Abstract
Reversible histone acetylation and deacetylation at the N-terminus of histone tails play a crucial role in regulating eukaryotic gene activity. Acetylation of core histones is associated with gene activation, whereas deacetylation of histone is often correlated with gene repression. The level of histone acetylation is antagonistically catalyzed by histone acetyltransferases citation(HATs) and histone deacetylases (HDACs). In this work, we examined the subcellular localization, expression pattern and function of HDT701, a member of the plant-specific HD2-type histone deacetylase in rice. HDT701 is localized at the subcellular level in the nucleus. Histochemical GUS-staining analysis revealed that HDT701 is constitutively expressed throughout the life cycle of rice. Overexpression of HDT701 in rice decreases ABA, salt and osmotic stress resistance during seed germination. Delayed seed germination of HDT701 overexpression lines is associated with decreased histone H4 acetylation and down-regulated expression of GA biosynthetic genes. Moreover, overexpression of HDT701 in rice enhances salt and osmotic stress resistance during the seedling stage. Taken together, our findings suggested that HDT701 may play an important role in regulating seed germination in response to abiotic stresses in rice.
Keywords: HDT701, histone deacetylase, seed germination, abiotic stress, gene expression
Introduction
In eukaryotic cells, gene activity is controlled not only by the DNA sequence but also by epigenetic events. Switch of the chromatin structure between the condensed status and the relaxed status plays an important role in the regulation of gene expression (Goldberg et al., 2007). Post-translational histone changes lead to an alteration of DNA activity by histone acetylation, methylation, phosphorylation, ubiquitination, and sumoylation (Berger, 2007). The reversible acetylation or deacetylation of specific lysine residues at the N-terminal of histone tails are catalyzed by histone acetyltransferases citation(HATs) and histone deacetylases citation(HDACs) (Pandey et al., 2002). In general, hyper-acetylation of the histones is associated with gene activation whereas the hypoacetylation of histones is correlated with transcriptional repression. Plant HDACs can be grouped into three major families, the RPD3/HDA1 superfamily, the SIR2 family, and the HD2 family (Pandey et al., 2002).
HD2 proteins are plant-specific HDACs. HD2 proteins were first identified in maize as an acidic nucleolar phosphoprotein in a high molecular weight complex (Brosch et al., 1996; Lusser et al., 1997). Four HD2 proteins, HD2A (also known as HDT1), HD2B (HDT2) (Wu et al., 2000), HD2C citation(HDT3) (Zhou et al., 2004), and HD2D (HDT4) have been identified in Arabidopsis (Dangl et al., 2001; Pandey et al., 2002). The functions of the HD2 family members have been well-studied in recent years. It was demonstrated that HD2A, HD2B, and HD2C are able to mediate transcriptional repression (Wu et al., 2000, 2003). Knockdown of HD2A in Arabidopsis results in seed abortion (Wu et al., 2003) and causes reduced silencing of Arabidopsis rDNA (Lawrence et al., 2004). HD2C is involved in abscisic acid (ABA) and salt and drought stress responses (Sridha and Wu, 2006; Luo et al., 2012a). Knockdown of HD2A and HD2B in the asymmetric leaf1 citation(as1) or as2 mutant background causes the formation of abaxialized and filamentous leaves, which suggests the involvement of HD2A and HD2B during leaf development in Arabidopsis (Ueno et al., 2007).
Phylogenic analysis of the rice genome suggests there are at least two members of HD2 genes, HDT701 and HDT702, in rice (Fu et al., 2007). Knockdown of HDT702 rice plants displayed severely narrowed leaves, suggesting that HDT702 might be involved in cell division or growth (Hu et al., 2009). Furthermore, overexpression of HDT701 in transgenic rice leads to decreased levels of histone H4 acetylation and enhanced susceptibility to rice pathogens Magnaporthe oryzae and Xanthomonas oryzae pv oryzae, while silencing of HDT701 in transgenic rice causes the levels of histone H4 acetylation and transcription pattern recognition of receptor and defense-related genes to become elevated, suggesting that HDT701 is a histone H4 deacetylase and acts as a negative regulator in plant innate immunity in rice (Ding et al., 2012).
In this study, we investigated the subcellular localization, expression pattern and function of HDT701 in rice. Our results suggested that HDT701 may play essential roles in the regulation of seed germination in response to abiotic stresses.
Results
Cis-acting element analysis of HDT701 and HDT702 promoter sequences
We analyzed the cis-acting elements of the promoter regions of HDT701 and HDT702. A 2000 bp DNA sequence before the start codon was used for the prediction using the PlantCARE database (Lescot et al., 2002). As shown in Supplemental Table 1, a large subset of stress-responsive cis-acting DNA regulatory elements was identified in HDT701 and HDT702 promoters. The following cis-acting elements were found in the promoters of HDT701 and HDT702, respectively: 18 and 14 dehydration-responsive, 8 and 22 early responsive to dehydration, 8 and 11 pathogenesis-related, 4 and 4 disease-resistant responsive, and 2 and 8 ABA-responsive. This suggests that HDT701 and HDT702 may play an important role in the response to abiotic and biotic stresses. Furthermore, 20 and 9 cytokinin-related (CK), 2 and 2 salicylic acid (SA)-induced, and 1 and 3 jasmonate acid (JA)-responsive elements were also identified in the promoters of HDT701 and HDT702, respectively. This indicates that HDT701 and HDT702 may be involved in multiple hormone signaling pathways.
Expression patterns of HDT701 and HDT702 under abiotic stresses
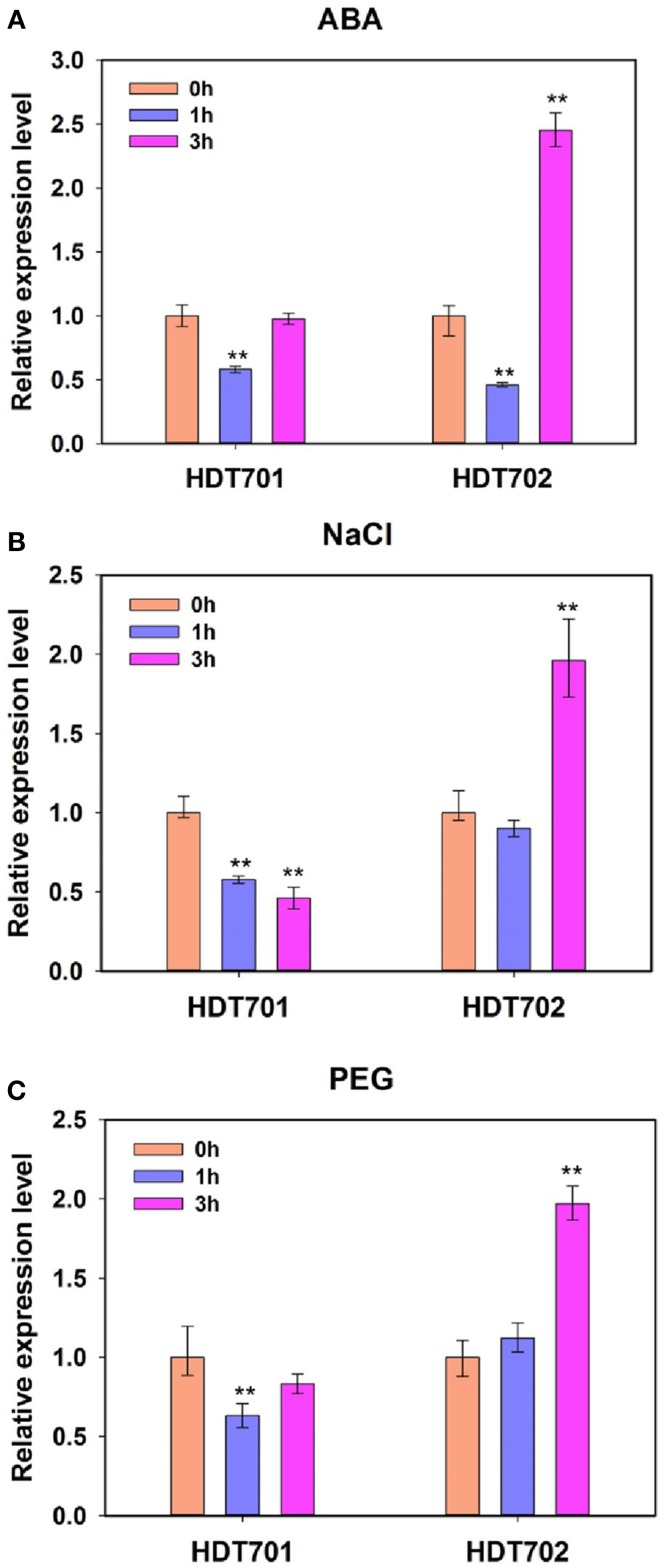
We further studied the expression patterns of HDT701 and HDT702 under ABA, salt and osmotic stresses. Here, 300 mM sodium chloride (NaCl) and 20% polyethylene glycol 6000 (PEG) were used to simulate salt and osmotic stress, respectively. The expression levels of HDT701 decreased significantly 1 h after ABA, NaCl and PEG treatments, but 3 h after ABA and PEG exposure, they recovered (Figure 1). Furthermore, the expression of HDT702 was down-regulated 1 h after ABA treatment, but 3 h after ABA, NaCl, and PEG exposure, the expression levels were significantly higher (Figure 1). Together, this data suggests that the expression patterns of HDT701 and HDT702 may be regulated by abiotic stresses.
Figure 1.
Expression patterns of rice HDT701 and HDT702 under ABA, salt, and PEG stresses. Two-week-old rice seedlings were treated with 100 μ M ABA (A), 300 mM NaCl (B), and 20% PEG (C) for 0, 1, and 3 h, respectively. ACTIN was used as the internal control. Eighteen seedlings were used for analysis for each treatment. Values are shown as means + SD (t-test: **P < 0.01, difference from WT).
GUS staining analysis of HDT701 expression patterns in rice organs
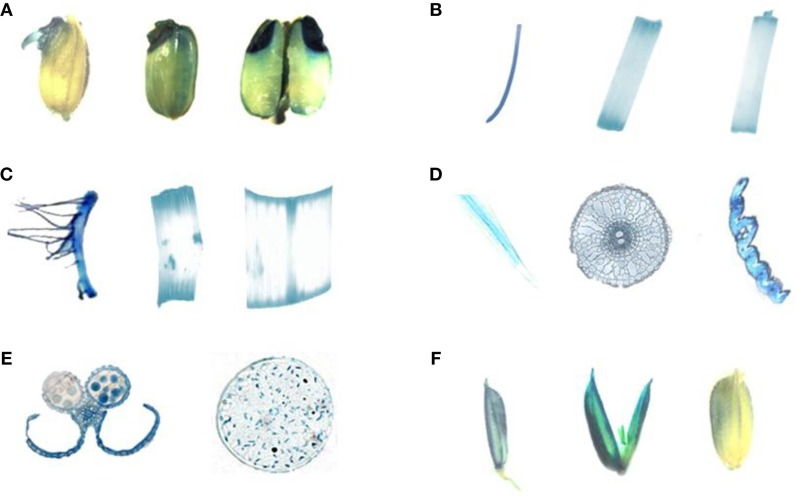
The expression patterns of HDT701 were then examined by using transgenic rice plants expressing β-glucuronidase (GUS) driven by the HDT701 promoter. A strong GUS signal was detected in germinating rice seeds (Figure 2A), 14 and 40-day-old roots, petioles and leaves (Figures 2B–D). GUS staining was also detected in anthers, pollen (Figure 2E), and in the pollen grains during the maturation stage (Figure 2F). These data suggest that HDT701 is constitutively expressed throughout the life cycle of rice.
Figure 2.
GUS staining of HDT701 expression at various growth stages in rice. (A) Germinating rice seeds. (B) Root, stem and leaf of 14-day-old seedling. (C) Root, stem, and leaf of 40-day-old rice plant. (D) Cross-section of root, stem, and leaf of 40-day-old rice plant. (E) Anther and pollen of 60-day-old rice plant. (F) Grains during maturation (85-day-old rice plant).
Subcellular localization of HDT701
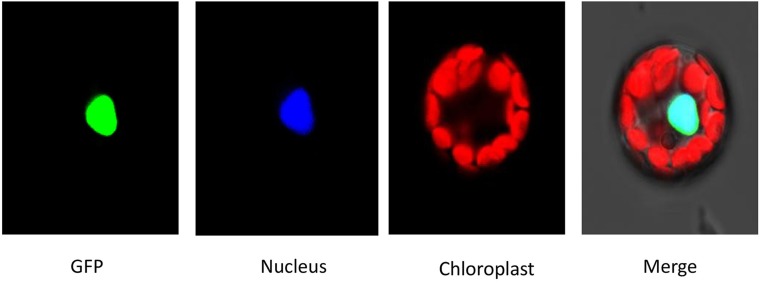
To investigate the subcellular localization of HDT701 proteins, the coding region of HDT701 fused with green fluorescent protein (GFP) was transformed into Arabidopsis protoplasts by PEG. Consistent with a previous report (Ding et al., 2012), we showed that HDT701-GFP is localized at a subcellular level in the nucleus of the protoplasts by a fluorescence microscopy assay (Figure 3).
Figure 3.
Subcellular localization analysis of HDA701. The constructs were transformed into Arabidopsis protoplast by PEG. mCherry was used as a nuclear marker.
Generation and identification of rice HDT701 overexpression and RNAi plants
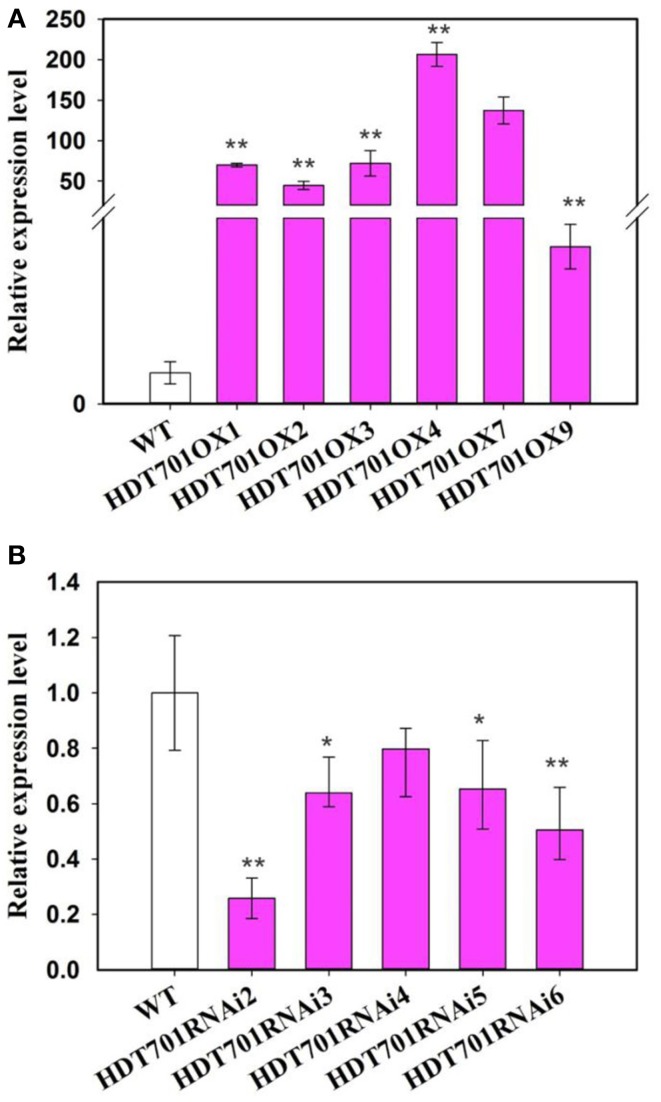
To elucidate the function of HDT701 in rice, an OX construct was made in which the full length HDT701 cDNA was expressed under the control of the maize ubiquitin promoter. The construct was transformed into rice via Agrobacterium tumefaciens-mediated transformation. Quantitative RT-PCR analysis confirmed increased expression of HDT701 in five independent HDT701OX homozygous transgenic lines (Figure 4A). Two transgenic lines (HDT701 OX4 and OX7) with high expression of HDT701 were selected for further studies. Furthermore, we also generated HDT701 RNAi transgenic plants using an RNAi construct, which was designed to target the specific 5' untranslated region (UTR) of HDT701 under the control of the maize ubiquitin promoter. Quantitative RT-PCR analysis showed that the expression of HDT701 was down-regulated in HDT701 RNAi2 and RNAi6 transgenic lines (Figure 4B).
Figure 4.
Transcription an alysis of HDT701OX plants (A) and RNAi plants (B) by quantitative RT-PCR assay. ACTIN was used as the internal control. At least 18 seedlings were used for analysis. Values shown as means + SD (t-test: *P < 0.05, **P < 0.01, difference from WT).
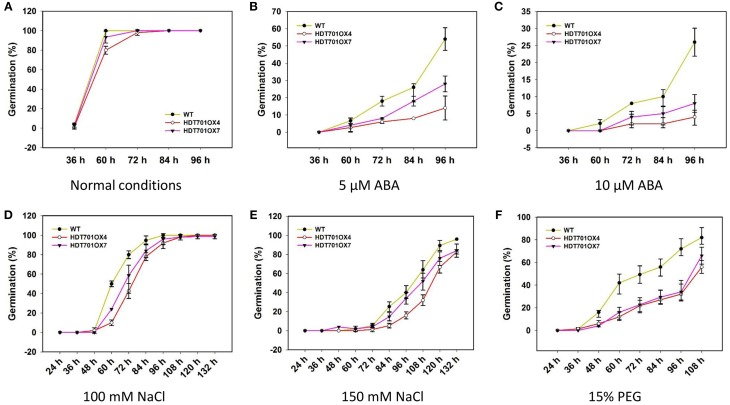
Overexpression of HDT701 in rice delays seed germination under ABA, salt, and osmotic stresses
Previous studies revealed that Arabidopsis HD2-type HDACs, namely HD2A, HD2B, HD2C and HD2D, are key regulators in stress responses (Liu et al., 2014), which prompted us to examine any possible role of rice HDT701 in responding to abiotic stresses. We compared the germination rates of HDT701 OX seeds with wild-type (WT) when separately treated with various concentrations of ABA, NaCl and PEG, as stressors. Relatively lower germination rates were detected for HDT701 OX4 and OX7 seeds 60 h after plating in medium compared to the WT under normal (control) conditions (Figure 5A), suggesting that overexpression of HDT701 in rice may delay seed germination. Furthermore, the germination of HDT701 OX4 and OX7 seeds was significantly delayed in response to 5 and 10 μ M ABA compared to the WT (Figures 5B,C). Similarly, the germination of HDT701 OX4 and OX7 seeds was also slower than the WT after treatment with 100 and 150 mM NaCl (Figures 5D,E). Upon exposure to 15% PEG, the germination rates of HDT701 OX4 and OX7 were obviously lower than the WT (Figure 5F). However, the germination rates of HDT701 RNAi2 and RNAi6 showed no obvious differences compared with the WT (Supplemental Figure 1). Collectively, these data indicate that overexpression of HDT701 in rice may delay seed germination under ABA, salt and osmotic stresses.
Figure 5.
Time course of seed germination of HDT701 OX4 and OX7 under normal conditions (A), 5 and 10 μM ABA (B,C), 100 and 150 mM NaCl (D,E), and 15% PEG (F). Hundred seeds were used for the analysis of each treatment.
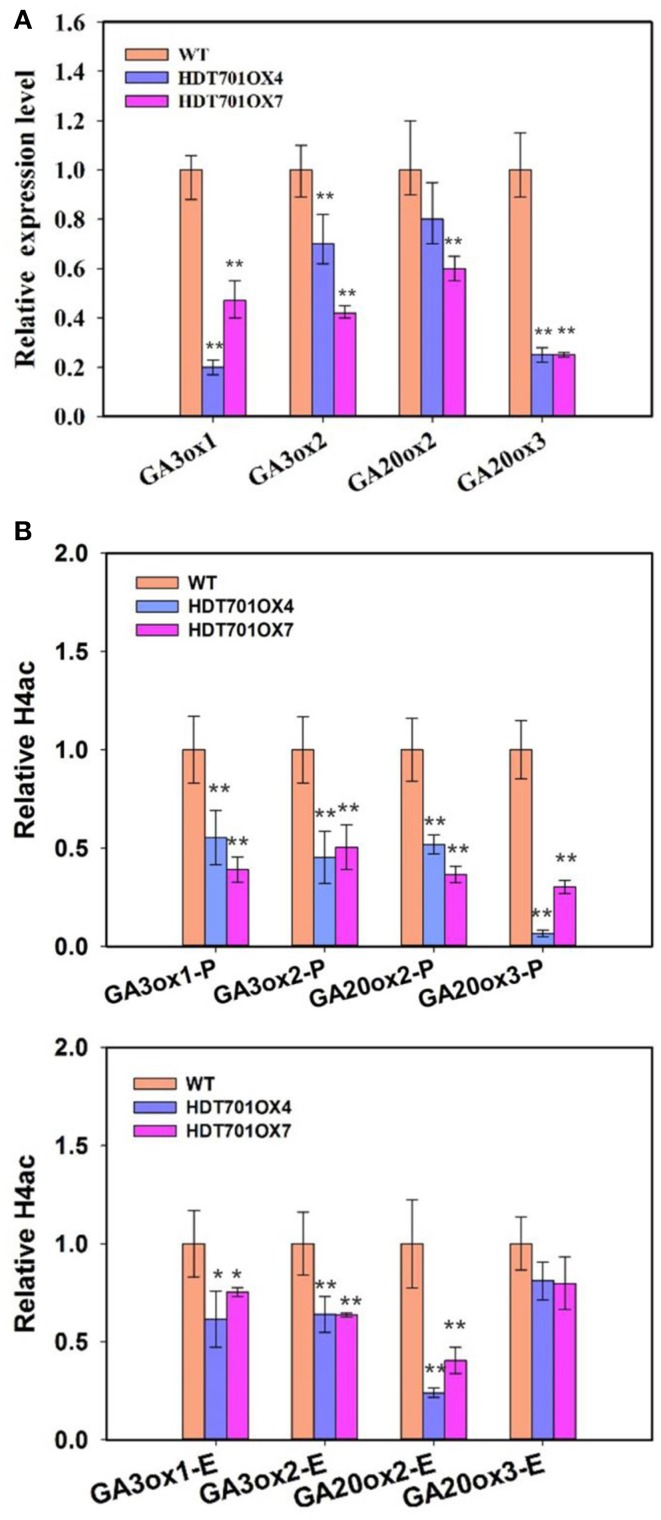
Overexpression of HDT701 decreases gene expression and histone H4 acetylation levels of GA biosynthetic genes during seed germination
Previous studies revealed that a plant hormone, gibberellin (GA), plays a key role in the control of seed germination (Ogawa et al., 2003). We further detected the transcription levels of the GA metabolic genes in germinating HDT701 OX seeds. The expression levels of GA biosynthetic genes, GA3ox1 and GA3ox2, GA20ox2 and GA20ox3, were significantly down-regulated in HDT701 OX4 and OX7 seeds (Figure 6A).
Figure 6.
Transcription and histone acetylation levels of GA biosynthesis genes in HDT701 overexpression lines during seed germination. (A) qRT-PCR analysis of the expression levels of GA biosynthetic genes, GA3ox1, GA3ox2, GA20ox2, and GA20ox3 in HDT701 OX4 and OX7. ACTIN was used as the internal control. (B) ChIP analysis of the histone H4 acetylation levels of the promoters and exons of GA biosynthetic genes in germinating HDT701 OX4 and OX7 seeds. P, promoter; E, exon. UBQ was used as the internal control. At least 30 seedlings of WT, HDT701 OX4, and OX7 were used for analysis. Values are shown as means + SD (t-test: *P < 0.05, **P < 0.01, difference from WT).
Recent work has shown that HDT701 is a histone H4 deacetylase (Ding et al., 2012). We further examined the histone H4 acetylation levels of these GA biosynthetic genes in germinating HDT701 OX seeds by chromatin immunoprecipitation (ChIP) assays. There was an obvious decrease in histone H4 acetylation levels at promoter and exon regions of GA3ox1, GA3ox2, and GA20ox2, and the promoter region of GA20ox3 in HDT701 OX4 and OX7 seeds compared with the WT (Figure 6B). This data suggests that HDT701 may repress the expression of GA biosynthetic genes by decreasing their histone H4 acetylation levels in germinating rice seeds.
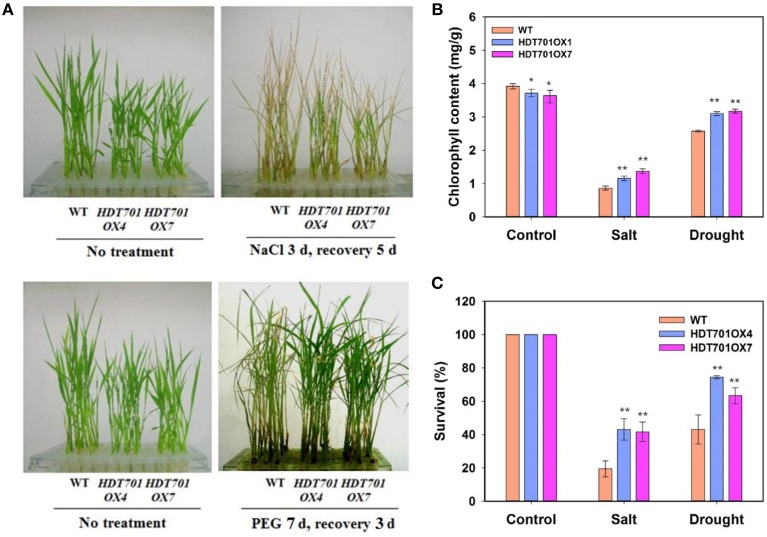
Overexpression of HDT701 in rice enhances salt and osmotic resistance during the seedling stage
We further examined the phenotype of HDT701 OX seedlings upon exposure to salt and osmotic stresses. Two-week-old HDT701 OX seedlings were treated with 150 mM NaCl or 20% PEG, respectively, and then recovered in MS medium for the indicated period. After recovery for 5 or 3 days, the NaCl- or PEG-treated HDT701 OX seedlings appeared less dehydrated than WT seedlings (Figure 7A). We also measured the chlorophyll content (Ming et al., 2007) of HDT701 OX seedlings, which was significantly higher than that of wild-type seedlings (Figure 7B). Furthermore, after recovery for 15 days, the survival rate of HDT701 OX seedlings was also higher than WT seedlings (Figure 7C). Taken together, this data indicates that overexpression of HDT701 in rice may enhance salt and osmotic resistance during the seedling stage.
Figure 7.
Overexpression of HDT701 enhances salt and osmotic stress tolerance. (A) Phenotype of HDT701 OX seedlings after PEG and NaCl treatment. Two-week-old WT, HDA701 OX4, and OX7 plants were treated with 150 mM NaCl or 20% PEG for indicated days and then recovered in MS medium. (B) Chlorophyll content of HDT701 OX4 and OX7 seedlings after NaCl and PEG treatments. (C) Survival rates of HDT701 OX4 and OX7 seedlings after NaCl and PEG exposure. Fifty seedlings were used for the analysis of each treatment. Values are shown as means + SD (t-test: *P < 0.05, **P < 0.01, difference from the WT).
Discussion
Previous studies have provided evidence for the involvement of Arabidopsis HDACs in ABA-related and other abiotic stresses (Yuan et al., 2013) In Arabidopsis, overexpression of HD2C plants demonstrated hyposensitivity to NaCl, in the form of inhibited seed germination and root growth (Sridha and Wu, 2006), while the loss of function of HDA6 and HDA19 mutants displayed a phenotype that is hypersensitive to NaCl and ABA stresses during seed germination in Arabidopsis (Chen and Wu, 2010; Chen et al., 2010). Down-regulation of SRT1, a SIR2 family HDAC, in rice enhanced tolerance to oxidative stress (Huang et al., 2007). In the present work, we showed that overexpression of HDT701 in rice displayed increased sensitivity to ABA, NaCl, and PEG during germination, and enhanced tolerance to NaCl and PEG stresses of two-week-old rice seedlings. Taken together, these findings indicate that HDACs could be essential players in the response of rice plants to abiotic stress.
The N-terminal tails of the histone proteins can undergo a variety of posttranslational modifications, including acetylation, methylation, ubiquitination, phosphorylation, and sumoylation (Yuan et al., 2013). Recent work noted that the histone arginine demethylases, JMJ20 and JMJ22, act positively in regulating seed germination in Arabidopsis. JMJ20 and JMJ22 increased the expression of GA biosynthetic genes, GA3ox1, and GA3ox2, by removing the repressive histone arginine methylation of these genes through histone arginine demethylation activity (Cho et al., 2012). In the present study, overexpression of HDT701 in rice decreased the expression levels of GA biosynthetic genes, GA3ox1, GA3ox2, GA20ox2 and GA20ox3, and delayed seed germination, suggesting that HDT701 may negatively regulate seed germination by decreasing the levels of GA in rice seeds. In Arabidopsis thaliana, HDACs interact with various transcription factors to repress gene expression in multiple developmental processes (Liu et al., 2014). HDA15 specifically interacts with Phytochrome-Interacting Factor 3 (PIF3), a key transcription factor involved in photomorphogenesis, to repress chlorophyll biosynthetic gene expression in Arabidopsis (Liu et al., 2013). Furthermore, HDA6 associates with Asymmetric Leaves 1 (AS1) in repression of KNOTTED-LIKE HOMOBOX (KNOX) genes expression in control of leaf development in Arabidopsis (Luo et al., 2012b). HDT701 may also be associated with specific transcription factors to repress GA biosynthetic gene expression during rice seed germination. HDT701 was previously identified as a histone H4 deacetylase which directly bound to promoter regions of defense-related genes, WRKY6 and WRKY5 (Ding et al., 2012). The decrease in histone H4 acetylation levels of the GA biosynthetic genes in HDT701 OX4 and OX7 suggests that HDT701 may repress their expression through histone deacetylation. However, it is still unclear whether HDT701 is able to directly regulate the expression of these genes. Further genome-wide ChIP analysis may identify more direct target genes of HDT701, which may help to elucidate the role of HDT701 in plant development and in response to abiotic stresses.
Materials and methods
Plant materials
Rice (Oryza sativa L. var. Japonica) cv. Zhonghua 11 was used in this study. Plants were grown in a greenhouse in South China Botanical Garden, Guangzhou, China, at 25°C (16-h photoperiod, with a light extensity of 60–80 μmol m−2 s−1) from March to July. After harvest, on the 15th July, rice seeds were stored at −20°C (with 15% humidity) for 2 months before they were used for analysis.
Bioinformatics analysis
The cis-acting elements in HDT701 and HDT702 promoters were analyzed using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002) and 2000-bp DNA sequences before the start codon were used for prediction.
Plasmid constructs and rice transformation
To generate GUS-HDT701 vectors, a 1884-bp fragment behind the transcriptional start site of HDT701 was amplified and subcloned into the pCAMBIA 1381Z binary vector (Cambia. Canberra, Australia). To generate the HDT701 OX vector, the cDNA fragment of HDT701 (accession number: AK072845) was amplified and then subcloned into binary vector pCU1301, which was modified based on pCAMIBA1301 (Cambia) and contained a maize (Zea mays) ubiquitin promoter. The primers used for vector construction are listed in Supplemental Table 2.
For transformation, at first, constructs were introduced into Agrobacterium tumefaciens strain EHA105 by freezing. Embryogenic callus from mature seeds of rice (“Zhonghua 11”) were used for transformation. The procedures for transformation, regeneration and transfer to soil were as described previously for rice (Hiei et al., 1994).
Detection of GUS expression
Histochemical GUS assays were carried out according to the procedure of Jefferson et al. (1987). The transgenic plants were grown in a greenhouse at 25°C. Various organs including leaves, roots, seeds, germinating seeds, spikes, and flowers of transgenic rice were collected to examine GUS expression. These organs were immerged in GUS staining buffer (1 mM 5-bromo-4-chloro-3-indolyl-β-D-glucuronide citation(X-gluc), 100 mM sodium phosphate buffer (pH 7.0), 2 mM potassium ferricyanide, 2 mM potassium ferrocyanide, and 0.1% Triton X-100) at 37°C overnight. The stained materials were washed in ethyl alcohol.
Salt and osmotic stress treatment of rice seedlings
For salt and osmotic stress treatment, two-week-old seedlings growing in MS (Murashige and Skoog, 2006) medium were transferred to MS medium supplemented with 150 mM NaCl for 3 days and then recovered in MS medium for 5 days, or transferred to MS medium to which 20% PEG was added for 7 days and then recovered in MS medium for 3 days.
Measurement of seed germination rate
For the germination assay, seeds were pre-soaked in a thermostatic culture box (Henan Brother Device and Equipment Co., Ltd., Henan, China) at 37°C for 3 days to break any possible dormancy. Seeds were surface-sterilized by incubating in 75% (v/v) ethanol for 1 min, treated with 0.1% (v/v) HgCl2 for 15 min, then washed with sterile distilled water three times. Surface-sterilized seeds were sown on MS plates supplemented with 5 and 10 μ M ABA, 100 and 150 mM NaCl, and 15% PEG, respectively. For normal conditions, the seeds were sown on MS plates and used as the control. Plates were placed in a greenhouse at 25°C under long-day conditions (16-h photoperiod). Each treatment was repeated at least three times.
Quantitative RT-PCR analysis
For gene expression analysis, after pre-soaking in sterile water for 3 d, the seeds were incubated on MS plates for 12 h then harvested for RNA extraction. Total RNA was extracted with TRIZOL reagent as described by the manufacturer (Invitrogen). After DNase treatment, the first strand of cDNA was synthesized by M-MLV reverse transcriptase (Promega) using 2 μg of total RNA as the template. Real-time PCR was performed on an optical 96-well plate with an ABI7500 real-time PCR system (Applied Biosystems). Each sample was quantified at least in triplicate. The rice Actin gene (accession no: NM_001057621) was used as the internal control. The gene-specific primers used for qRT-PCR are listed in Supplemental Table 2.
Chromatin immunoprecipitation assays
The chromatin immunoprecipitation assays were performed as described by Gendrel et al. (2005) with minor changes. Briefly, seeds were fixed with 1% formaldehyde in a vacuum for 1 h. Chromatin was extracted and sheared to an average length of 500 bp fragments by 6 repeats of 10 s sonication at 20% duty cycle and 1.5 power output using a Branson Sonifier 250, and then immunoprecipitated with an anti-acetyl-histone H4 antibody (Millipore; 06-866). Cross-linking was then reversed, and the amount of each precipitated DNA fragment was determined by quantitative PCR. UBQ (accession no: AC103891) was used as the internal control. Primers used for ChIP assays are listed in Supplemental Table 2.
Conflict of interest statement
The Associate Editor, Yuhai Cui, and the Review Editors, Steve Robinson and Ming-Jun Gao, declare that, despite being affiliated to the same institution as the author Lining Tian, the review process was handled objectively and no conflict of interest exists. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No. 31301056) and the Science and Technology Foundation of Guangdong Province (2011A020102008). The authors thank Dr. Jaime A. Teixeira da Silva for assistance with linguistic and scientific revisions.
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2014.00764/abstract
Seed germination rates of HDT701 RNAi2 and RNAi6 lines under normal conditions (A) 5 μM ABA (B) 100 mM NaCl (C), and 10% PEG (D).
References
- Berger S. L. (2007). The complex language of chromatin regulation during transcription. Nature 447, 407–412. 10.1038/nature05915 [DOI] [PubMed] [Google Scholar]
- Brosch G., Lusser A., Goralik-Schramel M., Loidl P. (1996). Purification and characterization of a high molecular weight histone deacetylase complex (HD2) of maize embryos. Biochemistry 35, 15907–15914. 10.1021/bi961294x [DOI] [PubMed] [Google Scholar]
- Chen L. T., Luo M., Wang Y. Y., Wu K. (2010). Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J. Exp. Bot. 61, 3345–3353. 10.1093/jxb/erq154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. T., Wu K. (2010). Role of histone deacetylases HDA6 and HDA19 in ABA and abiotic stress response. Plant Signal. Behav. 5, 1318–1320. 10.4161/psb.5.10.13168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J. N., Ryu J. Y., Jeong Y. M., Park J., Song J. J., Amasino R. M., et al. (2012). Control of seed germination by light-induced histone arginine demethylation activity. Dev. Cell 22, 736–748. 10.1016/j.devcel.2012.01.024 [DOI] [PubMed] [Google Scholar]
- Dangl M., Brosch G., Haas H., Loidl P., Lusser A. (2001). Comparative analysis of HD2 type histone deacetylases in higher plants. Planta 213, 280–285. 10.1007/s004250000506 [DOI] [PubMed] [Google Scholar]
- Ding B., Bellizzi Mdel R., Ning Y., Meyers B. C., Wang G. L. (2012). HDT701, a histone H4 deacetylase, negatively regulates plant innate immunity by modulating histone H4 acetylation of defense-related genes in rice. Plant Cell 24, 3783–3794. 10.1105/tpc.112.101972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W., Wu K., Duan J. (2007). Sequence and expression analysis of histone deacetylases in rice. Biochem. Biophys. Res. Commun. 356, 843–850. 10.1016/j.bbrc.2007.03.010 [DOI] [PubMed] [Google Scholar]
- Gendrel A. V., Lippman Z., Martienssen R., Colot V. (2005). Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods 2, 213–218. 10.1038/nmeth0305-213 [DOI] [PubMed] [Google Scholar]
- Goldberg A. D., Allis C. D., Bernstein E. (2007). Epigenetics: a landscape takes shape. Cell 128, 635–638. 10.1016/j.cell.2007.02.006 [DOI] [PubMed] [Google Scholar]
- Hiei Y., Ohta S., Komari T., Kumashiro T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282. 10.1046/j.1365-313X.1994.6020271.x [DOI] [PubMed] [Google Scholar]
- Hu Y., Qin F., Huang L., Sun Q., Li C., Zhao Y., et al. (2009). Rice histone deacetylase genes display specific expression patterns and developmental functions. Biochem. Biophys. Res. Commun. 388, 266–271. 10.1016/j.bbrc.2009.07.162 [DOI] [PubMed] [Google Scholar]
- Huang L., Sun Q., Qin F., Li C., Zhao Y., Zhou D. X. (2007). Down-regulation of a SILENT INFORMATION REGULATOR2-related histone deacetylase gene, OsSRT1, induces DNA fragmentation and cell death in rice. Plant Physiol. 144, 1508–1519. 10.1104/pp.107.099473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R. A., Kavanagh T. A., Bevan M. W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence R. J., Earley K., Pontes O., Silva M., Chen Z. J., Neves N., et al. (2004). A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol. Cell 13, 599–609. 10.1016/S1097-2765(04)00064-4 [DOI] [PubMed] [Google Scholar]
- Lescot M., Dehais P., Thijs G., Marchal K., Moreau Y., Van de Peer Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. 10.1093/nar/30.1.325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Chen C. Y., Wang K. C., Luo M., Tai R., Yuan L., et al. (2013). PHYTOCHROME INTERACTING FACTOR3 associates with the histone deacetylase HDA15 in repression of chlorophyll biosynthesis and photosynthesis in etiolated Arabidopsis seedlings. Plant Cell 25, 1258–1273. 10.1105/tpc.113.109710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Yang S., Zhao M., Luo M., Yu C. W., Chen C. Y., et al. (2014). Transcriptional repression by histone deacetylases in plants. Mol. Plant 7, 764–772. 10.1093/mp/ssu033 [DOI] [PubMed] [Google Scholar]
- Luo M., Wang Y. Y., Liu X., Yang S., Lu Q., Cui Y., et al. (2012a). HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J. Exp. Bot. 63, 3297–3306. 10.1093/jxb/ers059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Yu C. W., Chen F. F., Zhao L., Tian G., Liu X., et al. (2012b). Histone deacetylase HDA6 is functionally associated with AS1 in repression of KNOX genes in arabidopsis. PLoS Genet. 8:e1003114. 10.1371/journal.pgen.1003114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusser A., Brosch G., Loidl A., Haas H., Loidl P. (1997). Identification of maize histone deacetylase HD2 as an acidic nucleolar phosphoprotein. Science 277, 88–91. 10.1126/science.277.5322.88 [DOI] [PubMed] [Google Scholar]
- Ming H., Hu C. S., Zhang Y. M., Cheng Y. S. (2007). Improved extraction methods of chlorophyll from maize. J. Maize Sci. 15, 93–95 10.3969/j.issn.1005-0906.2007.04.025 [DOI] [Google Scholar]
- Murashige T., Skoog F. (2006). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–497 10.1111/j.1399-3054.1962.tb08052.x [DOI] [Google Scholar]
- Ogawa M., Hanada A., Yamauchi Y., Kuwahara A., Kamiya Y., Yamaguchi S. (2003). Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15, 1591–1604. 10.1105/tpc.011650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R., Muller A., Napoli C. A., Selinger D. A., Pikaard C. S., Richards E. J., et al. (2002). Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 30, 5036–5055. 10.1093/nar/gkf660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridha S., Wu K. (2006). Identification of AtHD2C as a novel regulator of abscisic acid responses in Arabidopsis. Plant J. 46, 124–133. 10.1111/j.1365-313X.2006.02678.x [DOI] [PubMed] [Google Scholar]
- Ueno Y., Ishikawa T., Watanabe K., Terakura S., Iwakawa H., Okada K., et al. (2007). Histone deacetylases and ASYMMETRIC LEAVES2 are involved in the establishment of polarity in leaves of Arabidopsis. Plant Cell 19, 445–457. 10.1105/tpc.106.042325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Tian L., Malik K., Brown D., Miki B. (2000). Functional analysis of HD2 histone deacetylase homologues in Arabidopsis thaliana. Plant J. 22, 19–27. 10.1046/j.1365-313x.2000.00711.x [DOI] [PubMed] [Google Scholar]
- Wu K., Tian L., Zhou C., Brown D., Miki B. (2003). Repression of gene expression by Arabidopsis HD2 histone deacetylases. Plant J. 34, 241–247. 10.1046/j.1365-313X.2003.01714.x [DOI] [PubMed] [Google Scholar]
- Yuan L., Liu X., Luo M., Yang S., Wu K. (2013). Involvement of histone modifications in plant abiotic stress responses. J. Integr. Plant Biol. 55, 892–901. 10.1111/jipb.12060 [DOI] [PubMed] [Google Scholar]
- Zhou C., Labbe H., Sridha S., Wang L., Tian L., Latoszek-Green M., et al. (2004). Expression and function of HD2-type histone deacetylases in Arabidopsis development. Plant J. 38, 715–724 10.1111/j.1365-313X.2004.02083.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Seed germination rates of HDT701 RNAi2 and RNAi6 lines under normal conditions (A) 5 μM ABA (B) 100 mM NaCl (C), and 10% PEG (D).