Significance
The present structure reveals the architecture of the Pseudomonas aeruginosa bacterial-type asparagine-transamidosome, the most common macromolecular assembly required for asparaginyl-tRNAAsn formation in bacteria. We show that the presence of an additional GAD domain in the aspartyl-tRNA synthetase, common in most bacteria but missing in the archaeal-type Thermus thermophilus transamidosome, results in a complex with a distinct architecture and stoichiometry. Furthermore, our kinetic studies reveal that bacterial transamidosomes have distinct kinetic properties compared with the archaeal complex, with rapid release of the Asn-tRNAAsn product, leading to improved turnover by the bacterial-type aspartyl-tRNA synthetase in the complex. Overall, our study provides a structural basis for understanding tRNA-dependent asparagine biosynthesis found in the in majority of bacterial species.
Keywords: transamidosome, asparagine biosynthesis, aspartyl-tRNA synthetase, GatCAB
Abstract
Many prokaryotes lack a tRNA synthetase to attach asparagine to its cognate tRNAAsn, and instead synthesize asparagine from tRNAAsn-bound aspartate. This conversion involves two enzymes: a nondiscriminating aspartyl-tRNA synthetase (ND-AspRS) that forms Asp-tRNAAsn, and a heterotrimeric amidotransferase GatCAB that amidates Asp-tRNAAsn to form Asn-tRNAAsn for use in protein synthesis. ND-AspRS, GatCAB, and tRNAAsn may assemble in an ∼400-kDa complex, known as the Asn-transamidosome, which couples the two steps of asparagine biosynthesis in space and time to yield Asn-tRNAAsn. We report the 3.7-Å resolution crystal structure of the Pseudomonas aeruginosa Asn-transamidosome, which represents the most common machinery for asparagine biosynthesis in bacteria. We show that, in contrast to a previously described archaeal-type transamidosome, a bacteria-specific GAD domain of ND-AspRS provokes a principally new architecture of the complex. Both tRNAAsn molecules in the transamidosome simultaneously serve as substrates and scaffolds for the complex assembly. This architecture rationalizes an elevated dynamic and a greater turnover of ND-AspRS within bacterial-type transamidosomes, and possibly may explain a different evolutionary pathway of GatCAB in organisms with bacterial-type vs. archaeal-type Asn-transamidosomes. Importantly, because the two-step pathway for Asn-tRNAAsn formation evolutionarily preceded the direct attachment of Asn to tRNAAsn, our structure also may reflect the mechanism by which asparagine was initially added to the genetic code.
Accurate translation of the genetic code into a protein sequence relies on a covalent attachment of amino acids to cognate tRNAs that are later used in protein synthesis (1). This attachment is catalyzed by aminoacyl-tRNA synthetases (aaRSs), each specific to one amino acid and a set of tRNA isoacceptors (2). However, the majority of prokaryotes lack several tRNA synthetases, particularly asparaginyl-tRNA synthetase (AsnRS), which ligates asparagine to tRNAAsn (3, 4). In these organisms, asparagine is synthesized in a two-step, tRNA-dependent pathway (5). First, a nondiscriminating aspartyl-tRNA synthetase (ND-AspRS) attaches aspartate to tRNAAsn to form Asp-tRNAAsn (6, 7). Then the tRNA-bound aspartate is converted to asparagine by the amidotransferase (AdT) GatCAB to yield the final product, Asn-tRNAAsn (6, 8–13). Likewise, in prokaryotes lacking glutaminyl-tRNA synthetase (GlnRS), Gln-tRNAGln is formed by the sequential actions of a nondiscriminating glutamyl-tRNA synthetase (ND-GluRS) (14) and an AdT (5). In bacteria, the role of AdT is played by GatCAB (15), whereas in archaea, it is played by GatDE (16, 17).
More than 25 years ago (18), it was proposed that ND-aaRSs and AdTs may form a complex—now called a transamidosome—to couple the two steps of Asn-tRNAAsn formation in space and time and allow efficient transfer of Asp-tRNAAsn from the aaRS to the AdT. The first characterized transamidosome was the Asn-transamidosome from Thermus thermophilus (TtAsn-transamidosome) (19, 20). This complex was identified as a tRNA-dependent association of AspRS2 (TtAspRS2) and GatCAB in a 2:2:2 ratio. It was shown that transamidosome formation stabilizes interactions between subunits of GatCAB (21) and protects Asn-tRNAAsn from hydrolysis, with product release being rate-limiting (19). In the complex, the AspRS forms a dimer with only one catalytic site active at a time (21). It was suggested that the key advantages of asparagine formation by the transamidosome compared with separate enzymes are enhanced aspartylation of tRNAAsn and better prevention of the misacylated Asp-tRNAAsn from use in translation, because this would compromise the fidelity of protein synthesis (19–21).
Importantly, TtAspRS2 was acquired through horizontal gene transfer from archaea (10), and lacks the GAD domain typical of bacterial AspRSs (22). The TtAsn-transamidosome crystal structure suggests that a complex between bacterial ND-AspRS and GatCAB should be less stable and more structurally distinct than the T. thermophilus complex, owing to the presence of the GAD domain in bacterial ND-AspRS (21, 23). Consistent with this notion, the stable association of the Helicobacter pylori ND-AspRS (HpND-AspRS) with GatCAB requires the presence of an auxiliary factor, Hp0100 (23, 24). In the complex, the activity of the HpND-AspRS is unchanged, although the activity of GatCAB increases (23); however, Hp0100 is phylogenetically limited to ε-proteobacteria (23). Therefore, most bacteria have a structurally and, possibly, functionally distinct class of transamidosomes than those described by the T. thermophilus and H. pylori complexes.
In the bacterium Pseudomonas aeruginosa, Asn-tRNAAsn formation is catalyzed by GatCAB and bacterial ND-AspRS, and thus represents the most common type of bacterial Asn-transamidosome (25). Here we report the crystal structure of the P. aeruginosa Asn-transamidosome (PaAsn-transamidosome), which represents the transamidation state of the Asn-tRNAAsn formation. The structure suggests that the additional GAD domain within the ND-AspRS changes the overall architecture of the complex relative to the previously described TtAsn-transamidosome. Consistent with the structure, our in vitro measurements show that PaAsn-transamidosome has unique kinetic properties and functions primarily to enhance tRNAAsn turnover and facilitate Asp-tRNAAsn handoff from AspRS to GatCAB.
Results
Overall Structure of the Bacterial Asn-Transamidosome.
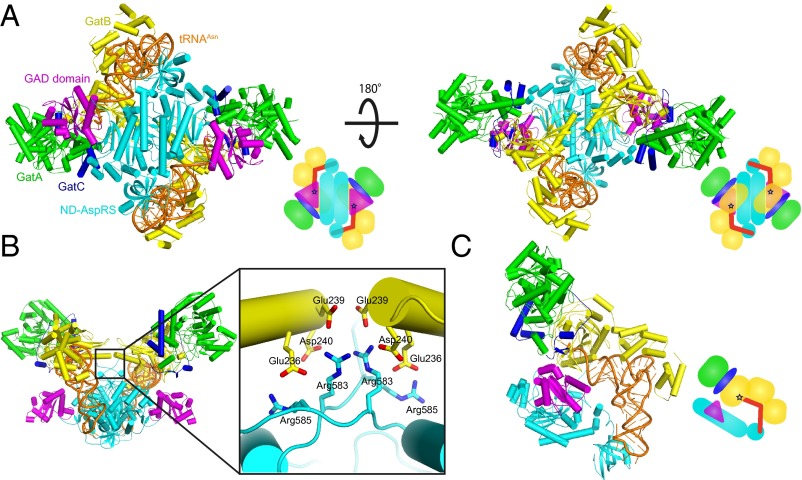
The structure of the PaAsn-transamidosome determined at 3.7-Å resolution (Table S1) reveals the architecture of a 413-kDa symmetric complex comprising ND-AspRS, GatCAB, and tRNAAsn in a 2:2:2 stoichiometry. Each GatCAB is bound to a different tRNAAsn and ND-AspRS, and all are related by a noncrystallographic twofold axis at the interface of the ND-AspRS monomers (Fig. 1A). The complex is stably formed without the auxiliary factor Hp0100 (Fig. S1), although an association of ND-AspRS with GatCAB is tRNA-dependent, as described previously (19, 24). Consistent with the crystal structure, an ∼400-kDa complex was detected in solution by gel filtration (Fig. S2).
Fig. 1.
(A) Overall structure of the Asn-transamidosome from P. aeruginosa. The catalytic and ABDs of ND-AspRS are shown in cyan, the insertion domain characteristic of bacterial AspRS (GAD domain) is in magenta, GatC is in blue, GatA is in green, GatB is in yellow, and tRNAAsn is in orange. The 3′ terminus of tRNAAsn positioned in the active site of GatB is represented by a dark-blue star in the schematic diagram. (B) Close-up view of the GatB and ND-AspRS interface region. The acidic and basic residues clustered around the interface region are shown as stick models. (C) A catalytic unit (one ND-AspRS, one GatCAB, and one tRNAAsn) of the PaAsn-transamidosome.
The structure reveals the interface between ND-AspRS and GatCAB. The α3 helix and β-hairpin connecting β-strands 13 and 14 in GatB contacts the C-terminal loop of ND-AspRS with an average interaction surface of ∼553 Ǻ2 (Fig. 1B, Fig. S3A, and Table S2). In addition, Arg583 and Arg585 in ND-AspRS are in a salt bridge distance from Glu236, Glu239, and Asp240 in GatB and seem to further stabilize the complex. Notably, these residues are largely conserved in organisms that use GatCAB (Fig. S4A) with a ND-AspRS (Fig. S4B), suggesting a common mode of GatB–ND-AspRS interaction across bacterial species.
The two tRNAAsn molecules are bound to the complex in an identical manner (Fig. 1C). The ND-AspRS anticodon-binding domain (ABD) recognizes the anticodon of the tRNA, whereas the 3′ CCA terminus of tRNAAsn is accommodated by the active site of GatCAB with the tRNAAsn U1-A72 base pair near the 310 turn in the GatB cradle domain. Consistent with previous studies, recognition of the U1-A72 base pair by the 310 turn enables the bacterial AdT to distinguish its tRNA substrates (tRNAAsn and tRNAGln) from tRNAAsp and tRNAGlu (21, 26–28). As seen in the Staphylococcus aureus GatCAB structure (27), the tail domain of GatB is positioned to size the D-loop of the tRNA, another major tRNA recognition element of bacterial GatCAB (26, 28). Accordingly, the structure likely represents the transamidation state of the bacterial Asn-transamidosome.
Unique Structural Features of the Bacterial Asn-Transamidosome.
The key difference between bacterial and archaeal-type transamidosomes is the presence of an additional GAD domain in the ND-AspRS proteins. The structural superposition of the PaAsn-transamidosome with the TtAsn-transamidosome (21) reveals how the GAD domain alters the overall architecture of the bacterial complex.
The previously described TtAsn-transamidosome comprises two dimers of the archaeal type TtAspRS2, two GatCABs, and four tRNAAsn molecules, although one TtAspRS2 dimer seems to dissociate from the complex in solution, according to small angle X-ray scattering analysis (21). The structure likely represents a transitional step in Asn-tRNAAsn formation. It was initially proposed that tRNA-dependent Asn biosynthesis in T. thermophilus requires binding of two tRNA molecules to the TtAspRS2 dimer (21). Two GatCAB molecules are then recruited and induce conformational changes in the TtAspRS2 dimer in such a way that makes only one TtAspRS2 monomer active at a time. In this complex, one tRNA molecule (cattRNAAsn) is bound to the active TtAspRS2 monomer to be aspartylated (Fig. 2A), whereas the other tRNA molecule (scaftRNAAsn) binds to the inactive monomer and plays the role of a scaffold that stabilizes the complex. In agreement with the structure, TtAspRS2 activity in the TtAsn-transamidosome has biphasic kinetics, with only one-half of the catalytic sites active at a time (19, 21).
Fig. 2.
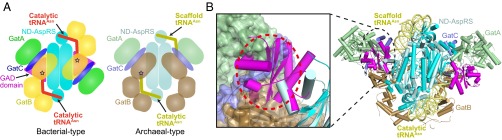
(A) Schematic comparison of the PaAsn-transamidosome (bacterial type) with the TtAsn-transamidosome (archaeal type) highlighting different architectures and tRNA-binding modes. The 3′ terminus of tRNAAsn positioned in the active site of GatB is represented by a dark-blue star in the schematic diagram. (B) Superposition of the ND-AspRS in the PaAsn-transamidosome on that in the TtAsn-transamidosome. (Left) Close-up view of the GatCAB-bound scaffold tRNAAsn in the TtAsn-transamidosome, represented as a surface model, and the GAD domain of the ND-AspRS in the PaAsn-transamidosome. The steric clash between the GAD domain of ND-AspRS in the PaAsn-transamidosome and the GatCAB bound scaffold tRNAAsn in the TtAsn-transamidosome is represented by a red dotted circle. The structure of the TtAsn-transamidosome is rotated 180° from A along the axis parallel to the paper surface.
We show that, unlike the TtAsn-transamidosome, the PaAsn-transamidosome comprises only one ND-AspRS dimer bound to two tRNAAsn molecules and two GatCAB molecules (Fig. 1A). Both tRNA molecules adopt a uniform cattRNAAsn conformation, suggesting that both ND-AspRS monomers can be active at the same time. The differences between the two Asn-transamidosomes likely are related to the bacteria-specific GAD insertion domain. Superposition of the ND-AspRS in the PaAsn-transamidosome and TtAsn-transamidosome reveals a steric clash between the GAD domain of the PaND-AspRS and the TtGatCAB (Fig. 2B). In the PaAsn-transamidosome, the PaGatCAB bends away from the rest of the complex to accommodate the PaND-AspRS GAD domain. This particular orientation of GatCAB in the PaAsn-transamidosome enables both tRNAAsn molecules bound to the ND-AspRS dimer to adopt cattRNAAsn conformations. The lack of a GAD insertion in the TtAspRS2 allows the TtGatCAB to bend toward the catalytic core of TtAspRS2, facilitating the binding of one tRNAAsn in the scaftRNAAsn confirmation (21).
Distinct Biochemical Properties of the Bacterial Asn-Transamidosome.
To test whether a distinct architecture of the complex results in distinct kinetic properties, we measured how the PaAsn-transamidosome protects Asn-tRNAAsn from hydrolysis. The lack of scaftRNAAsn to stabilize the complex suggests that the PaAsn-transamidosome can readily release Asn-tRNAAsn after product formation. Our measurements show that, consistent with that hypothesis and unlike the TtAsn-transamidosome, the P. aeruginosa complex does not protect Asn-tRNAAsn from deacylation (t1/2 of 29 min vs. 29 min with just GatCAB present). In a similar manner, the PaAsn-transamidosome does not protect Asp-tRNAAsn from hydrolysis better than GatCAB alone (t1/2 of 238 min and 246 min, respectively). However, PaGatCAB does protect Asp-tRNAAsn from deacylation (t1/2 of 246 min vs. 39 min with no enzyme present and 41 min with ND-AspRS present), but offers minimal protection of Asn-tRNAAsn (t1/2 of 29 min vs. 22 min without enzyme), consistent with GatCAB binding Asp-tRNAAsn to generate and release Asn-tRNAAsn for protein synthesis.
In agreement with the results of the protection assay, we did not detect burst phase kinetics with the PaAsn-transamidosome that had been observed with the TtAsn-transamidsome (19, 21). The association of GatCAB with ND-AspRS did increase ND-AspRS turnover by 3.2-fold; however, the presence of GatCAB also increased the Km of ND-AspRS for tRNAAsn by a similar factor, leading to no difference in ND-AspRS catalytic efficiency (Table 1). Taken together, the kinetic data show that the PaAsn-transamidosome can readily release Asn-tRNAAsn after its formation. This biochemical property may be a reflection of the GAD insertion rather than the lack of scaftRNAAsn to stabilize the complex.
Table 1.
Kinetic data for Asp-tRNAAsn formation by P. aeruginosa ND-AspRS
| Enzyme | KM, μM, mean ± SD | kcat, s−1, mean ± SD | kcat/KM, s−1/μM, mean ± SD |
| AspRS | 0.61 ± 0.07 | 0.111 ± 0.006 | 0.18 ± 0.02 |
| AspRS + GatCAB* | 2.0 ± 0.2 | 0.36 ± 0.01 | 0.18 ± 0.02 |
Measurements are from three separate experiments. Reactions with P. aeruginosa ND-AspRS (5 nM) were carried out at 37 °C in the presence of excess ATP (4 mM), Asp (3.3 mM), and Gln (2 mM), as described in Materials and Methods. The concentration of tRNAAsn varied between 0.1 and 10.1 μM.
GatCAB (2.0 μM) was added to the reaction mixture.
We attempted to determine whether the bacterial Asn-transamidosome can behave like the archaeal Asn-transamidsome when the GAD insertion domain is deleted. Unfortunately, none of the PaND-AspRS deletion mutant constructs (including a replacement of the GAD domain with the loop found in TtAspRS2) produced sufficient protein amounts for analysis.
Structural Basis of the CCA Terminus Translocation During the Transamidation Cycle.
The PaAsn-transamidosome structure likely represents the transamidation state of the complex; thus, we determined the cocrystal structure of PaND-AspRS with tRNAAsn at 3.3-Å resolution to provide insight into the aminoacylation state of PaND-AspRS with tRNAAsn (Fig. 3 A and B and Table S1). Overall, the PaND-AspRS:tRNAAsn binary complex is similar to the Escherichia coli D-AspRS (EcD-AspRS) enzyme bound to tRNAAsp (PDB ID code 1C0A) (29) and reveals subtle differences in the tRNA structures that may explain tRNA specificity of the enzyme (SI Text and Figs. S5–S7).
Fig. 3.
(A) Monomeric structure of the P. aeruginosa ND-AspRS:tRNAAsn (PaND-AspRS:tRNAAsn) binary complex in an asymmetric unit. The ABD (green), catalytic domain (cyan), GAD domain (magenta), and hinge region (blue) of the ND-AspRS are shown with tRNAAsn (orange). ND-AspRS-bound Asp is shown as a sphere model. (B) Dimeric structure of the PaND-AspRS:tRNAAsn binary complex. Each monomer is related by a crystallographic twofold axis.
The 3′ CCA ends of both tRNAAsn molecules bound to the PaAsn-transamidosome are positioned at the GatCAB transamidase active site. However, for tRNA-dependent Asn biosynthesis, the tRNA first must be aspartylated by ND-AspRS. Thus, the 3′ end must bind in the ND-AspRS active site and then flip ∼40 Å up into the GatCAB catalytic site after Asp-tRNAAsn formation. To clarify how the ND-AspRS accommodates the shifting of the CCA tRNAAsn terminus, we superposed the PaND-AspRS bound to tRNAAsn onto that in the PaAsn-transamidosome, with respect to the ABD (Fig. 4A). This superposition revealed that when the tRNA acceptor end flips from the active site of ND-AspRS to GatCAB, the catalytic domain shifts backward away from the tRNA. The shift is possible because of a hinge between the ABD and catalytic domain of ND-AspRS. In addition, the GAD insertion and helix bundle appends to the catalytic domain shift to open up the PaND-AspRS catalytic site (Fig. 4B). Presumably, this movement facilitates the flipping of the tRNA acceptor stem from ND-AspRS to GatCAB.
Fig. 4.

(A) Superposition of the PaND-AspRS:tRNAAsn structure in the binary complex onto that in the PaAsn-transamidosome with respect to the ABD. The black dotted circle represents the ABD, and the red arrow denotes the shift of the catalytic domain from the binary complex to the transamidosome. (B) Superposition of the PaND-AspRS:tRNAAsn binary complex on that in the PaAsn-transamidosome with respect to the catalytic core. The black dotted circle represents the catalytic core, and the red arrow denotes the shift of the GAD domain and helix bundle from the binary complex to the transamidosome. (C) Superposition of the tRNAAsn structure in the binary complex on that in the PaAsn-transamidosome with respect to the anticodon arm. The red arrow denotes the shift of the acceptor, D-arm, and T-arm from the binary complex to the transamidosome. (D) Superposition of the tRNAAsn structure in the binary complex onto that in the PaAsn-transamidosome with respect to the T-arms. Structurally different nucleotides between tRNAAsn in the binary complex and the Asn-transamidosome are shown as stick models.
These structural rearrangements in the ND-AspRS are accompanied by an ∼14-Å shift of the tRNAAsn elbow and an ∼40-Å movement of the tRNAAsn 3′ CCA end during the transition from the aminoacylation to transamidase state (Fig. 4C). Superposition of the T-arms of the tRNA in the two states revealed rearrangements within the tRNAAsn D-loop (Fig. 4D and Fig. S3 B and C). The shift results in C17 being flipped out of the D-loop and U20 pointing down toward the anticodon instead of up toward the TΨC-loop. Accordingly, G18 is shifted so it no longer hydrogen-bonds with U55. Similar structural changes are seen in the GatDE-tRNAGln structure (17).
Bacterial Asn-Transamidosome Structure Reveals the GatCAB Transamidation State.
With the tRNAAsn 3′ end bound in the GatCAB active site, the PaAsn-transamidosome likely represents the transamidation state of the complex. Apart from association with ND-AspRS, bacterial GatCAB can form a ternary complex with ND-GluRS and tRNAGln, known as a Gln-transamidosome and typical of organisms lacking a GlnRS (30, 31). In the Thermotoga maritima (Tm) Gln-transamidosome structure (30), the tRNAGln 3′ end is bound in the GluRS active site, and the GatCAB tail domain is associated with the D-loop of the tRNA. The structure likely represents the aminoacylation state of the bacterial GatCAB in a transamidosome.
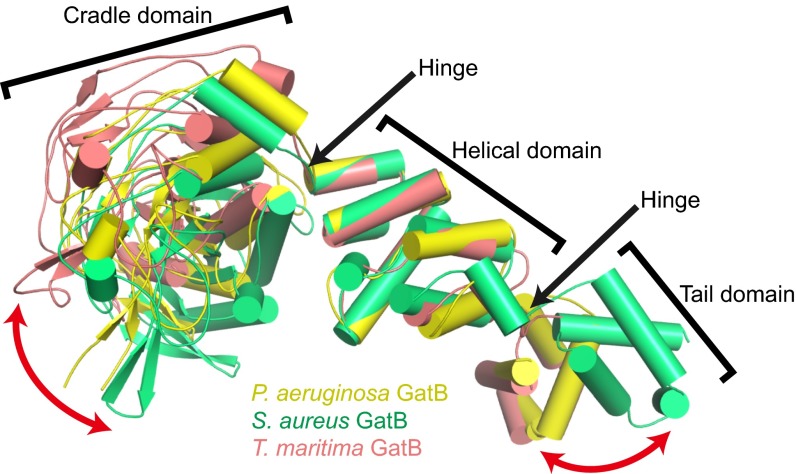
To reveal the structural differences between GatCAB in the aminoacylation and transamidation states, we superposed GatB in the PaAsn-transamidosome with the subunit in the TmGln-transamidosome (30) and the unbound S. aureus GatCAB structure (27, 28) with respect to the GatB helical domain. As was seen in the TmGln-transamidosome (30), we found two hinge regions in the PaGatB, between the helical domain and the cradle and tail domains (Fig. 5). In both transamidosomes, the GatB tail domain is curled toward the GatB helical and cradle domains compared with the unbound structure, enabling the AdT subunit to bind to the tRNA D-loop. The movement is facilitated by the flexibility of the hinge between the helical and tail domains. In the TmGln-transamidosome, the GatB cradle domain is turned up away from the other two domains to facilitate binding of the acceptor stem of the tRNA in the GluRS active site (30). Similar positioning by the PaGatB would help accommodate the tRNAAsn 3′ end bound in the ND-AspRS active site (Fig. 3A). In the transamidation state of the PaAsn-transamidosome, the GatB cradle domain is curled toward the helical domain, although not to the same extent as the unbound GatCAB. The positioning allows GatB to bind the tRNA acceptor stem.
Fig. 5.
Superposition of three GatB structures from P. aeruginosa (yellow), S. aureus (PDB ID code 3IP4; green), and T. maritima (PDB ID code 3ALO; pink) with respect to the helical domain showing two hinges in each domain interface. The movements of the cradle and tail domains are indicated by red arrows.
Discussion
Complexes of Translation Components for Improved Protein Synthesis.
Multi-aaRS complexes, or complexes of aaRSs with other translation machinery components, are known in all domains of life (32). Such higher-order structures have been shown to contribute to translational fidelity and to increased catalytic efficiency of aaRS reactions (33–35). The allure of substrate channeling (36), preventing the participation of misacylated aminoacyl-tRNA in protein synthesis, led to the suggestion of a multienzyme complex for tRNA-dependent Gln-tRNA synthesis (18). This has been borne out by detailed analyses of Gln-tRNA and Asn-tRNA formation by transamidosomes in bacteria and archaea (this work and refs. 19 and 30), although exceptions might exist (37). The transamidosome architecture nicely explains substrate channeling and efficient aa-tRNA formation by the transamidation route. Another multimeric complex (consisting of SepRS, SepCysS, SepCysE, and tRNACys) is essential for Cys-tRNA formation in methanogens (38). It is likely that the tRNA-dependent selenocysteine pathway also may involve a complex of tRNASec, SerRS, PSTK, and SepSecS (39).
Bacterial tRNA-Dependent Asn Biosynthesis.
Taken together, our data suggest that tRNA-dependent Asn biosynthesis in bacteria (Fig. 6) is overall analogous to Gln-tRNAGln formation by the TmGln-transamidosome (30). At the initial step, the PaND-AspRS binds tRNAAsn with its anticodon positioned in the enzyme’s binding domain and the tRNAAsn 3′ end positioned in the enzyme’s active site, as seen in the cocrystal structure of PaND-AspRS complexed with tRNAAsn. The GatB tail domain binds the tRNA’s D-loop to distinguish tRNAAsn from tRNAAsp, with the GatB cradle domain turned away from the complex to accommodate the tRNA acceptor stem bound in the ND-AspRS active site, as observed in the TmGln-transamidosome aminoacylation state (Fig. 6A). Aspartylation of the 3′-CCA terminus of tRNAAsn occurs in the aminoacylation state of the bacterial Asn-transamidosome. Asp-tRNAAsn formation triggers a conformational shift of the ND-AspRS GAD insertion and that catalytic domain and helix bundle appended to the catalytic domain. By opening up the ND-AspRS active site, this intermediate state facilitates the dissociation of the tRNA acceptor stem from ND-AspRS and a flipping up toward GatCAB (Fig. 6B). Finally, GatCAB, in particular the GatB cradle domain, moves into position to bind the tRNAAsn acceptor stem and adopt the transamidation state seen in the PaAsn-transamidosome crystal structure (Fig. 6C). The 310 turn in the GatB cradle domain selectively binds the tRNAAsn U1-A72 base pair; this provides a second check to ensure that Asp-tRNAAsn is amidated and not Asp-tRNAAsp (21, 26–28).
Fig. 6.
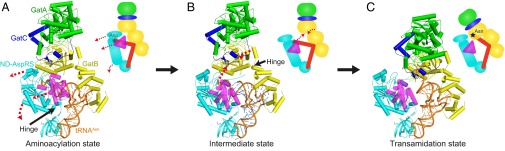
Proposed reaction model for Asn-tRNAAsn synthesis in the bacterial Asn-transamidosome. Each component is color-coded as in Fig. 1. Aminoacylation (A), intermediate (B), and transamidation (C) states of the Asn-transamidosome are shown. Movement from the aminoacylation to the transamidation state is indicated by red dotted arrows. In the schematic diagram, the amino acid ligated to the tRNAAsn is represented by a star (Asp, gray; Asn, dark blue).
Now the tRNAAsn-bound Asp is positioned in the GatCAB transamidase active site for amidation. Following Asn-tRNAAsn production, the complex dissociates, releasing Asn-tRNAAsn, consistent with the complex’s inability to protect Asn-tRNAAsn from hydrolysis. Given that the association of the PaGatCAB and PaND-AspRS is tRNA-dependent, GatCAB is also likely released from ND-AspRS when Asn-tRNAAsn dissociates.
Evolution of Asn-Transamidosomes.
In archaea, GatCAB is used only for Asn-tRNAAsn formation as GatDE forms Gln-tRNAGln (16, 17). Both AdTs likely were present in early archaea (40). The specificity of archaeal GatCAB for Asn-tRNAAsn synthesis might have enabled archaeal AspRS to coevolve with the AdT to form a thermostable complex in which GatCAB is not released following Asn-tRNAAsn synthesis. This stability likely was facilitated by the archaeal AspRS not retaining the GAD insertion domain found in bacterial-type AspRS. The thermostability of the archaeal complex may explain why T. thermophilus acquired an archaeal-type AspRS for tRNA-dependent Asn biosynthesis instead of using the bacterial-type AspRS that it also encodes in its genome (19, 21, 22, 41).
Early bacteria likely emerged from the last universal common ancestral state (LUCAS) with just one AdT—GatCAB—for both Gln-tRNAGln and Asn-tRNAAsn formation (40). After Asn-tRNAAsn formation in early bacteria, the release of GatCAB from bacterial ND-AspRS might be have been beneficial, because the free GatCAB could easily be repurposed for Gln-tRNAGln formation with GluRS. In that context, acquisition of the GAD insertion domain by bacterial AspRS might have facilitated GatCAB release from the Asn-transamidosome to better enable early bacteria to use GatCAB for both Gln-tRNAGln and Asn-tRNAAsn formation. Consistent with this idea, the GAD insertion was likely acquired early on in bacterial AspRS evolution and coevolved for an extended time with the rest of the enzyme (SI Text and Fig. S8).
In H. pylori, Hp0100 stabilizes GatCAB association with bacterial ND-AspRS even in the absence of tRNAAsn (23). H. pylori also encodes two GluRS enzymes, with the second (GluRS2) complexing with GatCAB for Gln-tRNAGln formation (31, 42, 43). Hp0100 and GluRS2 may be adaptations to better regulate the dual functions of HpGatCAB. In P. aeruginosa, which acquired GlnRS and uses GatCAB for only Asn-tRNAAsn synthesis (25), retention of the GAD insertion in AspRS may be a remnant the coevolution of the protein domains in early bacteria. The insertion domain may still be selected for to allow proper folding of PaND-AspRS and to enable simultaneous activity of both monomers of ND-AspRS in the complex, thereby improving ND-AspRS turnover of tRNAAsn. This coevolution may be preventing the generation of stable GAD domain deletion mutant PaND-AspRS enzymes.
Because the PaAsn-transamidosome allows Asp-tRNAAsn to be directly handed off from ND-AspRS to GatCAB, the misaminoacylated tRNA is protected from deacylation and use in protein synthesis, where it may compromise the fidelity of translation. Accordingly, under certain conditions, the complex of ND-AspRS with GatCAB may be less prone than AsnRS to mischarged tRNA formation (8). In addition, the Asn-transamidosome allows P. aeruginosa to directly and efficiently couple the biosynthesis of Asn with its use in translation. As such, a similar complex between ND-AspRS and ancestral GatCAB pre-LUCAS might have enabled the addition of Asn to the genetic code (44).
Materials and Methods
Preparation of the PaAsn-transamidosome and PaND-AspRS:tRNAAsn and the details of crystallization and structure determination are summarized in SI Materials and Methods. Structures of the PaAsn-transamidosome and PaND-AspRS:tRNAAsn were solved by molecular replacement methods. Atomic coordinates and structure factors have been deposited in the Protein Data Bank under ID codes 4WJ3 for PaAsn-transamidosome and 4WJ4 for PaND-AspRS:tRNAAsn. Detailed descriptions of the gel-shift assay, gel filtration analysis, kinetic analysis, protection assay, and phylogenetic analysis using established methods are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Keitaro Yamashita (RIKEN) and the beamline staff of BL41XU and BL44XU at SPring-8. We are grateful to Sergey Melnikov and Jiqiang Ling for their critical reading of the manuscript. This work was supported by a Grant-in-Aid for Scientific Research in a Priority Area (24567068, to M.Y.) and the Photon and Quantum Basic Research Coordinated Development Program (M.Y.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan; a grant from the US National Science Foundation (MCB-1244326, to K.S.); and a grant from the National Institute of General Medical Sciences (GM22854, to D.S.). A.N. has received a Japan Society for the Promotion of Science Postdoctoral Fellow for Research Abroad.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4WJ4 and 4WJ3).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1423314112/-/DCSupplemental.
References
- 1.Ibba M, Söll D. Quality control mechanisms during translation. Science. 1999;286(5446):1893–1897. doi: 10.1126/science.286.5446.1893. [DOI] [PubMed] [Google Scholar]
- 2.Ibba M, Söll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 3.Roy H, Becker HD, Reinbolt J, Kern D. When contemporary aminoacyl-tRNA synthetases invent their cognate amino acid metabolism. Proc Natl Acad Sci USA. 2003;100(17):9837–9842. doi: 10.1073/pnas.1632156100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sheppard K, Akochy PM, Salazar JC, Söll D. The Helicobacter pylori amidotransferase GatCAB is equally efficient in glutamine-dependent transamidation of Asp-tRNAAsn and Glu-tRNAGln. J Biol Chem. 2007;282(16):11866–11873. doi: 10.1074/jbc.M700398200. [DOI] [PubMed] [Google Scholar]
- 5.Yuan J, Sheppard K, Söll D. Amino acid modifications on tRNA. Acta Biochim Biophys Sin (Shanghai) 2008;40(7):539–553. doi: 10.1111/j.1745-7270.2008.00435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becker HD, Kern D. Thermus thermophilus: A link in evolution of the tRNA-dependent amino acid amidation pathways. Proc Natl Acad Sci USA. 1998;95(22):12832–12837. doi: 10.1073/pnas.95.22.12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cathopoulis T, Chuawong P, Hendrickson TL. Novel tRNA aminoacylation mechanisms. Mol Biosyst. 2007;3(6):408–418. doi: 10.1039/b618899k. [DOI] [PubMed] [Google Scholar]
- 8.Curnow AW, Ibba M, Söll D. tRNA-dependent asparagine formation. Nature. 1996;382(6592):589–590. doi: 10.1038/382589b0. [DOI] [PubMed] [Google Scholar]
- 9.Curnow AW, Tumbula DL, Pelaschier JT, Min B, Söll D. Glutamyl-tRNAGln amidotransferase in Deinococcus radiodurans may be confined to asparagine biosynthesis. Proc Natl Acad Sci USA. 1998;95(22):12838–12843. doi: 10.1073/pnas.95.22.12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Becker HD, et al. The heterotrimeric Thermus thermophilus Asp-tRNAAsn amidotransferase can also generate Gln-tRNAGln. FEBS Lett. 2000;476(3):140–144. doi: 10.1016/s0014-5793(00)01697-5. [DOI] [PubMed] [Google Scholar]
- 11.Salazar JC, et al. A dual-specific Glu-tRNAGln and Asp-tRNAAsn amidotransferase is involved in decoding glutamine and asparagine codons in Acidithiobacillus ferrooxidans. FEBS Lett. 2001;500(3):129–131. doi: 10.1016/s0014-5793(01)02600-x. [DOI] [PubMed] [Google Scholar]
- 12.Raczniak G, Becker HD, Min B, Söll D. A single amidotransferase forms asparaginyl-tRNA and glutaminyl-tRNA in Chlamydia trachomatis. J Biol Chem. 2001;276(49):45862–45867. doi: 10.1074/jbc.M109494200. [DOI] [PubMed] [Google Scholar]
- 13.Sheppard K, Sherrer RL, Söll D. Methanothermobacter thermautotrophicus tRNAGln confines the amidotransferase GatCAB to asparaginyl-tRNAAsn formation. J Mol Biol. 2008;377(3):845–853. doi: 10.1016/j.jmb.2008.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lapointe J, Duplain L, Proulx M. A single glutamyl-tRNA synthetase aminoacylates tRNAGlu and tRNAGln in Bacillus subtilis and efficiently misacylates Escherichia coli tRNAGln1 in vitro. J Bacteriol. 1986;165(1):88–93. doi: 10.1128/jb.165.1.88-93.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Curnow AW, et al. Glu-tRNAGln amidotransferase: A novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc Natl Acad Sci USA. 1997;94(22):11819–11826. doi: 10.1073/pnas.94.22.11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tumbula DL, Becker HD, Chang WZ, Söll D. Domain-specific recruitment of amide amino acids for protein synthesis. Nature. 2000;407(6800):106–110. doi: 10.1038/35024120. [DOI] [PubMed] [Google Scholar]
- 17.Oshikane H, et al. Structural basis of RNA-dependent recruitment of glutamine to the genetic code. Science. 2006;312(5782):1950–1954. doi: 10.1126/science.1128470. [DOI] [PubMed] [Google Scholar]
- 18.Schön A, Kannangara CG, Gough S, Söll D. Protein biosynthesis in organelles requires misaminoacylation of tRNA. Nature. 1988;331(6152):187–190. doi: 10.1038/331187a0. [DOI] [PubMed] [Google Scholar]
- 19.Bailly M, Blaise M, Lorber B, Becker HD, Kern D. The transamidosome: A dynamic ribonucleoprotein particle dedicated to prokaryotic tRNA-dependent asparagine biosynthesis. Mol Cell. 2007;28(2):228–239. doi: 10.1016/j.molcel.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 20.Bailly M, et al. tRNA-dependent asparagine formation in prokaryotes: Characterization, isolation and structural and functional analysis of a ribonucleoprotein particle generating Asn-tRNAAsn. Methods. 2008;44(2):146–163. doi: 10.1016/j.ymeth.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 21.Blaise M, et al. Crystal structure of a transfer-ribonucleoprotein particle that promotes asparagine formation. EMBO J. 2010;29(18):3118–3129. doi: 10.1038/emboj.2010.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delarue M, et al. Crystal structure of a prokaryotic aspartyl tRNA-synthetase. EMBO J. 1994;13(14):3219–3229. doi: 10.1002/j.1460-2075.1994.tb06623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silva GN, et al. A tRNA-independent mechanism for transamidosome assembly promotes aminoacyl-tRNA transamidation. J Biol Chem. 2013;288(6):3816–3822. doi: 10.1074/jbc.M112.441394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fischer F, et al. The asparagine-transamidosome from Helicobacter pylori: A dual-kinetic mode in non-discriminating aspartyl-tRNA synthetase safeguards the genetic code. Nucleic Acids Res. 2012;40(11):4965–4976. doi: 10.1093/nar/gks167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akochy PM, Bernard D, Roy PH, Lapointe J. Direct glutaminyl-tRNA biosynthesis and indirect asparaginyl-tRNA biosynthesis in Pseudomonas aeruginosa PAO1. J Bacteriol. 2004;186(3):767–776. doi: 10.1128/JB.186.3.767-776.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bailly M, et al. A single tRNA base pair mediates bacterial tRNA-dependent biosynthesis of asparagine. Nucleic Acids Res. 2006;34(21):6083–6094. doi: 10.1093/nar/gkl622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakamura A, et al. Two distinct regions in Staphylococcus aureus GatCAB guarantee accurate tRNA recognition. Nucleic Acids Res. 2010;38(2):672–682. doi: 10.1093/nar/gkp955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamura A, Yao M, Chimnaronk S, Sakai N, Tanaka I. Ammonia channel couples glutaminase with transamidase reactions in GatCAB. Science. 2006;312(5782):1954–1958. doi: 10.1126/science.1127156. [DOI] [PubMed] [Google Scholar]
- 29.Eiler S, Dock-Bregeon A, Moulinier L, Thierry JC, Moras D. Synthesis of aspartyl-tRNAAsp in Escherichia coli—a snapshot of the second step. EMBO J. 1999;18(22):6532–6541. doi: 10.1093/emboj/18.22.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ito T, Yokoyama S. Two enzymes bound to one transfer RNA assume alternative conformations for consecutive reactions. Nature. 2010;467(7315):612–616. doi: 10.1038/nature09411. [DOI] [PubMed] [Google Scholar]
- 31.Huot JL, et al. Gln-tRNAGln synthesis in a dynamic transamidosome from Helicobacter pylori, where GluRS2 hydrolyzes excess Glu-tRNAGln. Nucleic Acids Res. 2011;39(21):9306–9315. doi: 10.1093/nar/gkr619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hausmann CD, Ibba M. Aminoacyl-tRNA synthetase complexes: Molecular multitasking revealed. FEMS Microbiol Rev. 2008;32(4):705–721. doi: 10.1111/j.1574-6976.2008.00119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hausmann CD, Praetorius-Ibba M, Ibba M. An aminoacyl-tRNA synthetase:elongation factor complex for substrate channeling in archaeal translation. Nucleic Acids Res. 2007;35(18):6094–6102. doi: 10.1093/nar/gkm534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Godinic-Mikulcic V, Jaric J, Hausmann CD, Ibba M, Weygand-Durasevic I. An archaeal tRNA-synthetase complex that enhances aminoacylation under extreme conditions. J Biol Chem. 2011;286(5):3396–3404. doi: 10.1074/jbc.M110.168526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Das M, Vargas-Rodriguez O, Goto Y, Suga H, Musier-Forsyth K. Distinct tRNA recognition strategies used by a homologous family of editing domains prevent mistranslation. Nucleic Acids Res. 2014;42(6):3943–3953. doi: 10.1093/nar/gkt1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Srivastava DK, Bernhard SA. Metabolite transfer via enzyme-enzyme complexes. Science. 1986;234(4780):1081–1086. doi: 10.1126/science.3775377. [DOI] [PubMed] [Google Scholar]
- 37.Bhaskaran H, Perona JJ. Two-step aminoacylation of tRNA without channeling in Archaea. J Mol Biol. 2011;411(4):854–869. doi: 10.1016/j.jmb.2011.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y, et al. Ancient translation factor is essential for tRNA-dependent cysteine biosynthesis in methanogenic archaea. Proc Natl Acad Sci USA. 2014;111(29):10520–10525. doi: 10.1073/pnas.1411267111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuan J, et al. Distinct genetic code expansion strategies for selenocysteine and pyrrolysine are reflected in different aminoacyl-tRNA formation systems. FEBS Lett. 2010;584(2):342–349. doi: 10.1016/j.febslet.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sheppard K, Söll D. On the evolution of the tRNA-dependent amidotransferases, GatCAB and GatDE. J Mol Biol. 2008;377(3):831–844. doi: 10.1016/j.jmb.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Becker HD, et al. Thermus thermophilus contains an eubacterial and an archaebacterial aspartyl-tRNA synthetase. Biochemistry. 2000;39(12):3216–3230. doi: 10.1021/bi992573y. [DOI] [PubMed] [Google Scholar]
- 42.Skouloubris S, Ribas de Pouplana L, De Reuse H, Hendrickson TL. A noncognate aminoacyl-tRNA synthetase that may resolve a missing link in protein evolution. Proc Natl Acad Sci USA. 2003;100(20):11297–11302. doi: 10.1073/pnas.1932482100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salazar JC, et al. Coevolution of an aminoacyl-tRNA synthetase with its tRNA substrates. Proc Natl Acad Sci USA. 2003;100(24):13863–13868. doi: 10.1073/pnas.1936123100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sheppard K, et al. From one amino acid to another: tRNA-dependent amino acid biosynthesis. Nucleic Acids Res. 2008;36(6):1813–1825. doi: 10.1093/nar/gkn015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.