Significance
Understanding the structure and function of the cell division apparatus of Mycobacterium tuberculosis is crucial for advancing drug development against tuberculosis. Here, we report the solid-state NMR structure of a transmembrane protein, CrgA, that is a central component of the M. tuberculosis divisome. Small helical membrane protein structures are particularly sensitive to their environment, and consequently, we characterized CrgA in an environment that models well the biophysical properties of the native membrane. To determine the structure, both oriented sample and magic-angle spinning NMR data from liquid–crystalline lipid bilayer preparations were used along with refinement by restrained molecular dynamics simulations in the same lipid environment. The structure suggests how CrgA serves as a platform for binding and recruiting other proteins of the divisome.
Keywords: membrane protein structure, transmembrane helix binding motif, intrinsically disordered proteins, solid-state NMR, oriented samples
Abstract
The 93-residue transmembrane protein CrgA in Mycobacterium tuberculosis is a central component of the divisome, a large macromolecular machine responsible for cell division. Through interactions with multiple other components including FtsZ, FtsQ, FtsI (PBPB), PBPA, and CwsA, CrgA facilitates the recruitment of the proteins essential for peptidoglycan synthesis to the divisome and stabilizes the divisome. CrgA is predicted to have two transmembrane helices. Here, the structure of CrgA was determined in a liquid–crystalline lipid bilayer environment by solid-state NMR spectroscopy. Oriented-sample data yielded orientational restraints, whereas magic-angle spinning data yielded interhelical distance restraints. These data define a complete structure for the transmembrane domain and provide rich information on the conformational ensembles of the partially disordered N-terminal region and interhelical loop. The structure of the transmembrane domain was refined using restrained molecular dynamics simulations in an all-atom representation of the same lipid bilayer environment as in the NMR samples. The two transmembrane helices form a left-handed packing arrangement with a crossing angle of 24° at the conserved Gly39 residue. This helix pair exposes other conserved glycine and alanine residues to the fatty acyl environment, which are potential sites for binding CrgA’s partners such as CwsA and FtsQ. This approach combining oriented-sample and magic-angle spinning NMR spectroscopy in native-like lipid bilayers with restrained molecular dynamics simulations represents a powerful tool for structural characterization of not only isolated membrane proteins, but their complexes, such as those that form macromolecular machines.
Better understanding of cell division in Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis (TB), will generate new opportunities for pharmaceutical development. CrgA, a transmembrane (TM) protein, is a central component of the Mtb divisome (1). CrgA has homologs in other actinomycetes (2, 3), but not in the two bacteria, Escherichia coli and Bacillus subtilis, with better characterized cell division mechanisms. Conversely, many cell division proteins in the latter organisms, such as FtsA, FtsN, FtsL, and ZipA, appear to have no homologs in Mtb. CrgA is localized at the poles and septum, and interacts with multiple cell division proteins, including FtsZ, FtsQ, FtsI (PBPB), PBPA, and CwsA. One function of these interactions is to stabilize the divisome (1, 4). The interaction with CwsA, a protein that is unique to mycobacteria (5), might coordinate elongation at the poles and division at midcell (4). Moreover, CrgA appears to have an important role in peptidoglycan (PG) formation during cell division, by recruiting PG synthases to the divisome (4). Reduced production of CrgA results in elongated cells and reduced growth rate (1), and loss of CrgA impairs PG synthesis (5). In addition to CwsA, the Mtb divisome involves other atypical players such as FipA (FhaB), ChiZ, and MtrB (6–8), and thus there is much yet to be learned about the participants in mycobacterial cell division (9). Here, we determined the structure of CrgA in a lipid bilayer environment using solid-state NMR (ssNMR) spectroscopy.
TB is a devastating human disease that kills ∼1.3 million people each year with 8.6 million new cases diagnosed annually worldwide (10). Rising extreme drug-resistant Mtb strains do not succumb to the frontline antibiotics, generating a dire need for new drugs (11). Pathways critical for bacterial survival such as DNA replication and cell division include numerous potential drug targets and represent a major focus for structural biology. Also, TB treatment is expensive and significantly toxic and requires an extensive period caused by Mtb’s ability to exist in a latent state. Hence, there are additional motivations for characterizing the proteins associated with its survival in active and nonreplicative persistent states.
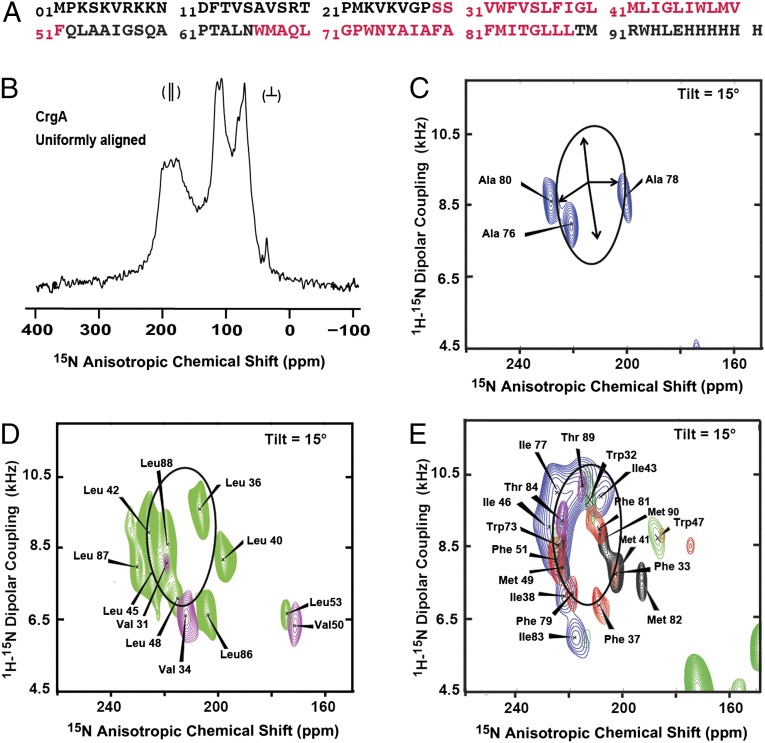
CrgA was first described from Streptomyces as being required for sporulation through coordinating several aspects of its reproductive growth (2, 3). The Mtb CrgA consists of 93 residues, with two predicted TM helices (12) (TM1: residues 29–51; and TM2: residues 66–88; Fig. 1A). The N-terminal 17 residues are predicted to be disordered by the software PONDR (13); the C terminus is predicted to be just five residues, whereas the loop between the TM helices is predicted to be just 14 residues. The predicted TM1 sequence contains a pair of conserved tryptophan residues (W32 and W47) that appear from the sequence to be positioned for anchoring the helix to the membrane interfacial regions. A second pair of conserved tryptophan residues is at positions 73 and 92. Because the TM2 prediction has W73 eight residues into the helix and W92 four residues beyond the end of the predicted helix, this prediction may not be as accurate. Both predicted helices contain a number of other conserved residues, whereas the loop between the helices is much more variable both in length and in composition (SI Appendix, Fig. S1).
Fig. 1.
Amino acid sequence and ssNMR spectra of full-length Mtb CrgA membrane protein. (A) The sequence of the expressed CrgA with a C-terminal His6 tag. The predicted transmembrane (TM) helical residues (TMHMM, version 2.0) are indicated by red lettering. (B) One-dimensional 15N cross-polarization spectrum of 15N uniformly labeled CrgA with the bilayer normal parallel to the magnetic field. The resonance intensities near 200 ppm are from the TM helix residues, with their backbone amide NH bonds nearly parallel to the bilayer normal. The intensity near 120 ppm is from the highly dynamic sites, and most of the remaining intensity is from structured residues not in the TM domain. (C) Two-dimensional PISEMA spectra of 15N Ala-labeled CrgA with a superimposed PISA wheel calculated for an ideal helix [(φ, ψ) = (−60°, −45°)] with a 15° tilt angle relative to the bilayer normal. The set of four arrows in the middle of the PISA wheel define 90° rotational increments for the backbone nitrogen sites. Resonance assignments based on the PISA analysis are shown. (D) Superimposed PISEMA spectra of 15N Leu (green)- and 15N Val (purple)-labeled protein with resonance assignments. (E) Overlay of PISEMA spectra for 15N Ile (blue)-, 15N Met (black)-, 15N Thr (purple)-, 15N Phe (red)-, and 15N Trp (green)-labeled CrgA, with sequence-specific assignments shown. All spectra were collected at 60 MHz for 15N in oriented POPC/POPG (4:1 mol/mol) liquid–crystalline phospholipid bilayers, pH 7.0, 13 °C.
Surprisingly, this small membrane protein binds a large number of other proteins, all of which are transmembrane proteins except for FtsZ. In particular, FtsI, with a single TM helix, is a transpeptidase responsible for synthesis of the septal PG (1). A crgA-deletion mutant results in the loss of septal and polar localization of FtsI, suggesting the importance of CrgA for PG synthesis through its recruitment of FtsI. CwsA also contains a single TM helix. A crgA and cwsA double-deletion mutant showed the importance of the corresponding gene products for cell wall synthesis and cell shape maintenance (8).
The CrgA TM helices contain a number of conserved glycine and alanine residues (SI Appendix, Fig. S1). Although glycine residues are known to be helix breakers in water-soluble proteins, in TM helices, they may allow local helix bending in the low dielectric membrane environment where intrahelical hydrogen bonds are strengthened for maintaining the overall integrity of the helical structure. In addition, glycine and alanine residues permit close approach of adjacent helical backbones, resulting in backbone–backbone electrostatic and side-chain–side-chain van der Waals interactions that stabilize the tertiary structure. Therefore, glycines may allow helical membrane proteins to sacrifice secondary structural stability for tertiary structural stability (14–16). This is needed because the amino acid composition in the interior of membrane proteins is more hydrophobic than the interior of water-soluble proteins where there are more frequent tertiary hydrogen bonds than in TM domains (17). In addition, conserved glycine residues are rarely found on the fatty-acyl exposed surface of multihelix membrane proteins (16). In such a location, they would expose their hydrophilic backbone atoms to the low dielectric environment of the protein. If present, it is a strong indication that they are exposed for a required function such as binding another protein. Interestingly, E. coli FtsQ is thought to localize to the divisome through interactions with other components via its single TM helix (18).
Only a couple of full-length Mtb membrane protein structures have been determined. One is an X-ray structure of the mechanosensitive channel of large conductance, and the other is a single TM helix protein, Rv1761 (19, 20). In addition, water-soluble domains of other Mtb membrane proteins have been characterized such as those from PknB and FtsX (21–24). Although X-ray crystallographers have focused on large membrane proteins, the majority of the 1,162 ORFs of the Mtb genome code for small helical membrane proteins containing one to three TM helices with <40-kDa molecular weight (25). Structure–function studies of these small membrane proteins are essential for understanding Mtb cell division and other cellular processes. Small polytopic membrane protein structures are stabilized not just by interactions between their TM helices, but also by interactions with their membrane environment. Consequently, it is necessary to solve their structures in an appropriate membrane mimetic environment, one that possesses many of the restraining influences of the native membrane such as a relatively fixed hydrophobic thickness, a dramatic lateral pressure profile, and a hydrophobic core essentially devoid of water (26, 27).
For the structure determination of CrgA, here we used both oriented-sample (OS) and magic-angle spinning (MAS) ssNMR to characterize the full-length protein in lipid bilayers. All ssNMR spectroscopy was performed on fully hydrated liquid–crystalline lipid bilayer preparations of CrgA. The use of such bilayer preparations for supporting the native-like conformation of the M2 protein from Influenza A has been validated with the comparison of spectra from synthetic bilayers and from cellular membranes where the protein has been inserted by the cellular machinery and never removed from this environment or exposed to a detergent environment (28). Multiple recent membrane protein structures have now been determined by OS ssNMR (29–34), and the first membrane protein structure has been obtained from MAS ssNMR (35). OS ssNMR generates information on the orientations of peptide planes with respect to the bilayer normal, and for a TM helix it yields the tilt angle of the helix relative to the lipid bilayer normal and rotational orientation about the helical axis along the entire length of helix. However, it does not directly provide information on the helix–helix packing interface. The latter information can be ascertained by relatively few distance restraints between the helices, as the degrees of freedom for packing the helices have been minimized by the orientational restraints, to just the relative rotation around the bilayer normal and relative translation in the bilayer plane. The combination of OS and MAS ssNMR thus allows the complete determination of the helical TM domain structure.
Based on the OS and MAS data, we refined the structure using restrained molecular dynamics simulations in an all-atom representation of the same lipid bilayer environment as in the protein samples. The two TM helices both have a tilt angle of 13° but are tilted in nearly opposite directions such that they form a left-handed packing arrangement with a crossing angle of 24° at the conserved Gly39 residue. The two-helix TM domain exposes other conserved glycine and alanine residues that potentially form binding sites for TM helices of CrgA binders. Much of the N-terminal region is disordered, but a nine-residue motif therein appears to form an amphipathic helix. In the interhelical loop, a short segment appears to be disordered while a 12-residue motif appears to form a β-hairpin in the membrane interface. The C terminus comprises just two residues. Overall, the structure suggests how CrgA serves as a platform where other proteins of the divisome assemble.
Results
Sample Characterization.
15N, 13C uniform-labeled and amino acid-specific–labeled full-length CrgA was expressed with a C-terminal His6 tag through an LE linker (Fig. 1A). In 12% (wt/vol) SDS/PAGE gel, the purified protein appeared as a single band at 12 kDa (SI Appendix, Fig. S2A). The purified protein was then reconstituted in 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC)/1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol (POPG) (4:1 mol/mol) liposomes for structural characterization. As an initial secondary structure characterization, the circular dichroism (CD) spectrum of the reconstituted proteoliposomes was obtained suggesting 49% helix, 12% β-sheet, 17% turn, and 22% random coil [CD Pro analysis (36); SI Appendix, Fig. S2B].
OS ssNMR.
The orientation of full-length CrgA in glass-slide–supported POPC/POPG lipid bilayers (with a gel-to-liquid crystalline phase transition temperature of −2 °C) was first assessed using the one-dimensional (1D) 15N ssNMR chemical shift spectrum of uniform 15N-labeled protein at 13 °C, pH 8.0 (Fig. 1B). The spectrum is dominated by the amide backbone resonances. Approximately 10% of the intensity is at the isotropic frequency (110–130 ppm) indicating structural disorder for a significant portion of the amino acid sequence in addition to the His-tag, considering that cross-polarization of the isotropic intensity is often weak. Here, the B0 field direction was parallel to the bilayer normal, and thus the considerable intensity near the parallel edge of the anisotropic chemical shift tensor (∼230–180 ppm) suggests that the TM helices account for most but maybe not all of the 49% α-helical content indicated by CD spectroscopy. The rest of the spectral intensity accounts for the loops and terminal regions.
Fig. 1C shows the 15N Ala polarization inversion spin exchange at magic angle (PISEMA) (37) spectrum for three alanine residues (at positions 76, 78, 80) predicted to be in TM2. Similar to helical wheels, there are 3.6 resonances per turn in a polarity index slant angle (PISA) wheel for an α-helix (i.e., 100° separation between adjacent resonances) (38, 39), as shown superimposed on the spectrum. The tilt of the helix controls the position and size of the PISA wheel. This wheel, for a 15° tilt of the helix axis relative to the bilayer normal, was chosen for best alignment with all of the spectral data (not just the alanine data) and provides an initial assessment of helix tilt. The assignments for these alanine residues were based on the fact that residues i and i + 2 should be on opposite sides of the wheel (∼200° apart) and residues i to i + 4 should be near each other (∼40° apart). The unique assignment of these residues also provided an initial assessment of the rotational orientation of the helix. The full 15N Ala spectrum shows resolved resonances for all nine alanine residues in CrgA (SI Appendix, Fig. S3) including resonances from the disordered regions of the protein and the structured interfacial regions. Although a bacterial two-hybrid assay suggested that CrgA forms a dimer (1), the single resonances observed here for all of the residues indicate a monomer (or symmetric dimer) in these preparations.
Fig. 1D shows the superimposed PISEMA spectra of 15N Val- and 15N Leu-labeled CrgA obtained from separate samples. All three valine residues predicted to be in the TM helices are in TM1 (at positions 31, 34, and 50), but residue 50, due to its predicted position in the last turn of TM1 has a potentially deformed orientation for its peptide plane (40), and consequently it was excluded from initial assignment. Residues 31 and 34, corresponding to i and i + 3 positions, should be on one side of the wheel (∼60° apart). The initial tilt assignment of TM1, again by alignment of the corresponding PISA wheel with all of the data, is ∼15°, similar to that of TM2. Via the assignment of the two valine resonances, an initial assessment of the rotational orientation for TM1 was also made. Based on the tilt and rotational characterizations for the two TM helices, the remaining resonance assignments were made.
The leucine residues in TM1 (at positions 36, 40, 42, 45, and 48) are distributed almost uniformly around the helical and PISA wheels, whereas those in TM2 (at positions 86, 87, and 88) are on one-half of its wheel. Fig. 1E shows five additional superimposed PISEMA spectra for 15N Phe (residues 33, 37, 51, 79, and 81), Met (residues 41, 49, 82, and 90), Trp (residues 32, 47, and 73), Ile (residues 38, 43, 46, 77, and 83), and Thr (residues 84 and 89). The vast majority of the resonances for the TM helices (Fig. 1E) fall in close proximity to the 15° PISA wheels for both TM helices, with the exception of the resonances from residues in the first or last turns of the helices. All of the helical backbone sites in the TM helices were observed except for Gly44. The anisotropic chemical shifts and dipolar couplings for the TM helices are listed in Table 1. In addition, the conformity of the data to PISA wheels permits the use of typical α-helical restraints associated with hydrogen bonding distances and membrane protein helical torsion angle (ϕ = −60° and ψ = −45°) restraints (41). Most of the resonances in the N-terminal region, the two-residue C terminus, and interhelical loop were observed, and many indicate significant structural disorder by displaying very little anisotropy in their oriented sample resonance frequencies.
Table 1.
CrgA 1H-15N dipolar coupling (DC) and 15N anisotropic chemical shift (ACS) data from the PISEMA spectra
| TM1 (residue Val31–Gln52) | TM2 (residue Trp73–Arg91) | ||||
| Residues | DC, kHz | ACS, ppm | Residues | DC, kHz | ACS, ppm |
| Val31 | 8.07 | 221.3 | Trp73 | 8.40 | 224.6 |
| Trp32 | 9.23 | 210.3 | Asn74 | 9.57 | 208.9 |
| Phe33 | 7.84 | 203.5 | Tyr75 | 4.28 | 189.7 |
| Val34 | 6.5 | 211.5 | Ala76 | 7.80 | 220.9 |
| Ser35 | 9.07 | 227.5 | Ile77 | 10.07 | 224.8 |
| Leu36 | 9.57 | 206.9 | Ala78 | 8.75 | 200.8 |
| Phe37 | 6.96 | 209.4 | Phe79 | 7.20 | 219.1 |
| Ile38 | 7.23 | 222.0 | Ala80 | 8.70 | 228.5 |
| Gly39 | 9.66 | 220.4 | Phe81 | 9.10 | 210.0 |
| Leu40 | 8.20 | 198.3 | Met82 | 7.80 | 193.7 |
| Met41 | 7.90 | 202.9 | Ile83 | 6.06 | 218.1 |
| Leu42 | 8.93 | 226.0 | Thr84 | 9.17 | 223.0 |
| Ile43 | 9.90 | 208.6 | Gly85 | 9.77 | 203.3 |
| Gly44 | NA | NA | Leu86 | 6.62 | 203.8 |
| Leu45 | 7.59 | 224.6 | Leu87 | 7.95 | 230.4 |
| Ile46 | 9.02 | 225.3 | Leu88 | 8.57 | 219.1 |
| Trp47 | 8.75 | 188.6 | Thr89 | 10.35 | 215.8 |
| Leu48 | 7.09 | 215.8 | Met90 | 8.79 | 207.4 |
| Met49 | 8.07 | 222.5 | Arg91 | 4.73 | 177.3 |
| Val50 | 6.42 | 171.3 | |||
| Phe51 | 8.23 | 225.1 | |||
| Gln52 | 6.64 | 174.3 | |||
The orientational restraints for the TM helices (Table 1) when plotted as a function of residue number, known as dipolar and chemical shift waves (42, 43), provide additional structural detail for the experimentally characterized helices consisting of residues 31–52 and residues 73–91 (SI Appendix, Fig. S4). There appear to be some local perturbations to the helices in addition to the first and last turns, such as the anisotropic chemical shifts observed at Met41 in TM1 and Tyr75 in TM2, where the data lie significantly off the waves. Perturbations primarily associated with the anisotropic chemical shifts may reflect differences in their chemical shift tensors. Perturbations in the first or last turn are likely to reflect the interaction of this helical turn with the membrane interface through amphipathic side chains, anchoring and stabilizing the TM domain in the lipid bilayer (44, 45), and hence the helical backbone may be somewhat perturbed at these sites. However, it is clear that the tilt and rotational orientation of the helices remain constant throughout their lengths.
Most of the remaining PISEMA resonances could not be sequence-specifically assigned except for Ile56 and Trp66 to a structured portion of the interhelical loop and Phe12 to the disordered N-terminal region. The other resonances were amino acid-specifically assigned because of the isotopic labeling strategy. For methionine, there are two residues in addition to those in the TM helices. Met67 is in the interhelical loop, next to Trp66, and could be assigned to a Met resonance similar to that of Trp66. That left Met22 with a resonance indicating a structured portion in the N-terminal region. The rest of the observed resonances were tentatively assigned according to proximity in the spectra (sequentially neighboring residues were assigned to nearby resonances, i.e., those having similar dynamics or conformation; SI Appendix, Table S1).
MAS ssNMR.
Although OS ssNMR characterizes the helical backbone structure and the orientations of the helices relative to the bilayer normal, distances between the helices are valuable for unique determination of the tertiary structure. We obtained the latter data using 13C-13C correlation [dipolar-assisted rotational resonance (DARR)] spectra. Before distances could be obtained, sequence-specific assignments were made for the isotropic 13C resonances. This was complicated by the uniformity of the helical structures resulting from the hydrophobic amino acid content and the low dielectric environment that strengthens intrahelical hydrogen bonds (41, 46). However, we were able to make unique assignments for segments of the helical backbone and some of the side chains from 3D MAS experiments [including NCACX, NCOCX, and CAN(CO)CX experiments (47, 48)] of uniformly 15N and 13C-labeled CrgA samples (SI Appendix, Fig. S5). The resonance assignments for residues 38–43 and 48–50 of TM1 and 75–85 of TM2 are listed in SI Appendix, Table S2, representing enough assignments to obtain a set of unique interhelical distances. Fig. 2 A and B shows the superposition of 2D DARR spectra (49, 50) with different mixing times from a 13C, 15N reverse-labeled sample in which all of the residues were labeled except for Thr, Ile, Phe, Ser, and Trp. These spectra resulted in interhelical cross peaks between Met49 and both Tyr75 and Ala78 as well as a cross peak between Leu42 and Ala80 (distances listed in SI Appendix, Table S3). In a second 13C, 15N reverse-labeled sample, all of the amino acid residues except for Ile, Leu, Phe, Tyr, and Ser were isotopically labeled, and another interhelical resonance between Gly39 and Thr84 was observed in the DARR spectra (Fig. 2C).
Fig. 2.
Two-dimensional 13C-13C DARR-MAS spectra of 13C, 15N reverse-labeled CrgA. (A and B) DARR spectra of CrgA with all but Thr, Ile, Phe, Ser, and Trp labeled, using mixing times of 1,000 ms (red), 700 ms (green), and 300 ms (blue). Cross peaks corresponding to three interhelical distance restraints are indicated. (C) Superimposed DARR spectra of CrgA with unlabeled Ile, Leu, Phe, Tyr, and Ser at 100-ms mixing time (blue) and 13C, 15N uniform-labeled CrgA at 50-ms mixing time (red). The Gly39–Thr84 cross peak represents an interhelical distance restraint. All spectra were collected at 600 MHz proton frequency in liquid–crystalline POPC/POPG liposomes at 13 °C and 12-kHz spinning rate.
CrgA Structure.
Based on the OS and MAS ssNMR data described above, we calculated the structure of the CrgA TM domain (34). First, a set of 960 initial conformations was generated by simulated annealing (51), with the dipolar couplings and anisotropic chemical shifts of helical residues restrained to the values observed in the OS ssNMR experiments (Table 1) and the interhelical distances observed in the MAS ssNMR experiments restrained to a conservative upper bound of 8 Å (SI Appendix, Table S3). After filtering by TM1 and TM2 tilt angles and clustering, the average pairwise backbone root-mean-square deviation among 10 selected conformations is 0.38 Å (see SI Appendix, Table S4, for other statistics of the structure calculation). A representative conformation was then further refined by restrained molecular dynamics simulations, again with the restraints from the ssNMR observables, but now with an all-atom representation of the same lipid bilayer environment used for collecting the ssNMR data. ssNMR parameters calculated on the final structure are in close agreement with the experimental values (SI Appendix, Fig. S6).
The refined structure has residues 31–49 forming TM1, two residues shorter on both termini of the helix compared with the prediction (Fig. 1A). These terminal residues are likely to be part of the helix, but their peptide plane orientations appear to deviate substantially from those in the helix core (Table 1), and consequently their PISEMA resonances fall outside of the PISA wheel, but still in close proximity. TM2 is formed by residues 73–91, considerably different from the predicted residues 66–88 (Fig. 1A). Residues 66, 67, and 68 were labeled, but their resonances did not appear in the vicinity of the PISA wheel, and therefore these residues are unlikely to be part of TM2. Gly71 appears to be disordered, and the next residue is a proline, a known helix breaker, leaving Trp73 as the starting residues of TM2. It is thus important to experimentally characterize these helices and not depend solely on prediction tools.
The side view of the refined structure of CrgA TM domain shows a pair of antiparallel α-helices with a tilt angle of 13° with respect to the bilayer normal for both TM1 and TM2 (Fig. 3A). These tilt values are close to those obtained initially from the PISA wheel and wave analysis of the PISEMA spectra. The four interhelical distances from MAS ssNMR define the packing arrangement of the two helices (Fig. 3B), in which the two TM helices tilt in nearly opposite directions, resulting in a left-handed bundle with a crossing angle of 24° and a crossing midpoint near Gly39 and Leu42 of TM1 and Phe81 and Thr84 of TM2, positioned near the center of the membrane (Fig. 3).
Fig. 3.
Structure of CrgA transmembrane domain determined in lipid bilayers. (A) TM1 (cyan) and TM2 (pink) both have a tilt angle of 13°, and cross each other at a 24° angle near Gly39 and Leu42 of TM1 and Phe81 and Thr84 of TM2. (B) A rotated view showing the four interhelical distance restraints (dashed red lines) determined from the MAS spectra.
The interhelical interface can be well described as “knobs in holes” (52). Close van der Waals interactions extend over the entire length of the helix pair, involving 11 residues from TM1 (at positions 31, 32, 35, 36, 38, 39, 42, 43, 45, 46, and 49) and 9 residues from TM2 (at positions 74, 77, 78, 80, 81, 84, 87, 88, and 91), and contribute significantly to the tertiary structural stability (Fig. 4A). Position 39 in the middle of the interface is conserved as a small residue, either glycine or alanine (SI Appendix, Fig. S1). One of the primary reasons for the conservation of Gly/Ala in TM domains is for forming stable helical complexes (53, 54). Two more conserved small residues, A78 and A80, also contribute to the interhelical interface. Interestingly, two highly conserved glycines, Gly44 and Gly85, are not part of the interface and can potentially form interaction sites for other TM proteins or as a CrgA dimer (Fig. 5A; Discussion).
Fig. 4.
Interhelical and intrahelical interactions in CrgA. (A) Interhelical interface with a “knobs-in-holes” pattern involving complementary large and small side chains on the two helices. (B) Interhelical and intrahelical side-chain–backbone hydrogen bonds.
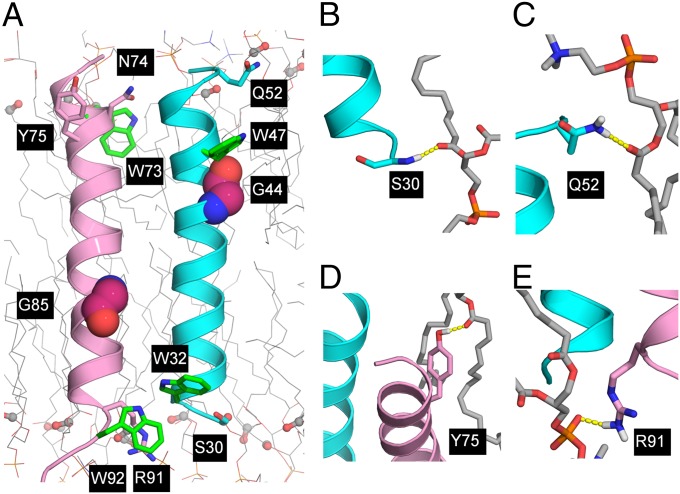
Fig. 5.
Interactions of CrgA with its environment. (A) Positioning of the transmembrane helices in a lipid bilayer. Conserved Trp side chains are shown as green sticks for carbon atoms; neighboring polar and charged side chains are shown as cyan (TM1) or pink (TM2) sticks for carbon atoms. Conserved outward-facing Gly residues are shown as magenta spheres for carbons. Lipid carbonyls are shown as ball-and-stick. In all cases, oxygen atoms are in red and nitrogen atoms are in blue. (B–E) Hydrogen bonds of Ser30, Gln52, Tyr75, and Arg91 with lipid phosphate and carbonyl groups.
Gly39 CαH potentially forms a hydrogen bond across the interface with the side-chain hydroxyl of Thr84 (Fig. 4B), a type of interaction known to stabilize helix–helix packing (33, 55). Besides Thr84, there are two more polar side chains (Ser35 and Thr89) within the helices that form intrahelical hydrogen bonds with neighboring backbone carbonyls. Ser30 is in a helix-capping position and its side chain hydrogen bonds with a backbone amide. In addition, the side chain of Asn74 reaches across the interhelical interface to hydrogen bond with the backbone carbonyl of Leu53. This variety of side-chain–backbone hydrogen bonding is typical of polar residues in TM helices (56).
The structure is defined not only internally but also with respect to its environment (Fig. 5A). These helices span nearly 30 Å of the POPC/POPG bilayer hydrophobic thickness. Two tryptophan residues at the helix termini (Trp47 on TM1 and Trp73 on TM2) have their indole N–H oriented toward the membrane interfacial regions and serve as anchors (44, 57); two other tryptophans (Trp32 and Trp92) form an interhelical stacking interaction. Neighboring polar and charged residues (Ser30 and Gln52 for TM1 and Tyr75 and Arg91 for TM2) form hydrogen bonds with lipid phosphate and carbonyl groups (Fig. 5 B–E). All four tryptophans are absolutely conserved and somewhat less so for Tyr75. Clearly, these residues help to orient the protein in the membrane environment as has been noted for other proteins (45, 58). Interestingly, three consecutive polar residues at the N terminus of TM2 appear to interact with the membrane interface, something that is only possible when the helical tilt angle is small.
The OS ssNMR data outside TM1 and TM2, although sparsely sequence-specifically assigned, contain rich information for structural characterization of the full-length protein. They confirm the disorder of the N-terminal 18 residues predicted by PONDR and further reveal a short disordered segment (Ala68 to Pro72) at the C terminus of the interhelical loop. The sequences for both of these disordered segments are highly conserved among mycobacterial species (SI Appendix, Fig. S1). The structured portions in the N-terminal region (residues 19–27) and in the loop (residues 56–67) have remarkably uniform chemical shifts between 70 and 85 ppm and dipolar couplings between 4.3 and 5.5 kHz, with only a couple of outliers for each parameter. The immediate conclusion is these residues form α-helices or β-strands, with the peptide N–H bonds nearly parallel to the bilayer surface, such as an amphipathic helix or a β-sheet in the plane of the bilayer interface. Furthermore, the CD spectral analysis suggests that there are ∼10 residues of additional helix and ∼10 residues of β-strand, consistent with the preceding OS ssNMR data. Although both segments could form amphipathic helices, the segment in the loop has Pro61 in the middle, which potentially facilitates the formation of a γ-turn. Thereby, this segment can form a β-hairpin, with the terminal residues in close proximity, which is required by the close packing of the TM helices at the extracytoplasmic end (Fig. 3). Although these results are strongly suggestive for the structure of the full-length protein, these segments were not included in the refinement.
Discussion
Recently, general and robust protocols have been published for OS and MAS ssNMR sample preparation (59). Although the ssNMR structural approach demonstrated here for characterizing the structure of CrgA is relatively new, it has general applicability to a large segment of the proteome that has been largely inaccessible by other structural tools. This is because the crystallization of small helical membrane proteins is very difficult (60, 61) and the structures characterized in detergent micelles are often significantly distorted by the single hydrophilic surface of the micelle (27). Consequently, it is important to characterize these relatively small membrane proteins in lipid bilayer environments. As demonstrated here, the combination of OS and MAS ssNMR provides an ideal approach.
CrgA has the responsibility of coordinating the assembly of multiple membrane proteins for cell division (1). CrgA binds to FtsZ, the protein responsible for initiating the formation of the Z-ring when cells divide. Because FtsZ is a water-soluble protein, its interaction with CrgA, documented by bacterial two-hybrid assays, pull-down assays, and colocalization of fluorescent labeled proteins in vivo, must occur through the cytoplasmic N-terminal region of CrgA, which as shown here is largely disordered and consequently has certain advantages for binding other proteins (62). Two penicillin-binding proteins, the transpeptidases FtsI (PBPB) and PBPA, are single-TM helix membrane proteins responsible for synthesis of the septal PG. Based on crgA deletion experiments, this gene product appears to be responsible for the recruitment of FtsI to the divisome. Bacterial two-hybrid assays have demonstrated that the interaction between CrgA and FtsI occurs through the latter’s cytoplasmic domain (1). Similar assays have shown that CrgA also binds PBPA, but this protein has a minimalistic cytoplasmic N terminus of five or six residues that is unlikely to be responsible for the binding and its TM helix has no obvious helix binding motif. Therefore, we anticipate that PBPA binds to the interhelical loop of CrgA, which appears to form a β-hairpin followed by a disordered segment. The structure of CrgA determined here serves as an important platform for assembling these partner proteins (Fig. 6A).
Fig. 6.
The structure of Mtb CrgA as a platform for assembling other interacting proteins. (A) Schematic view of the likely binding sites for five cell division proteins known to interact with CrgA. FtsZ and FtsI interact with the cytoplasmic N terminus of CrgA. CwsA and FtsQ have binding motifs in their transmembrane (TM) helices and are therefore likely to interact with the CrgA TM helices. PBPA has no such binding motif in its TM helix and a minimalistic N-terminal sequence and therefore is hypothesized to interact with the CrgA interhelical loop. (B) A structural model of the CrgA–FtsQ complex. TM1 and TM2 of CrgA are shown as cyan and pink ribbons, respectively. FtsQ is shown as spheres. Residues involved in binding are shown as red spheres (Thr78 and Gly85) and blue ball-and-stick (Phe81 and Met82) on the CrgA side and as green spheres (Gly16, Gly12, and Ala9) on the FtsQ side.
Conserved residues are typically not observed on the lipid-facing surface of membrane proteins, where hydrophobic residues can replace each other without significantly affecting the tertiary stability of the protein in the lipid bilayer. Glycines on this surface, however, expose hydrophilic backbone atoms to the hydrophobic interstices of the bilayer, thereby reducing tertiary stability (16). Hence conserved glycine residues on the lipid-facing surface may indicate a functionally important role, such as stabilizing protein–protein interactions. For CrgA, although the highly conserved Gly39 residue is used in the intraprotein helix packing, the strictly conserved Gly44 and Gly85 residues are exposed to lipid acyl chains (Fig. 5A). Their positions strongly suggest that these are sites for the binding of additional proteins.
In addition to FtsZ, FtsI, and PBPA, CrgA interacts with FtsQ and CwsA, which are also associated with Mtb cell division and are TM helical proteins (1). FtsQ and CwsA both have helix-binding motifs in their single TM helices. The FtsQ TM helix has an AxxGxGxGxA sequence that translates into two helix-binding motifs on opposite sides of the FtsQ helix, an AxxGxxxG motif on one side and a GxxxA motif on the other side. Based on the CrgA TM domain structure, modeling suggests how the AxxGxxxG motif may take advantage of two exposed small residues in CrgA TM2, Ala78 and Gly85, in generating an extensive van der Waals binding interface between these two proteins (Fig. 6B). So we propose that the binding of CrgA to FtsQ is through the TM domain. This leaves CrgA G44 still exposed to the lipid interstices, and it could be a site for dimerization or binding CwsA. This small protein has a single TM helix, WxxAGxAAAxxAGGAxAxSxxR, that is rich in small residues. This sequence has a plethora of surfaces for protein interactions to the extent that large hydrophobic residues that provide the majority of the van der Waals interactions for helix–helix binding are scarce. A cwsA and crgA double-deletion mutant showed that these proteins are required for cell wall PG synthesis and cell morphology (5).
Fig. 6A summarizes the potential binding sites on CrgA for its important function in the divisome. CwsA and FtsQ may bind through the TM helices of CrgA, such as illustrated by the model in Fig. 6B for the FtsQ–CrgA complex. FtsZ and FtsI may bind to the cytoplasmic N-terminal region and PBPA may bind to the extracytoplasmic loop of CrgA. Although it is not clear how many of these proteins bind at the same time, there is potential through these multiple binding sites for CrgA to play the central role in recruitment of the PG synthesis machinery during cell division.
Methods
Methods used in this study are briefly summarized below. Full descriptions are given in SI Appendix.
CrgA Protein Expression, Purification, and Reconstitution.
The protocol for preparing the protein samples was similar to one described previously (1). For 13C, 15N uniformly labeled CrgA protein preparation, 1 L of M9 media was supplemented with 1 g of 15NH4Cl and 2 g of 13C-glucose. Media preparation for other types of labeling is found in SI Appendix. Details of the procedure for CrgA protein purification and of the buffer preparations were previously published (1, 59). Full-length CrgA was reconstituted in POPC/POPG (4:1 mol/mol) liposomes at pH 8.0 as previously described (59).
Circular Dichroism Spectroscopy.
Initial secondary structural analysis of CrgA in POPC/POPG (4:1 mol/mol) liposome was performed by CD spectroscopy at 25 °C. The secondary structure content was analyzed and calculated using the CDPro program (63) and the spectrum was plotted as molar ellipticity ([θ]m) (in degree⋅square centimeters per decimole) (SI Appendix, Fig. S2B).
ssNMR Spectroscopy.
Glass plate-supported OS preparation was described previously (59). OS ssNMR spectroscopy was performed on oriented lipid bilayer samples containing full-length CrgA with various labeling schemes (15N uniform labeling, 15N amino acid-specific labeling, and 15N reverse labeling). For MAS ssNMR spectroscopy, 2D DARR (49, 50) experiments and 3D NCACX, NCOCX (64), and CAN(CO)CX (48) experiments were performed on 13C, 15N uniform-labeled and two reverse-labeled 13C, 15N CrgA samples.
Structure Calculation.
Based on the OS and MAS ssNMR data (Table 1 and SI Appendix, Table S3), we calculated the structure of CrgA TM domain, largely following a previous protocol (34).
Supplementary Material
Acknowledgments
We thank Dr. Y. Tian and Dr. F. Marassi from Sanford–Burnham Medical Research Institute for providing the Xplor-NIH script. We also thank P. L. Gor’kov for his design of the OS sample holder and the sample transfer base. This work was supported, in part, by NIH Grants AI073891, AI074805, and AI023007. The NMR experiments were performed at the National High Magnetic Field Laboratory supported by a National Science Foundation Cooperative Agreement (DMR-1157490) with the State of Florida.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. H.L. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2mmu). Anisotropic chemical shifts and dipolar couplings from oriented-sample NMR, and isotropic chemical shifts from magic-angle spinning NMR, are deposited in the Biological Magnetic Resonance Bank, www.bmrb.wisc.edu (accession no. 19867).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1415908112/-/DCSupplemental.
References
- 1.Plocinski P, et al. Characterization of CrgA, a new partner of the Mycobacterium tuberculosis peptidoglycan polymerization complexes. J Bacteriol. 2011;193(13):3246–3256. doi: 10.1128/JB.00188-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Del Sol R, Pitman A, Herron P, Dyson P. The product of a developmental gene, crgA, that coordinates reproductive growth in Streptomyces belongs to a novel family of small actinomycete-specific proteins. J Bacteriol. 2003;185(22):6678–6685. doi: 10.1128/JB.185.22.6678-6685.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Del Sol R, Mullins JG, Grantcharova N, Flärdh K, Dyson P. Influence of CrgA on assembly of the cell division protein FtsZ during development of Streptomyces coelicolor. J Bacteriol. 2006;188(4):1540–1550. doi: 10.1128/JB.188.4.1540-1550.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kieser KJ, Rubin EJ. How sisters grow apart: Mycobacterial growth and division. Nat Rev Microbiol. 2014;12(8):550–562. doi: 10.1038/nrmicro3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plocinski P, et al. Mycobacterium tuberculosis CwsA interacts with CrgA and Wag31, and the CrgA-CwsA complex is involved in peptidoglycan synthesis and cell shape determination. J Bacteriol. 2012;194(23):6398–6409. doi: 10.1128/JB.01005-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sureka K, et al. Novel role of phosphorylation-dependent interaction between FtsZ and FipA in mycobacterial cell division. PLoS One. 2010;5(1):e8590. doi: 10.1371/journal.pone.0008590. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Chauhan A, et al. Interference of Mycobacterium tuberculosis cell division by Rv2719c, a cell wall hydrolase. Mol Microbiol. 2006;62(1):132–147. doi: 10.1111/j.1365-2958.2006.05333.x. [DOI] [PubMed] [Google Scholar]
- 8.Plocinska R, et al. Septal localization of the Mycobacterium tuberculosis MtrB sensor kinase promotes MtrA regulon expression. J Biol Chem. 2012;287(28):23887–23899. doi: 10.1074/jbc.M112.346544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hett EC, Rubin EJ. Bacterial growth and cell division: A mycobacterial perspective. Microbiol Mol Biol Rev. 2008;72(1):126–156. doi: 10.1128/MMBR.00028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.World Health Organization . Global Tuberculosis Report 2013. World Health Organization; Geneva: 2013. pp. 1–3. [Google Scholar]
- 11.Amaral L, Molnar J. Potential therapy of multidrug-resistant and extremely drug-resistant tuberculosis with thioridazine. In Vivo. 2012;26(2):231–236. [PubMed] [Google Scholar]
- 12.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol. 2001;305(3):567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 13.Li X, Romero P, Rani M, Dunker AK, Obradovic Z. Predicting protein disorder for N-, C-, and internal regions. Genome Inform Ser Workshop Genome Inform. 1999;10:30–40. [PubMed] [Google Scholar]
- 14.Senes A, Engel DE, DeGrado WF. Folding of helical membrane proteins: The role of polar, GxxxG-like and proline motifs. Curr Opin Struct Biol. 2004;14(4):465–479. doi: 10.1016/j.sbi.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 15.MacKenzie KR, Prestegard JH, Engelman DM. A transmembrane helix dimer: Structure and implications. Science. 1997;276(5309):131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- 16.Dong H, Sharma M, Zhou HX, Cross TA. Glycines: Role in α-helical membrane protein structures and a potential indicator of native conformation. Biochemistry. 2012;51(24):4779–4789. doi: 10.1021/bi300090x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou HX, Cross TA. Modeling the membrane environment has implications for membrane protein structure and function: Influenza A M2 protein. Protein Sci. 2013;22(4):381–394. doi: 10.1002/pro.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scheffers DJ, et al. Contribution of the FtsQ transmembrane segment to localization to the cell division site. J Bacteriol. 2007;189(20):7273–7280. doi: 10.1128/JB.00723-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang G, Spencer RH, Lee AT, Barclay MT, Rees DC. Structure of the MscL homolog from Mycobacterium tuberculosis: A gated mechanosensitive ion channel. Science. 1998;282(5397):2220–2226. doi: 10.1126/science.282.5397.2220. [DOI] [PubMed] [Google Scholar]
- 20.Page RC, Lee S, Moore JD, Opella SJ, Cross TA. Backbone structure of a small helical integral membrane protein: A unique structural characterization. Protein Sci. 2009;18(1):134–146. doi: 10.1002/pro.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mieczkowski C, Iavarone AT, Alber T. Auto-activation mechanism of the Mycobacterium tuberculosis PknB receptor Ser/Thr kinase. EMBO J. 2008;27(23):3186–3197. doi: 10.1038/emboj.2008.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mavrici D, et al. Mycobacterium tuberculosis FtsX extracellular domain activates the peptidoglycan hydrolase, RipC. Proc Natl Acad Sci USA. 2014;111(22):8037–8042. doi: 10.1073/pnas.1321812111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yao Y, Barghava N, Kim J, Niederweis M, Marassi FM. Molecular structure and peptidoglycan recognition of Mycobacterium tuberculosis ArfA (Rv0899) J Mol Biol. 2012;416(2):208–220. doi: 10.1016/j.jmb.2011.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wells RM, et al. Discovery of a siderophore export system essential for virulence of Mycobacterium tuberculosis. PLoS Pathog. 2013;9(1):e1003120. doi: 10.1371/journal.ppat.1003120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korepanova A, et al. Cloning and expression of multiple integral membrane proteins from Mycobacterium tuberculosis in Escherichia coli. Protein Sci. 2005;14(1):148–158. doi: 10.1110/ps.041022305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou HX, Cross TA. Influences of membrane mimetic environments on membrane protein structures. Annu Rev Biophys. 2013;42:361–392. doi: 10.1146/annurev-biophys-083012-130326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cross TA, Murray DT, Watts A. Helical membrane protein conformations and their environment. Eur Biophys J. 2013;42(10):731–755. doi: 10.1007/s00249-013-0925-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miao Y, et al. M2 proton channel structural validation from full-length protein samples in synthetic bilayers and E. coli membranes. Angew Chem Int Ed Engl. 2012;51(33):8383–8386. doi: 10.1002/anie.201204666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park SH, et al. Structure of the chemokine receptor CXCR1 in phospholipid bilayers. Nature. 2012;491(7426):779–783. doi: 10.1038/nature11580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verardi R, Shi L, Traaseth NJ, Walsh N, Veglia G. Structural topology of phospholamban pentamer in lipid bilayers by a hybrid solution and solid-state NMR method. Proc Natl Acad Sci USA. 2011;108(22):9101–9106. doi: 10.1073/pnas.1016535108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu GJ, Tian Y, Vora N, Marassi FM, Opella SJ. The structure of the mercury transporter MerF in phospholipid bilayers: A large conformational rearrangement results from N-terminal truncation. J Am Chem Soc. 2013;135(25):9299–9302. doi: 10.1021/ja4042115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahuja S, et al. A model of the membrane-bound cytochrome b5-cytochrome P450 complex from NMR and mutagenesis data. J Biol Chem. 2013;288(30):22080–22095. doi: 10.1074/jbc.M112.448225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jean-Francois FL, et al. Binding of MgtR, a Salmonella transmembrane regulatory peptide, to MgtC, a Mycobacterium tuberculosis virulence factor: A structural study. J Mol Biol. 2014;426(2):436–446. doi: 10.1016/j.jmb.2013.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharma M, et al. Insight into the mechanism of the influenza A proton channel from a structure in a lipid bilayer. Science. 2010;330(6003):509–512. doi: 10.1126/science.1191750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang S, et al. Solid-state NMR spectroscopy structure determination of a lipid-embedded heptahelical membrane protein. Nat Methods. 2013;10(10):1007–1012. doi: 10.1038/nmeth.2635. [DOI] [PubMed] [Google Scholar]
- 36.Sreerama N, Woody RW. On the analysis of membrane protein circular dichroism spectra. Protein Sci. 2004;13(1):100–112. doi: 10.1110/ps.03258404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramamoorthy A, Opella SJ. Two-dimensional chemical shift/heteronuclear dipolar coupling spectra obtained with polarization inversion spin exchange at the magic angle and magic-angle sample spinning (PISEMAMAS) Solid State Nucl Magn Reson. 1995;4(6):387–392. doi: 10.1016/0926-2040(95)00054-t. [DOI] [PubMed] [Google Scholar]
- 38.Wang J, et al. Imaging membrane protein helical wheels. J Magn Reson. 2000;144(1):162–167. doi: 10.1006/jmre.2000.2037. [DOI] [PubMed] [Google Scholar]
- 39.Marassi FM, Opella SJ. A solid-state NMR index of helical membrane protein structure and topology. J Magn Reson. 2000;144(1):150–155. doi: 10.1006/jmre.2000.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murray DT, Hung I, Cross TA. Assignment of oriented sample NMR resonances from a three transmembrane helix protein. J Magn Reson. 2014;240:34–44. doi: 10.1016/j.jmr.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Page RC, Kim S, Cross TA. Transmembrane helix uniformity examined by spectral mapping of torsion angles. Structure. 2008;16(5):787–797. doi: 10.1016/j.str.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mesleh MF, et al. Dipolar waves map the structure and topology of helices in membrane proteins. J Am Chem Soc. 2003;125(29):8928–8935. doi: 10.1021/ja034211q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kovacs FA, Denny JK, Song Z, Quine JR, Cross TA. Helix tilt of the M2 transmembrane peptide from influenza A virus: An intrinsic property. J Mol Biol. 2000;295(1):117–125. doi: 10.1006/jmbi.1999.3322. [DOI] [PubMed] [Google Scholar]
- 44.Braun P, von Heijne G. The aromatic residues Trp and Phe have different effects on the positioning of a transmembrane helix in the microsomal membrane. Biochemistry. 1999;38(30):9778–9782. doi: 10.1021/bi990923a. [DOI] [PubMed] [Google Scholar]
- 45.von Heijne G. Membrane-protein topology. Nat Rev Mol Cell Biol. 2006;7(12):909–918. doi: 10.1038/nrm2063. [DOI] [PubMed] [Google Scholar]
- 46.Kim S, Cross TA. Uniformity, ideality, and hydrogen bonds in transmembrane alpha-helices. Biophys J. 2002;83(4):2084–2095. doi: 10.1016/S0006-3495(02)73969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Igumenova TI, et al. Assignments of carbon NMR resonances for microcrystalline ubiquitin. J Am Chem Soc. 2004;126(21):6720–6727. doi: 10.1021/ja030547o. [DOI] [PubMed] [Google Scholar]
- 48.Franks WT, Kloepper KD, Wylie BJ, Rienstra CM. Four-dimensional heteronuclear correlation experiments for chemical shift assignment of solid proteins. J Biomol NMR. 2007;39(2):107–131. doi: 10.1007/s10858-007-9179-1. [DOI] [PubMed] [Google Scholar]
- 49.Takegoshi K, Nakamura S, Terao T. C-13-H-1 dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem Phys Lett. 2001;344(5-6):631–637. [Google Scholar]
- 50.Takegoshi K, Nakamura S, Terao T. C-13-H-1 dipolar-driven C-13-C-13 recoupling without C-13 rf irradiation in nuclear magnetic resonance of rotating solids. J Chem Phys. 2003;118(5):2325–2341. [Google Scholar]
- 51.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160(1):65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 52.Walther D, Eisenhaber F, Argos P. Principles of helix-helix packing in proteins: The helical lattice superposition model. J Mol Biol. 1996;255(3):536–553. doi: 10.1006/jmbi.1996.0044. [DOI] [PubMed] [Google Scholar]
- 53.Lemmon MA, Flanagan JM, Treutlein HR, Zhang J, Engelman DM. Sequence specificity in the dimerization of transmembrane alpha-helices. Biochemistry. 1992;31(51):12719–12725. doi: 10.1021/bi00166a002. [DOI] [PubMed] [Google Scholar]
- 54.Javadpour MM, Eilers M, Groesbeek M, Smith SO. Helix packing in polytopic membrane proteins: Role of glycine in transmembrane helix association. Biophys J. 1999;77(3):1609–1618. doi: 10.1016/S0006-3495(99)77009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Senes A, Ubarretxena-Belandia I, Engelman DM. The Cα—H⋅⋅⋅O hydrogen bond: A determinant of stability and specificity in transmembrane helix interactions. Proc Natl Acad Sci USA. 2001;98(16):9056–9061. doi: 10.1073/pnas.161280798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vijayakumar M, Qian H, Zhou HX. Hydrogen bonds between short polar side chains and peptide backbone: Prevalence in proteins and effects on helix-forming propensities. Proteins. 1999;34(4):497–507. [PubMed] [Google Scholar]
- 57.Killian JA, von Heijne G. How proteins adapt to a membrane-water interface. Trends Biochem Sci. 2000;25(9):429–434. doi: 10.1016/s0968-0004(00)01626-1. [DOI] [PubMed] [Google Scholar]
- 58.de Planque MR, et al. Interfacial anchor properties of tryptophan residues in transmembrane peptides can dominate over hydrophobic matching effects in peptide-lipid interactions. Biochemistry. 2003;42(18):5341–5348. doi: 10.1021/bi027000r. [DOI] [PubMed] [Google Scholar]
- 59.Das N, Murray DT, Cross TA. Lipid bilayer preparations of membrane proteins for oriented and magic-angle spinning solid-state NMR samples. Nat Protoc. 2013;8(11):2256–2270. doi: 10.1038/nprot.2013.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Höfer N, Aragão D, Caffrey M. Crystallizing transmembrane peptides in lipidic mesophases. Biophys J. 2010;99(3):L23–L25. doi: 10.1016/j.bpj.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Separovic F, Killian JA, Cotten M, Busath DD, Cross TA. Modeling the membrane environment for membrane proteins. Biophys J. 2011;100(8):2073–2074, author reply 2075. doi: 10.1016/j.bpj.2011.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oldfield CJ, Dunker AK. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu Rev Biochem. 2014;83:553–584. doi: 10.1146/annurev-biochem-072711-164947. [DOI] [PubMed] [Google Scholar]
- 63.Sreerama N, Woody RW. Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal Biochem. 2000;287(2):252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 64.Igumenova TI, Wand AJ, McDermott AE. Assignment of the backbone resonances for microcrystalline ubiquitin. J Am Chem Soc. 2004;126(16):5323–5331. doi: 10.1021/ja030546w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.