Summary
Somatic mutations occur during brain development and are increasingly implicated as a cause of neurogenetic disease. However, the patterns in which somatic mutations distribute in the human brain are unknown. We used high-coverage whole-genome sequencing of single neurons from a normal individual to identify spontaneous somatic mutations as clonal marks to track cell lineages in human brain. Somatic mutation analyses in >30 locations throughout the nervous system identified multiple lineages and sub-lineages of cells marked by different LINE-1 (L1) retrotransposition events and subsequent mutation of poly-A microsatellites within L1. One clone contained thousands of cells limited to the left middle frontal gyrus, whereas a second distinct clone contained millions of cells distributed over the entire left hemisphere. These patterns mirror known somatic mutation disorders of brain development, and suggest that focally distributed mutations are also prevalent in normal brains. Single-cell analysis of somatic mutation enables tracing of cell lineage clones in human brain.
Introduction
Somatic mutations, occurring during or after the mitotic cell divisions that generate the body, cause not only cancer, but also diverse neurologic diseases, including cortical malformations, epilepsy, intellectual disability, and neurodegeneration (Poduri et al., 2013). Somatic mutations also remain an important, unexplored possible etiology of other neuropsychiatric diseases (Insel, 2014). In contrast to inherited mutations, somatic mutations cause disease depending not only on their effects on gene function, but also on the time, place, and cell lineage during development at which they occur (Frank, 2010). Therefore, pathogenic somatic mutations pose a challenge because of the variety of ways their effects are shaped by normal development. Systematic tracing of the patterns of distribution of clonally related cells in human brain has not been possible, relying instead on extrapolation from animal models and in vitro studies (Clowry et al., 2010). Knowledge of these patterns, in conjunction with systematic measurement of somatic mutation rates in the brain (Evrony et al., 2012; McConnell et al., 2013; Cai et al., 2014), is crucial to understand how somatic mutations might cause disease by impairing circuit function and their potential role in the large unexplained burden of neuropsychiatric disease.
Somatic mutations also present an opportunity to study the developmental processes that create the human brain. Marking all progeny of a specific cell or population of cells is a central tool of developmental biology, revealing patterns of progenitor proliferation, migration, and differentiation (Kretzschmar and Watt, 2012). Existing tools to mark cell lineages, such as retroviral tracers and genetic and fluorescent markers, have uncovered key aspects of brain development in model organisms (Franco and Muller, 2013; Marin and Muller, 2014), but are invasive and cannot be applied to human tissue in vivo. Somatic mutations, however, occur spontaneously and possess the key features required of lineage markers: a) they are inherited by all descendant cells; and b) they are not transferred between cells. Retrotransposon mutations in particular have been shown to occur in mouse brain in vivo (Muotri et al., 2005) and human neuronal progenitors in vitro (Coufal et al., 2009), and are detectable in human brain (Baillie et al., 2011; Evrony et al., 2012; Reilly et al., 2013). Retrotransposons also have unique sequence structures that make each insertion differentiable from other insertions (Goodier and Kazazian, 2008), enabling detection even at low mosaicism and suggesting they could be used as non-invasive cell lineage markers in human brain.
Here we show that single-neuron, high-coverage whole-genome sequencing (WGS), along with profiling of all active retrotransposon families and further single-molecule somatic mutation analyses, can identify and leverage somatic mutations as tags to identify unexpected spatial patterns of cell lineages in the human brain. Our data show a proof of principle that clonal patterns defined by somatic retroelement insertions and mutations of associated repeat sequences delineate patterns of lineage resembling those defined in animal models while enabling study of human-specific features, and suggest that deep analysis of the gamut of somatic mutations will allow a systematic reconstruction of key features of lineage patterns in the human brain.
Results
High-coverage whole-genome sequencing of single neuronal genomes
We selected 16 single neuronal genomes for high-coverage WGS from a population of large neuronal nuclei from the left middle frontal gyrus of the dorsolateral prefrontal cortex of a neurologically normal individual (UMB1465). These genomes were amplified by multiple-displacement amplification (MDA) (Dean et al., 2002) as part of a prior targeted study of LINE-1 (L1) retrotransposition (Evrony et al., 2012). WGS at a genome-wide average read depth of 42x achieved coverage of 98±0.5% of the genome at ≥1x and 81±2% at ≥10x read depth on average (±SD) across all single neurons (Figures 1A–B; Tables S1–S2), consistent with prior estimates of MDA locus dropout measured by targeted genotyping (Evrony et al., 2012) and WGS of MDA-amplified single cancer cells (Hou et al., 2012). Single neurons showed highly consistent sequencing quality, genome read alignment, and genome coverage (Figure S1; Tables S1–S2). Sequencing and alignment metrics were generally similar to WGS of unamplified bulk DNA from cortex and heart, although, as seen in prior single-cell studies (Evrony et al., 2012; Hou et al., 2012; Voet et al., 2013), MDA samples showed systematic and mostly correctable biases in genome coverage due to GC-sequence content (Figure 1C and S2–S5; Table S2;Supplemental Note 1). Compared to single cells amplified by the MALBAC method in a prior study (Zong et al., 2012), MDA achieves improved overall genome coverage, as well as more even amplification at smaller scales (< 50 kb) necessary for reliable detection of sequence variants such as retrotransposons insertions (Figures S1, S6, S7; Table S2; Supplemental Note 1). On the other hand, MALBAC shows more even and reproducible coverage at larger scales (Figures S6–S7; Supplemental Note 1), consistent with its better performance in detecting large copy-number variants (Hou et al., 2013). Our high-coverage single-cell WGS dataset, the most extensive to date, provided an opportunity for additional detailed analyses of single-cell MDA performance, including in-depth investigation of genome coverage, GC-sequence bias, comparisons to other publicly available single-cell datasets, and MDA chimeras (stochastic false positive structural variants created during amplification). These comprehensive analyses are presented in Supplemental Note 1 (see also Figures S1–S9; Tables S1–S2) to aid future single-cell genomics research in understanding mechanisms of single-cell genome amplification and developing improved amplification methods (Blainey and Quake, 2014).
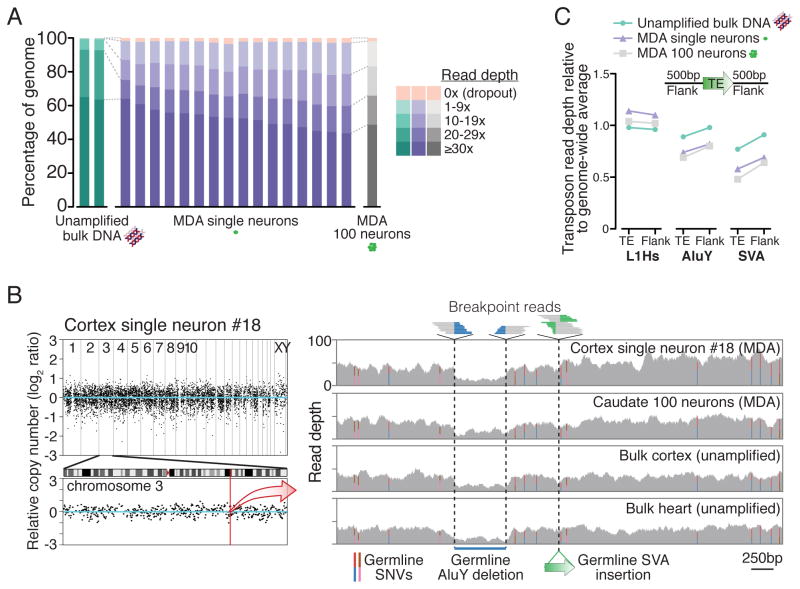
Figure 1. Single-neuron WGS genome coverage.
(A) Percentage of genome covered at specified read depths for all samples in this study. (B) Representative genome-wide coverage plot of single neuron #18 in ~500kb bins, with enlargement of chromosome 3 in which L1#2 was identified (Figure 2). See Figure S5 for plots of all samples. Right panel zoom of a 5kb region (chr3:147,605,344–147,610,345; hg19) shows single-neuron WGS simultaneous detection of 3 types of germline mutations, all concordant with 100-neuron and bulk samples: single nucleotide variants (SNVs, colored bars represent ratios of allele reads), a deletion (578bp AluY and additional flanking sequence), and an SVA retrotransposon insertion. All mutations have been previously identified in public polymorphism databases. (C) Average read depth of retrotransposon insertions annotated in the human genome reference and their 500bp flanks, relative to the genome-wide average read depth. Relative read depths correlate with average GC-content of each retrotransposon family (L1Hs: 42%; AluY: 54%; SVA: 63% GC content), due to GC amplification bias of MDA (see Table S2 and Supplemental Note 1).
See also Figures S1–S9 and Tables S1–S2.
Somatic retrotransposon insertion analysis with the single-cell Transposable element analyzer
We searched for somatic retrotransposon insertions deriving from all major active retrotransposon families (AluY, L1Hs, and SVA) using scTea (single-cell Transposable element analyzer), a pipeline based on the Tea method originally developed for detection of somatic insertions in tumor samples (Lee et al., 2012). scTea incorporates significant additional features and improvements for single-cell analysis including identification of true insertions with high sensitivity and specificity (Figures 2A–B and S10–S11; see Supplemental Experimental Procedures for details). scTea achieves sensitivity of 95%, 96%, and 86% in detection of AluY, L1Hs, and SVA insertions, respectively, that are absent from the human genome reference (non-reference insertions) in simulations generated from the only Sanger-sequenced diploid genome (HuRef) (Figure S10C). Specificity of AluY, L1Hs, and SVA bulk DNA insertion calls estimated by PCR and Sanger sequencing validation of 80 randomly selected insertion candidates from bulk DNA WGS of individual 1465 was 97%, 100%, and 100%, respectively (Table S3). In single-neuron genomes, scTea detected an average of 805 AluY, 131 L1Hs, and 17 SVA germline non-reference insertions (i.e. insertions also found in bulk samples of the individual), of which 708, 117, and 9, on average were ‘known’ insertions independently detected by prior population studies of retrotransposon polymorphism (Figure 2A). scTea achieved a single-neuron sensitivity of 74%, 79%, and 62% for AluY, L1Hs, and SVA, respectively, using the high-confidence ‘known’ germline insertions of the individual as a reference (Figures S10D–E; see Supplemental Experimental Procedures for details).
Figure 2. Single-neuron WGS analysis identifies somatic retrotransposon insertions.
(A) Average number of germline non-reference insertions (i.e. present in either bulk cortex or heart) detected per sample with an scTea-calculated score ≥9 (see Supplemental Experimental Procedures for details on score calculation). Error bars (SD). The number of ‘known’ insertions (reported in public databases and prior population studies of retrotransposon polymorphism) are shaded in darker color. (B) Score distribution of all AluY, L1Hs, and SVA germline known non-reference insertions (upper panel, n=13,849 insertions) and somatic calls (lower panel, i.e. calls absent in bulk heart) detected across all 16 single neurons. Dashed line indicates score threshold ≥9 used to call somatic candidates, with bars below threshold drawn in lighter color. Note the distribution of known insertions at higher scores compared to somatic calls at lower scores that arise from low-level MDA chimeras below threshold. (C) Whole-genome sequencing reads detecting L1#1. Main panel shows fully-aligned reads whose pairs aligned to L1, with darker and lighter colors indicating plus and minus orientations, respectively. Smaller panels on right show partially-aligned (breakpoint) reads spanning the insertion breakpoint, detecting the TSD and poly-A tail. Mismatched bases relative to the human genome reference are colored. (D) Whole-genome sequencing reads detecting L1#2. A few partially-aligned reads distant from the insertion site are chimeras. (E) Schematic of L1#2 and its source L1. (F) Full-length PCR validation and cloning of L1#2. (G) Representative gel from a 3′PCR screen for L1#2 in single neurons.
See also Figures S10–S12 and Table S3.
Analysis of the 16 single-neuron genomes with scTea identified 18 somatic insertion candidates (Figure 2B; Table S3). The 4 highest scoring candidates were 2 L1Hs insertions, each identified in 2 neurons: L1#1 in neurons 2 and 77 and L1#2 in neurons 6 and 18 (Figures 2B–D), and were the only candidates with convincing in silico evidence on manual review of WGS data (Table S3). Follow-up evaluation of all 18 candidates by independent PCR assays validated only these 4 candidates. Remarkably, L1#1 was the same somatic insertion on chromosome 15 previously identified by targeted L1 insertion profiling (L1-IP) in the same 2 neurons (2 and 77) (Evrony et al., 2012). This represents important validation of L1-IP (Evrony et al., 2012) by an entirely independent sequencing method, dataset, and analysis pipeline.
Full-length cloning of L1#2 revealed that like L1#1, it showed all the hallmarks of a bona fide retrotransposition event (target site duplication [TSD] and poly-A tail), but also showed truncation, inversion, and a long 3′ transduction (614 bp) identifying the source L1 on chromosome 13 (Figures 2E–F and S12A–B; Table S3). The site of insertion was in an intergenic region far away from any obvious transcribed gene, strongly suggesting that this L1 does not alter the function of any nearby gene. Its long 3′ transduction, which occurs infrequently during retrotransposition (< 5% of insertions transduce >500 bp) (Goodier et al., 2000; Pickeral et al., 2000; Xing et al., 2006), is longer than the DNA fragments amplified by L1-IP, explaining why the insertion was not identified by L1-IP. Additional 3′-junction PCR (3′PCR) screening of a large set of single cells from the individual identified L1#2 in 13 of 587 single cortical neuron genomes, but not in 59 single caudate neuron genomes or 68 single cerebellar neuron genomes (Figures 2G and S12C–D). Intriguingly, the source element for L1#2 on chromosome 13 was not active in previous in vitro assays (Brouha et al., 2003), suggesting that in vivo retrotransposon activity may differ from in vitro estimates and highlighting how single-cell studies can reveal in vivo activity of source elements. WGS analysis of single neurons was consistent with our previous targeted L1-IP (Evrony et al., 2012) and our prior estimate of low rates of L1 retrotransposition in the cerebral cortex (with 12/16 single neurons lacking validated insertions and two validated insertions each shared by two clonally related cells), and extends these results to find no evidence of Alu and SVA retrotransposition in the 16 sequenced single neurons from this normal individual. These results illustrate the advantage of single-cell WGS by its ability to analyze all retrotransposon families simultaneously and to recover somatic insertions that elude targeted sequencing approaches.
Tracing spatial distributions of progenitor lineages in human brain
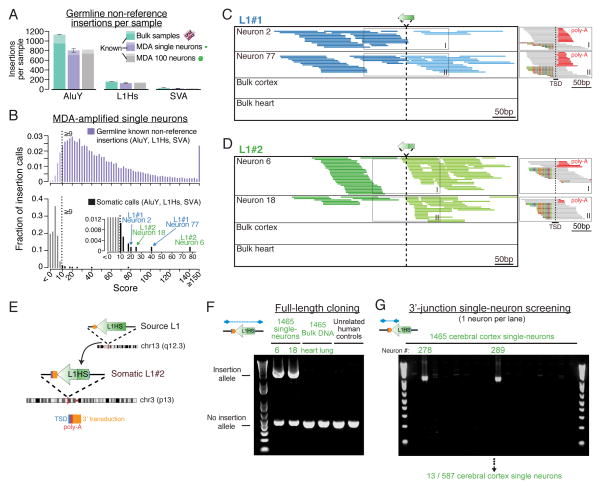
A custom droplet digital PCR (ddPCR) assay with single-copy sensitivity (Figure S13) allowed quantification of the mosaicism (% of cells) and distribution of the two somatic L1s in unamplified (‘bulk’) DNA extracted from frozen tissues from 32 regions across the left cerebral cortical hemisphere, left caudate, left cerebellum, and spinal cord (Figure S14; Table S4); the right hemisphere was formalin-fixed and therefore studied by a different nested PCR assay (see Supplemental Experimental Procedures). Remarkably, L1#1 was detected only in 5 adjacent locations in the left middle frontal gyrus of the cortex, spanning a region ≈ 2×1cm in size, and showing an average mosaicism of 0.09% (range: 0.04%–0.22%) (Figures 3 and S14–S15; Table S4). Absolute copy number quantification by ddPCR further estimated that at least 2,200 cells harbored L1#1 in our DNA samples, extrapolating to likely no more than fifty thousand cells total in the cortex (see Supplemental Experimental Procedures). L1#1 was not detected in non-neuronal cells sorted from the left middle frontal gyrus, nor in multiple caudate, cerebellum, spinal cord, right cortex, heart, lung, and liver samples, illustrating the assay’s specificity and ability to detect ultra-low mosaicism. The localized spatial distribution and very low mosaicism strongly suggest that the insertion marked a neocortical progenitor of the left middle frontal gyrus giving rise to mostly if not exclusively neurons.
Figure 3. Mosaicism of somatic L1 insertions measured by ddPCR.
(A) Representative ddPCR plots of L1#1 and L1#2. L1+ droplets are plotted with larger points for better visualization. Reduced L1 signal in double-positive versus single-positive droplets is due to relatively higher PCR efficiency of RNaseP amplicons. (B) Mosaicism levels measured in individual UMB1465, plotted on a representative brain, using logarithmic box plots to indicate level of mosaicism. Empty rectangles indicate no detection. Blue shading indicates the estimated distribution of L1#1 in the middle frontal gyrus. The 16 WGS single neurons were originally obtained from location D (underlined). (C) Lucida tracings of cortex sections 2, 3, and 4 in which L1#1 was found, traced from photographs of sections. Dashed lines indicate regions that were not present in photographs of sections due to sampling prior to this study. Anatomy of these regions was extrapolated based on records of sampled locations, adjacent sections, photographs of right hemisphere formalin-fixed sections, and atlases of normal brain anatomy. Locations in which L1#1 was detected are highlighted in blue. See Figure S14 for diagrams of all sampled brain sections.
See also Figures S13–S15 and Table S4.
In contrast to L1#1, L1#2 was detected in every sample of the left cerebral cortex and caudate nucleus tested, though at very low and highly variable mosaicism (cortex average 0.4%, range 0.01–1.7%) (Figures 3 and S15; Table S4). While it is not possible to estimate the total number of cells harboring L1#2 without assaying the entire brain, extrapolation from assayed regions suggests that L1#2’s lineage encompasses tens to hundreds of millions of cells. L1#2 was also detected in sorted non-neuronal cells and at extremely low levels in left cerebellum, but not in formalin-fixed tissue of the right hemisphere, nor in spinal cord, heart, lung or liver (Figures 3 and S15; Table S4), suggesting it mobilized considerably earlier in nervous system development than L1#1 and in a progenitor for both neurons and glia.
Poly-A tails of somatic insertions are highly mutable and mark sub-lineages
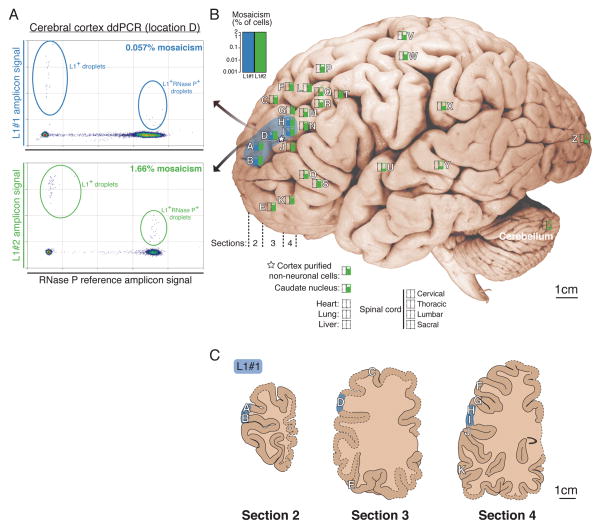
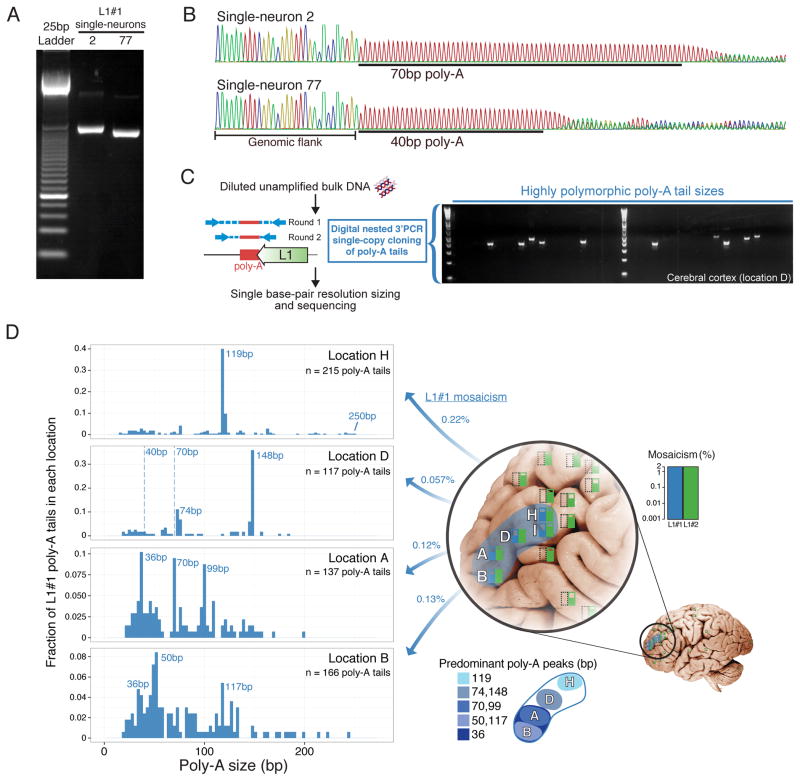
3′PCR validation data for L1#1 in neurons 2 and 77 suggested that the insertion was slightly different in size in each neuron (Figure 4A), which reflected unexpected secondary mutations in the poly-A tail of the L1 sequence. The difference in size was initially surprising, as L1#1 was inherited by both neurons from a single event in a shared progenitor, as confirmed by identical breakpoints, TSD, and transduction sequences in both neurons (Evrony et al., 2012). Comparison of L1#1’s sequence in the two neurons revealed that the poly-A tail, which was reverse transcribed into the genome from the original retrotransposon transcript’s poly-A tail and shown before to be a highly mutable sequence element (Grandi et al., 2012), was longer in neuron 2 (70 bp) compared to neuron 77 (40 bp) (Figure 4B), fully accounting for the difference in insertion size. This suggested that the poly-A sequence of L1#1 underwent somatic mutation in descendant cells after the original insertion event. Using a digital nested 3′-junction PCR assay (dnPCR) with near 100% sensitivity and specificity in cloning single copies of L1#1’s poly-A tail directly from unamplified bulk DNA (Figure S16A), we found that the poly-A tail was highly polymorphic (Figure 4C), indicating it mutated somatically many times.
Figure 4. Somatic retrotransposon insertion poly-A tail polymorphism reveals sub-lineages across brain regions.
(A) L1#1 3′PCR products differ in size between single-neurons 2 and 77. (B) Sanger sequencing of L1#1 in single neurons 2 and 77 reveals poly-A tail polymorphism. (C) Brief schematic of dnPCR single-copy cloning of poly-A tails directly from unamplified bulk DNA, and a representative gel showing highly polymorphic product sizes. Poly-A tail lengths were precisely measured by capillary electrophoresis. (D) L1#1 poly-A tail size distributions (in 3 bp bins) obtained by dnPCR shows peaks representing sub-lineages and marked variability within and between locations. Dashed lines mark poly-A tail sizes of single-neurons 2 and 77 sorted from location D. Location I not shown, since due to extremely low mosaicism only 4 poly-A tails were cloned.
See also Figures S16–S19 and Table S5.
Profiling the lengths of many L1#1 poly-A tails (n=639) from locations where L1#1 was found revealed striking differences in poly-A size distributions between locations, including distinct peaks marking subset lineages of cells, as well as additional highly variable poly-A tail lengths at lower levels indicating frequent somatic mutation (Figure 4D; Table S5). Importantly, dnPCR of control poly-A tails of known lengths (Figure S16B) and reproducibility of peaks across tissues (Figure S17) shows that dnPCR reliably measures poly-A tail lengths with a precision up to ±1bp (see Supplemental Experimental Procedures for details). Overall, these results suggest that the poly-A tail of L1#1’s originating retrotransposition event may have been >200bp long (Figure 4D; and see Supplemental Note 2 for further discussion), and that in subsequent descendant cells, in vivo somatic mutation generated widely varying poly-A lengths marking distinct sub-lineages — a striking example of ‘nested somatic mutation’. Moreover, the distinct distributions of poly-A tails in each location (Figure 4D) is consistent with migration of a subset of progenitors each with a different distribution of poly-A tails.
L1#2’s poly-A tail showed less polymorphism. Only 1 of the 13 single neurons with the L1#2 insertion showed a large difference in poly-A size (Table S5), and dnPCR profiling of >1,500 poly-A tails across 12 locations in the cortex, caudate, and cerebellum showed some variability, though much less than L1#1 (Figure S17; Table S5). The different poly-A mutation rates of L1#1 and L1#2 may reflect a difference in poly-A size of the original insertion, regional genomic variability in mutation rates, timing of the insertion during development, or epigenetic effects on microsatellite and somatic mutation rates (Kim et al., 2013).
Notably, smaller clonal sets of cells carrying L1#1 with similar poly-A lengths appear to occupy smaller zones of the middle frontal gyrus (Figure 4D; Table S5). For example, cells carrying L1#1 marked by poly-A tails 118–120bp in length (110/639 cells) were limited to location H of the middle frontal gyrus (104/110 = 95% of cells). Similarly, cells carrying 147–149bp poly-A tails (46/639 cells) were found predominantly in location D (42/46 = 91% of cells), with the remaining cells in adjacent locations, while 99–100bp poly-A tails (13/639 cells) were found predominantly in location A (12/13 = 92% of cells; 1/13 cells in adjacent location B). We interpret cells carrying the same poly-A length as sub-lineages, defined by poly-A mutations, that are offspring of the original progenitor in which L1#1 inserted. The distribution of these “sub-lineages” suggests that tangential dispersion becomes progressively restricted in later generations of neocortical progenitor lineages, though even these sub-lineages show remarkable intermingling with cells from distinct clonal origins. Larger scale single-cell analyses of somatic mutations will be necessary to study the generalizability of these patterns across different progenitor types and anatomic locations.
Discussion
Here, we show how single-neuron WGS and in-depth characterization of somatic mutations can reveal spatial patterns of cell lineages in normal human brain. We were able to take advantage of somatic mutations for this purpose by our ability to definitively validate them and recover their full sequences from single-neurons, a level of validation not routinely performed in single-cell studies. Although here we focused on somatic retrotransposition and the highly mutable poly-A microsatellites they create, potentially any type of somatic mutation that can similarly be definitively validated could be used for this purpose (Shapiro et al., 2013). Indeed, prior studies have found diverse types of somatic mutation in human brain, including copy-number variants (Cai et al., 2014; McConnell et al., 2013), point mutations (Poduri et al., 2012), and other microsatellite polymorphisms (Gonitel et al., 2008). Since our single-neuron WGS captures most of the genome at high read depth, our methods may be extended to examine nearly all types of somatic mutation in one experiment. Further single-cell WGS studies of all classes of mutation simultaneously may achieve high-resolution tracing of lineages in human brain.
One limitation of retrotransposons for lineage tracing is our prior (Evrony et al., 2012) and current finding that at least in the cerebral cortex somatic insertions are relatively infrequent, being undetectable in 12/16 single-neuron genomes. Nonetheless, they offer important advantages as lineage markers, relative to other mutation types: a) they possess characteristic sequence signatures confirming they were created in vivo and not by MDA; b) their breakpoints enable ultra-sensitive assays; c) each insertion is unique so that homoplasy (occurrence of identical independent mutations) does not confound analysis. Spontaneous somatic retrotransposition as a tool to study brain development is compellingly analogous to classical retroviral labeling used to study cortical development in other mammals (Walsh and Cepko, 1992; Ware et al., 1999); in fact, retrotransposons and retroviruses are evolutionarily related (Eickbush and Jamburuthugoda, 2008). Identification of genetic backgrounds more permissive for retrotransposition (Muotri et al., 2010; Zhao et al., 2013), or individuals with a higher load of active elements, may identify brains with more spontaneously labeled lineages.
Notably, a recent study with a transgenic synthetic L1 mouse model found significant rates of somatic truncation of long (>100bp) L1 poly-A tails (Grandi et al., 2012), consistent with our findings with endogenous human L1 elements. We also profiled single poly-A tails of a tumor-specific somatic L1 insertion we identified in a breast cancer (Figures S18A–C; Table S3) and found distinct poly-A size distributions in a metastasis of the cancer compared to the primary tumor (Figures S18D–E; Table S5), consistent with most of the metastasis deriving from likely one or at most a few cells. The significant somatic mutation of retrotransposon poly-A tails (see Supplemental Note 2 for discussion) supports the potential of high-throughput microsatellite analysis for systematic lineage tracing (Naxerova et al., 2014; Shapiro et al., 2013).
The somatic mutations we studied exhibited distinct spatial patterns of mosaicism, resembling patterns of clonal dispersion previously seen only in animal models and suggesting that focal patches of somatic mutation are prevalent throughout normal brains. The detection of L1#1 only in the middle frontal gyrus suggests it occurred in a neocortical progenitor relatively late in cortical development. Its isolation from a population of neurons with the largest nuclear size also suggests that it is likely present in pyramidal neurons (Evrony et al., 2012). Moreover, the focal spatial distributions of the L1#1 lineage and its sub-lineages imply radial ontogenetic units (Rakic, 2009). On the other hand, the dispersion of the L1#1 lineage at very low mosaicism (≈0.1%) across at least 2 cm of cortex supports the existence of clonal heterogeneity among neocortical progenitor-derived cells within any given cortical column, consistent with lineage tracing studies in other mammals (Gao et al., 2014; Kriegstein and Noctor, 2004; Reid et al., 1997; Torii et al., 2009; Walsh and Cepko, 1988; Ware et al., 1999). Importantly, this implies additional complexity in the possible ways different somatic mutations may overlap spatially and interact to affect cortical circuits.
L1#2 marks a distinct lineage with a much wider geographic distribution than L1#1, suggesting it arose earlier in development. However, L1#2’s low mosaicism (<2%) spanning the entire rostrocaudal length of the brain (from forebrain to hindbrain) implies surprising intermingling of clones in the early central nervous system (CNS). Remarkably, genetic fate-mapping of CNS clones in mouse (Mathis and Nicolas, 2000) revealed the same unexpected finding of significant rostrocaudal dispersion, with evidence suggesting this results from intermixing along the rostrocaudal axis among the earliest CNS progenitors. Therefore, L1#2 likely inserted into one of the earliest progenitors of the CNS in the anterior (rostral) epiblast or early neural plate, prior to the transition to coherent growth when clonally related cells have more restricted rostrocaudal dispersion. It is possible that some L1#2-containing cells derived from ventral telencephalon progenitors, which give rise to interneurons that disperse across the cortex (Marin, 2013), though proving this would require new phenogenomic technologies combining single-cell genomics with broader single-cell phenotyping. In situ hybridization methods, such as High-Definition DNA Fluorescence In Situ Hybridization (HD-FISH), offer one possible route for phenogenomic study of somatic mutations (Bienko et al., 2013) to resolve cell-type, morphology, and layer distributions of cells within a lineage. However, attempts so far to detect somatic insertions shorter than 1 kb (such as the L1 transduction sequences available as targets in the current study) directly in brain tissue sections using this approach pose formidable challenges due to the sensitivity limits of current probe designs (M. Bienko, personal communication). We provide a developmental model for the somatic mutation events we identified in Figure 5. Overall, these results illustrate how somatic mutations can yield important insight into clonal dispersion patterns in human brain development and point to the potential of future systematic study of large numbers of mutations and single cells to delineate lineages in the human nervous system.
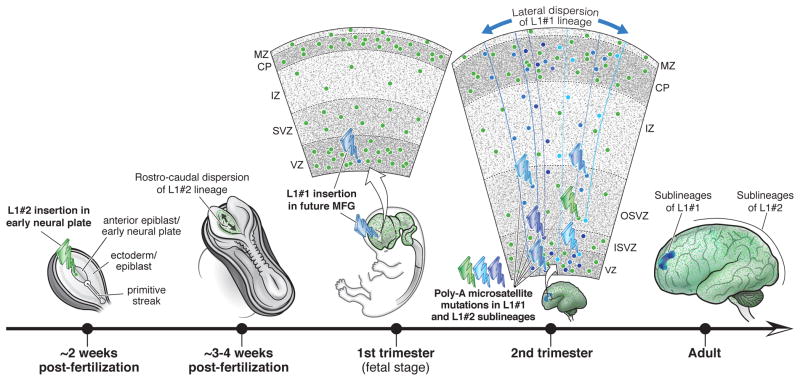
Figure 5. Developmental model of mutation events and lineages in this study.
L1#1 is illustrated as occurring in the 1st trimester, though its exact timing is unknown. MFG, middle frontal gyrus; VZ; SVZ, subventricular zone; ISVZ, inner SVZ; OSVZ, outer SVZ; IZ, intermediate zone; CP, cortical plate; MZ, marginal zone.
The two L1 clones and smaller sub-lineages also match patterns of known somatic mutation disorders of human brain development, and predict the existence of additional types of somatic lesions. Deleterious somatic mutations in mTOR pathway genes cause hemimegalencephaly and show wide dispersion throughout an entire hemisphere (Poduri et al., 2012; http://www.ncbi.nlm.nih.gov/pubmed/22729223), similar to L1#2, while focal cortical dysplasias, the most common cause of intractable epilepsy, are generally limited to smaller areas of cortex, remarkably similar to L1#1 (Poduri et al., 2013). Other deleterious mutations with such restricted distributions could potentially impact cortical areas strikingly unequally, affecting small regions of cortex (L1#1) or only one hemisphere (L1#2), providing a possible mechanism to generate selective and unpredictable disorders of cognition. Focal lesions of unknown etiology have been described in histology of brains of patients with autism spectrum disorder (Stoner et al., 2014), however, many autism brains do not have structural or radiographic findings. Focal mutation of genes involved in synaptic function, for example, may impair neuronal function locally without being structurally evident. Comprehensive single-cell sequencing and somatic mutation analyses across all cell types, brain regions, and timepoints in development will inform an understanding of normal human brain development and the role of somatic mutation in neuropsychiatric disease.
Experimental Procedures
See ‘Supplemental Experimental Procedures’ in the Supplemental Information for full method details.
Human tissues and DNA samples
Post-mortem tissues from individual UMB1465 were obtained from the NIH NeuroBioBank at the University of Maryland. UMB1465 was a 17 year-old male and one of the individuals profiled in our previous single-neuron L1 insertion-profiling (L1-IP) study (Evrony et al., 2012). All UMB1465 tissues were frozen and stored at −80°C without fixation within 4 hours of death, except for the right cerebral hemisphere which was formalin-fixed. Coronal sections of the frozen left cerebral cortex were photographed before and after sampling. Sampled locations were mapped to a representative brain using measured section thicknesses and anatomy of gyri. Since an image of the complete brain of individual 1465 prior to sectioning was not available, sampled locations are illustrated on a representative brain image from the University of Wisconsin and Michigan State Comparative Mammalian Brain Collection (http://brainmuseum.org).
Bulk DNA was extracted from tissues with the QIAamp DNA Mini or QIAamp DNA FFPE Tissue kits (Qiagen). Genomes of the 16 cerebral cortex single neuron samples and the caudate nucleus 100-neuron sample were amplified by MDA (Dean et al., 2002) as part of our previous targeted L1 insertion-profiling (L1-IP) study (Evrony et al., 2012). The 16 single neurons were originally sorted from location D of the left middle frontal gyrus. Unamplified bulk DNA from a breast cancer primary tumor, lymph node metastasis, and normal blood from an individual (ID: TCGA-E1-A15E) were obtained with permission from The Cancer Genome Atlas (TCGA) project.
Whole-genome sequencing and read alignment
Paired-end whole-genome sequencing libraries were prepared with the NEXTflex DNA Sequencing Kit (Bioo Scientific) from 500ng of DNA. Paired-end sequencing (100bp × 2 or 101bp × 2) was performed on HiSeq 2000 sequencers (Illumina). High coverage whole-genome sequencing data from prior studies of MALBAC-amplified single cancer cells (SW480 cancer cell line) (Zong et al., 2012) and MDA-amplified single lymphoblastoid cells (YH cell line) (Hou et al., 2012), and corresponding unamplified bulk DNA, were obtained from the NCBI Sequence Read Archive (SRA). High-coverage whole-genome sequencing data for breast cancer primary tumor, metastasis, and normal blood samples from individual TCGA-E1-A15E were obtained from CGHub. Sequencing reads were aligned to hs37d5 (1000 Genomes Project human genome reference based on the GRCh37 primary assembly) using bwa (Li and Durbin, 2009). Single-neuron whole-genome sequencing data are deposited in the NCBI SRA with accession SRP041470.
Single-cell analysis of somatic retrotransposition
Somatic retrotransposon insertion analysis was performed with scTea (Single-cell Transposable element analyzer). scTea is based on the Tea pipeline originally developed to detect somatic insertions of transposable elements in cancer genomes (Lee et al., 2012), with additional significant modifications for single-neuron whole genome analysis, including: a) a scoring scheme assigning a score to each call, taking into account MDA and library preparation amplification noise; b) improved handling of poly-A signals; c) copy number genotyping of insertion calls; d) local read assembly to detect transduced sequences; e) a revised transposable element sequence library using only known active retrotransposon subfamilies; f) rigorous sensitivity analyses to establish call criteria; and g) specificity analyses using independent PCR validation.
Validation and cloning of retrotransposon candidates
Validation of germline and somatic insertion candidates predicted by scTea was attempted by: 1) full-length PCR (FL-PCR) with genomic primers flanking the candidate (for Alu and L1 candidates), and 2) 3′-junction PCR (3′PCR) with a primer designed downstream of the 3′-end of the candidate paired with an internal primer specific to the 3′ sequence of the retrotransposon (for L1 and SVA candidates). Primer design and full-length cloning were performed as previously described (Evrony et al., 2012). Sequences of validation primers used for each candidate insertion can be found in Table S3. Positive validation reactions were confirmed by Sanger sequencing.
Droplet digital PCR (ddPCR)
Custom ddPCR assays for L1#1 and L1#2 were performed with the QX100 Droplet Digital PCR System (Bio-Rad). L1 assays were multiplexed with an assay for RNaseP serving as a genomic copy number reference for calculation of mosaicism. Multiple unrelated human control samples confirmed assay specificity (Table S4), and the presence or absence of L1#1 and L1#2 in unamplified bulk DNA from every location and tissue was independently verified by a bulk nested 3′-junction PCR (Figure S15).
Poly-A tail cloning and sizing
Poly-A tail lengths of somatic retrotransposon insertions were measured using a digital nested 3′PCR approach (dnPCR) in which single copies of poly-A tails are cloned directly from unamplified bulk DNA, thereby avoiding potential MDA artifacts. Single-copy (digital) cloning by dnPCR also recovers the true poly-A tail distribution in tissues, which is not possible with bulk (non-digital) PCR since PCR amplification efficiency varies with poly-A tail length (data not shown). dnPCR is performed by diluting DNA to a target retrotransposon insertion concentration of 0.3 copies/reaction based on the absolute concentration measured by ddPCR, such that there would be < 5% chance that the diluted DNA input into a dnPCR reaction would contain >1 poly-A tail. A two-round nested PCR targeting the 3′ junction (containing the poly-A tail) of the somatic retrotransposon insertion is then performed on the diluted DNA, using a FAM-labeled primer in the second-round PCR. dnPCR reactions are screened by agarose gel electrophoresis to identify reactions yielding a product. dnPCR products are then sized by capillary electrophoresis on 3130 or 3730 DNA Analyzers (Life Technologies) to obtain the poly-A tail length. A subset of positive dnPCR reactions from each tissue and location were Sanger sequenced (Genewiz, Inc.) and confirmed that dnPCR amplifies the targeted retrotransposon insertion with 100% specificity (data not shown). dnPCR results across tissues shows that dnPCR measures poly-A tails with a precision of ±1 bp across a wide range of poly-A tail sizes (see Supplemental Experimental Procedures for details).
Supplementary Material
Highlights.
High-coverage whole-genome sequencing of single neurons from human brain
Spatial tracing of cell lineages in human brain using somatic retrotransposon insertions
Highly dynamic mutation of microsatellite repeats within insertions marks sub-lineages
Somatic mutations reveal patterns of clonal dispersion and focal mutation in normal brain
Acknowledgments
We thank Aldo Rozzo for logistical assistance, the Orchestra research computing team at Harvard Medical School for assistance with computing resources, Magda Bienko and Alexander van Oudenaarden for helpful discussions and pilot attempts to detect the somatic insertions in brain tissue using HD-FISH, Alexander Subtelny for assistance analyzing transcriptome poly-A length profiling data (Supplemental Note 2) (Subtelny et al., 2014), and Haig Kazazian and Tara Doucet for helpful discussions regarding activities of source retroelements. We also thank Kim Petro (Bio-Rad) for assistance with droplet digital PCR, and Dick Bennett of the Boston Children’s Hospital Molecular Biology core for assistance with capillary electrophoresis. Representative brain image in Figures 3B, 4D, and S14A was adapted with permission from the University of Wisconsin and Michigan State Comparative Mammalian Brain Collections (http://brainmuseum.org), supported by the National Science Foundation. Figure 5 was illustrated by Ken Probst (Xavier Studio). Sequencing was performed with support of the Research Connection of Boston Children’s Hospital. G.D.E. is supported by NIH MSTP grant T32GM007753 and the Louis Lange III Scholarship in Translational Research. E.L. is supported in part by the Eleanor and Miles Shore Fellowship. C.A.W. is supported by the Manton Center for Orphan Disease Research and grants from the NINDS (R01 NS079277 and R01 NS032457). C.A.W. is a Distinguished Investigator of the Paul G. Allen Family Foundation, and an Investigator of the Howard Hughes Medical Institute.
Footnotes
Author contributions
G.D.E. and B.K.M. performed all wet-lab experiments, with input from X.C. and H.S.L. General and retrotransposition analyses of WGS data were performed by G.D.E. and E.L., respectively, with assistance from Y.B., L.Y, and P.H. Y.B. performed GC-content bias analyses. R.M.J. procured human tissues. G.D.E., E.L., P.J.P, and C.A.W. conceived and designed the project, and wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baillie JK, Barnett MW, Upton KR, Gerhardt DJ, Richmond TA, De Sapio F, Brennan PM, Rizzu P, Smith S, Fell M, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479:534–537. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienko M, Crosetto N, Teytelman L, Klemm S, Itzkovitz S, van Oudenaarden A. A versatile genome-scale PCR-based pipeline for high-definition DNA FISH. Nat Methods. 2013;10:122–124. doi: 10.1038/nmeth.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blainey PC, Quake SR. Dissecting genomic diversity, one cell at a time. Nat Methods. 2014;11:19–21. doi: 10.1038/nmeth.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH., Jr Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci USA. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Evrony GD, Lehmann HS, Elhosary PC, Mehta BK, Poduri A, Walsh CA. Single-Cell, Genome-wide Sequencing Identifies Clonal Somatic Copy-Number Variation in the Human Brain. Cell reports. 2014;8:1280–1289. doi: 10.1016/j.celrep.2014.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowry G, Molnar Z, Rakic P. Renewed focus on the developing human neocortex. J Anat. 2010;217:276–288. doi: 10.1111/j.1469-7580.2010.01281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, Morell M, O’Shea KS, Moran JV, Gage FH. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean FB, Hosono S, Fang L, Wu X, Faruqi AF, Bray-Ward P, Sun Z, Zong Q, Du Y, Du J, et al. Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci USA. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickbush TH, Jamburuthugoda VK. The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res. 2008;134:221–234. doi: 10.1016/j.virusres.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrony GD, Cai X, Lee E, Hills LB, Elhosary PC, Lehmann HS, Parker JJ, Atabay KD, Gilmore EC, Poduri A, et al. Single-Neuron Sequencing Analysis of L1 Retrotransposition and Somatic Mutation in the Human Brain. Cell. 2012;151:483–496. doi: 10.1016/j.cell.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco SJ, Muller U. Shaping our minds: stem and progenitor cell diversity in the Mammalian neocortex. Neuron. 2013;77:19–34. doi: 10.1016/j.neuron.2012.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank SA. Somatic evolutionary genomics: mutations during development cause highly variable genetic mosaicism with risk of cancer and neurodegeneration. Proc Natl Acad Sci USA. 2010;107:1725–1730. doi: 10.1073/pnas.0909343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Postiglione MP, Krieger TG, Hernandez L, Wang C, Han Z, Streicher C, Papusheva E, Insolera R, Chugh K, et al. Deterministic progenitor behavior and unitary production of neurons in the neocortex. Cell. 2014;159:775–788. doi: 10.1016/j.cell.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonitel R, Moffitt H, Sathasivam K, Woodman B, Detloff PJ, Faull RL, Bates GP. DNA instability in postmitotic neurons. Proc Natl Acad Sci U S A. 2008;105:3467–3472. doi: 10.1073/pnas.0800048105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL, Kazazian HH. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- Goodier JL, Ostertag EM, Kazazian HH., Jr Transduction of 3′-flanking sequences is common in L1 retrotransposition. Hum Mol Genet. 2000;9:653–657. doi: 10.1093/hmg/9.4.653. [DOI] [PubMed] [Google Scholar]
- Grandi FC, Rosser JM, An W. LINE-1 Derived Poly(A) Microsatellites Undergo Rapid Shortening and Create Somatic and Germline Mosaicism in Mice. Mol Biol Evol. 2012;30:503–512. doi: 10.1093/molbev/mss251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Fan W, Yan L, Li R, Lian Y, Huang J, Li J, Xu L, Tang F, Xie XS, Qiao J. Genome analyses of single human oocytes. Cell. 2013;155:1492–1506. doi: 10.1016/j.cell.2013.11.040. [DOI] [PubMed] [Google Scholar]
- Hou Y, Song L, Zhu P, Zhang B, Tao Y, Xu X, Li F, Wu K, Liang J, Shao D, et al. Single-Cell Exome Sequencing and Monoclonal Evolution of a JAK2-Negative Myeloproliferative Neoplasm. Cell. 2012;148:873–885. doi: 10.1016/j.cell.2012.02.028. [DOI] [PubMed] [Google Scholar]
- Insel TR. Brain somatic mutations: the dark matter of psychiatric genetics? Mol Psychiatry. 2014;19:156–158. doi: 10.1038/mp.2013.168. [DOI] [PubMed] [Google Scholar]
- Kim TM, Laird PW, Park PJ. The landscape of microsatellite instability in colorectal and endometrial cancer genomes. Cell. 2013;155:858–868. doi: 10.1016/j.cell.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar K, Watt FM. Lineage tracing. Cell. 2012;148:33–45. doi: 10.1016/j.cell.2012.01.002. [DOI] [PubMed] [Google Scholar]
- Kriegstein AR, Noctor SC. Patterns of neuronal migration in the embryonic cortex. Trends Neurosci. 2004;27:392–399. doi: 10.1016/j.tins.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Lee E, Iskow R, Yang L, Gokcumen O, Haseley P, Luquette LJ, 3rd, Lohr JG, Harris CC, Ding L, Wilson RK, et al. Landscape of Somatic Retrotransposition in Human Cancers. Science. 2012;337:967–971. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin O. Cellular and molecular mechanisms controlling the migration of neocortical interneurons. Eur J Neurosci. 2013;38:2019–2029. doi: 10.1111/ejn.12225. [DOI] [PubMed] [Google Scholar]
- Marin O, Muller U. Lineage origins of GABAergic versus glutamatergic neurons in the neocortex. Curr Opin Neurobiol. 2014;26C:132–141. doi: 10.1016/j.conb.2014.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis L, Nicolas JF. Different clonal dispersion in the rostral and caudal mouse central nervous system. Development. 2000;127:1277–1290. doi: 10.1242/dev.127.6.1277. [DOI] [PubMed] [Google Scholar]
- McConnell MJ, Lindberg MR, Brennand KJ, Piper JC, Voet T, Cowing-Zitron C, Shumilina S, Lasken RS, Vermeesch JR, Hall IM, Gage FH. Mosaic copy number variation in human neurons. Science. 2013;342:632–637. doi: 10.1126/science.1243472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muotri AR, Chu VT, Marchetto MCN, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- Muotri AR, Marchetto MC, Coufal NG, Oefner R, Yeo G, Nakashima K, Gage FH. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468:443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naxerova K, Brachtel E, Salk JJ, Seese AM, Power K, Abbasi B, Snuderl M, Chiang S, Kasif S, Jain RK. Hypermutable DNA chronicles the evolution of human colon cancer. Proc Natl Acad Sci U S A. 2014;111:E1889–1898. doi: 10.1073/pnas.1400179111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickeral OK, Makalowski W, Boguski MS, Boeke JD. Frequent human genomic DNA transduction driven by LINE-1 retrotransposition. Genome Res. 2000;10:411–415. doi: 10.1101/gr.10.4.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poduri A, Evrony GD, Cai X, Elhosary PC, Beroukhim R, Lehtinen MK, Hills LB, Heinzen EL, Hill A, Hill RS, et al. Somatic Activation of AKT3 Causes Hemispheric Developmental Brain Malformations. Neuron. 2012;74:41–48. doi: 10.1016/j.neuron.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poduri A, Evrony GD, Cai X, Walsh CA. Somatic mutation, genomic variation, and neurological disease. Science. 2013;341:1237758. doi: 10.1126/science.1237758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. Evolution of the neocortex: a perspective from developmental biology. Nat Rev Neurosci. 2009;10:724–735. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid CB, Tavazoie SF, Walsh CA. Clonal dispersion and evidence for asymmetric cell division in ferret cortex. Development (Cambridge, England) 1997;124:2441–2450. doi: 10.1242/dev.124.12.2441. [DOI] [PubMed] [Google Scholar]
- Reilly MT, Faulkner GJ, Dubnau J, Ponomarev I, Gage FH. The role of transposable elements in health and diseases of the central nervous system. J Neurosci. 2013;33:17577–17586. doi: 10.1523/JNEUROSCI.3369-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro E, Biezuner T, Linnarsson S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat Rev Genet. 2013;14:618–630. doi: 10.1038/nrg3542. [DOI] [PubMed] [Google Scholar]
- Stoner R, Chow ML, Boyle MP, Sunkin SM, Mouton PR, Roy S, Wynshaw-Boris A, Colamarino SA, Lein ES, Courchesne E. Patches of disorganization in the neocortex of children with autism. N Engl J Med. 2014;370:1209–1219. doi: 10.1056/NEJMoa1307491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subtelny AO, Eichhorn SW, Chen GR, Sive H, Bartel DP. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature. 2014;508:66–71. doi: 10.1038/nature13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii M, Hashimoto-Torii K, Levitt P, Rakic P. Integration of neuronal clones in the radial cortical columns by EphA and ephrin-A signalling. Nature. 2009;461:524–528. doi: 10.1038/nature08362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voet T, Kumar P, Van Loo P, Cooke SL, Marshall J, Lin ML, Zamani Esteki M, Van der Aa N, Mateiu L, McBride DJ, et al. Single-cell paired-end genome sequencing reveals structural variation per cell cycle. Nucleic Acids Res. 2013;41:6119–6138. doi: 10.1093/nar/gkt345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C, Cepko CL. Clonally related cortical cells show several migration patterns. Science. 1988;241:1342–1345. doi: 10.1126/science.3137660. [DOI] [PubMed] [Google Scholar]
- Walsh C, Cepko CL. Widespread dispersion of neuronal clones across functional regions of the cerebral cortex. Science. 1992;255:434–440. doi: 10.1126/science.1734520. [DOI] [PubMed] [Google Scholar]
- Ware ML, Tavazoie SF, Reid CB, Walsh CA. Coexistence of widespread clones and large radial clones in early embryonic ferret cortex. Cereb Cortex. 1999;9:636–645. doi: 10.1093/cercor/9.6.636. [DOI] [PubMed] [Google Scholar]
- Xing J, Wang H, Belancio VP, Cordaux R, Deininger PL, Batzer MA. Emergence of primate genes by retrotransposon-mediated sequence transduction. Proc Natl Acad Sci U S A. 2006;103:17608–17613. doi: 10.1073/pnas.0603224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Du J, Han X, Goodier JL, Li P, Zhou X, Wei W, Evans SL, Li L, Zhang W, et al. Modulation of LINE-1 and Alu/SVA retrotransposition by Aicardi-Goutieres syndrome-related SAMHD1. Cell reports. 2013;4:1108–1115. doi: 10.1016/j.celrep.2013.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338:1622–1626. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.