Significance
Epstein–Barr Virus (EBV)-infected lymphoblasts can give rise to non-Hodgkin’s lymphomas, Hodgkin’s disease, and lymphoproliferative disorders, especially in immunosuppressed and HIV-infected individuals. EBV-driven lymphoblast growth requires EBV nuclear antigen 3A (EBNA3A) for suppression of CDKN2A-mediated cell senescence responses. We have described the EBNA3A genome-wide landscape in EBV-infected human lymphoblasts. EBNA3A was found mostly at strong enhancers, colocalized with BATF, ETS, IRF4, and RUNX3. EBNA3A was tethered to DNA through BATF protein complexes.
Keywords: EBNA3A, CDKN2A, RUNX3, BATF, IRF4
Abstract
Epstein–Barr Virus (EBV) conversion of B-lymphocytes to Lymphoblastoid Cell Lines (LCLs) requires four EBV nuclear antigen (EBNA) oncoproteins: EBNA2, EBNALP, EBNA3A, and EBNA3C. EBNA2 and EBNALP associate with EBV and cell enhancers, up-regulate the EBNA promoter, MYC, and EBV Latent infection Membrane Proteins (LMPs), which up-regulate BCL2 to protect EBV-infected B-cells from MYC proliferation-induced cell death. LCL proliferation induces p16INK4A and p14ARF-mediated cell senescence. EBNA3A and EBNA3C jointly suppress p16INK4A and p14ARF, enabling continuous cell proliferation. Analyses of the EBNA3A human genome-wide ChIP-seq landscape revealed 37% of 10,000 EBNA3A sites to be at strong enhancers; 28% to be at weak enhancers; 4.4% to be at active promoters; and 6.9% to be at weak and poised promoters. EBNA3A colocalized with BATF-IRF4, ETS-IRF4, RUNX3, and other B-cell Transcription Factors (TFs). EBNA3A sites clustered into seven unique groups, with differing B-cell TFs and epigenetic marks. EBNA3A coincidence with BATF-IRF4 or RUNX3 was associated with stronger EBNA3A ChIP-Seq signals. EBNA3A was at MYC, CDKN2A/B, CCND2, CXCL9/10, and BCL2, together with RUNX3, BATF, IRF4, and SPI1. ChIP-re-ChIP revealed complexes of EBNA3A on DNA with BATF. These data strongly support a model in which EBNA3A is tethered to DNA through a BATF-containing protein complexes to enable continuous cell proliferation.
Epstein–Barr Virus (EBV) was discovered 50 years ago in the search for a virus in Burkitt’s lymphoma cells (1). EBV is highly prevalent in all populations and persists in almost all people as a lifelong symbiont. Nevertheless, primary infection with EBV is the usual cause of infectious mononucleosis (IM), and adolescent IM increases the risk of subsequent Hodgkin’s disease (2). EBV also causes lymphoma and Hodgkin’s disease in people with HIV infection or T-cell immune deficiency. Moreover, EBV causes nasopharyngeal carcinoma and some gastric carcinomas (3, 4). EBV infection of primary human B-lymphocytes in vitro results in continuous infected B-cell proliferation and immortal Lymphoblastoid Cell Lines (LCLs). This process is mediated by the expression of EBNAs and LMP1. In addition, EBV also expresses more than 20 microRNAs. Genetic experiments indicate that EBNA2, EBNA3A, EBNA3C, EBNALP, and LMP1 are essential for LCL proliferation (3–7).
EBNA3A, EBNA3B, and EBNA3C are the consequence of a partially divergent gene triplication and encode proteins that regulate virus and cell gene expression (4). The EBNA3s have short 5′ and long 3′ coding exons. Their transcription is driven by the 5′ distal EBNA Cp-promoter. The EBNA3s have a homologous N-terminal domain that binds to the cell DNA sequence-specific transcription factor (TF) RBPJ, which mediates EBNA2 and Notch binding to DNA (8–13). Although EBNA2 interaction with RBPJ on DNA displaces repressors and activates transcription (14), EBNA3A or EBNA3C binding to RBPJ substantially decreases RBPJ binding to DNA in electrophoretic mobility shift assays in vitro (8–11). EBNA3A and EBNA3C can also repress EBNA2 activation of the EBNA2- and RBPJ-dependent EBV Cp-promoter, indicative of EBNA3A and EBNA3C inhibition of RBPJ binding to cognate DNA (8–11). Genome-wide data on EBNA3C DNA binding found low EBNA3C and RBPJ co-occupancy at the same DNA site in LCLs, indicating that EBNA3C is not mostly tethered to DNA through RBPJ (15). Conditional EBNA3A overexpression in LCLs inhibits EBNA2-affected cell gene transcription, including MYC or CD21 (16). When tethered to DNA by the Gal4 DNA-binding domain, EBNA3 proteins repress transcription by recruiting histone deacetylases and transcription repressor CtBP (17–21).
Despite sequence and functional similarities, the EBNA3 proteins each have unique functions. Although EBNA3B is entirely dispensable for LCL growth, EBNA3A and EBNA3C are essential for immortal LCL proliferation (5–7, 22–25). Overexpression of EBNA3A cannot rescue LCLs from growth arrest caused by EBNA3C deficiency and vice versa, suggesting that the unique functions are required for LCL growth and survival (5, 23).
Both EBNA3A and EBNA3C are required to repress p16INK4A and p14ARF expression (23, 25, 26). Knockdown of both p16INK4A and p14ARF, p16INK4A null mutations or HPV E6 and E7 expression can restore LCL growth in the absence of EBNA3A or EBNA3C (27, 28). EBNA3A and EBNA3C repression of p16INK4A and p14ARF is linked to repressive histone modifications at these loci (26, 27). EBNA3C ChIP-seq finds EBNA3C binding strongly to the p14ARF promoter through SPI1, IRF4, and BATF and recruits the transcription repressor Sin3A. Conditional EBNA3C inactivation significantly decreases Sin3A binding at the p14ARF promoter (15). Joint EBNA3A and EBNA3C repression of MYC-induced senescence responses allows continuous MYC-driven LCL proliferation. EBNA3A can also repress CDKN2B p15INK4B expression through MIZ1 and CDKN1A p21WAF1/CIP1 (29, 30).
EBNA3A initiates and maintains polycomb repressive signatures at the CXCL9 and CXCL10 loci (31), and EBNA3A expression in BL cells represses proapoptotic Bim expression (32). EBNA3A specifically induces transcription of Hsp70, Hsp70B/B′, Bag3, and DNAJA1/Hsp40 chaperones (33). The EBNA3s have also been implicated in chromatin looping (34).
The essential EBNA3A role in suppressing p14ARF and p16INK4A expression to enable continuous LCL proliferation is evident from conditional EBNA3A studies (27). We therefore undertook a genome-wide approach to further elucidate the role of EBNA3A in cell gene transcription, including p14ARF and p16INK4A. We discovered that EBNA3A binds to LCL enhancer sites with high H3K4me1 and H3K27ac marks. EBNA3A, like EBNA3C, frequently colocalized with RUNX3, BATF, IRF4, and SPI1, TFs that are critical for lymphocyte transcription, differentiation, and development. Surprisingly, EBNA3A did not recruit Sin3A to repress p14ARF and p16INK4A. Indeed, EBNA3A and EBNA3C had distinctly different binding sites at CDKN2A/B and repress these loci through different mechanisms.
Results
EBNA3A Binding Sites in LCLs.
Two EBNA3A ChIP-seq biological replicates were done from LCLs transformed by a recombinant EBV BACmid, wherein EBNA3A was C-terminally tagged with Flag and HA epitopes (EBNA3AFHA). These LCLs were similar in EBNA3A expression to wild-type LCLs. EBNA3A ChIP-seq reads were mapped to human genome version hg18, using Bowtie, allowing two mismatches. Both EBNA3A ChIP-seq replicas had quality tags >1 (Fig. S1), indicative of high quality, as defined by the Encyclopedia of DNA Elements (ENCODE). The ChIP-seq processing pipeline (SPP) identified the top 10,000 EBNA3A sites to have Irreproducible Discovery Rates (IDR) < 0.01 (35, 36). EBNA3A sites were assigned to functional genome domains, as defined by ENCODE GM12878 LCL epigenetic landscapes (37), which divide the human genome into seven epigenetic domains: strong enhancers, weak enhancers, active promoters, weak promoters, poised promoters, heterochromatin, and other domains.
EBNA3A was 37% at strong enhancers with high H3K4me1 and H3K27ac signals; 28% at weak enhancers with intermediate H3K4me1 and weak H3K27ac signals; 4.4% at active promoters with high H3K4me3 and H3K9ac signals; 6.9% at weak or poised promoters with high H3K4me3 and H3K27me3 signals; 18% at sites that lacked predictive histone modifications; and 5.8% at other sites including insulators, transcription elongation, and transcription transition (Fig. 1A).
Fig. 1.
Genome-wide distribution of EBNA3A sites and percentage overlap with EBNA2, EBNA3C, RBPJ, BATF, IRF4, and RUNX3. (A) EBNA3A sites are annotated to ENCODE GM12878 LCL chromatin states. EBNA3A sites localize to six chromatin state domains defined by their unique histone modification profiles: strong enhancers, weak enhancers, active promoters, weak and poised promoters, heterochromatin, and other sites, such as insulators. (B) Venn diagrams indicating that 50% of EBNA3A sites overlap (±250 bp) with EBNA3C, 8% with EBNA2, 16% with RBPJ, 63% with RUNX3, 65% with BATF, 44% with IRF4, and 42% with both BATF and IRF4 (numbers in Venn diagrams indicate the percentage of EBNA3A peaks overlapping with the indicated TFs).
EBNA3A Sites Coincide 50% with EBNA3C Sites, 8% with EBNA2/RBPJ Sites, and 8% with RBPJ Sites Without EBNA2 (±250 bp).
Comparison of the EBNA3A and EBNA3C ChIP-seq landscapes (15) revealed a 50% coincidence of EBNA3A and EBNA3C sites, likely related to EBNA3A and EBNA3C binding to BATF/IRF4 (AP-1–IRF Composite Elements, AICE) and ETS/IRF4 (ETS–IRF composite elements, EICEs) sites (Fig. 1B) (15). EBNA3A sites also overlapped 8% with EBNA2/RBPJ sites and 8% with RBPJ sites without EBNA2 (38). As expected, the 16% coincidence of EBNA3A with RBPJ excluded RBPJ as a major TF that tethers EBNA3A to DNA (Fig. 1B).
EBNA3A Sites Coincide with RUNX3, BATF, and IRF4.
To identify cell TFs that likely tether EBNA3A to DNA or modify EBNA3A effects, ENCODE GM12878 cell TF ChIP-seq data were reanalyzed, and EBNA3A-associated cell TFs were identified. EBNA3A sites highly overlapped with RUNX3 (63%), BATF (65%), and IRF4 (44%), positioning these TFs as modifiers of EBNA3A binding to DNA and transcription effects (Fig. 1B and Table S1).
Nonheterochromatic EBNA3A Sites Are Divided into Seven Clusters, Dependent on Cell TF Co-Occupancies.
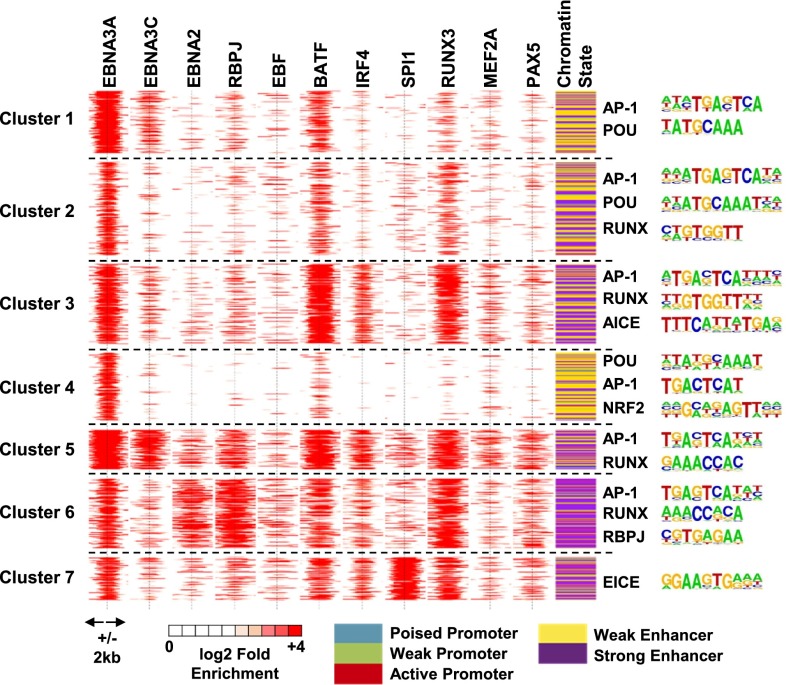
EBNA3A, EBNA3C, EBNA2, and multiple-cell TF ChIP-seq signals within ±2 kb of EBNA3A sites were normalized against matching inputs, and log2 enrichment over input was calculated. Partitioning Around Medoids (PAM) was used to cluster EBNA3A sites into seven distinct clusters, differing in EBNA3C, EBNA2, RBPJ, EBF, BATF, IRF4, SPI1, RUNX3, MEF2A, and PAX5 enrichment (Fig. 2). EBF, SPI1, and PAX5 are early B-cell lineage factors that maintain open chromatin sites for B-cell TF access at subsequent stages of B-cell development. The chromatin states of each cluster are indicated in the last column of Fig. 2. DNA sequences embedded in each EBNA3A site (±250 bp) were extracted, and HOMER was used to identify cell TF motifs enriched in each cluster (Fig. 2, Right).
Fig. 2.
EBNA3A-associated cell TF clusters, chromatin states, and embedded motifs at each cluster. PAM is used to divide nonheterochromatin EBNA3A sites into seven unique clusters with different EBNA3C, EBNA2, RBPJ, EBF, BATF, IRF4, SPI1, RUNX3, MEF2A, and PAX5 signals. All TF ChIP-seq signals at ±2 kb of EBNA3A sites are shown on a red (high ChIP-seq signal) to white (no ChIP-seq signal) scale. EBNA3A site chromatin states are indicated in the right most column. Enriched motifs at each cluster are shown next to the clusters.
Cluster 1 EBNA3A sites were most highly enriched for EBNA3A, EBNA3C, and BATF signals and were substantially less enriched for EBNA2, RBPJ, EBF, IRF4, SPI1, RUNX3, MEF2A, and PAX5 signals. Cluster 1 EBNA3A sites were also highly enriched for embedded AP-1 and POU motifs and were at strong (purple) and weak (yellow) enhancers.
Cluster 2 EBNA3A sites had weaker signals than cluster 1, but still had strong EBNA3A, BATF, and RUNX3 signals. Cluster 2 was highly enriched for embedded AP-1, POU, and RUNX motifs, as well as for strong and weak enhancers.
Cluster 3 EBNA3A sites had strong EBNA3A, BATF, IRF4, and RUNX3 signals, as well as weak MEF2A and PAX5 signals. Cluster 3 sites were enriched for embedded AP-1, RUNX, and AICE motifs and were mostly at strong or weak enhancers.
Cluster 4 EBNA3A sites had strong EBNA3A signals, weak BATF signals, and embedded POU, AP-1, and NRF2 TF motifs. Cluster 4 sites were mostly at weak enhancers.
Cluster 5 EBNA3A sites had strong EBNA3A and EBNA3C signals, weak EBNA2 and strong RBPJ signals, weak EBF signals, strong BATF and IRF4 signals, weak SPI1 signals, strong RUNX3 signals, moderate MEF2A and PAX5 signals, and embedded AP1 and RUNX motifs. Cluster 5 sites were mostly at strong enhancers.
Cluster 6 EBNA3A sites had strong EBNA3A signals, weak EBNA3C signals, strong EBNA2 and RBPJ signals, moderate EBF signals, strong BATF signals, moderate IRF4 signals, weak SPI1 signals, strong RUNX3 signals, weak MEF2A, moderate PAX5 signals, and embedded AP-1, RUNX, and RBPJ motifs. Cluster 6 sites were mostly at strong enhancers.
Cluster 7 EBNA3A sites had strong EBNA3A signals, weak EBNA3C signals, weak EBNA2 and RBPJ signals, weak EBF signals, strong BATF, moderate IRF4, strong SPI1 and RUNX3 signals, weak MEF2A and PAX5 signals, and embedded EICE motifs. Cluster 7 sites were mostly at strong enhancers.
EBNA3A ChIP-seq Signals at EBNA3A Sites Alone or Co-Occupied with Other Cellular and Viral TFs.
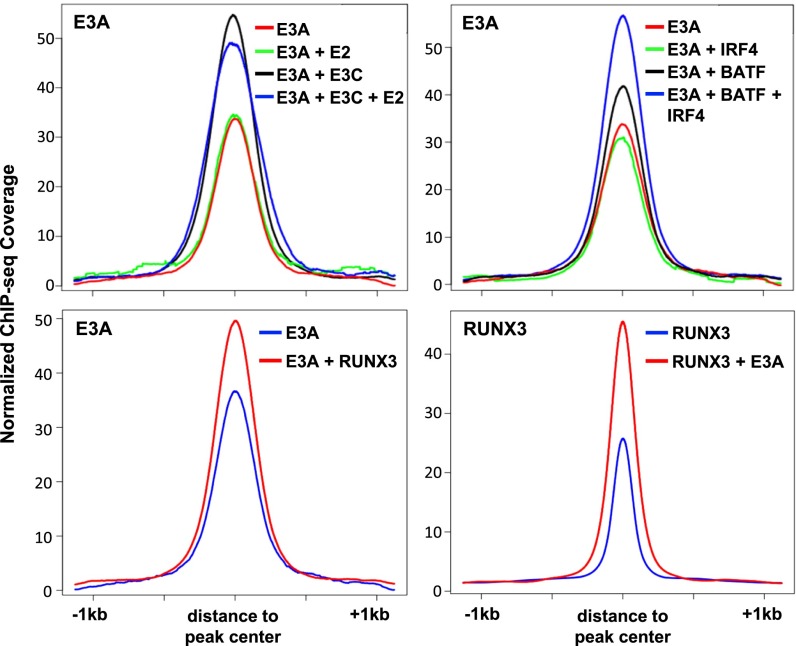
Because EBNA3A, EBNA3C, and EBNA2 co-occur with common B-cell TFs, including RBPJ and RUNX3, the effect of their co-occurrence on DNA binding was examined. EBNA3A ChIP-seq signals for EBNA3A alone sites or EBNA3A sites with EBNA3C, EBNA2, or both were evaluated. Sites for EBNA3A alone or with EBNA2 had the lowest EBNA3A signals. However, EBNA3A sites had the highest ChIP-seq signals when co-occurring with EBNA3C or with both EBNA3C and EBNA2, suggesting that EBNA3A preferred co-occupancy with EBNA3C or EBNA3C/EBNA2 (Fig. 3, Upper Left). Similarly, EBNA3C had higher signals when co-occurring with EBNA3A and EBNA2 (Fig. S2). EBNA3A ChIP-Seq signals at BATF/IRF4 sites were higher, whereas EBNA3A sites co-occurring with IRF4 had lower EBNA3A signals than EBNA3A sites co-occurring with BATF (Fig. 3, Upper Right). BATF and IRF4 signals were also higher when EBNA3A, BATF, and IRF4 co-occurred (Fig. S2). Also, EBNA3A co-occurrence with RUNX3 had higher EBNA3A signals (Fig. 3, Lower Left), whereas RUNX3 had higher signals at sites co-occurring with EBNA3A (Fig. 3, Lower Right; Fig. S3). The EBNA3A signals were also higher when co-occurring with RBPJ, NFκB(p65), MEF2A, POU2F2 (Oct2), and PAX5 (Fig. S3). Surprisingly, MEF2A and PAX5 signals were higher when co-occurring with EBNA3A, whereas EBNA3A co-occurrence had no effect on RBPJ, POU2F2, or NFκB(p65) binding (Fig. S3). SPI1 co-occurrence did not alter the EBNA3A ChIP-seq signal, but SPI1 signals were much higher when co-occurring with EBNA3A (Fig. S3).
Fig. 3.
ChIP-seq signals at EBNA3A sites with other EBV and cell TFs. Anchor plots of EBNA3A (E3A) sites showing the effect of co-occupancy with EBNA3C and EBNA2 (Upper Left) and AICEs (BATF and IRF4, Upper Right) on EBNA3A ChIP-seq signals. Anchor plots of EBNA3A (E3A) (Lower Left) or RUNX3 sites (Lower Right) showing the impact of co-occupancy on ChIP-seq signals. ChIP-seq signals were plotted in a ±1-kb window.
EBNA3A ChIP-seq Signals at MYC, CDKN2A/B, CCND2, BCL2, and CXCL9/10 Loci.
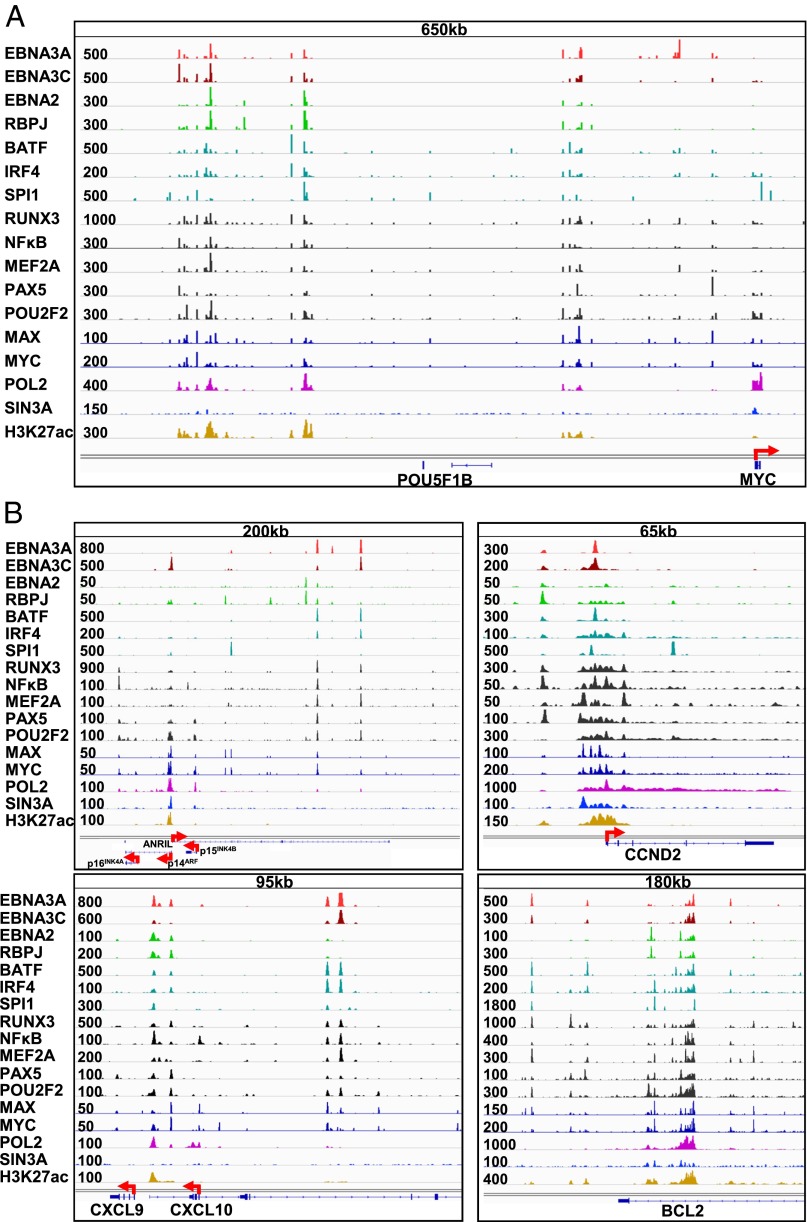
EBNA2 and EBNALP are the first EBV genes expressed after initial EBV infection of resting B-lymphocytes. EBNA2 binds to enhancers from 428 to 525 kb upstream of the MYC TSS and loops to the MYC promoter to activate MYC expression (Fig. 4A) (38). EBNA3A and EBNA3C have strong signals at the −428 to −525 kb MYC enhancer sites, which were co-occupied by EBNA2, RBPJ, BATF, IRF4, SPI1, MEF2A, PAX5, POU2F2, and POLII (Fig. 4A). EBNA3A was also at multiple sites 220 kb upstream of the MYC TSS. However, the strongest EBNA3A peak was at 70 kb upstream of the MYC TSS. Intriguingly, this site lacked EBNA3C, EBNA2, H3K27ac, and POLII signals. Other EBNA3A sites had high H3K27ac and POLII signals, indicative of strong enhancers (Fig. 4A). The MYC and MAX heterodimers cause cell cycle entry and induce p16INK4A- and p14ARF-mediated cell senescence (Fig. 4B, Upper Left). However, sustained LCL growth requires EBNA3A and EBNA3C repression of p16INK4A and p14ARF, as well as EBNA3A, EBNA3C, EBNA2, RBPJ, BATF, IRF4, SPI1, RUNX3, NFκB, and MEF2A-driven CCND2 and BCL2 expression (Fig. 4B, Upper and Lower Right).
Fig. 4.
EBNA3A ChIP-seq signals at MYC, CDKN2A/B, CCND2, CXCL9/CXCL10, and BCL2 loci. (A) ChIP-seq tracks and overall signal strength for EBV, cell TFs, and histone marks within 1 Mb 5′ of the MYC TSS. EBNA3A, EBNA3C, EBNA2, RBPJ, BATF, IRF4, SPI1, RUNX3, NFκB, MEF2A, PAX5, POU2F2 (OCT2), MAX, MYC, POLII, Sin3A, and H3K27ac tracks are shown. (B) ChIP-seq tracks and overall signal strength for EBV, cell TFs, and histone marks at CDKN2A/B (Upper Left), CCND2 (Upper Right), CXCL9/CXCL10 (Lower Left), and BCL2 (Lower Right) loci. EBNA3A, EBNA3C, EBNA2, RBPJ, BATF, IRF4, SPI1, RUNX3, NFκB, MEF2A, PAX5, POU2F2 (OCT2), MAX, MYC, POLII, Sin3A, and H3K27ac signals are shown.
Although EBNA3C’s effects are mediated by Sin3A recruitment to the p14ARF promoter, the molecular mechanisms underlying EBNA3A’s repressive effects are not fully evident. EBNA3A ChIP-seq places EBNA3A at three sites upstream of the p14ARF promoter and within the ANRIL noncoding RNA gene body (Fig. 4B, Upper Left). ANRIL has been implicated in negative control of the CDKN2A locus (39). These sites lack strong EBNA2, RBPJ, or Sin3A signals. Furthermore, EBNA3A signals were low at the p14ARF promoter site, indicating that EBNA3A’s repressive effects on CDKN2A differ from EBNA3C’s repressive effects.
EBNA3A binds to the CXCL9/CXCL10 loci in LCLs by ChIP quantitative PCR (qPCR) (31). Our ChIP-seq data confirmed this finding in another LCL line (Fig. 4B, Lower Left). Additional strong EBNA3A-binding sites that may regulate CXCL9/CXCL10 expression were also evident further upstream of this locus. EBNA3A signals were also evident at MED26 and IGF2R loci (Fig. S4).
EBNA3A Binds to DNA Through a BATF-Containing Protein Complex.
EBNA3A localized with B-cell TFs that substantially increased EBNA3A signals on DNA (Fig. 3). These B-cell TFs are likely candidates that mediate EBNA3A DNA binding. The frequent co-occurrence of EBNA3A with BATF (Fig. 1B and Fig. S2) at LCL genome DNA sites and the higher EBNA3A signals at these sites implicate BATF as one of the factors tethering EBNA3A to DNA.
ChIP-re-ChIP was used to further characterize EBNA3A DNA complexes with BATF. The SMG6 and CCR6 intron enhancer sites had high ChIP-seq signals for EBNA3A, EBNA2, RBPJ, BATF, IRF4, and RUNX3 (Fig. 5A and Fig. S4). HA-tagged EBNA3A was immunoprecipitated with anti-HA antibody. Protein–DNA complexes were eluted and re-immunoprecipitated with anti-HA– and anti-BATF–specific antibodies. A no-antibody control was used to assess whether the anti-HA antibodies used in the first immunoprecipitation had been efficiently removed and not carried over to the second immunoprecipitation. qPCR was used to quantitate the immunoprecipitated DNA. As expected, re-ChIP with anti-HA antibody was enriched for the SMG6 and CCR6 EBNA3A sites ∼20-fold over a no-Ab control, and re-ChIP with BATF antibody was significantly enriched over the no-Ab controls (P < 0.01) (Fig. 5B). Coimmunoprecipitation experiments, in contrast, showed that EBNA3A did not directly interact with BATF without cross-linking. Together, these data indicate that both EBNA3A and BATF are part of a ternary protein complex on DNA in which EBNA3A may bind to BATF via additional cell proteins or DNA.
Fig. 5.
EBNA3A is tethered to enhancers through BATF containing protein complex. (A) EBNA3A site at the SMG6 and CCR6 loci. Normalized EBNA3A, EBNA2, RBPJ, RUNX3, BATF, and IRF4 tag density signals are shown. The peak heights are indicated at the left of each track. (B) ChIP-re-ChIP analyses with antibodies against HA and BATF and subsequent qPCR analyses showing a high fold enrichment of both samples over negative control. (C) EBNA3A binds to enhancers through a complex containing BATF, IRF4, and RUNX3 to recruit factors involved in gene expression to activate transcription of target genes.
Discussion
Genetic and biochemical evidence indicate that EBNA3A and EBNA3C bind similarly to RBPJ and CtBP (8–10, 18, 40, 41). We now find that 50% of EBNA3A genome-wide binding sites co-occur with EBNA3C. EBNA3A and EBNA3C ChIP-seq signals are similarly dependent on co-occupying B-cell lineage TFs, notably, BATF/IRF4 AICE and SPI1/IRF4 EICE composite sites, which strongly affect EBNA3A and EBNA3C DNA binding (15). The strong positive effects of AICE, EICE, and RUNX3 on EBNA3A signals were similar in magnitude to their effects on EBNA3C and were positively affected by co-occupancy with other B-cell TFs. We also find that EBNA3A and EBNA3C differ dramatically in their genome-wide DNA-binding landscape. Even though both EBNA3A and EBNA3C repress CDKN2A expression, they repress CDKN2A p14ARF- and p16INK4A-mediated cell senescence responses through distinct mechanisms. EBNA3C binds to the p14ARF promoter and recruits Sin3A repressive complexes to suppress CDKN2A expression (15). In contrast, EBNA3A binds to multiple enhancer sites upstream of the p14ARF/p15INK4B promoter and to the ANRIL long noncoding RNA gene body. EBNA3A and EBNA3C bind to common sites at −220, −428, and −525 kb of MYC, where strong EBNA2, RBPJ, BATF, IRF4, SPI1, MEF2A, PAX5, and POU2F2 signals, as well as high H3K27ac marks, are also evident. At these sites, EBNA3A is likely to be tethered to DNA through BATF, IRF4, and SPI1 and recruit POLII to enhance MYC expression through looping to the MYC TSS. In addition to these MYC enhancer sites, EBNA3A also binds strongly to unique enhancer sites ∼70 kb upstream of MYC. This EBNA3A site is different from other EBNA3A MYC sites in that there is no binding for other EBNAs. This site lacks POLII or H3K27ac signals, but the moderate H3K4me1 signals suggest enhancer activity. EBNA3C signals are almost identical when co-occurring with BATF or with IRF4 alone whereas EBNA3A signals are stronger when co-occurring with BATF alone than with IRF4.
Because EBNA3A and EBNA3C inactivation affect the expression of p14ARF, p16INK4A, and p15INK4B, EBNA3A and EBNA3C likely cause chromosome looping between the p14ARF, p16INK4A, or p15INK4B promoters. H3K27me3 methylase EZH2 can cause the p15INK4B promoter to loop to the p16INK4A promoter and coregulate their expression, leaving the p14ARF promoter unaffected (42). EBNA3A likely differs from EBNA3C in enhancer looping to the p14ARF or p16INK4A promoters because they bind differentially unique sites. EBNA3A and EBNA3C also have PXDLS sites that may bind CtBP and recruit repressive chromatin modifications to p14ARF or p16INK4A (18, 26, 41). However, null mutations in the EBNA3C PXDLS motifs have little effect on p14ARF or p16INK4A suppression (43, 44). The CDKN2A/B loci are affected by polycomb group proteins and the long noncoding RNA ANRIL (39). ANRIL recruits polycomb group proteins to the CDKN2A/B loci and increases the repressive H3K27me3 mark at the CDKN2A/B loci (45). EBNA3A binding to the ANRIL gene body may increase its expression and cause an increase in H3K27me3 signals at this locus.
EBNA3A can also displace EBNA2 from CXCL9 and CXCL10 promoter/enhancer sites, decreasing CXCL9 and CXCL10 expression and inducing PRC2-catalyzed H3K27me3 modifications (31), consistent with a two-step model in which EBNA3A binds and inactivates enhancers, followed by reduced POLII recruitment and PRC2-catalyzed H3K27me3 modifications across CXCL9 and CXCL10 (31). EBNA3A inactivation of intergenic enhancers may maintain polycomb signatures across the CXCL9 and CXCL10 chromatin domain. EBNA3A ChIP-seq data find EBNA3A tethered to these sites through BATF and IRF4/SPI1, possibly making the CXCL9/10 locus inaccessible to other activators and thus repressing CXCL9 and 10.
Approximately 16% of EBNA3A sites have co-occurring RBPJ signals. This seems to be contradictory to the previous in vitro and in vivo findings where EBNA3A prevents RBPJ binding to the viral Cp-promoter, and EBNA3A expression prevents EBNA2 from binding to the CXCL9/CXCL10 loci, where EBNA2 is likely be tethered to DNA through RBPJ (8, 10, 31, 46). However, it is also likely that, instead of being tethered to DNA through RBPJ, EBNA3A is tethered to neighboring sites through a BATF-containing protein complex or other cell TFs.
EBNA3C preferentially binds to IRF4 (47), whereas EBNA3A binding is mostly affected by BATF. Because EBNA3A does not directly interact with BATF, both proteins are likely part of a ternary complex on DNA in which EBNA3A associates with BATF via additional cell factors. BATF and IRF4 frequently form heterodimers and bind to AICE sites (48). Therefore, higher-order complexes of EBNA3A, EBNA3C, BATF, and IRF4 may be present in LCLs. This is supported by a 50% genome-wide co-occurrence of EBNA3A and EBNA3C in LCLs. However, EBNA3A or EBNA3C may also differentially bind to sites in different phases of the cell cycle.
In summary, EBNA3A is at >10,000 euchromatic sites and is extensively involved in cell gene regulation. EBNA3A preferentially binds to AICE and EICE sites with other important B-cell lineage TFs. EBNA3A is tethered to enhancer regions through complexes containing BATF, RUNX3, and IRF4/SPI1 and recruits POLII through bridging factors. Enhancer complexes can then be brought in close proximity to promoters to activate gene expression.
Materials and Methods
Cell Lines.
A recombinant EBV in which EBNA3A is C-terminally tagged with FLAG and HA epitope (EBNA3AFHA) was used to generate LCLs. LCLs were grown in RPMI 1640 medium supplemented with 10% (vol/vol) FBS (GIBCO) and 2 mM L-glutamine with penicillin and streptomycin.
ChIP-seq.
EBNA3AFHA LCLs were cross-linked with formaldehyde and EBNA3AFHA was immunoprecipitated using an anti-HA antibody (Abcam, ab9110) as previously described (38). Extensive washing was followed by the elution of the protein–DNA complexes (38). After reverse cross-linking, the purified DNA was sequenced using an Illumina HiSeq 2500.
ChIP-re-ChIP.
ChIP-re-ChIP was done using a Re-ChIP-IT kit (Active Motif) following the manufacturer’s instruction.
ChIP-Seq Data Analysis.
ChIP-seq reads were mapped to hg18, allowing for one alignment and two mismatches per read with Bowtie with the parameters −k 1 −m 1. Replicate 1 has 31.9 million reads and replicate 2 has 30.6 million reads with at least one alignment to the reference genome. We performed quality control of the ChIP-seq data using phantom peak calling in SPP (35). The top 10,000 peaks were then called using SPP with an IDR < 0.01 to ensure reproducibility between replicates. These 10,000 peaks were then mapped to the hg18 GM12878 chromatin states as defined by Ernst et al. (37).
ChIP-seq Signal Clustering Around EBNA3A Sites.
TF ChIP-seq visualization and clustering were performed as previously described (15). In short, SPP was used to calculate input-normalized ChIP-seq log2-fold enrichment of various transcription factors around EBNA3A promoter and enhancer peaks (±2 kb) for visualization purposes (20 bins) or clustering purposes (5 bins). PAM clustering was then performed with in-house R scripts.
Enriched Motif Calling.
Using the Bioconductor package (BSgenome.Hsapiens.UCSC.hg18), sequences ±250 bp of EBNA3A sites per cluster (from Fig. 2) were extracted for motif analysis. As a control for background sequences, random hg18 sites with an identical chromatin distribution for each EBNA3A cluster were used. The findMotifs.pl function in HOMER was used to find DNA-binding motifs for each EBNA3A cluster enriched over a chromatin distribution matched background.
Supplementary Material
Acknowledgments
We thank Ben Gewurz for helpful discussions. These experiments were supported by Grants R01CA047006, R01CA170023, and R01CA085180 from the National Cancer Institute (to E.K.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE59181).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1422580112/-/DCSupplemental.
References
- 1.Epstein MA, Achong BG, Barr YM. Virus particles in cultured lymphoblasts from Burkitt’s lymphoma. Lancet. 1964;1(7335):702–703. doi: 10.1016/s0140-6736(64)91524-7. [DOI] [PubMed] [Google Scholar]
- 2.Alexander FE, et al. Risk factors for Hodgkin’s disease by Epstein-Barr virus (EBV) status: Prior infection by EBV and other agents. Br J Cancer. 2000;82(5):1117–1121. doi: 10.1054/bjoc.1999.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Longnecker R, Kieff E, Cohen JI. Epstein-Barr Virus. 8th Ed. Lippincott Williams & Wilkins; Philadelphia: 2013. pp. 1898–1959. [Google Scholar]
- 4.Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4(10):757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 5.Maruo S, Johannsen E, Illanes D, Cooper A, Kieff E. Epstein-Barr Virus nuclear protein EBNA3A is critical for maintaining lymphoblastoid cell line growth. J Virol. 2003;77(19):10437–10447. doi: 10.1128/JVI.77.19.10437-10447.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maruo S, et al. Epstein-Barr virus nuclear protein 3A domains essential for growth of lymphoblasts: Transcriptional regulation through RBP-Jkappa/CBF1 is critical. J Virol. 2005;79(16):10171–10179. doi: 10.1128/JVI.79.16.10171-10179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tomkinson B, Robertson E, Kieff E. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J Virol. 1993;67(4):2014–2025. doi: 10.1128/jvi.67.4.2014-2025.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao B, Marshall DR, Sample CE. A conserved domain of the Epstein-Barr virus nuclear antigens 3A and 3C binds to a discrete domain of Jkappa. J Virol. 1996;70(7):4228–4236. doi: 10.1128/jvi.70.7.4228-4236.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robertson ES, et al. Epstein-Barr virus nuclear protein 3C modulates transcription through interaction with the sequence-specific DNA-binding protein J kappa. J Virol. 1995;69(5):3108–3116. doi: 10.1128/jvi.69.5.3108-3116.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robertson ES, Lin J, Kieff E. The amino-terminal domains of Epstein-Barr virus nuclear proteins 3A, 3B, and 3C interact with RBPJ(kappa) J Virol. 1996;70(5):3068–3074. doi: 10.1128/jvi.70.5.3068-3074.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalbiès-Tran R, Stigger-Rosser E, Dotson T, Sample CE. Amino acids of Epstein-Barr virus nuclear antigen 3A essential for repression of Jkappa-mediated transcription and their evolutionary conservation. J Virol. 2001;75(1):90–99. doi: 10.1128/JVI.75.1.90-99.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grossman SR, Johannsen E, Tong X, Yalamanchili R, Kieff E. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the J kappa recombination signal binding protein. Proc Natl Acad Sci USA. 1994;91(16):7568–7572. doi: 10.1073/pnas.91.16.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henkel T, Ling PD, Hayward SD, Peterson MG. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science. 1994;265(5168):92–95. doi: 10.1126/science.8016657. [DOI] [PubMed] [Google Scholar]
- 14.Hsieh JJ, Hayward SD. Masking of the CBF1/RBPJ kappa transcriptional repression domain by Epstein-Barr virus EBNA2. Science. 1995;268(5210):560–563. doi: 10.1126/science.7725102. [DOI] [PubMed] [Google Scholar]
- 15.Jiang S, et al. Epstein-Barr virus nuclear antigen 3C binds to BATF/IRF4 or SPI1/IRF4 composite sites and recruits Sin3A to repress CDKN2A. Proc Natl Acad Sci USA. 2014;111(1):421–426. doi: 10.1073/pnas.1321704111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooper A, et al. EBNA3A association with RBP-Jkappa down-regulates c-myc and Epstein-Barr virus-transformed lymphoblast growth. J Virol. 2003;77(2):999–1010. doi: 10.1128/JVI.77.2.999-1010.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cludts I, Farrell PJ. Multiple functions within the Epstein-Barr virus EBNA-3A protein. J Virol. 1998;72(3):1862–1869. doi: 10.1128/jvi.72.3.1862-1869.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hickabottom M, Parker GA, Freemont P, Crook T, Allday MJ. Two nonconsensus sites in the Epstein-Barr virus oncoprotein EBNA3A cooperate to bind the co-repressor carboxyl-terminal-binding protein (CtBP) J Biol Chem. 2002;277(49):47197–47204. doi: 10.1074/jbc.M208116200. [DOI] [PubMed] [Google Scholar]
- 19.Bain M, Watson RJ, Farrell PJ, Allday MJ. Epstein-Barr virus nuclear antigen 3C is a powerful repressor of transcription when tethered to DNA. J Virol. 1996;70(4):2481–2489. doi: 10.1128/jvi.70.4.2481-2489.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knight JS, Lan K, Subramanian C, Robertson ES. Epstein-Barr virus nuclear antigen 3C recruits histone deacetylase activity and associates with the corepressors mSin3A and NCoR in human B-cell lines. J Virol. 2003;77(7):4261–4272. doi: 10.1128/JVI.77.7.4261-4272.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Radkov SA, et al. Epstein-Barr virus nuclear antigen 3C interacts with histone deacetylase to repress transcription. J Virol. 1999;73(7):5688–5697. doi: 10.1128/jvi.73.7.5688-5697.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen A, Zhao B, Kieff E, Aster JC, Wang F. EBNA-3B- and EBNA-3C-regulated cellular genes in Epstein-Barr virus-immortalized lymphoblastoid cell lines. J Virol. 2006;80(20):10139–10150. doi: 10.1128/JVI.00854-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maruo S, et al. Epstein-Barr virus nuclear protein EBNA3C is required for cell cycle progression and growth maintenance of lymphoblastoid cells. Proc Natl Acad Sci USA. 2006;103(51):19500–19505. doi: 10.1073/pnas.0604919104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomkinson B, Kieff E. Second-site homologous recombination in Epstein-Barr virus: Insertion of type 1 EBNA 3 genes in place of type 2 has no effect on in vitro infection. J Virol. 1992;66(2):780–789. doi: 10.1128/jvi.66.2.780-789.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hertle ML, et al. Differential gene expression patterns of EBV infected EBNA-3A positive and negative human B lymphocytes. PLoS Pathog. 2009;5(7):e1000506. doi: 10.1371/journal.ppat.1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skalska L, White RE, Franz M, Ruhmann M, Allday MJ. Epigenetic repression of p16(INK4A) by latent Epstein-Barr virus requires the interaction of EBNA3A and EBNA3C with CtBP. PLoS Pathog. 2010;6(6):e1000951. doi: 10.1371/journal.ppat.1000951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maruo S, et al. Epstein-Barr virus nuclear antigens 3C and 3A maintain lymphoblastoid cell growth by repressing p16INK4A and p14ARF expression. Proc Natl Acad Sci USA. 2011;108(5):1919–1924. doi: 10.1073/pnas.1019599108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skalska L, et al. Induction of p16(INK4a) is the major barrier to proliferation when Epstein-Barr virus (EBV) transforms primary B cells into lymphoblastoid cell lines. PLoS Pathog. 2013;9(2):e1003187. doi: 10.1371/journal.ppat.1003187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tursiella ML, et al. Epstein-Barr virus nuclear antigen 3A promotes cellular proliferation by repression of the cyclin-dependent kinase inhibitor p21WAF1/CIP1. PLoS Pathog. 2014;10(10):e1004415. doi: 10.1371/journal.ppat.1004415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bazot Q, et al. Epstein-Barr virus nuclear antigen 3A protein regulates CDKN2B transcription via interaction with MIZ-1. Nucleic Acids Res. 2014;42(15):9700–9716. doi: 10.1093/nar/gku697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harth-Hertle ML, et al. Inactivation of intergenic enhancers by EBNA3A initiates and maintains polycomb signatures across a chromatin domain encoding CXCL10 and CXCL9. PLoS Pathog. 2013;9(9):e1003638. doi: 10.1371/journal.ppat.1003638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderton E, et al. Two Epstein-Barr virus (EBV) oncoproteins cooperate to repress expression of the proapoptotic tumour-suppressor Bim: Clues to the pathogenesis of Burkitt’s lymphoma. Oncogene. 2008;27(4):421–433. doi: 10.1038/sj.onc.1210668. [DOI] [PubMed] [Google Scholar]
- 33.Young P, Anderton E, Paschos K, White R, Allday MJ. Epstein-Barr virus nuclear antigen (EBNA) 3A induces the expression of and interacts with a subset of chaperones and co-chaperones. J Gen Virol. 2008;89(Pt 4):866–877. doi: 10.1099/vir.0.83414-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McClellan MJ, et al. Modulation of enhancer looping and differential gene targeting by Epstein-Barr virus transcription factors directs cellular reprogramming. PLoS Pathog. 2013;9(9):e1003636. doi: 10.1371/journal.ppat.1003636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol. 2008;26(12):1351–1359. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Landt SG, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22(9):1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473(7345):43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao B, et al. Epstein-Barr virus exploits intrinsic B-lymphocyte transcription programs to achieve immortal cell growth. Proc Natl Acad Sci USA. 2011;108(36):14902–14907. doi: 10.1073/pnas.1108892108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yap KL, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38(5):662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao B, et al. Transcriptional regulatory properties of Epstein-Barr virus nuclear antigen 3C are conserved in simian lymphocryptoviruses. J Virol. 2003;77(10):5639–5648. doi: 10.1128/JVI.77.10.5639-5648.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Touitou R, Hickabottom M, Parker G, Crook T, Allday MJ. Physical and functional interactions between the corepressor CtBP and the Epstein-Barr virus nuclear antigen EBNA3C. J Virol. 2001;75(16):7749–7755. doi: 10.1128/JVI.75.16.7749-7755.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kheradmand Kia S, et al. EZH2-dependent chromatin looping controls INK4a and INK4b, but not ARF, during human progenitor cell differentiation and cellular senescence. Epigenetics Chromatin. 2009;2(1):16. doi: 10.1186/1756-8935-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee S, et al. Epstein-Barr virus nuclear protein 3C domains necessary for lymphoblastoid cell growth: Interaction with RBP-Jkappa regulates TCL1. J Virol. 2009;83(23):12368–12377. doi: 10.1128/JVI.01403-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maruo S, et al. Epstein-Barr virus nuclear protein EBNA3C residues critical for maintaining lymphoblastoid cell growth. Proc Natl Acad Sci USA. 2009;106(11):4419–4424. doi: 10.1073/pnas.0813134106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aguilo F, Zhou MM, Walsh MJ. Long noncoding RNA, polycomb, and the ghosts haunting INK4b-ARF-INK4a expression. Cancer Res. 2011;71(16):5365–5369. doi: 10.1158/0008-5472.CAN-10-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waltzer L, Perricaudet M, Sergeant A, Manet E. Epstein-Barr virus EBNA3A and EBNA3C proteins both repress RBP-J kappa-EBNA2-activated transcription by inhibiting the binding of RBP-J kappa to DNA. J Virol. 1996;70(9):5909–5915. doi: 10.1128/jvi.70.9.5909-5915.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Banerjee S, et al. The EBV latent antigen 3C inhibits apoptosis through targeted regulation of interferon regulatory factors 4 and 8. PLoS Pathog. 2013;9(5):e1003314. doi: 10.1371/journal.ppat.1003314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ochiai K, et al. Transcriptional regulation of germinal center B and plasma cell fates by dynamical control of IRF4. Immunity. 2013;38(5):918–929. doi: 10.1016/j.immuni.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.