Abstract
ABA is a major plant hormone that plays important roles during many phases of plant life cycle, including seed development, maturity and dormancy, and especially the acquisition of desiccation tolerance. Understanding of the molecular basis of ABA-mediated plant response to stress is of interest not only in basic research on plant adaptation but also in applied research on plant productivity. Maize mutant viviparous-5 (vp5), deficient in ABA biosynthesis in seeds, is a useful material for studying ABA-mediated response in maize. Due to carotenoid deficiency, vp5 endosperm is white, compared to yellow Vp5 endosperm. However, the background difference at proteome level between vp5 and Vp5 seeds is unclear. This study aimed to characterize proteome alterations of maize vp5 seeds and to identify ABA-dependent proteins during seed maturation. We compared the embryo and endosperm proteomes of vp5 and Vp5 seeds by gel-based proteomics. Up to 46 protein spots, most in embryos, were found to be differentially accumulated between vp5 and Vp5. The identified proteins included small heat shock proteins (sHSPs), late embryogenesis abundant (LEA) proteins, stress proteins, storage proteins and enzymes among others. However, EMB564, the most abundant LEA protein in maize embryo, accumulated in comparable levels between vp5 and Vp5 embryos, which contrasted to previously characterized, greatly lowered expression of emb564 mRNA in vp5 embryos. Moreover, LEA proteins and sHSPs displayed differential accumulations in vp5 embryos: six out of eight identified LEA proteins decreased while nine sHSPs increased in abundance. Finally, we discussed the possible causes of global proteome alterations, especially the observed differential accumulation of identified LEA proteins and sHSPs in vp5 embryos. The data derived from this study provides new insight into ABA-dependent proteins and ABA-mediated response during maize seed maturation.
Keywords: abscisic acid (ABA), late embryogenesis abundant proteins (LEA proteins), maize ABA-deficient mutant vp5, mass spectrometry, protein abundance, proteome profiling, small heat shock proteins (sHSPs), 2-D electrophoresis (2-DE)
Introduction
Abscisic acid (ABA) is a major hormone that regulates a broad range of plant traits and is especially important for plant adaptation to environmental conditions. In seeds, ABA is thought to play a central role in many developmental stages, such as seed maturation and dormancy, the accumulation of nutritive reserves and the acquisition of desiccation tolerance (Quatrano, 1986). ABA-mediated plant response to stress has been extensively studied in different species ranging from Arabidopsis to crops, especially regarding ABA sensing, signaling, metabolism and transport (Umezawa et al., 2010). Knowledge about the complexity of ABA-mediated plant response to stress is still full of gaps, but the recent identification of ABA receptors (Ma et al., 2009; Santiago et al., 2009) and the key factors of the first step of ABA signal transduction (Park et al., 2009; Nishimura et al., 2010) in Arabidopsis provided an important insight into this mechanism.
Biosynthesis of ABA has been well characterized in Arabidopsis (Zeevaart and Creelman, 1988) and some data is available for other species, such as maize (Tan et al., 1997). Maize, viviparous-5 (vp5) is deficient in ABA biosynthesis with the first step catalyzed by phytoene desaturase being blocked, which results in the precursor phytoene accumulation and carotenoid deficiency (Robichaud et al., 1980; Hable et al., 1998). Previous studies reported that ABA content in vp5 embryos and endosperms was substantially reduced to 10 and 42% of the corresponding wild-type, respectively (Neill et al., 1986). The vp5 seeds exhibit a visible phenotypic difference: the endosperm of mutant vp5 seeds was white, while that of wild-type Vp5 seeds was yellow. Therefore, vp5 mutant is particularly useful not only for studies on the regulation of ABA-dependent maize genes, both in embryo and vegetative tissues, but also for studies of embryo development, seed germination and dormancy (Pla et al., 1989; Durantini et al., 2008).
Up to date, the expression of many individual gene/proteins has been studied using ABA-deficient mutant maize vp5 and wild-type Vp5 (Pla et al., 1989; Thomann et al., 1992). Williams and Tsang (1991) found emb564 mRNA is expressed at low level in vp5 embryos. The level of 3-hydroxy-3-methylglutaryl coenzyme A reductase activity, a rate-limiting enzyme of isoprenoid biosynthesis, is higher in vp5 endosperm (Moore and Oishi, 1994). Recently, we compared root and leaf proteome differences between vp5 and Vp5 seedlings with 2-D gel electrophoresis (2-DE) combined mass spectrometry (MS/MS) and found that many proteins accumulation in roots or leaves are differentially regulated by drought and light in an ABA-dependent way (Hu et al., 2011, 2012). However, protein accumulation alterations caused by ABA-deficient mutation in vp5 seeds are unclear at a proteome scale. Therefore, the characterization of seed proteome difference between vp5 and Vp5 is necessary for dissection of ABA-mediated maize response in the studies involved vp5 mutants.
2-DE-based proteomics approach provides a powerful tool to analyze the expression levels of proteins, distinguish varieties and genotypes and even to identify single mutations with multiple effects (Lehesranta et al., 2005). This study aimed to characterize proteome alterations due to ABA-deficient mutation and further to identify ABA-dependent protein accumulation during seed maturation. We found significant proteome differences between vp5 and Vp5 seeds, where 46 differentially accumulated proteins were successfully identified. Most notably, six out of eight late embryogenesis abundant (LEA) proteins and nine small heat shock proteins (sHSPs) were found to differentially accumulate in ABA-deficient vp5 embryos: six identified LEA proteins were repressed while nine sHSPs were induced.
Materials and methods
Plant materials
Maize (Zea mays L.) ABA-deficient mutant vp5 mutant was provided by Maize Genetics Cooperation Stock Center (Urbana, IL). The mutant vp5 was propagated in primarily W64 genetic backgrounds. The vp5 mutant was maintained as a heterozygote. Heterozygous seeds (Vp5/vp5) were planted under natural conditions at the farm of Henan Agricultural University (Zhengzhou, China). The homozygous vp5 kernels were identified on segregating ears by their lack of carotenoid pigments (Robichaud et al., 1980). The mutant vp5 kernels appear white, while Vp5 kernels are yellow. Mature vp5 and Vp5 seeds from the same ear were sampled (Presentation 1 in Supplementary Material) used in this study. Dry maize seeds were soaked in water for 2 h to soften starchy endosperm. For each biological replicate, the embryos and the endosperm of 20 maize seeds were manually separated and used for protein extraction, respectively.
Protein isolation
Embryos or endosperms were powdered in liquid N2 and further ground in a buffer containing 0.25 M Tris-HCl (pH 7.5), 1% SDS, 14 mM DTT and a cocktail of protease inhibitors. This slurry was heated to 65°C for 5 min, vortexed, and heated at 95°C for 2 min, vortexed again, and then centrifuged at 12,000 g for 10 min to remove cellular debris. The supernatant was recovered and subjected to phenol extraction as described (Wu et al., 2014). The protein pellet was dissolved in 2-DE rehydration buffer containing 7 M urea, 2 M thiourea, 2% (w/v) CHAPS, 20 mM DTT, 0.5% (v/v) IPG buffer (pH 4–7 or 7–10, GE Healthcare). The protein content was determined by Bradford microassay (Bio-Rad) with BSA standards.
SDS-PAGE and immunoblot
SDS-PAGE was performed in a Laemmli gel system (5% stacking gel and 12.5% resolving gel). After electrophoresis, proteins in gels were visualized with colloidal CBB R350 or electroblotted onto polyvinylidene difluoride membrane (Hybond-P, GE healthcare) in a transfer buffer (20% v/v methanol, 50 mM Tris, 40 mM glycine). For immunoblot analysis, protein blots were soaked in TBST buffer (50 mM Tris-HCl, pH 7.5, 0.15 M NaCl, 0.1% Tween-20) containing 5% low fat milk powder and gently shaken for 2 h at room temperature (RT). The blot was then incubated with anti-EMB564 polyclonal antibody (Wu et al., 2013a, 1: 5000 dilution) for 1 h. After washing with TBST, the blot was incubated in peroxidase-conjugated goat anti-rabbit IgG (1: 2000 dilution) at RT for 1 h. The blot was visualized with 0.08% 3,3′-diaminobenzidine tetrahydrochloride, 0.05% H2O2, 0.1 M Tris-HCl, pH 7.5.
2-DE, image and data analysis
Isoelectric focusing (IEF) was performed using 11-cm linear pH 4–7 IPG strips with the Ettan III system (GE Healthcare, USA). About 600 μg proteins were loaded into the strip by passive rehydration overnight at RT. The IEF voltage was set at 250 V for 1 h, 1000 V for 4 h, finally increasing to 8000 V for 4 h, and holding for 10 h (20°C). Focused strips were equilibrated in Buffer I (0.1 M Tris-HCl, pH 8.8, 2% SDS, 6 M urea, 30% glycerol, 0.1 M DTT) and then in Buffer II (same as Buffer I, but with 0.25 M iodoacetamide instead of DTT) for 15 min each. SDS-PAGE was run on a 13.5% gel with 0.1% SDS in the gel and the running buffer. The gels were stained with 0.1% CBB G-250 overnight and destained in 7% acetic acid until a clear background.
Protein gels were placed on a white plastic plate with transmission fluorescent lighting, and photographed using a DSLR camera (Nikon D7000) at an automatic mode. The digital images of the gels were analyzed by using PDQUEST 8.0 software (Bio-Rad). Protein spots were detected on scanned gels using the default spot detection setting. Gels of three biological replicates per genotype were analyzed. The spot intensities were normalized according to total intensity of valid spots to minimize possible errors due to differences in the amount of protein and staining intensity. Analysis of variance (ANOVA) was used to deal with protein spots quantification to identify individual protein spots with significantly different expression levels. Only those proteins with at least 1.5-fold quantitative variations in abundance were selected for mass spectrometry (MS) analysis.
MS/MS
Protein spots of interest were manually excised from the gels and digested using trypsin. Proteins were reduced (10 mM DTT), alkylated (50 mM iodoacetic acid) and then digested with 10 mg/ml trypsin for 16 h at 37°C in 50 mM ammonium bicarbonate. The supernatants were vacuum-dried and dissolved in 10 μl 0.1% trifluoroacetic acid and 0.5 μl added onto a matrix consisting of 0.5 μl of 5 mg/ml 2, 5-dihydroxybenzoic acid in water: acetonitrile (2:1). The digested fragments were analyzed using a MALDI-TOF/TOF analyzer (ultraflex III, Bruker, Germany). MALDI-TOF/TOF spectra were acquired in the positive ion mode and automatically submitted to Mascot 2.2 (http://www.matrixscience.com, Matrix Science) for peptide mass finger printings against the NCBInr 20131226 database (35149712 sequences; 12374887350 residues, http://www.ncbi.nlm.nih.gov/). The taxonomy was Viridiplantae (green plants, 1669695 sequences). The search parameters were as follows: type of search: MALDI-TOF ion search; enzyme: trypsin; fixed modifications: carbamidomethyl (C); variable modifications: acetyl (protein N-terminal) and oxidation (M); mass values: monoisotopic; protein mass: unrestricted; peptide mass tolerance: ±50 ppm; fragment mass tolerance: ±0.2 Da; max missed cleavages: 1; instrument type: MALDI-TOF. Only significant scores defined by Mascot probability analysis greater than “identity” were considered for assigning protein identity. All of the positive protein identification scores were significant (P < 0.05, score > 49).
Bioinformatics
To identify the sequences of all putative uncharacterized proteins, BLAST searches (http://www.expasy.org/tools/blast/) with these protein sequences were performed on the UniProt Knowledgebase (UniProtKB, http://www.uniprot.org/uniprot) to find their homologs. Functional categorization of the identified proteins was based on annotations in UniProtKB and previous studies on their homologs. Subcellular locations of the identified proteins were determined according to the annotation in UniProtKB or predicted at Plant-mPLoc server (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/). Theoretical Mr and isoelectric point of proteins were predicted at http://web.expasy.org/compute_pi/. ABA responsive element (ABRE) and dehydration responsive element (DRE) were analyzed using Plantcare (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) and PLACE (http://www.dna.affrc.go.jp/PLACE/signalscan.html).
Results
Maize seed consists of an embryo (a miniature plant), an endosperm (a nutrition provider for seed germination) and a seed coat (protecting structure). To reveal maize seeds proteome alterations due to ABA-deficient mutation and to identify ABA-dependent proteins during seed maturation, embryos and endosperms of the mutant vp5 and wild-type Vp5 were used for comparative proteomic analysis. In order to improve protein resolution, two kinds of IPG strips with a pH range of 4–7 and 7–10 were used in 2-DE.
Proteomic difference between vp5 and Vp5 embryos
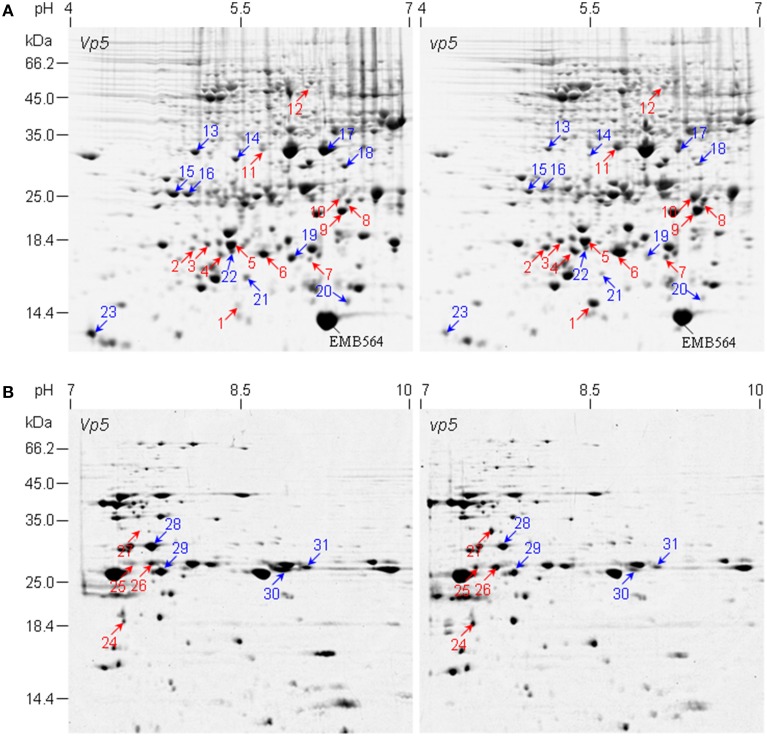
The embryo protein profiles between vp5 and Vp5 were compared by 2-DE. Approximately 780±10 and 130±5 CBB-stained protein spots were reproducibly detected using pH 4–7 and 7–10 IPG gels, respectively (Figure 1). PDQUEST analysis indicated that 96% of total protein spots were matched, unchanged in abundance between vp5 and Vp5 embryos across all the gels (Figure 1, Presentations 2, 3 in Supplementary Material). Spot-to-spot comparison revealed that 31 protein spots, i.e., 23 in pH 4–7 gels (Figure 1A and Presentation 2 in Supplementary Material) and 8 in pH 7–10 gel (Figure 1B and Presentation 3 in Supplementary Material), showed a minimum of a 1.5-fold difference in spot volume (Tables 1, 2). Those differentially accumulated embryo protein spots are mainly in the range of 10–35 kDa, with a consistent change in three biological replicates, except for three spots in two biological replicates (spot 5, 17, and 22) (Presentation 2 in Supplementary Material). Sixteen embryo protein spots (spots 1–12 and 24–27) accumulated in higher abundance in vp5 compared to Vp5 and 15 embryo protein spots (spots 13–23 and 28–31) accumulated in lower abundance in vp5. In particular, the abundance of spots 1, 10, and 27 were 13.71, 7.31, and 9.28 folds higher in vp5 embryos than in Vp5 embryos, respectively, whereas the abundance of spots 19 and 23 was 6.76 and 6.52 folds higher in Vp5 than in vp5 embryos, respectively.
Figure 1.
2-DE comparison of embryo protein profiles between maize mutant vp5 and wild type Vp5. (A,B) are representative 2-DE gels of pH 4–7 and pH 7–10, respectively. Differentially accumulated protein spots between vp5 and Vp5 embryos are indicated by arrows. Red arrow, increased abundance in vp5; blue arrow, increased abundance in Vp5. Embryo protein was extracted and resolved in 2-DE with IEF in the first dimension and 13.5% SDS-PAGE gel in the second dimension. Gels were stained with colloidal CBB G250. The identified proteins were listed in Tables 1, 2.
Table 1.
The differentially accumulated proteins identified by MS/MS between maize vp5 and Vp5 embryos.
| Spot | Protein name | NCBI accession | pI Theor/exp. | MW (kDa) Theor/exp. | Mascot score | Coverage (%) | Matched peptide sequences |
|---|---|---|---|---|---|---|---|
| 1 | Late embryogenesis abundant protein EMB564 | gi|162463828 | 6.60/5.47 | 9.68/14.9 | 172 | 15 | R.EQLGQQGYSEMGK.K |
| R.EQLGQQGYSEMGK.K R.REQLGQQGYSEMGK.K | |||||||
| R.REQLGQQGYSEMGK.K | |||||||
| 2 | 17.8 kDa class II heat shock protein | gi|162459174 | 5.33/5.09 | 17.85/17.9 | 124 | 23 | K.FVLPDNADVDK.V K.FVLPDNADVDKVAAVCR.D K.ELPGAYAFVVDMPGLGTGDIR.V |
| 3 | 16.9 kDa class I heat shock protein 1 | gi|226508268 | 5.82/5.21 | 17.28/18.2 | 248 | 22 | R.VDWKETPEAHVFK.A |
| R.SIVPSAASSGGGSETAAFANAR.V | |||||||
| 4 | 17.4 kDa class I heat shock protein 3 | gi|226530365 | 5.80/5.32 | 17.93/17.6 | 408 | 24 | R.FRLPENAR.T |
| R.VDWKETPEAHVFK.T K.VELEDGNVLQISGER.S | |||||||
| K.EEVKVELEDGNVLQISGER.S | |||||||
| 5 | Heat shock protein 17.9 | gi|1122317 | 5.55/5.41 | 17.78/18.1 | 506 | 32 | R.FRLPENAK.T |
| R.TSSETAAFAGAR.I | |||||||
| R.IDWKETPEAHVFK.A R.IDWKETPEAHVFK.A | |||||||
| K.VEVEDGNVLQISGER.N | |||||||
| K.EEVKVEVEDGNVLQISGER.N | |||||||
| 6 | Heat shock protein 17.2 | gi|162459222 | 5.54/5.68 | 17.15/17.6 | 474 | 38 | K.VEVEDGNVLVISGQR.S |
| R.SIVPSATSTNSETAAFASAR.I | |||||||
| K.EEVKVEVEDGNVLVISGQR.S | |||||||
| R.SNVFDPFSMDLWDPFDTMFR.S | |||||||
| R.SNVFDPFSMDLWDPFDTMFR.S | |||||||
| 7 | 17.0 kDa class II heat shock protein | gi|162458291 | 7.85/6.09 | 17.09/17.3 | 136 | 24 | K.FVLPDNADVDKVAAVCR.D |
| K.ELAGAYAFVVDMPGLSTGDIR.V | |||||||
| K.ELAGAYAFVVDMPGLSTGDIR.V | |||||||
| 8 | 22.0 kDa class IV heat shock protein | gi|226501206 | 6.72/6.41 | 25.22/24.9 | 792 | 33 | R.EDLKIEVEDYSR.V R.ETPDAHEIVVDVPGMR.R |
| R.ETPDAHEIVVDVPGMR.R | |||||||
| R.LPENADLDSVGASLDNGVLTVR.F | |||||||
| R.GGDEAAAAAASPLSGPGVGLLADPFR.I | |||||||
| R.RGGDEAAAAAASPLSGPGVGLLADPFR.I | |||||||
| 9 | 22.0 kDa class IV heat shock protein | gi|226509936 | 6.01/6.35 | 22.93/23.0 | 1063 | 43 | K.LAPEQIKGPR.V R.ETPDAHEIVVDVPGMR.R |
| R.ETPDAHEIVVDVPGMR.R | |||||||
| R.GLDEAAVSDVGLLAADPFR.I | |||||||
| RGLDEAAVSDVGLLAADPFR.I | |||||||
| R.LPENADLDSVAASLDSGVLTVR.F | |||||||
| R.ILEHVPFGFDRDDVAMVSMAR.V | |||||||
| R.FRLPENADLPSVAASLDSGVLTVR.F | |||||||
| R.FRLPENADLDSVAASLDSGVLTVR.F | |||||||
| 10 | 1-Cys peroxiredoxin PER1 | gi|162460575 | 6.31/6.31 | 25.06/25.0 | 350 | 21 | K.VTFPILADPAR.D |
| K.LSFLYPATTGR.N | |||||||
| K.LLGISCDDVESHR.Q R.QLNMVDPDEKDAAGR.S | |||||||
| R.QLNMVDPDEKDAAGR.S | |||||||
| 11 | Short-chain dehydrogenase/reductase SDR family protein | gi|226500748 | 5.78/5.67 | 33.15/34.9 | 525 | 27 | R.DIGSETGAR.E |
| R.ALSLQLADR.G | |||||||
| K.EGATVAFTFVR.G | |||||||
| R.GQEEKDAEETLR.A | |||||||
| K.VEQFGSQVPMKR.A R.IDVVVNNAAEQYER.E | |||||||
| R.EPMALPADLGYEANCR.E | |||||||
| 12 | UDP-glucose 6-dehydrogenase | gi|219885505 | 5.71/6.06 | 53.53/51.2 | 476 | 16 | K.LAANAFLAQR.I |
| K.AADLTYWESAAR.M | |||||||
| K.DVYAHWVPEDR.I | |||||||
| K.AQISIYDPQVTEDQIQR.D | |||||||
| K.GINYQILSNPEFLAEGTAIEDLFKPDR.V | |||||||
| 13 | Late embryogenesis abundant protein D-34 | gi|226530343 | 5.41/5.11 | 27.28/34.8 | 303 | 19 | R.LNQERPRP.- |
| R.RVVTESVGGQVVGK.M R.LQAAEQSVLGGTQK.G | |||||||
| K.GGPAAVLQSAATVNAR.A | |||||||
| 14 | Late embryogenesis abundant protein D-34 | gi|226530343 | 5.41/5.45 | 27.28/34.2 | 72 | 7 | R.LNQERPRP.- |
| R.VVTESVGGQVVGK.M | |||||||
| 15 | Late embryogenesis abundant protein D-34 | gi|226493450 | 5.26/4.93 | 21.17/26.1 | 560 | 21 | K.GAGGGVAEAVVAAADMNEGR.M |
| K.IIEQGDQLGGALHVDQTDLPAGR.R | |||||||
| R.DKIIEQGDQLGGALHVDQTDLPAGR.R | |||||||
| 16 | Late embryogenesis abundant protein D-34 | gi|226501566 | 5.23/5.05 | 21.29/26.2 | 88 | 16 | R.GGALHVDQTDLPAGR.R |
| K.GAGGGVAEAVVAAADMNEGR.M | |||||||
| 17 | Short-chain dehydrogenase/reductase SDR family protein | gi|226500748 | 5.78/6.21 | 33.15/34.9 | 491 | 24 | R.ALSLQLADR.G |
| K.EGATVAFTFVR.G | |||||||
| R.GQEEKDAEETLR.A | |||||||
| K.VEQFGSQVPMKR.A R.IDVVVNNAAEQYER.E | |||||||
| R.EPMALPADLGYEANCR.E | |||||||
| R.EPMALPADLGYEANCR.E | |||||||
| 18 | Short-chain dehydrogenase/reductase SDR family protein | gi|226500748 | 5.78/6.38 | 33.15/32.5 | 492 | 27 | R.DIGSETGAR.E |
| R.ALSLQLADR.G | |||||||
| K.EGATVAFTFVR.G | |||||||
| R.GQEEKDAEETLR.A | |||||||
| K.VEQFGSQVPMKR.A R.IDVVVNNAAEQYER.E | |||||||
| R.EPMALPADLGYEANCR.E | |||||||
| R.EPMALPADLGYEANCR.E | |||||||
| 19 | Hypothetical protein ZEAMMB73_326753 | gi|413941572 | 5.93/5.95 | 11.50/17.4 | 511 | 54 | R.AGPEEQTGR.G |
| R.QGMAEPTAGGR.R | |||||||
| R.TGFFDGTPLEGGK.I K.NAVIAESEPVDLPASAR.G | |||||||
| R.VADAPYSSQQEGPHEEGTNK.N | |||||||
| 20 | Thioredoxin | gi|194694706 | 6.19/6.04 | 13.25/15.2 | 555 | 67 | K.FTQVVFLK.V |
| R.KDELLAQIEK.H | |||||||
| R.AIAPLFVEHAK.K | |||||||
| M.ASEQGVVIACHSK.A K.LVVIDFTAAWCGPCR.A | |||||||
| K.VDVDEVKEVTAAYEVEAMPTFHFVK.N | |||||||
| K.VDVDEVKEVTAAYEVEAMPTFHFVK.N | |||||||
| 21 | Glyoxalase family protein superfamily | gi|226532762 | 5.47/5.53 | 15.13/16.4 | 463 | 51 | K.AASFYDAAFGYTVR.R |
| R.ETDELSGAVQLPDSSAAGR.G | |||||||
| R.GSVEVCFAYADVDAAYKR.A | |||||||
| R.KWAELESGATTIAFTPLHQR.E | |||||||
| 22 | Late embryogenesis abundant protein Lea14-A | gi|226491145 | 5.64/5.41 | 16.14/17.7 | 458 | 38 | R.TVASGTVPDPGSLAGDGATTR.L |
| R.NPYSHAIPVCEVTYTLR.S | |||||||
| K.LANIQKPEAELADVTVGHVGR.D | |||||||
| 23 | Uncharacterized protein | gi|223945515 | 5.73/4.23 | 22.86/13.7 | 27 | R.YRGDPGPR.C | |
| K.TYPLTCSCFDR.V | |||||||
| K.TYPLTCSCFDRVER.C R.CSDACKECVETEDSR.H | |||||||
| R.DEERPWGECCDLAVCVK.T | |||||||
| 24 | 17.4 kDa class I heat shock protein 3 | gi|212276212 | 6.86/7.48 | 17.87/19.1 | 603 | 49 | R.FRLPDNAK.A |
| R.TSSETAAFAGAR.I | |||||||
| R.IDWKETPEAHVFK.A K.VEVEDGNVLQISGER.N | |||||||
| K.EEVKVEVEDGNVLQISGER.N | |||||||
| R.GNAFDPFSLDLWDPFEGFFPFGSGGVR.S | |||||||
| 25 | Uncharacterized protein | gi|194693636 | 6.90/7.55 | 27.88/27.9 | 492 | 26 | R.YGLSTEELR.A |
| K.FWCTWQVDR.G | |||||||
| K.IFDSLPAEEQR.L | |||||||
| R.KIFDSLPAEEQR.L | |||||||
| R.ADVEAPAEEHPGQADYWLR.H | |||||||
| R.LNQDFLQCAVYDSDKADAR.L | |||||||
| 26 | Uncharacterized protein | gi|194693636 | 6.90/7.73 | 27.88/27.9 | 790 | 47 | R.YGLSTEELR.A |
| R.LIGVEYIVSR.K | |||||||
| R.QVETHHYVSR.L | |||||||
| K.FWCTWQVDR.G | |||||||
| K.IFDSLPAEEQR.L | |||||||
| R.KIFDSLPAEEQR.L K.SGLWTSPHVAGLLEK.A | |||||||
| K.VLDMGAAAMQSLRPVK.Q | |||||||
| R.ADVEAPAEEHPGQADYWLR.H | |||||||
| R.LNQDFLQCAVYDSDKADAR.L | |||||||
| 27 | Uncharacterized protein | gi|413955864 | 6.60/7.63 | 38.12/33.3 | 57 | 12 | R.GAAGGGGILESVQEGAR.S |
| R.ETASTHDTDREQGQGLLGALGNVTGAIK.E | |||||||
| 28 | Uncharacterized protein | gi|194693636 | 6.90/7.73 | 27.88/30.4 | 923 | 47 | R.YGLSTEELR.A |
| R.LIGVEYIVSR.K | |||||||
| R.QVETHHYVSR.L | |||||||
| K.FWCTWQVDR.G | |||||||
| K.IFDSLPAEEQR.L | |||||||
| R.KIFDSLPAEEQR.L K.SGLWTSPHVAGLLEK.A | |||||||
| K.VLDMGAAAMQSLRPVK.Q | |||||||
| R.ADVEAPAEEHPGQADYWLR.H | |||||||
| R.LNQDFLQCAVYDSDKADAR.L | |||||||
| 29 | Late embryogenesis abundant protein, group 3 | gi|162463970 | 8.80/7.81 | 22.75/26.4 | 164 | 10 | K.AAEAGQYAK.D |
| K.SGGVIQQATEQVK.S K.DKSGGVIQQATEQVK.S | |||||||
| 30 | Globulin-1 S allele | gi|195658011 | 6.16/8.87 | 50.28/26.8 | 207 | 8 | R.LLDMDVGLANIAR.G R.LLDMDVGLANIAR.G |
| K.LLAFGADEEQQVDR.V | |||||||
| R.HYEITGDECPHLR.L | |||||||
| 31 | Globulin-1 S allele | gi|195658011 | 6.16/9.09 | 50.28/27.2 | 353 | 14 | R.GSMMAPSYNTR.A |
| R.LLDMDVGLANIAR.G R.LLDMDVGLANIAR.G | |||||||
| K.LLAFGADEEQQVDR.V R.HYEITGDECPHLR.L | |||||||
| K.GQGYFEMACPHVSGGR.S | |||||||
| K.GQGYFEMACPHVSGGR.S |
Table 2.
The differentially accumulated proteins identified in maize vp5 and Vp5 embryos associated to putative functions.
| Spot | Protein name | UniProtKB accession | Protein homology by Blast (Score/Identity) | Abundance change folds | Subcellular location | Molecular function |
|---|---|---|---|---|---|---|
| PROTEINS IN INCREASED ABUNDANCE IN vp5 COMPARED TO Vp5 | ||||||
| LEA proteins | ||||||
| 1 | Late embryogenesis abundant protein EMB564 | P46517_MAIZE | 13.1 | Nucleus(Wu et al., 2013a) | Stress response | |
| 27 | Uncharacterized protein | K7VM99_MAIZE | Group 3 LEA protein (1170/ 68.0%) | 9.3 | Cell wallb | Stress response |
| HSPs | ||||||
| 2 | 17.8 kDa class II heat shock protein | P24632_MAIZE | 2.4 | Cytoplasma | Stress response; protein folding | |
| 3 | 16.9 kDa class I heat shock protein 1 | B6T6N6_MAIZE | 2.3 | Nucleusb | Stress response; protein processing in ER | |
| 4 | 17.4 kDa class I heat shock protein 3 | B6TLK8_MAIZE | 2.4 | Nucleusb | Stress response; protein folding; protein oligomerization | |
| 5 | Heat shock protein 17.9 | B6TDB5_MAIZE | 2.5 | Nucleusb | Stress response | |
| 6 | Heat shock protein 17.2 | Q43701_MAIZE | 2.5 | Nucleusb | Stress response; protein processing in ER | |
| 7 | 17.0 kDa class II heat shock protein | Q08275_MAIZE | 2.1 | Cytoplasma | Stress response; protein processing in ER | |
| 8 | 22.0 kDa class IV heat shock protein | B6TG53_MAIZE | 2.4 | Plastidb | Stress response; protein processing in ER | |
| 9 | 22.0 kDa class IV heat shock protein | B6TXB5_MAIZE | 1.5 | Plastidb | Stress response; protein processing in ER | |
| 24 | 17.4 kDa class I heat shock protein 3 | B4F976_MAIZE | 2.8 | Nucleusb | Stress response; protein processing in ER | |
| Oxidoreductase | ||||||
| 10 | 1-Cys peroxiredoxin PER1 | A2SZW8_MAIZE | 7.3 | Nucleusa | Phenylalanine metabolism; biosynthesis of other secondary metabolites | |
| 11 | Short-chain dehydrogenase/reductase SDR family protein | B4FNZ9_MAIZE | 3.6 | Plastidb | Oxidation-reduction process | |
| 12 | UDP-glucose 6-dehydrogenase | B7ZYX8_MAIZE | 2.2 | Plastidb | Cell wall pectin metabolic process | |
| Others | ||||||
| 25 | Uncharacterized protein | B4FFK9_MAIZE | Lipoprotein-like (1104/ 81.0%) | 3.1 | Plastidb | Unknown |
| 26 | Uncharacterized protein | B4FFK9_MAIZE | Lipoprotein-like (1104/ 81.0%) | 2.1 | Plastidb | Unknown |
| PROTEINS IN REDUCED ABUNDANCE IN vp5 COMPARED TO Vp5 | ||||||
| LEA proteins | ||||||
| 13 | Late embryogenesis abundant protein D-34 | B6UH67_MAIZE | 2.2 | Nucleusb | Stress response | |
| 14 | Late embryogenesis abundant protein D-34 | B6UH67_MAIZE | 3.9 | Nucleusb | Stress response | |
| 15 | Late embryogenesis abundant protein D-34 | B6SNS4_MAIZE | 3.1 | Nucleusb | Stress response | |
| 16 | late embryogenesis abundant protein D-34 | B6SN63_MAIZE | 2.4 | Nucleusb | Stress response | |
| 22 | Late embryogenesis abundant protein Lea14-A | B6UH99_MAIZE | 1.8 | Plastidb Nucleusb Golgi apparatusb | Stress response | |
| 29 | Late embryogenesis abundant protein, group 3 | Q42376_MAIZE | 2.2 | Cell wallb | Stress response | |
| Enzymes | ||||||
| 17 | Short-chain dehydrogenase/reductase SDR family protein | B4FNZ9_MAIZE | 1.7 | Plastidb | Oxidation-reduction process | |
| 18 | Short-chain dehydrogenase/reductase SDR family protein | B4FNZ9_MAIZE | 2.0 | Plastidb | Oxidation-reduction process | |
| 20 | Thioredoxin | B4FH44_MAIZE | 2.3 | Cytoplasmb | Cell redox homeostasis; glycerol ether metabolic process; oxidation-reduction process | |
| 21 | Glyoxalase family protein superfamily | B6SGF3_MAIZE | 3.4 | Cytoplasmb | Methylglyoxal metabolic process | |
| 23 | Uncharacterized protein | C0HII8_MAIZE | Bowman-Birk serine protease inhibitor family protein (365/ 40.0%) | 6.5 | Extracellular regiona | Negative regulation of endopeptidase activity |
| Storage proteins | ||||||
| 30 | Globulin-1 S allele | B6UGJ0_MAIZE | 3.6 | Cell walla | Seed maturation | |
| 31 | Globulin-1 S allele | B6UGJ0_MAIZE | 2.7 | Cell walla | Seed maturation | |
| Others | ||||||
| 19 | Hypothetical protein ZEAMMB73_326753 | K7UCT3_MAIZE | Seed maturation protein (152/ 52.0%) | 6.8 | Nucleusb | Unknown |
| 28 | Uncharacterized protein | B4FFK9_MAIZE | Lipoprotein-like (1104/ 81.0%) | 2.7 | Plastidb | Unknown |
Subcellular location of proteins was annotated in UniProtKB/Swiss-Prot (http://www.expasy.org/).
Subcellular location of proteins was predicted using the online Plant-mPLoc server (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/).
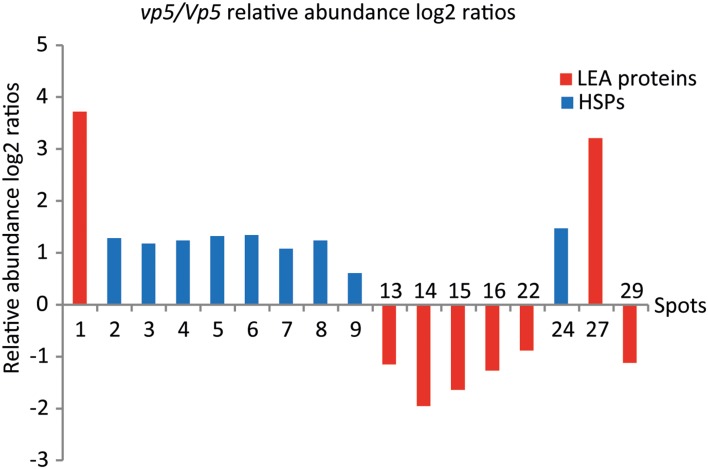
The 31 embryo proteins were successfully identified by MS/MS analysis, representing 26 distinct proteins in NCBI or SWISS-PROT protein databases (Tables 1, 2). In several cases, proteins were identified as uncharacterized proteins. We searched their homologous proteins in other plant species by BLAST, such as spot 19 homologous to seed maturation protein of Glycine latifolia (identity of 40%) and spots 25, 26, and 28 homologous to lipoprotein-like of Oryza sativa (rice, identity of 81%), or were based on family and domain databases, such as spot 23 belonging to Bowman-Birk serine protease inhibitor family, and spot 27 belonging to group 3 LEA protein. These differentially accumulated protein spots were assigned to various LEA family proteins (spots 1, 13–16, 22, 27, and 29), HSP20 family proteins (spots 2–9 and 24), 1-Cys peroxiredoxin PER1 (spot 10), short-chain dehydrogenase/reductase SDR family protein (spot 11, 17, and 18), UDP-glucose 6-dehydrogenase (spot 12), seed maturation protein (spot 19), thioredoxin (spot 20), glyoxalase family protein (spot 21), Bowman-Birk serine protease inhibitor family protein (spot 23), lipoprotein-like (spot 25, 26, and 28), and globulin-1 S allele (spots 30 and 31). Nine sHSPs accumulated in higher abundance in vp5 embryos, while six out of eight LEA proteins (except for spots 1 and 27) accumulated in higher abundance in Vp5 embryos (Figure 2).
Figure 2.
The relative abundance log2 ratios of LEA proteins and sHSPs between vp5 and Vp5 embryos. Log2 ratio > 1 represents abundance change more than two fold. Spot volumes were determined using the PDQuest software. Histograms full in red and blue represent LEA proteins and sHSPs, respectively.
In the present study, two identified proteins belong to group 3 LEA protein (spot 27, K7VM99, spot 29 Q42376), but they exhibit opposite accumulation between vp5 and Vp5 embryos. In vp5 embryos, Q42376, like other five identified LEA proteins, decreased in abundance, while K7VM99, like nine identified sHSPs, increased in abundance. This discrepancy may be explained by protein and gene sequence differences. K7VM99 and Q42376 share only a 35% identity in protein sequence alignment by BLAST.
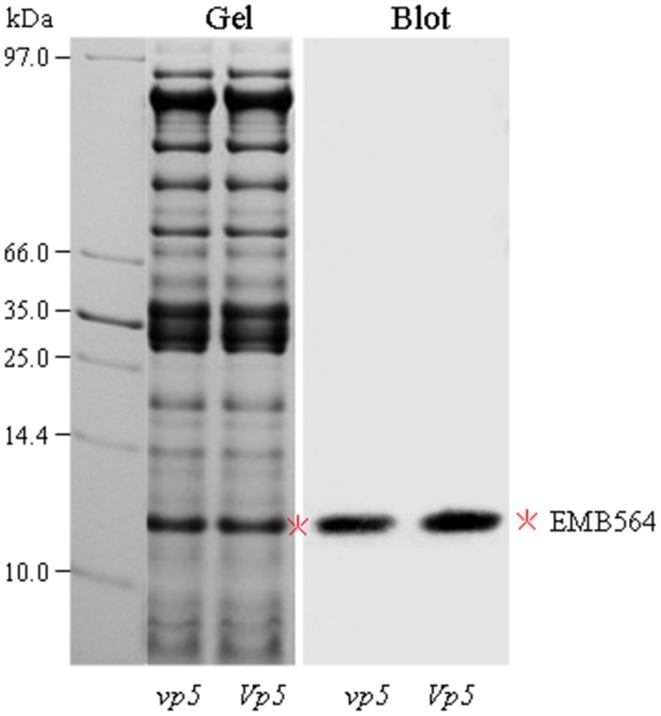
In addition, we found EMB564 existed in two isoforms: a weak spot 1 and a most predominant spot (indicated in Figure 1A). These two isoforms displayed a contrast accumulation in vp5 compared to Vp5: the weak isoform (spot 1) increased greatly whereas the other decreased a little. However, the total abundance of EMB564 was comparable between vp5 and Vp5. This result contrasted to previously characterized, greatly lowered expression of emb564 mRNA in vp5 embryos (Williams and Tsang, 1991). In order to further confirm the accumulation level of EMB564 in the two genotypes, we examined the abundance of EMB564 using immunoblot analysis (Figure 3). The specificity of the antibody has been recently characterized, and it specifically reacts with EMB564 (Wu et al., 2013a). Obviously, EMB564 existed in comparable levels between vp5 and Vp5 embryos, with a slightly reduced level in vp5.
Figure 3.
Immunoblot detection of EMB564 in vp5 and Vp5 embryos. Embryo protein (30 μg/lane) was resolved by 12.5% SDS-PAGE. Gel was stained with CBB. Blot was probed with an antibody specifically against EMB564 (1:5000 dilution). EMB564 was indicated by asterisk.
Proteomic difference between vp5 and Vp5 endosperms
In preliminary 2-DE experiments, none obviously differentially accumulated endosperms protein spots were observed to exist when pH 7–10 IPG gels were used (data not shown). Therefore, endosperm proteome analysis was performed only using pH 4–7 IPG gels.
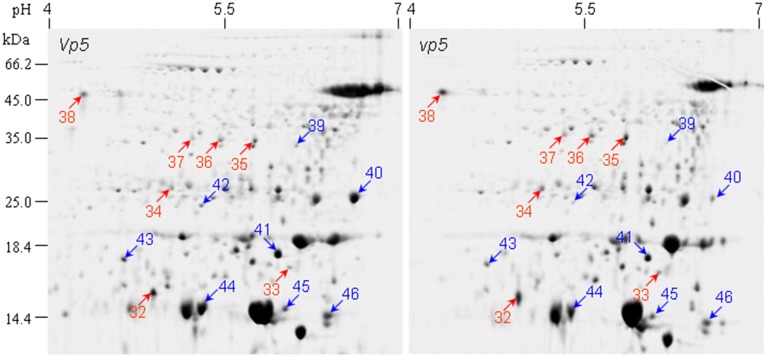
The endosperm protein profiles between vp5 and Vp5 were compared by 2-DE. Approximately 380±10 CBB-stained protein spots were detected in endosperms, which was much less compared to embryos and most of proteins existed in low abundance (Figure 4). PDQUEST analysis indicated that 15 protein spots, especially spot 40, showed an obvious difference between vp5 and Vp5 endosperms, with a consistent change in three biological replicates (Figure 4, Presentation 4 in Supplementary Material). Seven spots (32–38) existed in higher abundance and eight spots (39–46) in lower abundance in vp5. These differentially accumulated protein spots were successfully identified by MS/MS (Tables 3, 4), representing 11 unique protein species.
Figure 4.
2-DE comparison of endosperm protein profiles between maize vp5 mutant and its wild type Vp5. Endosperm proteins were resolved by 2-DE as described in Figure 1.
Table 3.
The differentially accumulated proteins identified by MS/MS between maize vp5 and Vp5 endosperms.
| Spot | Protein name | NCBI accession | pI Theor/exp. | MW (kDa) Theor/exp. | Mascot score | Coverage (%) | Matched peptide sequences |
|---|---|---|---|---|---|---|---|
| 32 | Glyoxalase family protein superfamily | gi|226532762 | 5.47/4.90 | 15.13/15.7 | 783 | 51 | K.AASFYDAAFGYTVR.R R.ETDELSGAVQLPDSSAAGR.G |
| R.GSVEVCFAYADVDAAYKR.A K.WAELESGATTIAFTPLHQR.E | |||||||
| R.KWAELESGATTIAFTPLHQR.E R.KWAELESGATTIAFTPLHQR.E | |||||||
| 33 | 50 kD gamma zein | gi|162458022 | 6.83/6.06 | 35.40/17.2 | 459 | 23 | K.NCHEFLR.Q |
| R.CSYNYYSSSSNLK.N | |||||||
| R.QQCSPLVMPFLQSR.L | |||||||
| R.QQCSPLVMPFLQSR.L K.SQQQQCHCQEQQQTTR.C | |||||||
| R.LIQPSSCQVLQQQCCHDLR.Q | |||||||
| 34 | Uncharacterized protein | gi|223944797 | 6.05/5.06 | 26.45/27.1 | 594 | 27 | K.VSFGENFSPAR.A |
| K.VKEGVEVAQLVEK.V | |||||||
| R.SPAAEALGPTHLLHTR.Y R.LRSPAAEALGPTHLLHTR.Y | |||||||
| K.GYQFGMVAVFDSVEELDAVEGDGKVEEAK.A | |||||||
| 35 | Lactoylglutathione lyase | gi|195639070 | 6.62/5.76 | 35.31/34.5 | 352 | 24 | R.MLHAVYR.V |
| K.YYTECFGMK.L | |||||||
| K.IASFVDPDGWK.V | |||||||
| R.ADTPEPLCQVMLR.V | |||||||
| R.ADTPEPLCQVMLR.V | |||||||
| K.VVLVDNTDFLKELH.- | |||||||
| K.GGSTVIAFAQDPDGYMFELIQR.A | |||||||
| 36 | Lactoylglutathione lyase | gi|195639070 | 6.62/5.47 | 35.31/34.6 | 438 | 24 | R.MLHAVYR.V |
| K.YYTECFGMK.L | |||||||
| K.IASFVDPDGWK.V | |||||||
| K.VVLVDNTDFLK.E | |||||||
| R.ADTPEPLCQVMLR.V | |||||||
| K.VVLVDNTDFLKELH.- K.GGSTVIAFAQDPDGYMFELIQR.A | |||||||
| 37 | Lactoylglutathione lyase | gi|195639070 | 6.62/5.23 | 35.31/35.0 | 391 | 24 | R.MLHAVYR.V |
| K.YYTECFGMK.L | |||||||
| K.IASFVDPDGWK.V | |||||||
| K.VVLVDNTDFLK.E | |||||||
| R.ADTPEPLCQVMLR.V | |||||||
| K.VVLVDNTDFLKELH.- | |||||||
| K.GGSTVIAFAQDPDGYMFELIQR.A | |||||||
| 38 | Maize protease inhibitor | gi|75994143 | 5.63/4.32 | 7.49/48.8 | 462 | 75 | R.IFVDIVAQTPHIG.- |
| K.TSWPEVVGLSVEDAK.K K.TSWPEVVGLSVEDAKK.V | |||||||
| K.TSWPEVVGLSVEDAKK.V K.DKPDADIVVLPVGSVVTADYRPNR.V | |||||||
| 39 | Short-chain dehydrogenase/reductase SDR family protein | gi|226500748 | 5.78/6.12 | 33.15/33.7 | 575 | 29 | R.ALSLQLADR.G |
| K.EGATVAFTFVR.G | |||||||
| R.ALRDIGSETGAR.E | |||||||
| R.GQEEKDAEETLR.A | |||||||
| K.VEQFGSQVPMKR.A | |||||||
| R.IDVVVNNAAEQYER.E R.VASAYGGRIDVVVNNAAEQYER.E | |||||||
| R.IDVVVNNAAEQYERESIGDVTEADLER.V | |||||||
| 40 | 1-Cys peroxiredoxin PER1 | gi|162460575 | 6.31/6.62 | 25.06/25.5 | 941 | 48 | K.VTFPILADPAR.D |
| K.LSFLYPATTGR.N | |||||||
| K.LLGISCDDVESHR.Q | |||||||
| R.QLNMVDPDEKDAAGR.S, K.MFPQGFETADLPSKK.G | |||||||
| M.PGLTIGDTVPNLELDSTHGK.I | |||||||
| K.VATPANWKPGECAVIAPGVSDEEAR.K | |||||||
| K.VATPANWKPGECAVIAPGVSDEEARK.M | |||||||
| 41 | Superoxide dismutase [Cu-Zn] 4AP | gi|13431904 | 5.65/5.96 | 15.22/17.9 | 315 | 21 | K.AVAVLGSSEGVK.G |
| R.AVVVHADPDDLGK.G R.AVVVHADPDDLGKGGHELSK.T | |||||||
| 42 | Globulin-1 S allele | gi|195658011 | 6.16/5.30 | 50.28/24.4 | 448 | 16 | R.VAELEAAPR.T |
| K.EGEGVIVLLR.G | |||||||
| K.GEITTASEEQIR.E | |||||||
| R.FTHELLEDAVGNYR.V | |||||||
| K.QSKGEITTASEEQIR.E R.EGDVMVIPAGAVVYSANTHQSEWFR.V | |||||||
| 43 | Superoxide dismutase [Cu-Zn] | gi|167860184 | 5.45/4.66 | 21.00/17.6 | 515 | 25 | K.GGHELSLSTGNAGGR.L K.GASEVEGVVTLTQDDDGPTTVNVR.I |
| R.AFVVHELEDDLGKGGHELSLSTGNAGGR.L | |||||||
| 44 | Trypsin/factor XIIA inhibitor | gi|157830250 | 8.07/5.30 | 16.30/14.8 | 479 | 46 | R.LPWPELKR.R |
| R.LEDLPGCPR.E | |||||||
| R.ELADIPAYCR.C -.SAGTSCVPGWAIPHNPLPSCR.W | |||||||
| R.CTALSILMDGAIPPGPDAQLEGR.L | |||||||
| R.CTALSILMDGAIPPGPDAQLEGR.L | |||||||
| 45 | Glycine-rich RNA-binding protein 2 | gi|195612516 | 6.10/6.02 | 15.48/14.9 | 652 | 54 | M.AASDVEYR.C, |
| R.NITVNEAQSR.G, | |||||||
| K.IILDRETQR.S, | |||||||
| R.GGGYGNSDGNWRN.- | |||||||
| R.GFGFVTFSTEEAMR.N | |||||||
| R.GFGFVTFSTEEAMR.N | |||||||
| R.DGGGGYGGGGGYGGGGGYGGGGGGYGGGNR.G | |||||||
| 46 | Trypsin/factor XIIA inhibitor | gi|157830250 | 8.07/6.39 | 16.30/14.5 | 455 | 46 | R.LPWPELKR.R |
| R.LEDLPGCPR.E | |||||||
| R.ELADIPAYCR.C -.SAGTSCVPGWAIPHNPLPSCR.W | |||||||
| R.CTALSILMDGAIPPGPDAQLEGR.L |
Table 4.
The differentially accumulated proteins identified in maize vp5 and Vp5 endosperms associated to putative functions.
| Spot | Protein name | UniProtKB accession | Protein homology by Blast (Score/Identity) | Abundance change folds | Subcellular location | Molecular function |
|---|---|---|---|---|---|---|
| PROTEINS IN INCREASED ABUNDANCE IN vp5 COMPARED TO Vp5 | ||||||
| Enzymes | ||||||
| 32 | Glyoxalase family protein superfamily | B6SGF3_MAIZE | 1.5 | Cytoplasmb | Methylglyoxal metabolic process | |
| 35 | Lactoylglutathione lyase | B6TPH0_MAIZE | 1.6 | Cytoplasmb | Metabolic process | |
| 36 | Lactoylglutathione lyase | B6TPH0_MAIZE | 1.5 | Cytoplasmb | Metabolic process | |
| 37 | Lactoylglutathione lyase | B6TPH0_MAIZE | 1.7 | Cytoplasmb | Metabolic process | |
| 38 | Maize protease inhibitor | Q2XX01_MAIZE | 1.9 | Plastidb Nucleusb | Proteolysis; response to wounding | |
| Storage proteins | ||||||
| 33 | 50 kD gamma zein | Q946W1_MAIZE | 2.1 | Nucleusb | Seed maturation | |
| Stress-responsive protein | ||||||
| 34 | Uncharacterized protein | C0HHH9_MAIZE | Stress responsive alpha-beta barrel domain protein (371/42.0%) | 1.5 | Plastidb | Stress response |
| PROTEINS IN REDUCED ABUNDANCE IN vp5 COMPARED TO Vp5 | ||||||
| Enzymes | ||||||
| 39 | Short-chain dehydrogenase/reductase SDR family protein | B4FNZ9_MAIZE | 2.3 | Plastidb | Oxidation-reduction process | |
| 40 | 1-Cys peroxiredoxin PER1 | A2SZW8_MAIZE | 5.2 | Nucleusa | Phenylalanine metabolism; biosynthesis of other secondary metabolites | |
| 41 | Superoxide dismutase [Cu-Zn] 4AP | P23346_MAIZE | 2.0 | Cytoplasma | Superoxide metabolic process | |
| 43 | Superoxide dismutase [Cu-Zn] | B1PEY4_MAIZE | 1.7 | Plastidb | Superoxide metabolic process | |
| 44 | Trypsin/factor XIIA inhibitor | P01088_MAIZE | 2.0 | Extracellular regiona | Negative regulation of endopeptidase activity | |
| 46 | Trypsin/factor XIIA inhibitor | P01088_MAIZE | 1.5 | Extracellular regiona | Negative regulation of endopeptidase activity | |
| Storage proteins | ||||||
| 42 | Globulin-1 S allele | B6UGJ0_MAIZE | 2.5 | Cell walla | Seed maturation | |
| Other | ||||||
| 45 | Glycine-rich RNA-binding protein 2 | B6STA5_MAIZE | 1.7 | Plastidb Nucleusb | DNA duplex unwinding; RNA secondary structure unwinding; mRNA export from nucleus; Stress response | |
Subcellular location of proteins was annotated in UniProtKB/Swiss-Prot (http://www.expasy.org/).
Subcellular location of proteins was predicted using the online Plant-mPLoc server (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/).
Discussion
Global proteome alterations in embryo and endosperm of maize vp5 and Vp5
Maize viviparous mutants, germinating directly on the ear (McCarty et al., 1991), are widely used for studying maize seed maturation, dormancy, and germination. Various viviparous mutants have been identified, such as vp1、vp2、vp5、vp7、vp8、 and vp9. Among them, vp5 mutant is deficient in ABA biosynthesis with the first step catalyzed by phytoene desaturase being blocked, resulting in the precursor phytoene accumulation and carotenoid deficiency (Robichaud et al., 1980). However, the background difference at proteome level between vp5 and Vp5 seeds is still unclear.
In the present study, comparative proteomics was used to determine the variation of protein expression at the proteome level between maize mutant vp5 and its wild type Vp5. There are great differences in the structure, composition and function between embryo and endosperm of maize seeds. Compared to endosperm, embryo is more active in nucleic acid, protein, and lipid metabolism. In total, 46 seed protein spots were found to be exhibited a differential change in abundance between these two genotypes, 31 spots in embryo and 15 spots in endosperm. Obviously, proteome alterations caused by ABA-deficient mutation are more significant in embryos than in endosperms in vp5 seeds. This may be explained by two possible causes: Firstly, ABA deficiency mainly takes place in vp5 embryos. Due to deficient in ABA biosynthesis, maize vp5 embryos contains a low ABA content (about 10% of Vp5) throughout seed maturation, whereas 42% ABA in vp5 endosperms (Neill et al., 1986). ABA in endosperm can be from maternal organs (Ober and Setter, 1992). Secondly, maize endosperms act mainly as starch storage tissue and contain fewer proteins in low amounts in mature seeds. Although ABA-deficient mutation affects the phenotype of vp5 endosperm (carotenoid deficiency), proteins or enzymes involved in the related pathways were not detected, probably due to their low abundance and low sensitivity of CBB staining.
In the present study, four proteins, i.e., 1-Cys peroxiredoxin PER1 (spot 10 in embryo, spot 40 in endosperm), globulin-1 s allele (spot 30 and 31 in embryo, spot 42 in endosperm), glyoxalase family protein (spots 21 in embryo and 32 in endosperm) and short-chain dehydrogenase/reductase SDR family protein (spot 11, 17, and 18 in embryo, spot 39 in endosperm) were the same as identified embryo proteins. Among them, globulin-1 s allele (spot 30, 31, and 42) and short-chain dehydrogenase/reductase SDR family protein (spot 17, 18, and 39) decreased both in vp5 embryo and endosperm compared to Vp5.
It is worth to note that EMB564, the most abundant LEA protein in maize embryos, was found to exist in comparable levels between vp5 and Vp5 embryos. EMB564 consists of two isoforms with obvious differences in apparent and theoretical values of sizes and isoelectric points, indicating a post-translational modification of this protein (e.g., phosphorylation, Amara et al., 2012). Williams and Tsang (1991) first cloned and characterized an embryo-specific recombinant, termed emb564 [recently renamed as embryo specific protein 1 (esp1) in NCBI database]. The emb564 mRNA is expressed at low level in ABA-deficient (e.g., vp5) but not in ABA-insensitive (e.g., vp1) embryos during embryogenesis, and exogenous ABA has little effect on the accumulation of emb564 mRNA in more mature embryos (Williams and Tsang, 1991). Therefore, there is a discrepancy between EMB564 protein and mRNA levels.
The transcript/protein discordance has been well documented in mammals, yeasts and plants (mainly Arabidopsis and maize). It is largely of biological origin (“true discordance”) and of post-transcriptional regulation (Vélez-Bermúdez and Schmidt, 2014). For example, in Arabidopsis roots, a lack of correlation between down-regulated transcripts and the amount of their corresponding proteins was observed in response to phosphate deficiency, whereas for induced genes changes in the levels of mRNAs and proteins were reasonably well correlated (Lan et al., 2012). In the ABA-deficient vp5 embryos, EMB564 can accumulate to a similar level as its wild type. Thus, this transcript/protein discordance is possibly caused by post-transcriptional regulation (via unknown factors) in an ABA-independent way.
Recently, we found that EMB564 is associated with maize seed vigor (Wu et al., 2011) and is highly thermal stable (Wang et al., 2012). By immunoelctron microscopy, we showed that EMB564 locates preferentially in the nucleus of maize embryonic cells (Wu et al., 2013a). Likewise, Amara et al. (2012) demonstrated that EMB564-green fluorescent protein fusions are expressed in the cytosol and nucleus in the agroinfiltrated leaves of Nicotiana bentamiana. Based on bioinformatics analysis, we proposed that EMB564 may function within the nucleus by binding DNA (Wu et al., 2013b).
In addition, we also noticed that some spots are not consistent in abundance between the biological replicates. This inconsistence results mainly from inherit drawbacks of 2-DE (e.g., poor reproducibility between gels) and from minor changes in manipulation during protein extraction among independent biological experiments. However, only those reproducibly, differentially accumulated proteins between vp5 and Vp5 are subjected to MS/MS analysis.
LEA proteins and sHSPs displayed differential accumulations in ABA-deficient embryos
The most interesting finding in this study is that most LEA proteins and sHSPs displayed differential accumulations in ABA-deficient vp5 embryos: six out of eight identified LEA proteins decreased while nine identified sHSPs increased in abundance, compared to Vp5.
LEA proteins are characterized by high hydrophilicity and accumulate to high levels during the last stage of seed maturation (Dure et al., 1989; Espelund et al., 1992). Although the roles of LEA proteins remain speculative, there is evidence supporting their participation in acclimation and/or in the adaptive response to dehydration, low temperature, salinity, or exogenous ABA treatment stress (Battaglia et al., 2008). sHSPs are produced in seeds during maturation and under various stress conditions. The synthesis of sHSPs during seed maturation indicates their probable role in cell component protection mechanisms. Mutants sensitive to desiccation contain smaller amounts of sHSPs during maturation (Wehmeyer and Vierling, 2000).
We tried to explain the possible causes of the observed differential accumulation of six identified LEA proteins and nine identified sHSPs between vp5 and Vp5 seeds. Firstly, the threshold content of ABA inducing the accumulation of LEA proteins and sHSPs may differ: some LEA proteins may require higher ABA content than sHSPs during seed maturation. ABA-deficiency in vp5 embryos greatly inhibited the accumulation of most LEA proteins, but promoted the accumulation of sHSPs. In another words, compared to developmentally specific accumulation of LEA proteins, sHSPs seemed to be less strictly ABA-dependent. In the ABA-deficient vp5 embryos, EMB564 accumulates to a similar level as its wild type, implying an ABA-independent accumulation of EMB564 in maize embryos. This deduction will be examined by measuring the ABA content and the time-course of accumulation of LEA proteins and sHSPs during vp5 seed development in future. Previously, differential regulation of ABA-induced 23–25 kDa proteins in embryo and vegetative tissues of maize vp mutants was reported (Pla et al., 1989). In a recent study, we identified several proteins in maize Vp5 and vp5 seedlings whose syntheses are ABA-independent, such as ADP-dependent malic enzyme and fructose-bisphosphate aldolase (Hu et al., 2012). Maize seeds undergo dehydration process during maturation. ABA mutant vp5 seeds contain low ABA content and lack obvious dehydration process; therefore, the accumulation of strict ABA-dependent protein (e.g., LEA proteins in this study) may reduce, and not strict ABA-dependent proteins (e.g., sHSPs in this study) may increase to enhance seed dehydration/drought tolerance.
We checked whether there was any difference in the promoter region, especially regarding ABRE and DRE, between differentially accumulated proteins identified here (Tables 5, 6). Most LEA genes are sensitive to ABA because of the presence of ABRE, i.e., regulatory elements in promoter regions, which contain the ACGT sequence called cassette G. Sensitivity to ABA also depends on the presence of MYC elements that include the sequence CACCTG, and MYB elements that include the TAACTG motive (Kalemba and Pukacka, 2007). Dehydration responsive element, DRE, is 9-bp consensus sequence, TACCGACAT, was first identified in the promoter of Arabidopsis rd29A/lti78 and shown to be essential for drought induction in the absence of ABA (Yamaguchi-Shinozaki and Shinozaki, 1994).
Table 5.
Cis-acting elements of LEA and sHSPs proteins analyzed by Plantcare.
| Spot | Accession | Protein | ABRE (Plantcare) | Other (Plantcare) |
|---|---|---|---|---|
| 1 | P46517 | EMB564 | AGTTCGTGGC; GCCACGCACA; CGCACGCGTC; CCGCGTCGGC | |
| 27 | K7VM99 | Group 3 LEA protein | CCCACGTGTC; CGTACGTGTC; GTCACGTACGT; TACGTG; CACGTG | CAACTG (MBS: MYB binding site involved in drought-inducibility) |
| 2 | P24632 | HSP 17.8 | CACGTG; ACGTGGC; CCAACGTGGC | |
| 4 | B6TLK8 | HSP 17.4 | NO | |
| 5 | B6TDB5 | HSP 17.4 | CCGCGTAGGC; CCGCGTCGGC;CACGTG | TGGCCGAC(C-repeat/DRE) |
| 7 | Q08275 | HSP 17.0 | TACGTG; CACGTG | |
| 8 | B6TG53 | HSP 22.0 | No | CGGGAAGCTTCCAG(HSE); AAAAAATTTA(HSE); CAACTG(MBS: MYB binding site involved in drought-inducibility) |
| 9 | B6TXB5 | HSP 22.0 | TACGTG | CAACTG(MBS: MYB binding site involved in drought-inducibility) |
| 24 | B4F976 | HSP 17.4 | No | CAACTG(MBS: MYB binding site involved in drought-inducibility) |
| 13,14 | B6UH67 | LEA D34 | No | No |
| 22 | B6UH99 | LEA 14-A | CCAACGTGTC | |
| 29 | Q42376 | Group 3 LEA protein | GCTACGTGGC; TACGTG; ACGTGGC; CGTTCGTGCA |
Table 6.
Cis-acting elements of LEA and sHSPs proteins analyzed by Place.
| Spot | Accession | Protein | ABRE (Place) | DRE (Place) |
|---|---|---|---|---|
| 1 | P46517 | EMB564 | CACGCGT; CACGCGC | GCCGAC(DRECRTCOREAT) |
| 27 | K7VM99 | Group 3 LEA protein | ACGTG;TACGTGTC;CACGCGT | ACCGAGA(DRE1COREZMRAB17); ACCGAC(DRE2COREZMRAB17); ACCGAC(DRECRTCOREAT) |
| 2 | P24632 | HSP 17.8 | ACGTG | ACCGAGA(DRE1COREZMRAB17) |
| 4 | B6TLK8 | HSP 17.4 | CACGCGGT | GCCGAC(DRECRTCOREAT) |
| 5 | B6TDB5 | HSP 17.4 | ACGTG | GCCGAC(DRECRTCOREAT) |
| 7 | Q08275 | HSP 17.0 | ACGTG;CACGTGT | ACCGAGA(DRE1COREZMRAB17); ACCGAC(DRE2COREZMRAB17); ACCGAC(DRECRTCOREAT) |
| 8 | B6TG53 | HSP 22.0 | ACGTG | GCCGAC(DRECRTCOREAT) |
| 9 | B6TXB5 | HSP 22.0 | ACGTG;TACGTGTC | ACCGAGA((DRE1COREZMRAB17); ACCGAC(DRE2COREZMRAB17); ACCGAC(DRECRTCOREAT) |
| 24 | B4F976 | HSP 17.4 | NO | NO |
| 13,14 | B6UH67 | LEA D34 | NO | NO |
| 22 | B6UH99 | LEA 14-A | ACGTG; AACGTGT | GCCGAC(DRECRTCOREAT) |
| 29 | Q42376 | Group 3 LEA protein | TACGTGGC; ACGTG; ACGTGGCG; ACGTGGCC; AACGTGG | ACCGAC(DRE2COREZMRAB17); ACCGAC(DRECRTCOREAT) |
In addition, the accumulation pattern of group 3 LEA protein (spot 27, K7VM99) in vp5 embryo is different with other five identified LEA proteins. The group 3 LEA proteins are characterized by a repeating motif of 11 amino acids (TAQAAKEKAGE) (Dure et al., 1989), which are quite diverse in sequence structure compared with other LEA groups (Battaglia et al., 2008). In maize vp5 seeds, group 3 LEA protein accumulation is dependent upon ABA (Thomann et al., 1992), whereas corresponding mRNA has no response to exogenous ABA (White and Rivin, 2000). In maize leaves, group 3 LEA gene can be induced by ABA (Liu et al., 2013). Cis-acting elements analysis showed that group 3 LEA protein (K7VM99) shares more identical ABRE and/or DRE with sHSPs (e.g., HSP 17.0) than other LEA proteins (Tables 5, 6). This may explain why group 3 LEA protein (K7VM99) differentially accumulates in vp5 embryos, like sHSPs but unlike other LEA proteins.
In conclusion, comparative gel-based proteomics revealed significant proteome difference between vp5 and Vp5 seeds. Most notably, LEA proteins and sHSPs displayed a differential accumulation pattern in ABA-deficient vp5 embryos. The data derived from the present study is highly applicable to other crops. The characterization of proteome difference between vp5 and Vp5 seeds is necessary for dissection of ABA-mediated maize response in the studies involved vp5 mutants. The data derived from this study provides insight into ABA-dependent proteins and ABA-mediated response during seed maturation in maize.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by National Natural Science Foundation of China (grant 31371543 and 31171470), Plan for Scientific Innovation Talent of Henan Province (grant 144200510012) and State Key Laboratory of Crop Biology (2012KF01) at Shandong Agricultural University, China.
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2014.00801/abstract
References
- Amara I., Odena A., Oliveira E., Moreno A., Masmoudi K., Pagès M., et al. (2012). Insights into maize LEA proteins: from proteomics to functional approaches. Plant Cell Physiol. 53, 312–329. 10.1093/pcp/pcr183 [DOI] [PubMed] [Google Scholar]
- Battaglia M., Olvera-carrillo Y., Garciarrubio A., Campos F., Covarrubias A. A. (2008). The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 148, 6–24. 10.1104/pp.108.120725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durantini D., Giulini A., Malgioglio A., Pilu R., Tuberosa R., Sanguineti C., et al. (2008). Vivipary as a tool to analyze late embryogenic events in maize. Heredity 101, 465–470. 10.1038/hdy.2008.29 [DOI] [PubMed] [Google Scholar]
- Dure L., Crouch M., Harada J. J., Ho T. H. D., Mundy J., Quatrano R., et al. (1989). Common amino acid sequence domains among the LEA proteins of higher plants. Plant Mol. Biol. 12, 475–486. 10.1007/BF00036962 [DOI] [PubMed] [Google Scholar]
- Espelund M., Saeboe-Larssen S., Hughes D. W., Galau G. A., Larsen F., Jakobsen K. S. (1992). Late embryogenesis-abundant genes encoding proteins with different numbers of hydrophilic repeats are regulated differentially by abscisic acid and osmotic stress. Plant J. 2, 241–252. [PubMed] [Google Scholar]
- Hable W. E., Oishi K. K., Schumaker K. S. (1998). Viviparous-5 encodes phytoene desaturase, an enzyme essential for abscisic acid (ABA) accumulation and seed development in maize. Mol. Gen. Genet. 257, 167–176. 10.1007/s004380050636 [DOI] [PubMed] [Google Scholar]
- Hu X. L., Lu M. H., Li C. H., Liu T. X., Wang W., Wu J., et al. (2011). Differential expression of proteins in maize roots in response to abscisic acid and drought. Acta Physiol. Plant. 33, 2437–2446. 10.1007/s11738-011-0784-y21659145 [DOI] [Google Scholar]
- Hu X. L., Wu X. L., Li C. H., Lu M. H., Liu T. X., Wang Y., et al. (2012). Abscisic acid refines the synthesis of chloroplast proteins in maize (Zea mays) in response to drought and light. PLoS ONE 7:e49500. 10.1371/journal.pone.0049500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalemba E. M., Pukacka S. (2007). Possible roles of LEA proteins and sHSPs in seed protection: a short review. Biol. Lett. 44, 3–16. [Google Scholar]
- Lan P., Li W., Schmidt W. (2012). Complementary proteome and transcriptome profiling in phosphate-deficient Arabidopsis roots reveals multiple levels of gene regulation. Mol. Cell. Proteomics 11, 1156–1166. 10.1074/mcp.M112.020461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehesranta S. J., Davies H. V., Shepherd L. V. T., Nunan N., McNicol J. W., Auriola S., et al. (2005). Comparison of tuber proteomes of potato varieties, Landraces, and genetically modified lines. Plant Physiol. 138, 1690–1699. 10.1104/pp.105.060152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Wang L., Xing X., Sun L. P., Pan J. W., Kong X. P., et al. (2013). ZmLEA3, a multifunctional group 3 LEA protein from maize (Zea mays L.), is involved in biotic and abiotic stresses. Plant Cell Physiol. 54, 944–959. 10.1093/pcp/pct047 [DOI] [PubMed] [Google Scholar]
- Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., et al. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068. 10.1126/science.1172408 [DOI] [PubMed] [Google Scholar]
- McCarty D. R., Hattori T., Carson C. B., Vasil V., Lazar M., Vasil I. K. (1991). The Viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell 66, 895–905. 10.1016/0092-8674(91)90436-3 [DOI] [PubMed] [Google Scholar]
- Moore K. B., Oishi K. K. (1994). 3-Hydroxy-3-methylglutaryl coenzyme A reductase activity in the endosperm of maize vivipary mutants. Plant Physiol. 105, 119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill S. J., Horgan R., Parry A. D. (1986). The carotenoid and abscisic acid content of viviparous kernels and seedlings of Zea mays L. Planta 169, 87–96. 10.1007/BF01369779 [DOI] [PubMed] [Google Scholar]
- Nishimura N., Sarkeshi A., Nito K., Park S. Y., Wang A., Carvalho P. C., et al. (2010). PYR/PYL/RACR family members are major in vivo ABI1 protein phosphatase 2c interacting proteins in Arabidopsis. Plant J. 61, 290–299. 10.1111/j.1365-313X.2009.04054.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober E. S., Setter T. L. (1992). Water deficit induces abscisic acid accumulation in endosperm of maize viviparous mutants. Plant Physiol. 98, 353–356. 10.1104/pp.98.1.353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. Y., Fung P., Nishimura N., Jensen D. R., Fujii H., Zhao Y., et al. (2009). Abscisic acid inhibits type 2c protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071. 10.1126/science.1173041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pla M., Goday A., Vilardell J., Gomez. J., Pages M. (1989). Differential regulation of the ABA-induced 23-25 kD proteins in embryos and vegetative tissues of the viviparous mutants of maize. Plant Mol. Biol. 13, 385–394. 10.1007/BF00015550 [DOI] [PubMed] [Google Scholar]
- Quatrano R. S. (1986). Regulation of gene expression by abscisic acid during angiosperm embryo development. Oxford Surveys Plant Mol. Cell Biol. 3, 467–477. [Google Scholar]
- Robichaud C. S., Wong J., Sussex I. M. (1980). Control of in vitro growth of viviparous embryo mutants of maize by abscisic acid. Dev. Genet. 1, 325–330 10.1002/dvg.1020010405 [DOI] [Google Scholar]
- Santiago J., Dupeux F., Round A., Antoni R., Park S. Y., Jamin M., et al. (2009). The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 462, 665–668. 10.1038/nature08591 [DOI] [PubMed] [Google Scholar]
- Tan B. C., Schwartz S. H., Zeevaart J. A., McCarty D. R. (1997). Genetic control of abscisic acid biosynthesis in maize. Proc. Natl. Acad. Sci. U.S.A. 94, 12235–12240. 10.1073/pnas.94.22.12235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomann E. B., Sollinger J., White C., Rivin C. J. (1992). Accumulation of group 3 late embryogenesis abundant proteins in Zea mays embryos: roles of abscisic acid and the Viviparous-1 gene product. Plant Physiol. 99, 607–614. 10.1104/pp.99.2.607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T., Nakashima K., Miyakawa T., Kuromori T., Tanokura M., Shinozaki K., et al. (2010). Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol. 51, 1821–1839. 10.1093/pcp/pcq156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vélez-Bermúdez I. C., Schmidt W. (2014). The conundrum of discordant protein and mRNA expression. Are plants special? Front. Plant Sci. 5:619. 10.3389/fpls.2014.00619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Wu X. L., Xiong E. H., Tai F. J. (2012). Improving gel-based proteome analysis of soluble protein extracts by heat prefractionation. Proteomics 12, 938–943. 10.1002/pmic.201100475 [DOI] [PubMed] [Google Scholar]
- Wehmeyer N., Vierling E. (2000). The expression of small heat shock proteins in seeds responds to discrete development signals and suggest a general protective role in desiccation tolerance. Plant Physiol. 122, 1099–1108. 10.1104/pp.122.4.1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White C. N., Rivin C. J. (2000). Gibberellins and seed development in maize. ii. gibberellin synthesis inhibition enhances abscisic acid signaling in cultured embryos. Plant Physiol. 122, 1089–1097. 10.1104/pp.122.4.1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B., Tsang A. (1991). A maize gene expressed during embryogenesis is abscisic acid-inducible and highly conserved. Plant Mol. Biol. 16, 919–923. 10.1007/BF00015086 [DOI] [PubMed] [Google Scholar]
- Wu X. L., Gong F. P., Wang W. (2013b). Functional assignment to maize group 1 LEA protein EMB564 within the cell nucleus using computational analysis. Bioinformation 9, 276–279. 10.6026/97320630009276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X. L., Liu H. Y., Wang W., Chen S. N., Hu X. L., Li C. H. (2011). Proteomic analysis of seed viability in maize. Acta Physiol. Plant. 33, 181–191 10.1007/s11738-010-0536-4 [DOI] [Google Scholar]
- Wu X. L., Scali M., Faleri C., Wang W. (2013a). Polyclonal antibody preparation and immunolocalization of maize (Zea mays) seed protein. Plant Omics J. 6, 359–363. [Google Scholar]
- Wu X. L., Xiong E. H., Wang W., Scali M., Cresti M. (2014). Universal sample preparation method integrating trichloroacetic acid/acetone precipitation with phenol extraction for crop proteomic analysis. Nat. Protoc. 9, 362–374. 10.1038/nprot.2014.022 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6, 251–264. 10.1105/tpc.6.2.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart J. A. D., Creelman R. A. (1988). Metabolism and physiology of abscisic acid. Annu. Rev. Plant Physiol. Plant Mol. Biol. 39, 439–473 10.1146/annurev.pp.39.060188.002255 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.