Multiple sequence aligners typically work by progressively aligning the most closely related sequences or group of sequences according to guide trees. In PNAS, Boyce et al. (1) report that alignments reconstructed using simple chained trees (i.e., comb-like topologies) with random leaf assignment performed better in protein structure-based benchmarks than those reconstructed using phylogenies estimated from the data as guide trees. The authors state that this result could turn decades of research in the field on its head. In light of this statement, it is important to check immediately whether their result holds under evolutionary criteria: recovery of homologous sequence residues and inference of phylogenetic trees from the alignments (2). We have done this and the results are entirely opposed to Boyce et al.’s findings (1).
Simulation entails simplifying assumptions, but provides a baseline for which the truth is known with certainty. Using ALF (3), we simulated over 100 different evolutionary scenarios, each containing 1,024 homologous sequences evolved along trees generated from birth–death processes. We then applied the same aligners as Boyce et al. (1) (ClustalOmega, Mafft, Muscle) and additionally Prank (4), using as guide trees: (i) chained tree with random leaf assignment of Boyce et al. (1); (ii) balanced tree with leaf assignment optimized using the traveling salesman problem heuristic as tested by Boyce et al. (1); (iii) default tree estimated by each aligner; (iv) least-squares distance tree estimated using specialized phylogenetic software; and (v) the true tree, known from simulation.
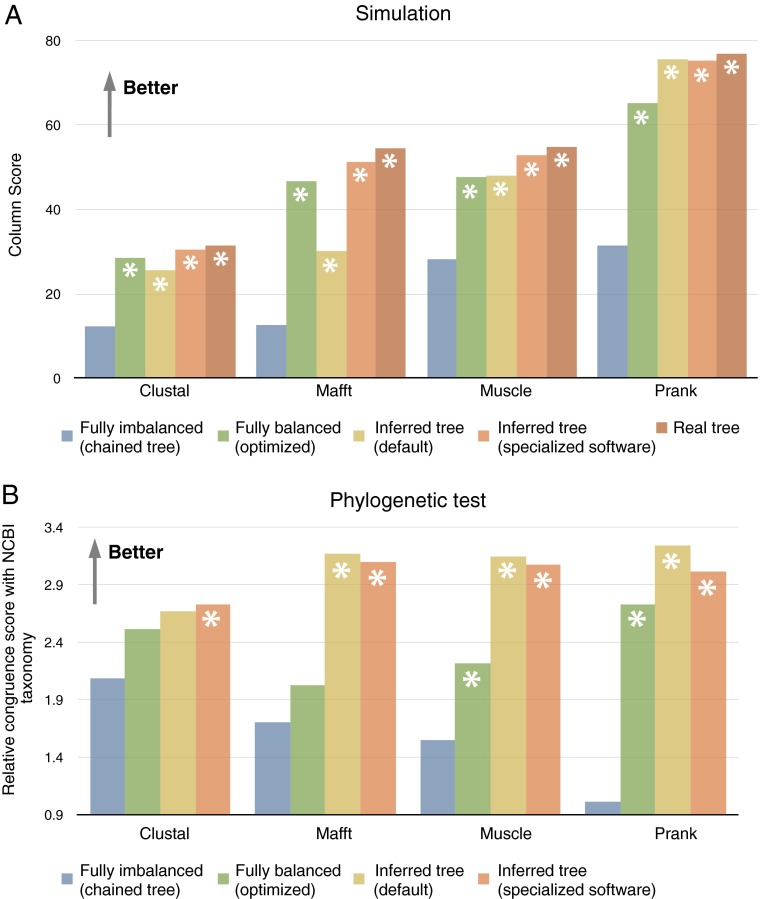
With all aligners, using better trees consistently yielded alignments with more homologous columns (Fig. 1A). In particular, chained trees with random leaf assignments yielded the worst alignments under that measure, with only about half as many correct alignment columns.
Fig. 1.
Evaluation of alignments reconstructed with various aligners and guide tree methods. (A) Average true column score over 113 simulated datasets of 1,024 sequences. (B) Average consistency with the National Center for Biotechnology Information taxonomy over 106 sets of 1,024 biological sequences. Note that the real tree is unknown for empirical data. With fully imbalanced trees as input guide tree, Prank failed to reconstruct alignments in 38 empirical data problem instances; results reported in B are thus based on the remaining 68 alignments. Significant difference from fully imbalanced guide trees is indicated with an asterisk (Wilcoxon double-sided test, P < 0.001). All data available at dx.doi.org/10.5061/dryad.4r5b8.
To confirm these results on empirical data, we performed a similar analysis on gene families of 1,024 homologous sequences each, sampled from the OMA (Orthologous Matrix) database. Based on the alignments obtained with the various guide trees, we inferred trees and compared their congruence with the National Center for Biotechnology Information taxonomy, assuming that more accurate alignments should yield more accurate trees, which in turn should have a higher congruence with the known biology (5). Here too, there is a clear correlation between the accuracy of the input guide trees and that of the resulting trees (Fig. 1B).
So why can the structure-based benchmark used by Boyce et al. (1) yield results that are so diametrically at odds with simulation-based and phylogeny-based ones? One clue may be that structural benchmarks exclusively consider highly compact, highly conserved core regions, which are atypical outside of structural contexts. In Balibase, used by Boyce et al. (1), the core regions constitute only 18.8% of all alignment columns; the benchmark is thus uninformative about the alignment of the vast majority of the protein sequences. In these conserved regions—50,787 columns in total—only four columns contain gaps; the benchmark provides virtually no information about the placement of insertions and deletions either.
For evolutionary analyses, the conclusion is clear: guide trees closer to the correct evolutionary history of the sequences result in better alignments.
Footnotes
The authors declare no conflict of interest.
References
- 1.Boyce K, Sievers F, Higgins DG. Simple chained guide trees give high-quality protein multiple sequence alignments. Proc Natl Acad Sci USA. 2014;111(29):10556–10561. doi: 10.1073/pnas.1405628111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iantorno S, Gori K, Goldman N, Gil M, Dessimoz C. Who watches the watchmen? An appraisal of benchmarks for multiple sequence alignment. Methods Mol Biol. 2014;1079:59–73. doi: 10.1007/978-1-62703-646-7_4. [DOI] [PubMed] [Google Scholar]
- 3.Dalquen DA, Anisimova M, Gonnet GH, Dessimoz C. ALF—A simulation framework for genome evolution. Mol Biol Evol. 2012;29(4):1115–1123. doi: 10.1093/molbev/msr268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Löytynoja A, Goldman N. Phylogeny-aware gap placement prevents errors in sequence alignment and evolutionary analysis. Science. 2008;320(5883):1632–1635. doi: 10.1126/science.1158395. [DOI] [PubMed] [Google Scholar]
- 5.Dessimoz C, Gil M. Phylogenetic assessment of alignments reveals neglected tree signal in gaps. Genome Biol. 2010;11(4):R37. doi: 10.1186/gb-2010-11-4-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]