Abstract
Bispecific antibodies were constructed using genetically encoded unnatural amino acids with orthogonal chemical reactivity. A two-step process afforded homogeneous products in excellent yield. Using this approach, we synthesized an anti-HER2/anti-CD3 bispecific antibody, which efficiently crosslinked HER2+ cells and CD3+ cells. In vitro effector-cell mediated cytotoxicity was observed at picomolar concentrations.
Recently, there has been considerable interest in the generation of bispecific antibodies that simultaneously bind two different antigens1-4. Indeed, several clinical successes of bispecific antibody therapies have been reported and in 2009 catumaxomab (Removab) became the first approved bispecific antibody drug for the treatment of malignant ascites5. A number of recombinant strategies have been developed to synthesize bispecific antibodies, which include single chain variable fragment (scFv)-derived formats such as diabodies6, tandem diabodies7, BiTEs (bispecific T-cell engager)8, and DARTs (Dual Affinity Re-Targeting)9, as well as immunoglobulin G (IgG)-based formats such as Triomab10, DVD-Ig (Dual Variable Domain antibodies)11, and two-in-one antibody12. In addition, a number of chemical approaches have been developed which largely exploit the reactivity of lysine or cysteine residues within the antibody13,14. However, lysine modification often yields heterogeneous products due to multiple reactive surface lysines in an antibody; and while cysteine-based approaches are more selective, the reaction is complicated due to multiple disulfide bonds in the antibody molecule15. Recently a novel chemical strategy has been reported in which heterodimeric peptides with a branched azetidinone linker were fused to the antibody in a site-specific manner16,17. In this approach, however, antigen-specific ligands must first be developed, rather than directly utilizing the diverse pool of existing selective, high-affinity monoclonal antibodies. Herein we report a simple, high yielding, and general method to generate chemically-defined homogeneous bispecific antibodies.
Our strategy takes advantage of genetically encoded unnatural amino acids with orthogonal chemical reactivity relative to the canonical twenty amino acids to site-specifically modify antibody fragments18. Specifically, we used an evolved tRNA/aminoacyl-tRNA synthetase pair to site-specifically incorporate p-acetylphenylalanine (pAcF, Figure 1A) at defined sites in each of two Fab fragments19 in response to an amber nonsense codon. The mutant Fab fragments were then selectively coupled by a stable oxime bond to bifunctional linkers with an alkoxy-amine on one terminus and an azide or cyclooctyne group at the other (Figure 1B). In a second step, the two Fab-linker conjugates were linked to obtain the heterodimer through a copper-free [3+2] Huisgen-cycloaddition (“Click” reaction) (Figure 1C)20-22. This approach has a number of advantages over recombinant technologies and conventional coupling chemistries. For example, the use of bioorthogonal chemistries produces homogeneous, chemically-defined products; variable linker lengths and conjugation sites on the antibody can be easily optimized to ensure flexibility and good efficacy for each specific application; sequences from existing monoclonal antibodies can be directly adopted; and the modular approach easily and rapidly allows for combinatorial generation of diverse heterodimers (antibodies, enzymes, cytokines, etc.).
Figure 1.
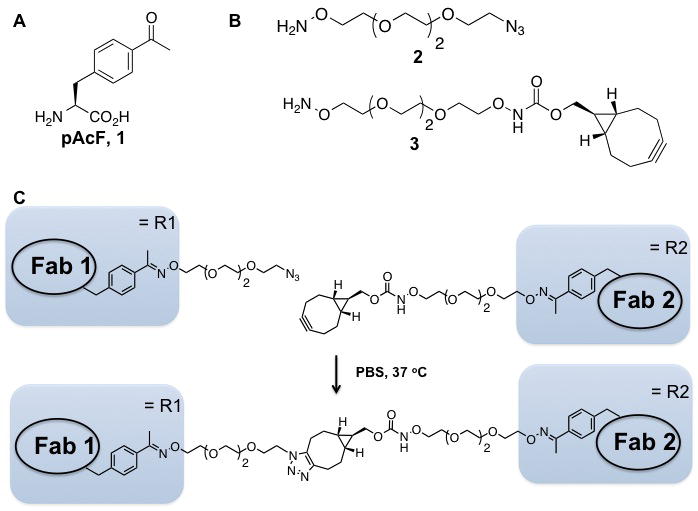
A. Structure of p-acetylphenylanine (pAcF, 1). B. Structure of bifunctional ethylene glycol linkers. C. General scheme for the generation of bispecific antibodies.
To test the feasibility of this approach, we first synthesized a homodimer of the Fab fragment of the HER2-specific antibody, trastuzumab (Herceptin; Roche/Genentech)23. The surface exposed residue Ser202 in the anti-HER2 Fab light chain (LC) was selected for pAcF incorporation because this residue is distal to the antigen-binding site and is not expected to affect binding or crosslinking. A LC-Ser202 amber mutant of the anti-HER2 Fab gene was inserted into the pBAD plasmid and cotransformed into E. coli DH10B strain with a plasmid containing a M. jannaschii mutant tRNA/aminoacyl-tRNA synthetase pair specific for pAcF (pEVOL-pAcF). Cells were grown in Luria-Bertani (LB) medium supplemented with 1 mM pAcF at 37 °C, and induced with 0.2 % arabinose.
The Fab mutants were purified by protein G chromatography (GE Healthcare) and analyzed by SDS-PAGE and ESI-MS (expected 47858 Da; observed 47855 Da. See Supporting Information for detailed expression and purification procedures). The mutant protein yielded 2 mg/L in shake flasks and 500 mg/L by high-density fermentation. Binding activity was comparable to wild type Fab as confirmed by enzyme-linked immunosorbent assay (ELISA) with the extracellular domain of HER2 (Fc conjugate) and detection with anti-human-kappa-HRP. Water soluble, flexible bifunctional crosslinkers were designed and synthesized to provide up to 40 Å distance between two covalently linked Fab fragments.
The bifunctional linkers (50-fold molar equivalents) were then coupled to the pAcF-containing anti-HER2 Fab (5 mg/mL) in excellent yields (>95 % by ESI-MS, See supporting information) in 100 mM acetate buffer pH 4.5 at 37 °C for 16 hours (Supplementary Figure 1). Excess linker was removed by an Amicon 10K concentrator column (Millipore) or by size exclusion chromatography (Superdex-75, GE Healthcare). The two Fab-linker conjugates were separately buffer exchanged into PBS, pH 7.4, then mixed at a 1:1 ratio at 10 mg/mL and incubated at 37 °C for the copper-free Click reaction. The reaction was monitored by SDS-PAGE, and a band at ∼100 kDa was observed, corresponding to the molecular weight of the Fab dimer. After 48 hours, about 70 % of starting material was consumed (Figure 2A, Supplementary Figure 2), and the homodimer was easily purified by sizeexclusion chromatography (Superdex-200, GE Healthcare) from the unreacted Fabs (Supplementary Figure 2).
Figure 2.
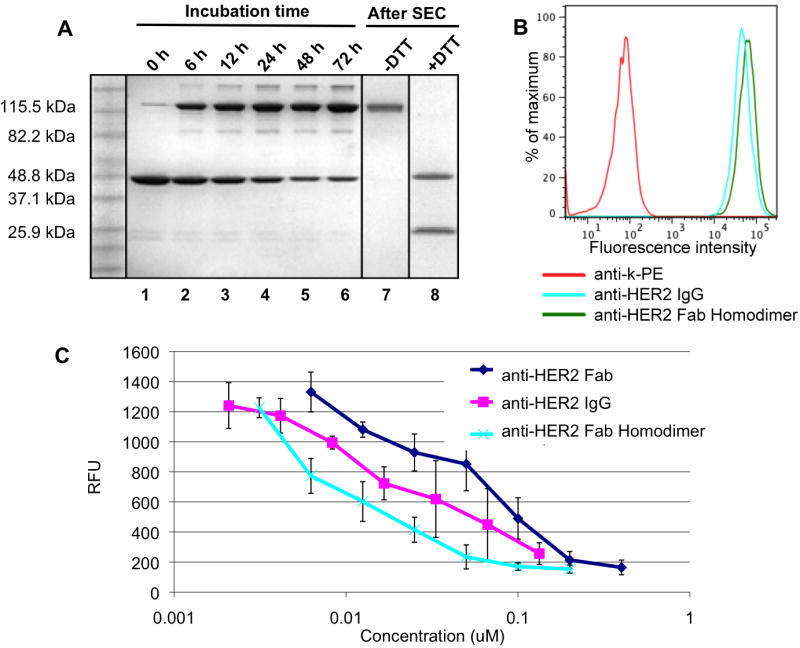
Time-course analysis of the ligation reaction and functional assays of the anti-HER2 Fab homodimer. A. Two anti-HER2 Fab fragments modified with distinct bifunctional linkers (azide or cyclooctyne) were co-incubated in PBS (10 mg/ml, 100 μL) at 37 °C. Aliquots were taken at different timepoints and analyzed by SDS-PAGE (lanes 1 - 6). After 72 hours, the reaction mixture was purified by size exclusion chromatography (SEC) and analyzed under non-reducing (lane 7) and reducing (lane 8) conditions, revealing bands at 25 kDa (free heavy chain) and 50 kDa (light chain dimer). All samples were analyzed by SDS-PAGE gel and stained with Coomassie brilliant blue. B. FACS-based binding assay of anti-HER2 Fab homodimer (100 nM, green), and IgG (100 nM, blue) to SK-BR-3 (HER2+) cells. Antihuman kappa-PE was used as secondary antibody. C. ELISA analysis of phosphorylated HER2 levels in SK-BR-3 cells using Human Phospho-ErBB2 DuoSet IC kit (R&D Systems).
We then assessed binding of the anti-HER2 homodimer to a HER2-positive breast-cancer cell line, SK-BR-3, by FACS (fluorescence-activated cell sorting) analysis and found the affinity to be comparable to full-length trastuzumab (Figure 2B). We further analyzed the ability of the homodimer to block HER2 receptor signaling in an ELISA-based HER2 phosphorylation assay (Human Phospho-ErbB2 DuoSet IC, R&D Systems) 24. The anti-HER2 Fab homodimer inhibits HER2 receptor phosphorylation more potently than anti-HER2 Fab alone and comparably to the bivalent full-length trastuzumab (Figure 2C).
We next applied this strategy to the synthesis of a heterodimeric bispecific antibody consisting of the anti-HER2 Fab linked to the Fab of monoclonal antibody UCHT1, which specifically binds human CD3 (cluster of differentiation 3) on CD8/CD3 positive cytotoxic T lymphocytes25. Such a construct is expected to recruit T lymphocytes to tumor cells and form a pseudo-immunological synapse that results in the activation of T lymphocytes and subsequent lysis of the target cells26-28. Indeed a genetically fused anti-CD19/anti-CD3 scFv Blinatumomab (MT103) has been generated by MicroMet and had shown efficacy in the treatment of patients with relapsed/refractory B-precursor acute lymphoblastic leukemia (ALL) in phase II clinical trials29. However, genetically fused scFvs can have short serum half lives and stability issues30. The ability to link two Fabs at any exposed amino acid position should allow one to optimize binding characteristics, synapse formation, stability, and pharmacokinetics of bispecific antibodies 31.
pAcPhe was substituted at HC-K138 in the anti-CD3 antibody, UCHT1. This site is distal to the antigen binding site and, when conjugated to the LC-S202 mutant anti-HER2 Fab with the same polyethylene glycol linker used above, should be long and flexible enough to allow the resulting bispecific antibody to productively bind both a CD3 positive T-cell and the HER2 positive target cell simultaneously. UCHT1 Fab was expressed in E. coli by the same method as described for anti-HER2 Fab, and the cyclooctyne linker-modified anti-CD3 was prepared as described above in >95 % yield as confirmed by ESI-MS (Supplementary Figure 1). The anti-CD3-Fabcyclooctyne conjugate was then coupled to anti-HER2-Fabazide conjugate as described above in 70 % percent yield (determined by SDS-PAGE and chromatographic separation) and purified from unreacted Fab monomers by Superdex-200 size exclusion column (Supplementary Figure 3). FACS analysis of the Fab heterodimer showed affinity for both HER2-positive (SK-BR-3) and CD3-positive (Jurkat) cells with no cross-reactivity of anti-CD3 Fab and anti-HER2 Fab, respectively (Figure 3A). To demonstrate the simultaneous binding of the heterodimer to the two antigens, we visualized cross-linking of fluorescently labeled SK-BR-3 and Jurkat cells. Specifically, SK-BR-3 cells and Jurkat cells were first stained with Mito Tracker Red (Invitrogen) and carboxyfluorescein succinimidyl ester (CFSE, Invitrogen), respectively. The labeled Jurkat cells were incubated with 100 nM of the anti-HER2/anti-CD3 heterodimer in RPMI media supplemented with 10% FBS (fetal bovine serum) at 4 °C for 30 minutes, and excess conjugate was washed away before mixing the Jurkat cells with SK-BR-3 cells. A 1:1 mixture of unconjugated anti-CD3 and anti-HER2 Fabs was used as a negative control under the same labeling conditions. The cells were incubated at 37 °C for 6 hours, allowing the adherent SK-BR-3 cells to attach to the plate; excess Jurkat cells were removed by gentle washing with media. In the sample with the heterodimer, significantly more Jurkat cells were bound to the SK-BR-3 cells after washing compared to cells incubated with the mixture of unconjugated Fabs (Figure 3B), confirming heterodimer-mediated cell-cell interaction.
Figure 3.
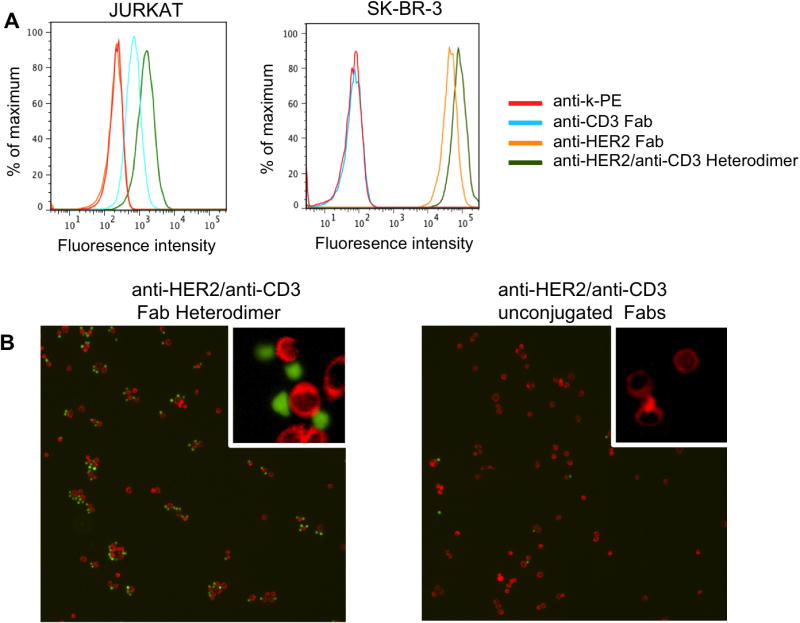
Bispecific binding of anti-HER2/anti-CD3 Fab heterodimer. A. FACS-based binding assay. Primary antibodies (100nM) were incubated with Jurkat (CD3+) and SK-BR-3 (HER2+) cell lines in PBS, and then detected by the secondary antibody (anti-human kappa-PE). B. (Left) Fluorescence microscope images of the interaction between SK-BR-3 (red) cells and Jurkat cells (green) in the presence of the conjugated anti-HER2/anti-CD3 heterodimer. (Right) A mixture of unconjugated anti-HER2 Fab and anti-CD3 Fab at 1:1 ratio was used as a negative control.
Next, we demonstrated that the heterodimer could recruit T cells to kill the target cancer cells in an in vitro cytotoxicity assay. Human PBMCs (peripheral blood mononuclear cells) were purified with Ficoll (GE Healthcare) from fresh blood of healthy donors and mixed with target cells, HER2-transfected MDA-MB-43532, at an effector to target cell ratio of 10 to 1 (1×105 to 1×104 cells) in RPMI media supplemented with 10 % FBS. Non-transfected MDA-MB-435 cells were used as an isogenic negative control. A 1:1 mixture of unconjugated anti-HER2 and anti-CD3 Fabs were used as an additional negative control to further demonstrate that cytotoxicity was due to bispecific antibody-based interactions. After incubation for 16 hours at 37 °C, the amount of LDH (lactate dehydrogenase) was measured from lysed cells as an indicator of cytotoxicity9. As shown in Figure 4, lysis of HER2 positive target cells was observed in a dose dependent manner only when the conjugated anti-HER2/anti-CD3 heterodimer was present; the half maximal effective concentration (EC50) was ∼20 pM. HER2-negative MDA-MB-435 cells were not affected by the hetero-dimer, and neither cell line was affected by the unconjugated Fab mixture. Additionally, cell lysis and formation of aggregates could be directly observed under a microscope in the heterodimer-present HER2 positive cell sample, but not in any of the controls (Supplementary Figure 4).
Figure 4.
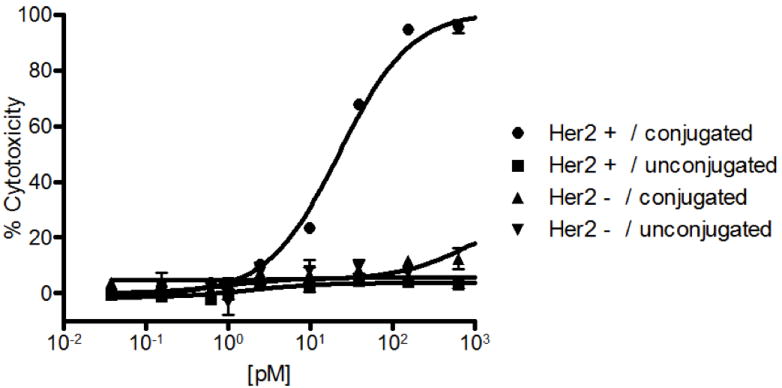
Dose-dependent cytotoxicity with MDA-MB-435/HER2+ cells in the presence of human PBMCs and antibody heterodimer. Different concentrations of anti-HER2/anti-CD3 Fab heterodimer (circles), or a 1:1 mixture of unconjugated anti-HER2 Fab and anti-CD3 Fab (square) were incubated with MDA-MB-435/HER2+ cells. Heterodimer (upward triangle) or a mixture (downward triangle) of anti-HER2 Fab and anti-CD3 Fab were incubated with MDA-MB-435/HER2- cells as negative controls. After 17 hours of incubation at 37 °C and 5 % CO2, cytotoxicity was measured by levels of LDH (lactate dehydrogenase) release from lysed cells using the Cytotox 96 Nonradioactive Cytotoxicity Assay Kit (Promega). In separate wells, MDA-MB-435/HER2+ or MDA-MB-435/HER2- cells with no PBMCs were incubated and lysed using the lysis buffer (provided in the assay kit) as maximum cytotoxicity controls. The absorbance at 490 nm was recorded using SpectraMax 250 plate reader (Molecular Devices Corp.). Percent cytotoxicity was calculated by: % Cytotoxicity = (Absorbanceexpt − Absorbancespontaneous average)/(Absorbancemax average − Absorbancespontaneous average).
In conclusion, we have demonstrated the rapid construction of bispecific antibodies by the site-specific incorporation of unnatural amino acids with orthogonal chemical reactivity. Two different conjugates, a bivalent anti-HER2-Fab homodimer and a bispecific anti-HER2/anti-CD3 Fab heterodimer, were synthesized and shown to have excellent in vitro activity. This method provides highly homogeneous products after two simple purification steps in good overall yield. Moreover, because each antibody fragment was directly derived from parent monoclonal antibodies, they retain the original immunoglobulin fold without loss of binding affinity or stability. Finally, we have shown that an anti-HER2/anti-CD3 bispecific antibody can induce targeted cell lysis in the presence of effector cells at picomolar concentrations in vitro, which is comparable to the most potent bispecific formats reported to date2.
Supplementary Material
Acknowledgments
This work was supported by the Skaggs Institute for Chemical Biology and NIH grant R01 GM062159 (P. G. S) and National Research Foundation of Korea Grant NRF-2009-352-C00079 (C. H. K). This is manuscript no. 21767 of The Scripps Research Institute.
Footnotes
Supporting Information. Additional experimental methods and Figure S1-S4 as mentioned in text. This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Holmes D. Nat Rev Drug Discov. 2011;10:798. doi: 10.1038/nrd3581. [DOI] [PubMed] [Google Scholar]
- 2.Chames P, Baty D. MAbs. 2009;1:539. doi: 10.4161/mabs.1.6.10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan AC, Carter PJ. Nat Rev Immunol. 2010;10:301. doi: 10.1038/nri2761. [DOI] [PubMed] [Google Scholar]
- 4.Carter P. Nat Rev Cancer. 2001;1:118. doi: 10.1038/35101072. [DOI] [PubMed] [Google Scholar]
- 5.Heiss MM, Murawa P, Koralewski P, Kutarska E, Kolesnik OO, Ivanchenko VV, Dudnichenko AS, Aleknaviciene B, Razbadauskas A, Gore M, Ganea-Motan E, Ciuleanu T, Wimberger P, Schmittel A, Schmalfeldt B, Burges A, Bokemeyer C, Lindhofer H, Lahr A, Parsons SL. Intl J Cancer. 2010;127:2209. doi: 10.1002/ijc.25423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holliger P, Winter G. Cancer Immunol Immunother. 1997;45:128. doi: 10.1007/s002620050414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cochlovius B, Kipriyanov SM, Stassar MJ, Schuhmacher J, Benner A, Moldenhauer G, Little M. Cancer Res. 2000;60:4336. [PubMed] [Google Scholar]
- 8.Baeuerle PA, Reinhardt C. Cancer Res. 2009;69:4941. doi: 10.1158/0008-5472.CAN-09-0547. [DOI] [PubMed] [Google Scholar]
- 9.Moore PA, Zhang W, Rainey GJ, Burke S, Li H, Huang L, Gorlatov S, Veri MC, Aggarwal S, Yang Y, Shah K, Jin L, Zhang S, He L, Zhang T, Ciccarone V, Koenig S, Bonvini E, Johnson S. Blood. 2011;117:4542. doi: 10.1182/blood-2010-09-306449. [DOI] [PubMed] [Google Scholar]
- 10.Jager M, Schoberth A, Ruf P, Hess J, Lindhofer H. Cancer Res. 2009;69:4270. doi: 10.1158/0008-5472.CAN-08-2861. [DOI] [PubMed] [Google Scholar]
- 11.Wu C, Ying H, Grinnell C, Bryant S, Miller R, Clabbers A, Bose S, McCarthy D, Zhu RR, Santora L, Davis-Taber R, Kunes Y, Fung E, Schwartz A, Sakorafas P, Gu J, Tarcsa E, Murtaza A, Ghayur T. Nat Biotechnol. 2007;25:1290. doi: 10.1038/nbt1345. [DOI] [PubMed] [Google Scholar]
- 12.Schaefer G, Haber L, Crocker LM, Shia S, Shao L, Dowbenko D, Totpal K, Wong A, Lee CV, Stawicki S, Clark R, Fields C, Lewis Phillips GD, Prell RA, Danilenko DM, Franke Y, Stephan JP, Hwang J, Wu Y, Bostrom J, Sliwkowski MX, Fuh G, Eigenbrot C. Cancer Cell. 2011;20:472. doi: 10.1016/j.ccr.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 13.Graziano RF, Guptill P. Methods Mol Biol. 2004;283:71. doi: 10.1385/1-59259-813-7:071. [DOI] [PubMed] [Google Scholar]
- 14.Brennan M, Davison PF, Paulus H. Science. 1985;229:81. doi: 10.1126/science.3925553. [DOI] [PubMed] [Google Scholar]
- 15.Junutula JR, Bhakta S, Raab H, Ervin KE, Eigenbrot C, Vandlen R, Scheller RH, Lowman HB. J Immunol Methods. 2008;332:41. doi: 10.1016/j.jim.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 16.Gavrilyuk JI, Wuellner U, Salahuddin S, Goswami RK, Sinha SC, Barbas CF., 3rd Bioorg Med Chem Lett. 2009;19:3716. doi: 10.1016/j.bmcl.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doppalapudi VR, Huang J, Liu D, Jin P, Liu B, Li L, Desharnais J, Hagen C, Levin NJ, Shields MJ, Parish M, Murphy RE, Del Rosario J, Oates BD, Lai JY, Matin MJ, Ainekulu Z, Bhat A, Bradshaw CW, Woodnutt G, Lerner RA, Lappe RW. Proc Nat Acad Sci USA. 2010;107:22611. doi: 10.1073/pnas.1016478108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L, Xie J, Schultz PG. Annu Rev Biophys Biomol Struct. 2006;35:225. doi: 10.1146/annurev.biophys.35.101105.121507. [DOI] [PubMed] [Google Scholar]
- 19.Wang L, Zhang Z, Brock A, Schultz PG. Proc Nat Acad Sci USA. 2003;100:56. doi: 10.1073/pnas.0234824100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dommerholt J, Schmidt S, Temming R, Hendriks LJA, Rutjes FPJT, van Hest JCM, Lefeber DJ, Friedl P, van Delft FL. Angew Chem Int Ed Engl. 2010;49:9422. doi: 10.1002/anie.201003761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bundy BC, Swartz JR. Bioconjug Chem. 2010;21:255. doi: 10.1021/bc9002844. [DOI] [PubMed] [Google Scholar]
- 22.Hudak JE, Barfield RM, de Hart GW, Grob P, Nogales E, Bertozzi CR, Rabuka D. Angew Chem Int Ed Engl. 2012;51:4161. doi: 10.1002/anie.201108130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho HS, Mason K, Ramyar KX, Stanley AM, Gabelli SB, Denney DW, Leahy DJ. Nature. 2003;421:756. doi: 10.1038/nature01392. [DOI] [PubMed] [Google Scholar]
- 24.Mukherji M, Brill LM, Ficarro SB, Hampton GM, Schultz PG. Biochemistry. 2006;45:15529. doi: 10.1021/bi060971c. [DOI] [PubMed] [Google Scholar]
- 25.Zhu Z, Carter P. J Immunol. 1995;155:1903. [PubMed] [Google Scholar]
- 26.Staerz UD, Kanagawa O, Bevan MJ. Nature. 1985;314:628. doi: 10.1038/314628a0. [DOI] [PubMed] [Google Scholar]
- 27.Offner S, Hofmeister R, Romaniuk A, Kufer P, Baeuerle PA. Mol Immunol. 2006;43:763. doi: 10.1016/j.molimm.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 28.Haas C, Krinner E, Brischwein K, Hoffmann P, Lutterbüse R, Schlereth B, Kufer P, Baeuerle PA. Immunobiology. 2009;214:441. doi: 10.1016/j.imbio.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 29.Bargou R, Leo E, Zugmaier G, Klinger M, Goebeler M, Knop S, Noppeney R, Viardot A, Hess G, Schuler M, Einsele H, Brandl C, Wolf A, Kirchinger P, Klappers P, Schmidt M, Riethmüller G, Reinhardt C, Baeuerle PA, Kufer P. Science. 2008;321:974. doi: 10.1126/science.1158545. [DOI] [PubMed] [Google Scholar]
- 30.Worn A, Pluckthun A. J Mol Biol. 2001;305:989. doi: 10.1006/jmbi.2000.4265. [DOI] [PubMed] [Google Scholar]
- 31.Hutchins BM, Kazane SA, Staflin K, Forsyth JS, Felding-Habermann B, Schultz PG, Smider VV. J Mol Biol. 2011;406:595. doi: 10.1016/j.jmb.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hutchins BM, Kazane SA, Staflin K, Forsyth JS, Felding-Habermann B, Smider VV, Schultz PG. Chem biol. 2011;18:299. doi: 10.1016/j.chembiol.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


