Significance
Human B cells secrete highly diverse antibody molecules to recognize and defend against infectious agents. Developing B cells independently rearrange their genomes to produce antibody-encoding sequences. It is uncertain to what degree genetic factors control antibody repertoires and the antibodies elicited by defined antigenic stimuli. Analysis of 134,000 antibody heavy chain sequences from genetically identical twins vaccinated with varicella-zoster vaccine indicates that twins show increased correlation in antibody gene segment usage, junctional features, and mutation rates in their antibody pools but show little similarity in clonal responses to an acute stimulus. Therefore, a shared germ-line genome sequence is correlated with overall convergence of antibody repertoires, but the particular antibody response to a given vaccination is less predictable.
Keywords: B cell repertoire, Varicella Zoster vaccine, twin, antibody, convergent
Abstract
Adaptive immune responses in humans rely on somatic genetic rearrangements of Ig and T-cell receptor loci to generate diverse antigen receptors. It is unclear to what extent an individual’s genetic background affects the characteristics of the antibody repertoire used in responding to vaccination or infection. We studied the B-cell repertoires and clonal expansions in response to attenuated varicella-zoster vaccination in four pairs of adult identical twins and found that the global antibody repertoires of twin pair members showed high similarity in antibody heavy chain V, D, and J gene segment use, and in the length and features of the complementarity-determining region 3, a major determinant of antigen binding. These twin similarities were most pronounced in the IgM-expressing B-cell pools, but were seen to a lesser extent in IgG-expressing B cells. In addition, the degree of antibody somatic mutation accumulated in the B-cell repertoire was highly correlated within twin pair members. Twin pair members had greater numbers of shared convergent antibody sequences, including mutated sequences, suggesting similarity among memory B-cell clonal lineages. Despite these similarities in the memory repertoire, the B-cell clones used in acute responses to ZOSTAVAX vaccination were largely unique to each individual. Taken together, these results suggest that the overall B-cell repertoire is significantly shaped by the underlying germ-line genome, but that stochastic or individual-specific effects dominate the selection of clones in response to an acute antigenic stimulus.
Human responses to infectious diseases or vaccinations rely on many different cell populations, soluble mediators, and interactions between cells. Prior studies of identical twins have highlighted aspects of human immunity that are heavily influenced by the germ-line genome, such as the proportions of particular leukocyte subsets (1). In addition to the germ line-encoded genes that affect the responses of immune cells, mammalian immune systems also make use of somatic genetic rearrangements to produce diverse repertoires of immunoglobulins (Igs) and T-cell receptors (TCRs) for specific recognition of foreign antigens. Antibody and TCR sequences are generated through the combinatorial use of a set of predetermined gene segments in the genome, as well as by more random exonuclease digestion of the ends of the gene segments, and addition of nontemplated bases at the junctions between gene segments. Clonal expansion of the populations of cells that recognize particular pathogens provides immune system memory of prior exposures.
The germ-line genome sequence plays a role in the initial generation and selection of antibody and T-cell receptor repertoires in each individual, as demonstrated in prior work (2–6). Whether such genetic effects have a prominent effect on the clonal B-cell responses to particular pathogens or vaccinations is much less clear. In adult humans, it is possible that the accumulation of the effects of responses to prior antigenic exposures in the lymphocyte memory compartments could greatly decrease the importance of an individual's germ-line genome on subsequent responses to pathogens or vaccines.
Convergent antibodies (i.e., antibodies with highly similar sequences observed in different individuals after vaccination or infection with the same agents) have been identified by deep sequencing of antibody genes in the context of Dengue virus and HIV infection and influenza vaccination and in earlier studies of polysaccharide vaccines (7–13), providing evidence that some antigenic stimuli can provoke relatively predictable responses even in genetically diverse human populations. It is not known to what extent convergent responses are influenced by germ-line genomic variation between individuals.
We studied a cohort of four pairs of identical twin adults undergoing vaccination with live attenuated varicella-zoster virus (VZV) vaccine (ZOSTAVAX) to more fully address which aspects of human B-cell responses are most influenced by the germ-line genome. We found that the composition of the antibody heavy chain repertoire at baseline was highly similar in identical twins, as assessed by V (variable), D (diversity), and J (joining) segment use and complementarity-determining region 3 (CDR3) lengths. These effects were especially prominent in the IgM repertoires whereas IgG sequence pools were more divergent. Surprisingly, the levels of mutation achieved by antibodies from the antigen-experienced B cells in the repertoires of twins were also highly similar. Convergent antibody sequences were enriched in twin pairs, including mutated sequences likely derived from memory B cells. In contrast, twins showed no correlation in the magnitude of the B-cell clonal expansions to VZV vaccination. These results indicate that set points for features of both naive and memory B-cell repertoires in humans are influenced by the germ-line genome but that, despite this, the particular antibody response to a vaccination event is less predictable.
Results
B-Cell Clonal Expansion and Secreted Antibody Responses to ZOSTAVAX.
We vaccinated four pairs of healthy adult monozygotic twins and measured their serum antibody titers against VZV, prevaccination and 28 d after vaccination. As shown in Fig. 1A, increased anti-VZV antibodies were detected postvaccination in all participants, with a fold titer change ranging from 1.2 to 1.6. Increased numbers of plasmablast B cells were detected in the blood at day 8 postvaccination in all but one participant, A2 (Fig. 1B).
Fig. 1.
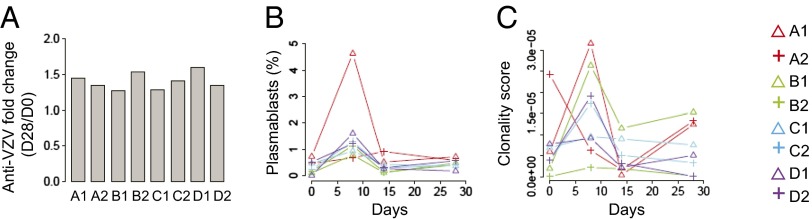
Antibody responses, plasmablast counts, and clonal expansion postvaccination. (A) The anti-VZV activity in the serum was expressed as a ratio of anti-VZV titer on day 28/day 0. (B) Plasmablasts as a percentage of total CD3− CD19+ B cells on days 0 (prevaccination), 8, 14, and 28 postvaccination. Samples from twin pair members are labeled with the same colors, with each individual within the twin pair labeled with a different symbol: twins A (red), twins B (green), twins C (blue), and twins D (purple). (C) Day 8 B-cell clonal expansion differs between twin pair members. Clonality score of gDNA sequences from each sample was calculated from six independent gDNA replicates. The x axis indicates the number of days postvaccination.
We then carried out high-throughput DNA sequencing of immunoglobulin heavy chain (IGH) variable regions, using replicate libraries from each time point prepared from genomic DNA (gDNA) and, for two of the twin pairs, cDNA template from peripheral blood mononuclear cells (PBMCs). Libraries were sequenced with the 454 high-throughput sequencing platform. The use of replicate gDNA libraries permits measurement of B-cell clonal expansions, quantified by a “clonality score” that can be considered as the probability that two B cells selected from the sample will belong to the same clone, and is not affected by differential expression of IGH mRNA by different B cells (8, 14). As shown in Fig. 1C, the clonality score increased on day 8 postvaccination for all but one participant and then decreased to baseline levels by day 14, consistent with detection of the clonal expansions of B cells induced by vaccination. Participant A2, whose plasmablasts did not peak on day 8, had a decreased clonality score on day 8 compared with day 0. Sequencing of cDNA-templated libraries from twin pairs C and D permitted analysis of antibody isotypes during the vaccination time course. Differential expression of IgG subtypes was observed during the vaccine response, with the proportion of IgG1 reads increasing transiently on day 8 whereas IgG2 showed a relative decrease on day 8 (SI Appendix, Fig. S1). These vaccine-induced B-cell responses did not show greater similarity between twin pair members compared with unrelated individuals and were a consistent feature of all vaccinated individuals.
B-Cell IGH V, D, J Segment Use and CDR3 Lengths Are Correlated in Twin Pairs.
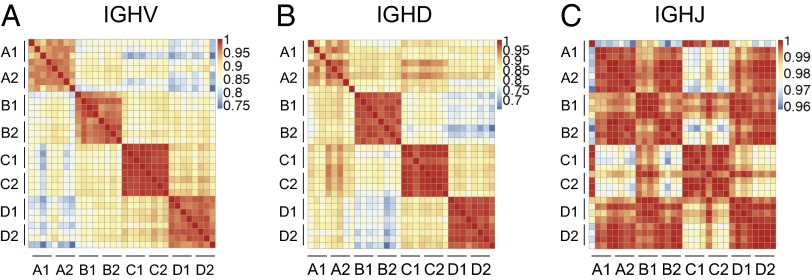
Duplicate sequences were removed from the total IGH sequence datasets, yielding ∼70,000 unique gDNA clone sequences for analysis of IGHV, IGHD, and IGHJ segment use. We found that each individual’s frequency of use of different gene segments over the vaccination time course was fairly stable, shown as high correlations in the heatmap of Fig. 2, suggesting that V, D, J segment use is not markedly altered by ZOSTAVAX (SI Appendix, Figs. S2 and S3). The high correlation within each individual was shared with their identical twin, as seen in each block of eight samples along the diagonal in the correlation heat map (four time points from each twin pair member) (Fig. 2). In contrast, V and D segment use frequencies are quite distinguishable between different twin pairs, due to differences in use frequencies for many segments (SI Appendix, Fig. S2). The six J segments in the human IGH locus provided less discrimination between twin pairs, with three twin pairs (twins A, B, and D) sharing high correlation with each other (Fig. 2C) whereas the fourth pair (twins C) used significantly less IGHJ6 and more IGHJ4 (SI Appendix, Fig. S2).
Fig. 2.
IGHV, IGHD, and IGHJ gene use is highly correlated within twin pairs. Correlation heatmap of use of V (A), D (B), and J (C) gene segments in the IgH of all samples, using gDNA template. Each square represents the correlation of gene segment use of two samples. Four samples from each individual are positioned sequentially in the order of time points (d0, d8, d14, and d28 from left to right in the x axis and from top to bottom in the y axis) for the eight individuals as labeled in the figure (A1, A2, B1, B2, C1, C2, D1, and D2 from left to right in the x axis and from top to bottom in the y axis). Correlations were calculated as Pearson correlations, using all V, D, or J gene segments identified in the data, without regard for allelic variants. The colored scale bar represents the strength of correlation of each sample pair.
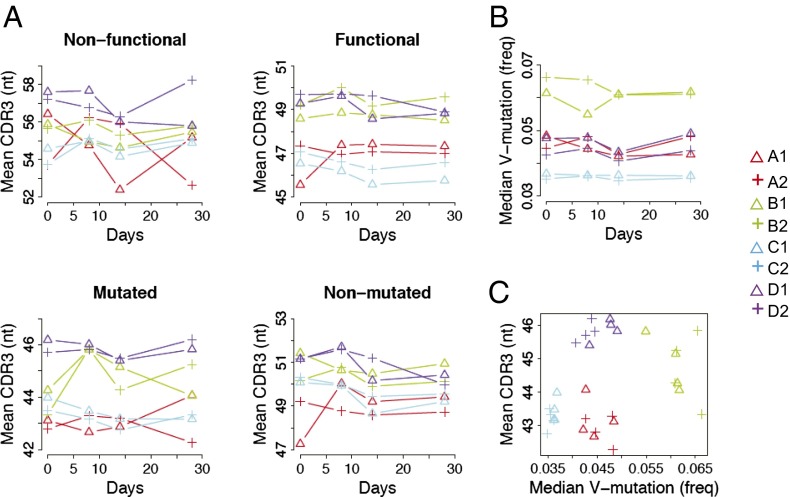
The CDR3 amino acids of the antibody heavy chain are often a key determinant of antigen binding specificity and are encoded by the region spanning the junctions of the joined V, D, and J gene segments. To analyze CDR3 regions amplified from gDNA, which would include unproductive rearrangements left over from initial attempts to rearrange the IGH locus in B-cell development, we first separated productive and unproductive rearrangements, as well as productive mutated and unmutated rearrangements. These categories of rearrangement are known to differ in CDR3 length, most likely as a result of selection processes acting on the expressed antibody proteins (14, 15). As shown in Fig. 3A, over the monitored vaccination time course, mean CDR3 length of the B-cell repertoire was relatively stable for all individuals. Twins shared a high correlation in CDR3 lengths of functional sequences in both the unmutated and mutated categories. CDR3 lengths of nonfunctional sequences were more variable, most likely as a result of the 5- to 10-fold decreased number of sequences available for analysis.
Fig. 3.
CDR3 length and IGHV mutation of functional gDNA is highly correlated within twin pairs. (A) Mean CDR3 of nonfunctional or functional sequences, and mutated or nonmutated sequences in the functional category on different days. (B) Median V-mutation frequency of the mutated sequences. (C) Samples from the same twin pairs of different time points form clusters on the 2D presentation of mean CDR3 and median V-mutation. Members of the same twin pair are labeled with the same color, and the two individuals from each twin pair are labeled with different symbols.
Mutation Levels in Antibody Heavy Chains Are Surprisingly Correlated in Twins.
Unmutated and mutated antibody genes are characteristic, respectively, of naive and antigen-experienced B cells such as memory B cells. Memory B cells accumulate somatic mutations over the affinity maturation process. The factors that control the levels of mutation reached in the memory B-cell pool are poorly understood and are only weakly associated with factors such as age in humans (14). We examined mutation levels in the IGHV genes of functional gDNA sequences in the twin subjects, all of whom are adults in the sixth decade of life, and observed similar distribution of IGHV-gene mutations within most twin pairs (SI Appendix, Fig. S4). The exception existed with twin pair B, one individual of which had unusually less mutated sequences than the other. On the other hand, both members of twin pair C had an unusually high proportion of mutated sequences. This phenomenon can also be seen in other healthy human subjects in an earlier study (14) and is now shown to be shared within twins, suggesting that it might be inherently controlled by genetic factors. The unusually low or high level of V-mutations in these individuals are not due to expansion of a few clones, as reflected in their clonality scores, which are similar to those of other individuals. Further examination of mutation levels in the mutated sequences revealed that mutation levels were tightly correlated in twin pair members (Fig. 3B). This finding suggests that either the rate of somatic mutation in stimulated B cells or the features of B cells that join the memory compartment may be under significant germ-line genetic control.
CDR3 Lengths and Mutation Frequency in IGH Are Under Independent Control.
The separation between different twin pairs on a plot of CDR3 length and IGHV segment mutation levels is very distinct (Fig. 3C). When the K-means nonsupervised learning method was used to cluster the samples with a cluster number of four (K = 4), only two out of 32 total samples were not clustered as expected with their twin partners (SI Appendix, Fig. S5). In an individual’s B-cell repertoire, there is typically a negative correlation between antibody mutation levels and CDR3 lengths (14–17). The mechanism underlying this phenomenon is not known. This relationship in antibody features within an individual was seen for all subjects in the current study. However, comparing the IGH CDR3 lengths and mutation levels between different twin pairs revealed the striking finding that these overall properties of a twin pair’s B-cell repertoire seem to be independently determined. In other words, although the antibodies within a particular individual show shorter CDR3 length for more mutated antibodies, some twin pairs, such as pair D, have relatively long CDR3s in proportion to the mutation levels of their antibodies (Fig. 3C). These observations suggest that CDR3 formation and IGH mutational storage are under separate kinds of genetically influenced control.
IgM Repertoires of Twins Are More Correlated than IgG Repertoires.
Isotype switching of activated B cells determines the effector functions that their antibodies can carry out, and some isotypes are stimulated more prominently in vaccine responses (SI Appendix, Fig. S1). Antibodies of different isotypes also differ in their mean CDR3 length and V-mutation levels (14). The above within-twin correlations of CDR3 length and IGHV mutation levels could just result from twin pair member correlation in isotype ratios. We therefore evaluated the antibodies of different expressed isotypes in two twin pairs from whom RNA samples were available (twin pairs C and D). For IgM, the mean CDR3 length of functional sequences expressed by B cells observed in the blood, mutated or not, was not altered by vaccination and was highly correlated within twin pairs (Fig. 4). IGHV mutation levels showed a similar correlation of IgM within twin pairs (SI Appendix, Fig. S6). In the case of IgG, changes in the CDR3 length and IGHV mutation levels at day 8 postvaccination were identified, consistent with the expected vaccine-induced plasmablast populations that typically peak in the blood 1 wk after vaccination. There were not enough sequences of other isotypes for analysis. These findings highlight differences between both the naive and mutated IgM- and IgG-expressing B-cell populations, suggesting that IgM-expressing memory B-cell repertoires are more similar in identical twins, compared with the IgG-expressing B-cell repertoires.
Fig. 4.
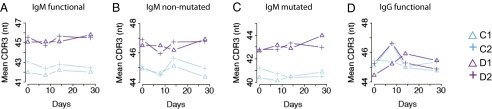
CDR3 lengths of IgM are correlated within twin pairs. Mean CDR3 length of IgM functional sequences (A), nonmutated (B) or mutated (C), and functional IgG (D) sequences. Twin pair C is blue and twin pair D is purple.
Using only the IgM sequences to reduce confounding variables, we further investigated the mechanisms responsible for the within-twins correlation in CDR3 length, in addition to the highly correlated use of IGHV, IGHD, and IGHJ segments. The human IGHJ6 gene segment contributes an unusually long sequence to the CDR3 of IGH rearrangements where it is used. Notably, the correlation of IGH CDR3 lengths within twin pairs remains, even if we split the sequences according to the IGHJ gene segment that they use (SI Appendix, Fig. S7), suggesting the involvement of other factors. Examination of the ways in which the CDR3 junctions were formed in each twin pair suggests that both the degree of exonuclease digestion of segment ends and the number of randomly added nucleotides during joining (SI Appendix, Fig. S8) contribute to the CDR3 length similarity within twin pair members.
Increased Convergence of Antibody Sequences Within Twins.
Given that twins’ B-cell receptor (BCR) repertoires share similar IGHV, IGHD, and IGHJ use profiles and CDR3 length, it seemed possible that, averaged over many different antigenic exposures, greater similarity in the antigen-stimulated B-cell clones might be observed within twin pairs. To test this hypothesis, we pooled all IGH sequences from the study and looked for convergent clones (defined as sequences from different samples having identical CDR3 length and >90% identity in CDR3 amino acid sequences) in all individuals and time points. To produce a normalized measure of the number of convergent sequences found between libraries, accounting for any differences in sequencing depth, we defined a convergence index as the ratio of the number of convergent clones to the product of the clone set sizes of the two libraries. As expected (SI Appendix, Fig. S9), many highly similar sequences are detected in different time points from the same person, owing to the presence of clonally expanded B cells within each individual. However, the number of convergent sequences found between any sample pair from different individuals was low, regardless of their immunization and twin status. We estimated the effect of immunization and twin status on the number of convergent lineages found between each pair of individuals using a Poisson regression model. We found that the number of convergent sequence lineages was significantly increased in twins by 1.5-fold (SI Appendix, Table S2, converted from the coefficient of 0.43 for twin status in the Poisson model). In the regression model, the influence of the immunization time point (comparing individuals at day 8) did not reach significance.
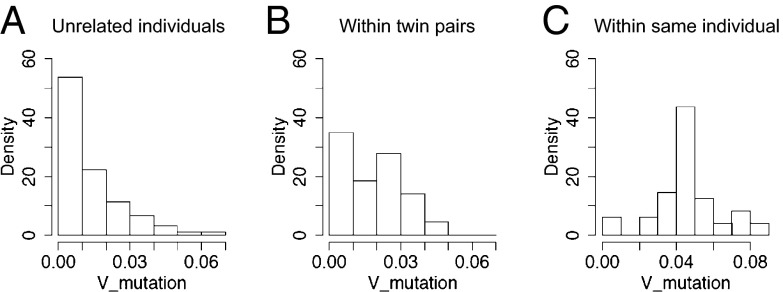
We further examined the mutation status of the convergent clones. We first calculated the average V-mutation rates of all convergent clones between any sample pair and then split these numbers to three categories depending on whether the sample pair came from unrelated individuals, from within twin pair members, or from different time points of the same individual. As summarized in Fig. 5, convergent clones between unrelated individuals were dominated by unmutated sequences likely representing similar rearrangements in the naive B-cell repertoire, before antigenic stimulation whereas those from within the same individual were enriched with mutated sequences, as one would expect from clonally expanded memory B cells in each subject. The convergent clones shared between twin pair members seem to be a mixture of these two patterns, having major populations of both germ-line and, to a lesser degree, mutated convergent antibodies, suggesting that the latter represent similar memory-cell populations elicited as a result of antigenic exposure. Therefore, although the particular B-cell clones stimulated by the single VZV vaccination event monitored in this study showed few similarities between twin pair members, it seems that the antigen-experienced mutated B-cell repertoires acquired over a lifetime of various antigenic stimuli do show greater convergence within twins.
Fig. 5.
IGHV mutation frequencies of convergent clones are affected by twin status. Average V-mutation frequencies of clones convergent between sample pairs from unrelated individuals (A), from within twin-pair members (B), and between different time point samples from the same individual (C).
Discussion
Examination of immune responses to ZOSTAVAX in identical twins provides a rare opportunity to evaluate the similarity of the serological and clonal B-cell responses to a known antigenic stimulus in genetically identical humans. All individuals recruited developed increased titers of anti-VZV antibodies, and seven out of eight had elevated plasmablasts at day 8 postvaccination, demonstrating detectable B-cell and serological responses to the vaccine. Clonal expansion of B cells determined by high-throughput DNA sequencing of IGH gene rearrangements similarly confirmed the vaccine responses. These serological and cellular aspects of the vaccine response were, however, variable between individuals and were unable to distinguish twins from nontwins.
In contrast, analysis of the Ig repertoire showed preferential and nonrandom use of V, D, and J gene segments in IGH rearrangements of identical twins, as suggested in earlier studies at the level of IGHV family use or in more limited sample numbers (3, 4). Perturbation of the immune system by VZV vaccination did not have a major impact on the gene segment use by circulating B cells. In our data, the similarity of gene segment use in twin IGH repertoires was greater in unmutated than mutated productive rearrangements, suggesting that the processes governing B-cell repertoire generation are more similar in twins compared with unrelated individuals, but that the subsequent selection processes and responses to environmental exposures are less genetically controlled.
Our results indicate that CDR3 length and the extent of somatic mutation in the IGHV segments, especially in antibodies of the IgM isotype, are both highly correlated within twin pairs but seem to be independent of each other, suggesting that they are affected by different kinds of control mechanisms. The twin correlation in CDR3 length shows the combined effects of higher similarity in V, D, and J segment use, as well as greater similarity of the extent of nucleotide trimming and nontemplated nucleotide addition at the segment junctions. The activity levels of exonucleases and TdT during VDJ rearrangement could account for the greater similarity in IGH CDR3 regions of twins, but it is also possible that selection acting on B cells leaving the bone marrow could contribute to the observed CDR3 lengths in functional rearrangements in peripheral blood B cells. One of the more surprising observations in the study was the close correlation between twins in the levels of mutation detected in their IGH sequences, presumably derived from memory B cells. The mechanisms controlling the extent of accumulation of mutations in memory B cells are not fully understood in humans, but our data indicate that different individuals preferentially generate or retain different levels of mutation in their memory B-cell compartments. Differences in the B-cell memory storage features between individuals could have major clinical implications for the capacity to recognize variant strains of a virus, in which case retaining less-mutated memory clones from prior exposures to similar viruses could be an advantage. On the other hand, storage of the highest affinity, and potentially most highly mutated, B-cell clones specialized toward binding a particular viral variant could accelerate the recall response if that particular virus is encountered at a future time. However, the within-twin correlation and intertwin differences in CDR3 length and V-mutations are not as obviously manifested in B cells expressing IgG, suggesting that the properties of these B cells are less controlled by genetic background and are potentially more strongly affected by particular antigenic exposures.
Convergent (or public) CDR3s in unrelated individuals are common in the IGK and IGL light chain repertoires, due to the limited number of gene segments and the single junction encoding them (18, 19). Low-frequency convergent IGH sequences have also been reported in patients with viral infections or recent vaccination, including responses to repetitive polysaccharide vaccine antigens (7, 8, 13, 20). We identified increased frequencies of antibodies with convergent CDR3 sequences in the overall B-cell repertoires of twin pairs. Notably, the convergent antibody sequences identified between twin pair members included both unmutated naive sequences, as well as mutated, presumably memory B cell-derived sequences, whereas convergent sequences between unrelated individuals were predominantly unmutated. Because the great majority of the B-cell clones stimulated by ZOSTAVAX vaccination were unique to each individual, stochastic events seem to play the greatest role in determining which VZV-reactive B-cell clones happen to be stimulated by a particular vaccination event, perhaps as a result of anatomic localization of individual naive and memory B cells relative to the site of vaccine delivery. Nonetheless, when averaged over the many antigenic exposures encountered by an immune system in five decades of life, the similarities in identical twins’ underlying B-cell repertoires do give rise to increased proportions of convergent IGH sequences in the mutated, antigen-experienced memory B-cell compartment.
In summary, our study with monozygotic twins vaccinated with live attenuated VZV vaccine suggests that genetic traits strongly influence some features of the B-cell repertoire, such as the IGHV, IGHD, and IGHJ gene use, as well as CDR3 length and IGHV mutation frequency. Increased numbers of convergent antibody heavy chains, including mutated sequences likely derived from memory B cells, are seen between twin pair members. Despite these overall repertoire similarities, however, the extent of the plasmablast response, the overall level of B-cell clonal expansion, and the serological response to vaccination are not well-correlated in twins, and the particular B-cell clones responding to the VZV vaccination event are, in a single vaccination episode, unique to each individual. The results highlight the relationship between the germ-line genomic locus that seems to set overall parameters for the IGH repertoire that will be generated in each individual, and the less predictable stimulation of particular B-cell clones that give rise to the immune response after a vaccination episode. We note that we cannot exclude some role for common antigenic exposures between twin pair members in the similarities of their B-cell repertoires, but the distinct responses each twin gave to the observed VZV vaccination, as well as the fact that the twins studied here were in their sixth decade of life, would suggest that shared antigen exposures are unlikely to account for our findings. In addition, the similar repertoires of naive, unmutated antibody genes shared between twins are unlikely to have been influenced by antigenic exposures. Importantly, the increased similarity of the mutated B-cell clones in identical twins’ repertoires gives reason to believe that statistical analysis of the clonal B-cell responses of much larger numbers of unrelated individuals will also begin to reveal predictable patterns of responses to vaccines and infectious agents, permitting better prediction of individual immune responses and effective vaccination strategies.
Materials and Methods
Specimen Collection.
Four pairs of monozygotic twins were recruited from the SRI twin registry. Demographic and recruitment details are available elsewhere (21). Zygosity was determined by a GoldenGate genotyping assay (Illumina Inc.) to interrogate 384 SNP loci, and assays were performed by IGenix. Twins were considered identical if DNA markers were above 99%. Inclusion criteria for this study included prior history of chicken pox. Exclusion criteria included an episode of shingles within 5 y of the study or prior vaccination with ZOSTAVAX. Demographic information is supplied in SI Appendix, Table S1. Twins were vaccinated with live attenuated ZOSTAVAX vaccine (Merck & Co. Inc.). Peripheral blood was collected on the day of vaccination (before vaccine delivery) and 8, 14, and 28 d after. Recruitment of patients, documentation of informed consent, collection of blood specimens, and experimental measurements were carried out with Institutional Review Board approval at Stanford University and SRI International.
Anti-VZV Antibody Titer Determination.
Anti-VZV IgG antibody titer was determined using a VZV IgG ELISA kit (Calbiotech). Before, and 28 d after, vaccination, serum samples from individuals were collected, frozen within 1 h of drawing, and stored at−80 °C. On the day of the assay, serum samples were thawed, diluted 1:21 in sample diluent, and added to ELISA plates in duplicate with positive and negative controls and the assay calibrator. After development with 3,3′,5,5′-tetramethylbenzidine (TMB) substrate, the reaction was stopped with stop solution, and plates were read at 450 nm within 15 min using an iMark Microplate Reader (Bio-Rad), with a reference filter of 655 nm. Duplicate readings were averaged to calculate antibody index and fold-change determined from day 0 to day 28.
Plasmablast Frequency Determination by Flow Cytometry.
Blood samples were taken before, and 8, 14, and 28 d after, vaccination. Peripheral blood mononuclear cells (PBMCs) were extracted by Ficoll density centrifugation and immediately stained with the following directly conjugated antibodies: anti-CD3 FITC, anti-CD38 APC (Biolegend), anti-CD20 PerCP-Cy5.5 (eBioscience), anti-CD19 PE-Cy7, and anti-CD27 V500 (BD Biosciences). Samples were run on a Fortessa flow cytometer (BD Biosciences) and analyzed using FlowJo (TreeStar). Plasmablasts were identified as CD3−, CD19+, CD20−, CD27+, and CD38+.
Genomic DNA and cDNA Template Preparation and PCR Amplification.
Library preparation of the IGH variable region followed the protocol as described (14). Briefly, gDNA and RNA were extracted from PBMCs, cDNA was synthesized with priming by random hexamers, and templates were amplified by PCR using Biomed primers (22), barcoded for sample identification. PCR was carried out in six independent replicates with the same sets of primers for gDNA and with different isotype primers separately for cDNA. High-throughput sequencing was performed on the 454 platform using Titanium chemistry (Roche).
Sequence Preprocessing.
Sequences were passed through a pipeline for sample decoding, annotation, and quality filtering (14). Briefly, V, D, and J gene and junctions were annotated based on alignment to germ-line sequences using iHMMune-align (23, 24), which rejected sequences with indels in the V segment. Non-IGH sequences and chimeric sequences were also removed. Substitution error rates from PCR errors and 454 sequencing are estimated to be less than 0.3% per base in data prepared with this protocol, as judged from constant region sequences. These levels are low enough that they are unlikely to influence the identification of the V, D, or J gene segments, and would very rarely cause a failure to recognize clonally related sequences in the IGH repertoires. We do not annotate the allelic variants for each V, D, and J gene segment because these are more sensitive to the error rates in current high-throughput sequencing data.
Sequence Analysis.
Candidate clonally related sequences were defined as sequences having the same V and J segment and identical CDR3 amino acid sequences. Clonality expansion in each repertoire was estimated using a metric called clonality score calculated as the following: sum of Nij × Nik (j!=k) over i, j, k, divided by the sum of Tj × Tk (j!=k) over j, k, where Nij and Nik are the copy numbers of clone i observed in independent replicate PCR libraries j and k generated from independent aliquots of template DNA, and Tj and Tk are total read numbers in the corresponding replicate libraries (for more details, see ref. 14). For analyses of V, D, and J gene use, CDR3, and V-D (N1) and D-J (N2) junction features, redundant sequences belonging to the same clone were collapsed to one sequence, resulting in 69,500 gDNA sequences and 64,700 cDNA sequences. Clones without D gene assignment were removed for V, D, and J use analysis. Sufficient cDNA/RNA sequences were available only for twin pairs C and D. For V-segment mutation analysis, we used a threshold of at least 1% mutation in the V segment when referring to “mutated sequences” to ensure that we did not overinterpret sequencing errors as somatic mutations.
K-Means Clustering of Samples Based on CDR3 Length and V-Mutation.
Mean CDR3 length and median V-mutation frequency of mutated gDNA sequences from each sample were centered to have a mean of zero and were scaled to have an SD of one. K-means clustering was performed with these two parameters using the “kmeans” command in the statistical package R. Within groups sum of squares of clusters was calculated to determine the appropriate number of clusters.
Convergence of Repertoire.
Converging clones were searched with the criteria of having >90% identity in their CDR3 amino acid sequences with the same CDR3 length, using program CD-HIT, version 4.6 (25, 26). Normalized convergence was defined as the number of converging clones between two samples divided by the product of their clone set sizes (total number of clones). The effect of twin- and immunization-status on convergence was estimated by Poisson regression with counts of converging clones between sample pairs against the log of product of their clone set sizes, immunization status, and twin status, including only the sample pairs from different individuals and from prevaccination or day 8 postvaccination. V-mutation extent of convergent clones was calculated as an average of all clones converged between any sample pair.
Statistical Analysis.
All statistical analyses and figures were generated using R.
Sequence Data.
Sequences from this study can be accessed via dbGaP with accession number phs000817.v1.p1.
Supplementary Material
Acknowledgments
We thank the following individuals: Mary McElroy, Lisa Jack, Ruth Krasnow, Jill Rubin, Dina Basin, Lucia Panini, Marty Ritchey, and Shilpa Joshi. Twins were recruited from the Twin Research Registry at SRI International, Menlo Park, CA. We thank the contributions and commitment to science provided by the twins through their ongoing participation in the Registry and various research studies. The Principal Investigator of the Twin Research Registry is Gary E. Swan (gswan@stanford.edu, 650-255-2448). The Stanford-LPCH Vaccine Program thanks our study volunteers for their participation; Sally Mackey for project, regulatory, and data management; research nurses Sue Swope and Nancy Mastman; research assistants Ashima Goel and Thu Quan; and phlebotomist Michele Ugur. This study was supported by grants from the NIH, including Grant U19 AI090019, an Ellison Medical Foundation New Scholar in Aging award (to S.D.B.), and Grants U19 AI090019, R01 AG015043, U19 AI057266, and R01 AI108891 (to J.J.G.). The project was supported by NIH/National Center for Research Resources Clinical Translational Sciences Award no. UL1 RR025744. Funds to support the development and maintenance of the Registry were provided through SRI’s Center for Health Sciences and through grants from the NIH, including Grants DA011170, DA023063, AI057229, AI090019, and ES022153.
Footnotes
Conflict of interest statement: Scott Boyd has consulted for Immumetrix, Inc., now part of CareDx, Inc., on topics distinct from those in the current manuscript.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the dbGaP database (accession no. phs000817.v1.p1).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1415875112/-/DCSupplemental.
References
- 1.Orrù V, et al. Genetic variants regulating immune cell levels in health and disease. Cell. 2013;155(1):242–256. doi: 10.1016/j.cell.2013.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyd SD, et al. Individual variation in the germline Ig gene repertoire inferred from variable region gene rearrangements. J Immunol. 2010;184(12):6986–6992. doi: 10.4049/jimmunol.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glanville J, et al. Naive antibody gene-segment frequencies are heritable and unaltered by chronic lymphocyte ablation. Proc Natl Acad Sci USA. 2011;108(50):20066–20071. doi: 10.1073/pnas.1107498108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kohsaka H, et al. The human immunoglobulin V(H) gene repertoire is genetically controlled and unaltered by chronic autoimmune stimulation. J Clin Invest. 1996;98(12):2794–2800. doi: 10.1172/JCI119106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fozza C, et al. T-cell receptor repertoire analysis in monozygotic twins concordant and discordant for type 1 diabetes. Immunobiology. 2012;217(9):920–925. doi: 10.1016/j.imbio.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 6.Zvyagin IV, et al. Distinctive properties of identical twins’ TCR repertoires revealed by high-throughput sequencing. Proc Natl Acad Sci USA. 2014;111(16):5980–5985. doi: 10.1073/pnas.1319389111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parameswaran P, et al. Convergent antibody signatures in human dengue. Cell Host Microbe. 2013;13(6):691–700. doi: 10.1016/j.chom.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson KJ, et al. Human responses to influenza vaccination show seroconversion signatures and convergent antibody rearrangements. Cell Host Microbe. 2014;16(1):105–114. doi: 10.1016/j.chom.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scott MG, et al. Clonal characterization of the human IgG antibody repertoire to Haemophilus influenzae type b polysaccharide. II. IgG antibodies contain VH genes from a single VH family and VL genes from at least four VL families. J Immunol. 1989;143(1):293–298. [PubMed] [Google Scholar]
- 10.Park MK, Sun Y, Olander JV, Hoffmann JW, Nahm MH. The repertoire of human antibodies to the carbohydrate capsule of Streptococcus pneumoniae 6B. J Infect Dis. 1996;174(1):75–82. doi: 10.1093/infdis/174.1.75. [DOI] [PubMed] [Google Scholar]
- 11.Silverman GJ, Lucas AH. Variable region diversity in human circulating antibodies specific for the capsular polysaccharide of Haemophilus influenzae type b: Preferential usage of two types of VH3 heavy chains. J Clin Invest. 1991;88(3):911–920. doi: 10.1172/JCI115394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ademokun A, et al. Vaccination-induced changes in human B-cell repertoire and pneumococcal IgM and IgA antibody at different ages. Aging Cell. 2011;10(6):922–930. doi: 10.1111/j.1474-9726.2011.00732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Binley JM, et al. Human antibody responses to HIV type 1 glycoprotein 41 cloned in phage display libraries suggest three major epitopes are recognized and give evidence for conserved antibody motifs in antigen binding. AIDS Res Hum Retroviruses. 1996;12(10):911–924. doi: 10.1089/aid.1996.12.911. [DOI] [PubMed] [Google Scholar]
- 14.Wang C, et al. Effects of aging, cytomegalovirus infection, and EBV infection on human B cell repertoires. J Immunol. 2014;192(2):603–611. doi: 10.4049/jimmunol.1301384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosner K, et al. Third complementarity-determining region of mutated VH immunoglobulin genes contains shorter V, D, J, P, and N components than non-mutated genes. Immunology. 2001;103(2):179–187. doi: 10.1046/j.1365-2567.2001.01220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Briney BS, Willis JR, Crowe JE., Jr Human peripheral blood antibodies with long HCDR3s are established primarily at original recombination using a limited subset of germline genes. PLoS ONE. 2012;7(5):e36750. doi: 10.1371/journal.pone.0036750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu YC, et al. High-throughput immunoglobulin repertoire analysis distinguishes between human IgM memory and switched memory B-cell populations. Blood. 2010;116(7):1070–1078. doi: 10.1182/blood-2010-03-275859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoi KH, Ippolito GC. Intrinsic bias and public rearrangements in the human immunoglobulin Vλ light chain repertoire. Genes Immun. 2013;14(4):271–276. doi: 10.1038/gene.2013.10. [DOI] [PubMed] [Google Scholar]
- 19.Jackson KJ, et al. Divergent human populations show extensive shared IGK rearrangements in peripheral blood B cells. Immunogenetics. 2012;64(1):3–14. doi: 10.1007/s00251-011-0559-z. [DOI] [PubMed] [Google Scholar]
- 20.Lucas AH, Reason DC. Polysaccharide vaccines as probes of antibody repertoires in man. Immunol Rev. 1999;171:89–104. doi: 10.1111/j.1600-065x.1999.tb01343.x. [DOI] [PubMed] [Google Scholar]
- 21.Krasnow RE, Jack LM, Lessov-Schlaggar CN, Bergen AW, Swan GE. The Twin Research Registry at SRI International. Twin Res Hum Genet. 2013;16(1):463–470. doi: 10.1017/thg.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Dongen JJ, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: Report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17(12):2257–2317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 23.Jackson KJ, Boyd S, Gaëta BA, Collins AM. Benchmarking the performance of human antibody gene alignment utilities using a 454 sequence dataset. Bioinformatics. 2010;26(24):3129–3130. doi: 10.1093/bioinformatics/btq604. [DOI] [PubMed] [Google Scholar]
- 24.Gaëta BA, et al. iHMMune-align: Hidden Markov model-based alignment and identification of germline genes in rearranged immunoglobulin gene sequences. Bioinformatics. 2007;23(13):1580–1587. doi: 10.1093/bioinformatics/btm147. [DOI] [PubMed] [Google Scholar]
- 25.Li W, Jaroszewski L, Godzik A. Clustering of highly homologous sequences to reduce the size of large protein databases. Bioinformatics. 2001;17(3):282–283. doi: 10.1093/bioinformatics/17.3.282. [DOI] [PubMed] [Google Scholar]
- 26.Li W, Jaroszewski L, Godzik A. Tolerating some redundancy significantly speeds up clustering of large protein databases. Bioinformatics. 2002;18(1):77–82. doi: 10.1093/bioinformatics/18.1.77. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.