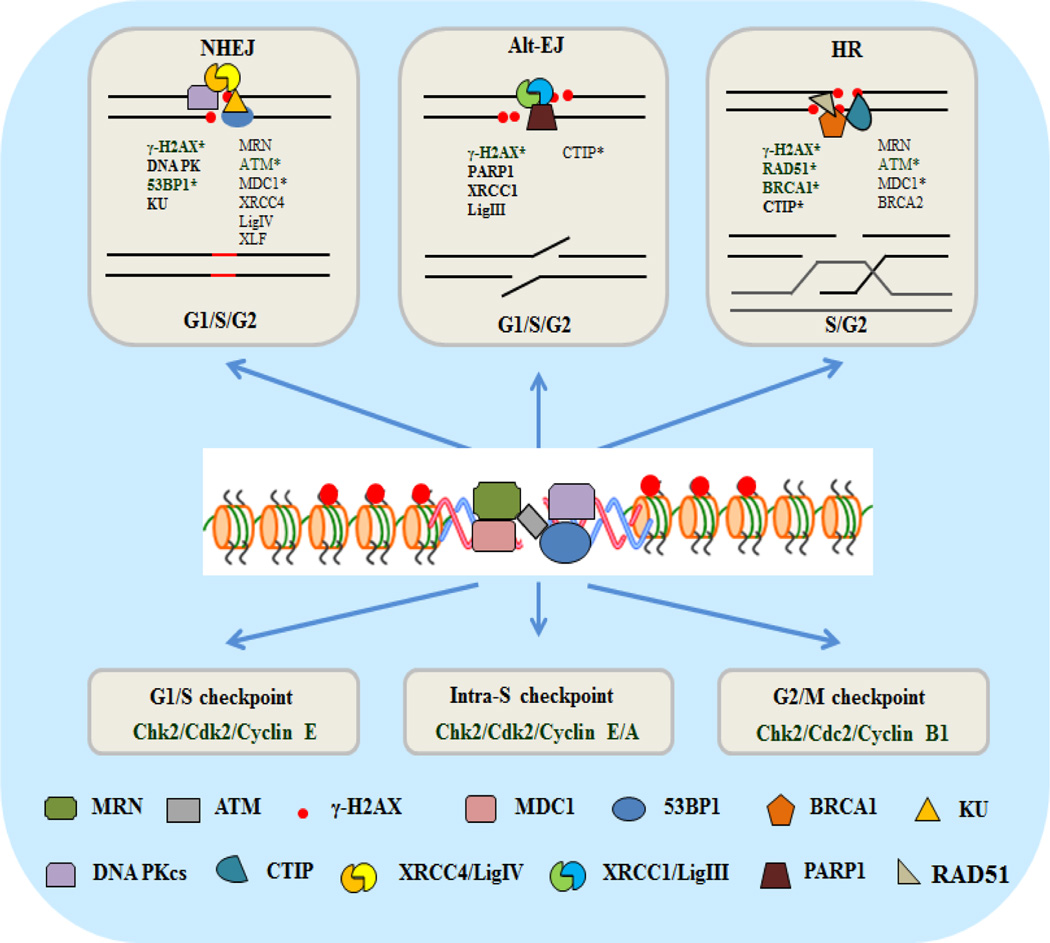
The DNA damage response (DDR) in mammalian cells is a complex and highly orchestrated signaling process that regulates the recruitment of specific DDR proteins to the DNA damage sites [1,2]. The most lethal type of DNA damage is the double-strand break (DSB), which is generated by ionizing radiation (IR), radiomimetic drugs and also drugs of DNA topoisomerase 2 poison family [3]. DSBs are also endogenously caused during replication of single-strand breaks generated by reactive oxygen species or at stalled replication forks [4,5]. While IR is extensively used in treating various cancers, IR-induced DSBs and other genomic lesions can cause mutations and gross genome rearrangements in normal cells as well, possibly leading to secondary leukemia and other cancers. Fortunately, multiple DSB repair (DSBR) systems, in coordination with cell cycle checkpoint control, have evolved to prevent both the cell lethality and mutations arising from unrepaired genome lesions. Activation of the master regulator ATM (Ataxia Telangiectasia Mutated) initiates the complex choreography of signaling required to interface with the repair pathways to protect cells after DSB induction [6]. In view of the critical role of DSBR in differential survival of healthy vs. tumor cells during radiotherapy, and its role in ageing and neurological diseases [7–9], reliable and sensitive biomarkers of DSBs and DSBR kinetics are necessary to monitor cellular responses. These would not only be useful for pre-clinical studies but also for evaluating the efficacy of radiotherapy in the clinical setting by assessing therapy-induced DNA damage or characterizing the inherent genomic instability of precancerous lesions. Radiation-induced DSBs in DNA mostly result from closely spaced (typically within 10 base pairs), bi-stranded single-strand breaks caused by ionization tracks. In response to DSB induction, the Mre11/Rad50/NBS1 (MRN) complex is initially recruited to damage sites to activate ATM, a serine/threonine kinase [10,11]. Activated ATM phosphorylates the H2AX variant of histone H2A at serine 139 at the DSB sites, which then triggers recruitment/accumulation of other DDR proteins including RNF8, RNF168, 53BP1 and BRCA1, at these damage sites [12]. Protein molecules localized in the vicinity of DSB termini, after conjugation with fluorescent antibodies, appear under a fluorescence microscope as bright dots termed foci. Many DDR proteins could be visualized as discrete foci at DSB sites by immuno-fluorescence (IF). However, it is important to note that not all proteins accumulating at damage sites are detectable as foci because accumulation of at least 100 molecules is needed to visualize a discrete focus [13]. While the mechanism underlying protein focus formation has begun to emerge only recently, the phenomenon is being extensively used.
Commonly used DSBR markers in Mammalian Cells
Phosphorylated H2AX
In principle, any DSBR protein that forms IF focus is a candidate for a DSB biomarker. However, phosphorylated H2AX [also known as gamma (γ)-H2AX] was first identified as a quantitative DSB marker due to its high sensitivity and almost immediate formation (within seconds) after DSB induction [14]. The γ-H2AX foci level is linearly related to the number of DSBs and IR dose in the range of 1.2 mGy and 2 Gy, as analyzed in primary human linearly related to the number of DSBs and IR dose in the range of 1.2 mGy and 2 Gy, as analyzed in primary human fibroblasts. Detection of DSBs by γ-H2AX foci formation is 100-fold more sensitive than by other available methods at clinically relevant radiation doses (1–10 Gy) [15,16]. The half-maximal number of foci is reached within 1 min and the maximum in 9 to 30min after irradiation [17]. Only few base pairs are involved in the initial DSB formation, commensurate with a small but distinct focus being formed initially at a DSB site. However, over time, the focus spreads to adjacent areas to cover up to 2 Mbp chromosomal DNA, and contains estimated 2000 γ-H2AX molecules. This suggests significant signal amplification, probably involving chromatin modification to facilitate the binding of a large number of DSBR components. Based on DSBR kinetics, 15–30min after irradiation appear to be the appropriate interval for precise and sensitive foci counting to quantitate DSBs [18]. However, spontaneous γ-H2AX foci are detectable in both normal and cancer cells, likely as a result of endogenous DSBs generated by DNA replication stress. The basal level of foci varies with the cell type, but commonly 1–2 foci/cell have been observed in normal tissues (colon, breast, ovary and human primary fibroblasts) while in cancer cell lines the number is larger,1–20 foci per cell, and more variable [19,20]. Although IF-based foci analysis is the most sensitive approach for DSB detection, it should be noted that the overlap of multiple foci due to high DSB density poses a major challenge for accurate manual or software-based quantitation, especially with high radiation doses [18].
The disappearance kinetics of the γ-H2AX foci, presumably linked to H2AX dephosphorylation, closely follows that of DSBR, irrespective of the repair pathway employed by the cell. However, the mechanism of de-phosphorylation is still controversial. Few studies reported the persistence of a small number of γ-H2AX foci even after apparent completion of DSB repair, particularly after formation of a high level of DSBs due to time required for resetting of chromatin after repair [16]. Nevertheless, the loss of γ-H2AX correlates well with DSBR at low-to-moderate levels of genome damage (<150 DSBs/genome) in repair proficient cells [21]. A unique advantage of using γ-H2AX foci as a DSB biomarker is that these foci are formed in all cell cycle phases. However, under certain circumstances, γ-H2AX foci may not exclusively reflect DSBs because dis-regulated DNA metabolism may also cause ATR-mediated H2AX phosphorylation in growing cells, which form DSB-independent background foci, in addition to heat mediated H2AX foci, that do not involve DSBs [22,23].
Phosphorylated ATM
ATM, which is activated via auto-phosphorylation at serine 1981, is recruited to DSB sites in the very early stage of DSBR and was proposed as an alternative DSB marker in growing cells [24]. Phospho-ATM foci formation follows a linear relationship with IR dose over the 10 mGy to 1 Gy range, and the number of foci is accurately correlated with the number of DSBs. However, one limitation of phospho-ATM as a DSB marker is that the senescent cells display phospho-ATM signal in the absence of DSBs [25,26].
Other DSBR pathway-specific proteins as DSB markers
In addition to phosphorylated H2AX and ATM, certain unmodified DSBR proteins also form foci at DSB sites, such as p53 binding protein 1 (53BP1), MDC1, RAD50 and BRCA1, and are often used as DSB markers [27,28]. 53BP1 co-localizes with γ-H2AX at DSB sites and the number of foci reaches a maximum at 15–30 min after IR treatment, followed by steady decrease to the background level after 10–16h. Foci disappearance matches the kinetics of DSBR occurring via non-homologous end joining (NHEJ); however, 53BP1 migrates or is released from the DSB sites in the case of other two modes of DSBR, namely, homologous recombination (HR) and alternative end joining (Alt-EJ) [13,29]. Furthermore, unlike phosphorylated H2AX/ATM, 53BP1 recruitment is specific to DSBs, except when DSBs occur during mitosis [30,31]. Other DSBR proteins including MDC1, RAD50, and BRCA1 also accumulate at DSB sites in sufficient numbers to form foci and were utilized as DSB markers in several studies [32]. However, their universal utilization is limited by their lack of correlation with DSBR kinetics in all cell types and all modes of DSB repair.
As already mentioned, not all DSBR proteins are capable of forming foci. For example, several key DSBR proteins like DNA-PK, Ku70/80, Smc1 and Smc3, although, recruited to DSBs, usually by direct binding to the damaged termini, do not form foci after irradiation [33]. Similarly, Chk2 (phosphorylated by ATM at threonine 68) and Chk1, critical DSB response elements in mammalian cells for damage-dependent cell cycle arrest, do not form foci at individual DSB sites after IR [34]. In fact, phospho-(Thr68)-Chk2 rapidly appears over the entire nucleus in laser microirradiated cells. Similarly, DNA-PK/Ku70 and Smc1 can be detected at laser microirradiated tracks after micro-laser irradiation at a substantial dose [33,34].
Other methods of quantitating DSBs
Complementary techniques used to measure DSBs include flow cytometry and Western blotting, which qualitatively (or semi-quantitatively at best) measure DSBs and their repair by monitoring covalent modification of DSBR proteins (e.g., γ-H2AX and phospho-ATM and also phospho-Chk2). In addition to the protein-based approaches, single cell electrophoresis to analyze damage-induced DNA fragments, named the comet assay, was developed some three decades ago and has been extensively utilized for the detection and quantitation of DNA single-strand and double-strand breaks. This assay method is based on the principle that fragmented DNA migrates out of the cell according to size during in situ electrophoresis, while intact nuclear DNA is too large to have detectable mobility. After staining with the dye acridine orange, the nucleus appears as a comet head with the smaller fragments migrating out of the cell appearing as a ‘tail’ under the microscope. The amount and size of the DNA fragments in the tail, measured as the ‘tail moment’, is proportional to the level of strand breaks [35]. A reasonably sensitive and rapid way to detect DNA damage, as was first introduced in 1984 [36]. The original comet assay was designed to measure single-strand breaks in the genome because electrophoresis was performed under alkaline conditions. The modified comet assay is able to detect cellular DSBs by carrying out electrophoresis at neutral pH. However, the sensitivity of the comet assay for detecting DSBs is significantly lower than that of γ-H2AX foci analysis [37–39].
Although not particularly sensitive, pulse field gradient electrophoresis (PFGE) and size-based DNA fractionation by sedimentation in an ultracentrifuge have also been used to monitor DNA fragmentation [40,41]. However, like comet analysis, these are not appropriate for quantitating small numbers of DSBs.
Perspective and future directions
A sensitive biomarker for chromosomal DSBs is a major prerequisite for analyzing genome damage and repair, which are important in both the laboratory and clinical settings. Some half dozen DSB markers and techniques have been developed for detection and quantification of the genome damage; the selection of the optimum marker largely depends on the objective of the research/diagnostic procedure, the cell types as well as the agents/drugs used to induce the DSBs. Although, γ-H2AX foci formation is not an exclusive indicator of DSBs, it is still the best marker based on its cell phase-independent formation, tight correlation with repair kinetics and repair pathway independence. Future studies should focus on defining distinct DSBR sub-pathways and identifying specific, quantitative biomarkers for analyzing each mode of DSB repair in cancer versus normal cell types. This may require a greater understanding of and range of DSB markers but could help identify and validate unique DSBR targets for cell/tissue type-specific cancer therapy.
Figure 1.
A schematic model of repair and damage response proteins recruited at chromosomal DSB sites in mammalian cells. The proteins that are commonly used as immune-fluorescence-based foci biomarkers are highlighted (*).
Table 1.
DSBR proteins that form immuno-fluorescence foci, which are commonly used as biomarker and their unique features.
| DSB foci-forming marker | Description/limitation |
|---|---|
| Gamma-H2AX |
|
| pATM 1981 |
|
| 53BP1 |
|
| BRCA1 |
|
| RAD-51 |
|
Acknowledgements
The research in authors’ laboratories is supported by grants from Muscular Dystrophy Association (MDA 294842; MLH), ALS Association (ALSA 15-IIP-204; MLH), Alzheimer’s Association (NIRG-12-242135; MLH), USPHS grants R01 CA158910 (SM and MLH), P01 CA92584; SM), R01CA129537 (TKP), R01CA154320; (TKP), R03 DA035193 (SA) and Houston Methodist Research Institute (MLH, TKP, SM). We thank Hegde lab members Joy Mitra, Erika Guerrero and Pavana Hegde for various help. This editorial commentary with a limited focus is not intended to provide a comprehensive review of literature on the topic, and hence, many citations may not be included, for which we sincerely apologize.
References
- 1.Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008;18(1):27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedberg EC, Aguilera A, Gellert M, Hanawalt PC, Hays JB, et al. DNA repair: from molecular mechanism to human disease. DNA Rep. 2006;5(8):986–996. doi: 10.1016/j.dnarep.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 3.Wilstermann AM, Osheroff N. Base excision repair intermediates as topoisomerase II poisons. J Biol Chem. 2001;276(49):46290–46296. doi: 10.1074/jbc.M105733200. [DOI] [PubMed] [Google Scholar]
- 4.Heller RC, Marians KJ. Replication fork reactivation downstream of a blocked nascent leading strand. Nature. 2006;439(7076):557–562. doi: 10.1038/nature04329. [DOI] [PubMed] [Google Scholar]
- 5.Hegde ML, Hegde PM, Bellot LJ, Mandal SM, Hazra TK, et al. Prereplicative repair of oxidized bases in the human genome is mediated by NEIL1 DNA glycosylase together with replication proteins. Proc. Natl Acad Sci USA. 2013;110(33):E3090–E3099. doi: 10.1073/pnas.1304231110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pandita TK. A multifaceted role for ATM in genome maintenance. Expert Rev Mol Med. 2003;5(16):1–21. doi: 10.1017/S1462399403006318. [DOI] [PubMed] [Google Scholar]
- 7.Hegde ML, Hegde PM, Rao KS, Mitra S. Oxidative genome damage and its repair in neurodegenerative diseases: function of transition metals as a double-edged sword. J Alzheimers Dis. 2011;24(Suppl 2):183–198. doi: 10.3233/JAD-2011-110281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitra J, Guerrero EN, Hegde PM, Wang H, Boldogh I, et al. New perspectives on oxidized genome damage and repair inhibition by pro-oxidant metals in neurological diseases. Biomolecules. 2014;4(3):678–703. doi: 10.3390/biom4030678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hegde ML, Gupta VB, Anitha M, Harikrishna T, Shankar SK, et al. Studies on genomic DNA topology and stability in brain regions of Parkinson’s disease. Arc Bio chem Biophys. 2006;449(1–2):143–156. doi: 10.1016/j.abb.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 10.Scott SP, Pandita TK. The cellular control of DNA double-strand breaks. J Cell Biochem. 2006;99(6):1463–1475. doi: 10.1002/jcb.21067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pandita TK, Richardson C. Chromatin remodeling finds its place in the DNA double-strand break response. Nucleic Acids Res. 2009;37(5):1363–1377. doi: 10.1093/nar/gkn1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar R, Horikoshi N, Singh M, Gupta A, Misra HS, et al. Chromatin modifications and the DNA damage response to ionizing radiation. Front Oncol. 2012;2:214. doi: 10.3389/fonc.2012.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta A, Hunt CR, Hegde ML, Chakraborty S, Udayakumar D, et al. MOF Phosphorylation by ATM Regulates 53BP1-Mediated Double-Strand Break Repair Pathway Choice. Cell Rep. 2014;8(1):177–189. doi: 10.1016/j.celrep.2014.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, et al. [gamma]H2AX and cancer. Nat Rev Cancer. 2008;8(12):957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rothkamm K, Kruger I, Thompson LH, Lobrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol Cell Biol. 2003;23(16):5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mah LJ, El-Osta A, Karagiannis TC. [gamma]H2AX: a sensitive molecular marker of DNA damage and repair. Leukemia. 2010;24(4):679–686. doi: 10.1038/leu.2010.6. [DOI] [PubMed] [Google Scholar]
- 17.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273(10):5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 18.Sak A, Stuschke M. Use of γH2AX and Other Biomarkers of Double-Strand Breaks During Radiotherapy. Seminars in radiation oncology. 2010;20(4):223–231. doi: 10.1016/j.semradonc.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 19.Banath JP, Macphail SH, Olive PL. Radiation sensitivity, H2AX phosphorylation, and kinetics of repair of DNA strand breaks in irradiated cervical cancer cell lines. Cancer Res. 2004;64(19):7144–7149. doi: 10.1158/0008-5472.CAN-04-1433. [DOI] [PubMed] [Google Scholar]
- 20.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146(5):905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bouquet F, Muller C, Salles B. The loss of gammaH2AX signal is a marker of DNA double strand breaks repair only at low levels of DNA damage. Cell cycle (Georgetown, Tex. 2006;5(10):1116–1122. doi: 10.4161/cc.5.10.2799. [DOI] [PubMed] [Google Scholar]
- 22.Ward JF. DNA damage produced by ionizing radiation in mammalian cells: identities, mechanisms of formation, and reparability. Prog Nucleic Acid Res Mol Biol. 1988;35:95–125. doi: 10.1016/s0079-6603(08)60611-x. [DOI] [PubMed] [Google Scholar]
- 23.Fernandez-Capetillo O, Mahadevaiah SK, Celeste A, Romanienko PJ, Camerini-Otero RD, et al. H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Developmental cell. 2003;4(4):497–508. doi: 10.1016/s1534-5807(03)00093-5. [DOI] [PubMed] [Google Scholar]
- 24.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421(6922):499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 25.Pospelova TV, Demidenko ZN, Bukreeva EI, Pospelov VA, Gudkov AV, et al. Pseudo-DNA damage response in senescent cells. Cell Cycle (Georgetown, Tex. 2009;8(24):4112–4118. doi: 10.4161/cc.8.24.10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pankotai T, Hoffbeck AS, Boumendil C, Soutoglou E. DNA damage response in the absence of DNA lesions continued. Cell Cycle Georgetown, Tex. 2009;8(24):4025–4026. [PubMed] [Google Scholar]
- 27.Xu X, Stern DF. NFBD1/KIAA0170 is a chromatin-associated protein involved in DNA damage signaling pathways. J Bio Chem. 2003;278(10):8795–8803. doi: 10.1074/jbc.M211392200. [DOI] [PubMed] [Google Scholar]
- 28.Yuan J, Chen J. MRE11-RAD50-NBS1 complex dictates DNA repair independent of H2AX. J Bio Chem. 2010;285(2):1097–1104. doi: 10.1074/jbc.M109.078436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gupta A, Hunt CR, Chakraborty S, Pandita RK, Yordy J, et al. Role of 53BP1 in the Regulation of DNA Double-Strand Break Repair Pathway Choice. Radiation Res. 2014;181(1):1–8. doi: 10.1667/RR13572.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giunta S, Belotserkovskaya R, Jackson SP. DNA damage signaling in response to double-strand breaks during mitosignaling in response to double-strand breaks during mitosis. J Cell Biol. 2010;190(2):197–207. doi: 10.1083/jcb.200911156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nelson G, Buhmann M, von Zglinicki T. DNA damage foci in mitosis are devoid of 53BP1. Cell Cycle (Georgetown, Tex. 2009;8(20):3379–3383. doi: 10.4161/cc.8.20.9857. [DOI] [PubMed] [Google Scholar]
- 32.Roukos V, Kinkhabwala A, Colombelli J, Kotsantis P, Taraviras S, et al. Dynamic recruitment of licensing factor Cdt1 to sites of DNA damage. J Cell Sci. 2011;124(Pt 3):422–434. doi: 10.1242/jcs.074229. [DOI] [PubMed] [Google Scholar]
- 33.Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan MB, et al. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J Cell Biol. 2006;173(2):195–206. doi: 10.1083/jcb.200510130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lukas C, Falck J, Bartkova J, Bartek J, Lukas J. Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat Cell Biol. 2003;5(3):255–260. doi: 10.1038/ncb945. [DOI] [PubMed] [Google Scholar]
- 35.Olive PL, Banath JP, Durand RE. Heterogeneity in radiation-induced DNA damage and repair in tumor and normal cells measured using the “comet” assay. Radiation Res. 1990;122(1):86–94. [PubMed] [Google Scholar]
- 36.Ostling O, Johanson KJ. Microelectrophoretic study of radiation-induced DNA damages in individual mammalian cells. Biochem Biophys Res Commun. 1984;123(1):291–298. doi: 10.1016/0006-291x(84)90411-x. [DOI] [PubMed] [Google Scholar]
- 37.Yu Y, Zhu W, Diao H, Zhou C, Chen FF, Yang J. A comparative study of using comet assay and gammaH2AX foci formation in the detection of N-methyl-N’-nitro-N-nitrosoguanidine-induced DNA damage. Toxicol In Vitro. 2006;20(6):959–965. doi: 10.1016/j.tiv.2006.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liao W, McNutt MA, Zhu WG. The comet assay: a sensitive method for detecting DNA damage in individual cells. Methods (San Diego, Calif) 2009;48(1):46–53. doi: 10.1016/j.ymeth.2009.02.016. [DOI] [PubMed] [Google Scholar]
- 39.Wood DK, Weingeist DM, Bhatia SN, Engelward BP. Single cell trapping and DNA damage analysis using microwell arrays. Proc. Natl Acad Sci USA. 2010;107(22):10008–10013. doi: 10.1073/pnas.1004056107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh SK, Bencsik-Theilen A, Mladenov E, Jakob B, Taucher-Scholz G, et al. Reduced contribution of thermally labile sugar lesions to DNA double strand break formation after exposure to heavy ions. Radiation Oncology (London, England) 2013;8:77. doi: 10.1186/1748-717X-8-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lobrich M, Ikpeme S, Kiefer J. Measurement of DNA double-strand breaks in mammalian cells by pulsed-field gel electrophoresis: a new approach using rarely cutting restriction enzymes. Radiation Res. 1994;138(2):186–192. [PubMed] [Google Scholar]