fever virus in Sudan outbreak, 2007. BMC Infect Dis. 2010;10:65. doi: 10.1186/1471-2334-10-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sabin LR, Hanna SL, Cherry S. Innate antiviral immunity in Drosophila. Curr Opin Immunol. 2010;22(1):4–9. doi: 10.1016/j.coi.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keene KM, et al. RNA interference acts as a natural antiviral response to O’nyong-nyong virus (Alphavirus; Togaviridae) infection of Anopheles gambiae. Proc Natl Acad Sci USA. 2004;101(49):17240–17245. doi: 10.1073/pnas.0406983101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell CL, Black WC, 4th, Hess AM, Foy BD. Comparative genomics of small RNA regulatory pathway components in vector mosquitoes. BMC Genomics. 2008;9:425. doi: 10.1186/1471-2164-9-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Myles KM, Wiley MR, Morazzani EM, Adelman ZN. Alphavirus-derived small RNAs modulate pathogenesis in disease vector mosquitoes. Proc Natl Acad Sci USA. 2008;105(50):19938–19943. doi: 10.1073/pnas.0803408105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siu RW, et al. Antiviral RNA interference responses induced by Semliki Forest virus infection of mosquito cells: Characterization, origin, and frequency-dependent functions of virus-derived small interfering RNAs. J Virol. 2011;85(6):2907–2917. doi: 10.1128/JVI.02052-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brackney DE, et al. C6/36 Aedes albopictus cells have a dysfunctional antiviral RNA interference response. PLoS Negl Trop Dis. 2010;4(10):e856. doi: 10.1371/journal.pntd.0000856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brackney DE, Beane JE, Ebel GD. RNAi targeting of West Nile virus in mosquito midguts promotes virus diversification. PLoS Pathog. 2009;5(7):e1000502. doi: 10.1371/journal.ppat.1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sánchez-Vargas I, et al. Dengue virus type 2 infections of Aedes aegypti are modulated by the mosquito’s RNA interference pathway. PLoS Pathog. 2009;5(2):e1000299. doi: 10.1371/journal.ppat.1000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khoo CC, Doty JB, Heersink MS, Olson KE, Franz AW. Transgene-mediated suppression of the RNA interference pathway in Aedes aegypti interferes with gene silencing and enhances Sindbis virus and dengue virus type 2 replication. Insect Mol Biol. 2013;22(1):104–114. doi: 10.1111/imb.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khoo CC, Piper J, Sanchez-Vargas I, Olson KE, Franz AW. The RNA interference pathway affects midgut infection- and escape barriers for Sindbis virus in Aedes aegypti. BMC Microbiol. 2010;10:130. doi: 10.1186/1471-2180-10-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xi Z, Ramirez JL, Dimopoulos G. The Aedes aegypti toll pathway controls dengue virus infection. PLoS Pathog. 2008;4(7):e1000098. doi: 10.1371/journal.ppat.1000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Souza-Neto JA, Sim S, Dimopoulos G. An evolutionary conserved function of the JAK-STAT pathway in anti-dengue defense. Proc Natl Acad Sci USA. 2009;106(42):17841–17846. doi: 10.1073/pnas.0905006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sim S, Dimopoulos G. Dengue virus inhibits immune responses in Aedes aegypti cells. PLoS ONE. 2010;5(5):e10678. doi: 10.1371/journal.pone.0010678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fragkoudis R, et al. Semliki Forest virus strongly reduces mosquito host defence signaling. Insect Mol Biol. 2008;17(6):647–656. doi: 10.1111/j.1365-2583.2008.00834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cirimotich CM, Dong Y, Garver LS, Sim S, Dimopoulos G. Mosquito immune defenses against Plasmodium infection. Dev Comp Immunol. 2010;34(4):387–395. doi: 10.1016/j.dci.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitri C, Vernick KD. Anopheles gambiae pathogen susceptibility: The intersection of genetics, immunity and ecology. Curr Opin Microbiol. 2012;15(3):285–291. doi: 10.1016/j.mib.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yassine H, Osta MA. Anopheles gambiae innate immunity. Cell Microbiol. 2010;12(1):1–9. doi: 10.1111/j.1462-5822.2009.01388.x. [DOI] [PubMed] [Google Scholar]
- 22.Pumpuni CB, Beier MS, Nataro JP, Guers LD, Davis JR. Plasmodium falciparum: Inhibition of sporogonic development in Anopheles stephensi by gram-negative bacteria. Exp Parasitol. 1993;77(2):195–199. doi: 10.1006/expr.1993.1076. [DOI] [PubMed] [Google Scholar]
- 23.Pumpuni CB, Demaio J, Kent M, Davis JR, Beier JC. Bacterial population dynamics in three anopheline species: The impact on Plasmodium sporogonic development. Am J Trop Med Hyg. 1996;54(2):214–218. doi: 10.4269/ajtmh.1996.54.214. [DOI] [PubMed] [Google Scholar]
- 24.Dong Y, Manfredini F, Dimopoulos G. Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog. 2009;5(5):e1000423. doi: 10.1371/journal.ppat.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frolet C, Thoma M, Blandin S, Hoffmann JA, Levashina EA. Boosting NF-kappaB-dependent basal immunity of Anopheles gambiae aborts development of Plasmodium berghei. Immunity. 2006;25(4):677–685. doi: 10.1016/j.immuni.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 26.Oduol F, Xu J, Niare O, Natarajan R, Vernick KD. Genes identified by an expression screen of the vector mosquito Anopheles gambiae display differential molecular immune response to malaria parasites and bacteria. Proc Natl Acad Sci USA. 2000;97(21):11397–11402. doi: 10.1073/pnas.180060997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodrigues J, Brayner FA, Alves LC, Dixit R, Barillas-Mury C. Hemocyte differentiation mediates innate immune memory in Anopheles gambiae mosquitoes. Science. 2010;329(5997):1353–1355. doi: 10.1126/science.1190689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waldock J, Olson KE, Christophides GK. Anopheles gambiae antiviral immune response to systemic O’nyong-nyong infection. PLoS Negl Trop Dis. 2012;6(3):e1565. doi: 10.1371/journal.pntd.0001565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sim C, et al. Modulation of Anopheles gambiae gene expression in response to o’nyong-nyong virus infection. Insect Mol Biol. 2005;14(5):475–481. doi: 10.1111/j.1365-2583.2005.00578.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brault AC, et al. Infection patterns of o’nyong nyong virus in the malaria-transmitting mosquito, Anopheles gambiae. Insect Mol Biol. 2004;13(6):625–635. doi: 10.1111/j.0962-1075.2004.00521.x. [DOI] [PubMed] [Google Scholar]
- 31.Pierro DJ, Myles KM, Foy BD, Beaty BJ, Olson KE. Development of an orally infectious Sindbis virus transducing system that efficiently disseminates and expresses green fluorescent protein in Aedes aegypti. Insect Mol Biol. 2003;12(2):107–116. doi: 10.1046/j.1365-2583.2003.00392.x. [DOI] [PubMed] [Google Scholar]
- 32.Gupta L, et al. The STAT pathway mediates late-phase immunity against Plasmodium in the mosquito Anopheles gambiae. Cell Host Microbe. 2009;5(5):498–507. doi: 10.1016/j.chom.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meister S, et al. Immune signaling pathways regulating bacterial and malaria parasite infection of the mosquito Anopheles gambiae. Proc Natl Acad Sci USA. 2005;102(32):11420–11425. doi: 10.1073/pnas.0504950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fraiture M, et al. Two mosquito LRR proteins function as complement control factors in the TEP1-mediated killing of Plasmodium. Cell Host Microbe. 2009;5(3):273–284. doi: 10.1016/j.chom.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 35.Povelones M, Waterhouse RM, Kafatos FC, Christophides GK. Leucine-rich repeat protein complex activates mosquito complement in defense against Plasmodium parasites. Science. 2009;324(5924):258–261. doi: 10.1126/science.1171400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitri C, et al. Fine pathogen discrimination within the APL1 gene family protects Anopheles gambiae against human and rodent malaria species. PLoS Pathog. 2009;5(9):e1000576. doi: 10.1371/journal.ppat.1000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hillyer JF, Barreau C, Vernick KD. Efficiency of salivary gland invasion by malaria sporozoites is controlled by rapid sporozoite destruction in the mosquito haemocoel. Int J Parasitol. 2007;37(6):673–681. doi: 10.1016/j.ijpara.2006.12.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.King JG, Hillyer JF. Spatial and temporal in vivo analysis of circulating and sessile immune cells in mosquitoes: hemocyte mitosis following infection. BMC Biol. 2013;11:55. doi: 10.1186/1741-7007-11-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Müller HM, Dimopoulos G, Blass C, Kafatos FC. A hemocyte-like cell line established from the malaria vector Anopheles gambiae expresses six prophenoloxidase genes. J Biol Chem. 1999;274(17):11727–11735. doi: 10.1074/jbc.274.17.11727. [DOI] [PubMed] [Google Scholar]
- 40.Christophides GK, et al. Immunity-related genes and gene families in Anopheles gambiae. Science. 2002;298(5591):159–165. doi: 10.1126/science.1077136. [DOI] [PubMed] [Google Scholar]
- 41.Osta MA, Christophides GK, Kafatos FC. Effects of mosquito genes on Plasmodium development. Science. 2004;303(5666):2030–2032. doi: 10.1126/science.1091789. [DOI] [PubMed] [Google Scholar]
- 42.Charroux B, Royet J. Gut-microbiota interactions in non-mammals: What can we learn from Drosophila? Semin Immunol. 2012;24(1):17–24. doi: 10.1016/j.smim.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 43.Myles KM, Morazzani EM, Adelman ZN. Origins of alphavirus-derived small RNAs in mosquitoes. RNA Biol. 2009;6(4):387–391. doi: 10.4161/rna.6.4.8946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cirimotich CM, Scott JC, Phillips AT, Geiss BJ, Olson KE. Suppression of RNA interference increases alphavirus replication and virus-associated mortality in Aedes aegypti mosquitoes. BMC Microbiol. 2009;9:49. doi: 10.1186/1471-2180-9-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Attarzadeh-Yazdi G, et al. Cell-to-cell spread of the RNA interference response suppresses Semliki Forest virus (SFV) infection of mosquito cell cultures and cannot be antagonized by SFV. J Virol. 2009;83(11):5735–5748. doi: 10.1128/JVI.02440-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saleh MC, et al. Antiviral immunity in Drosophila requires systemic RNA interference spread. Nature. 2009;458(7236):346–350. doi: 10.1038/nature07712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garver LS, et al. Anopheles Imd pathway factors and effectors in infection intensity-dependent anti-Plasmodium action. PLoS Pathog. 2012;8(6):e1002737. doi: 10.1371/journal.ppat.1002737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Avadhanula V, Weasner BP, Hardy GG, Kumar JP, Hardy RW. A novel system for the launch of alphavirus RNA synthesis reveals a role for the Imd pathway in arthropod antiviral response. PLoS Pathog. 2009;5(9):e1000582. doi: 10.1371/journal.ppat.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang Z, Kingsolver MB, Avadhanula V, Hardy RW. An antiviral role for antimicrobial peptides during the arthropod response to alphavirus replication. J Virol. 2013;87(8):4272–4280. doi: 10.1128/JVI.03360-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kane M, et al. Successful transmission of a retrovirus depends on the commensal microbiota. Science. 2011;334(6053):245–249. doi: 10.1126/science.1210718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuss SK, et al. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science. 2011;334(6053):249–252. doi: 10.1126/science.1211057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sim S, et al. Transcriptomic profiling of diverse Aedes aegypti strains reveals increased basal-level immune activation in dengue virus-refractory populations and identifies novel virus-vector molecular interactions. PLoS Negl Trop Dis. 2013;7(7):e2295. doi: 10.1371/journal.pntd.0002295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Povelones M, Upton LM, Sala KA, Christophides GK. Structure-function analysis of the Anopheles gambiae LRIM1/APL1C complex and its interaction with complement C3-like protein TEP1. PLoS Pathog. 2011;7(4):e1002023. doi: 10.1371/journal.ppat.1002023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Riehle MM, et al. Anopheles gambiae APL1 is a family of variable LRR proteins required for Rel1-mediated protection from the malaria parasite, Plasmodium berghei. PLoS ONE. 2008;3(11):e3672. doi: 10.1371/journal.pone.0003672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Riehle MM, et al. Natural malaria infection in Anopheles gambiae is regulated by a single genomic control region. Science. 2006;312(5773):577–579. doi: 10.1126/science.1124153. [DOI] [PubMed] [Google Scholar]
- 56.Baxter RH, et al. A heterodimeric complex of the LRR proteins LRIM1 and APL1C regulates complement-like immunity in Anopheles gambiae. Proc Natl Acad Sci USA. 2010;107(39):16817–16822. doi: 10.1073/pnas.1010575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harris C, et al. Polymorphisms in Anopheles gambiae immune genes associated with natural resistance to Plasmodium falciparum. PLoS Pathog. 2010;6(9):e1001112. doi: 10.1371/journal.ppat.1001112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kam YW, Ong EK, Rénia L, Tong JC, Ng LF. Immuno-biology of Chikungunya and implications for disease intervention. Microbes Infect. 2009;11(14-15):1186–1196. doi: 10.1016/j.micinf.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 59.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 60.Hombría JC, Brown S, Häder S, Zeidler MP. Characterisation of Upd2, a Drosophila JAK/STAT pathway ligand. Dev Biol. 2005;288(2):420–433. doi: 10.1016/j.ydbio.2005.09.040. [DOI] [PubMed] [Google Scholar]
- 61.Tauszig S, Jouanguy E, Hoffmann JA, Imler JL. Toll-related receptors and the control of antimicrobial peptide expression in Drosophila. Proc Natl Acad Sci USA. 2000;97(19):10520–10525. doi: 10.1073/pnas.180130797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cirimotich CM, et al. Natural microbe-mediated refractoriness to Plasmodium infection in Anopheles gambiae. Science. 2011;332(6031):855–858. doi: 10.1126/science.1201618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Team RC. 2012. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna). Available at www.R-project.org/. Accessed July 1, 2014.
- 64.Eklund A. 2012. Beeswarm: The bee swarm plot, an alternative to stripchart. R package version 0.1.5. Available at CRAN.R-project.org/package=beeswarm. Accessed July 1, 2014.
- 65.Blandin S, et al. Complement-like protein TEP1 is a determinant of vectorial capacity in the malaria vector Anopheles gambiae. Cell. 2004;116(5):661–670. doi: 10.1016/s0092-8674(04)00173-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.