Abstract
Neddylation is a post-translational protein modification that conjugates a ubiquitin-like protein NEDD8 to target proteins. Similar to ubiquitination, neddylation is mediated by a cascade of three NEDD8 specific enzymes, an E1 activating enzyme, an E2 conjugating enzyme and one of the several E3 ligases. Neddylation is countered by the action of deneddylases via a process termed deneddylation. By altering the substrate’s conformation, stability, subcellular localization or binding affinity to DNA or proteins, neddylation regulates diverse cellular processes including the ubiquitin-proteasome system-mediated protein degradation, protein transcription, cell signaling etc. Dysregulation of neddylation has been linked to cancer, neurodegenerative disorders, and more recently, cardiac disease. Here we comprehensively overview the biochemistry, the proteome and the biological function of neddylation. We also summarize the recent progress in revealing the physiological and pathological role of neddylation and deneddylation in the heart.
Keywords: Post-translational modification, NEDD8, neddylation, deneddylation, cardiac disease
Introduction
Protein post-translational modifications alter the target protein’s conformation, stability, subcellular localization, or binding affinity to protein or DNA partners, thereby largely expanding the functional diversity and dynamics of the proteome. Eukaryotic proteins can be covalently modified by chemical groups (phosphate, acetyl, methyl etc.) or certain proteins. The first and most studied protein-based modification is ubiquitination, which conjugates a 76-amino-acid protein ubiquitin to target protein via an ATP-dependent enzymatic pathway consisting of an E1 activating enzyme, an E2 conjugating enzyme, and an E3 ligase [1]. Although ubiquitination was first linked to signal the modified proteins for proteasomal degradation, it has since been shown to control many other cellular processes including receptor internalization, assembly of multiprotein complexes, regulation of enzyme activity, DNA repair, cell signaling, and vesicle trafficking [2,3].
Following the discovery of ubiquitin, more than a dozen of ubiquitin-like proteins have been uncovered to utilize ubiquitination-analogous but functionally distinct process to modify proteins, including NEDD8 (neural precursor cell expressed, developmentally down-regulated 8), SUMO-1, SUMO-2, SUMO-3, ISG15, UFM1, FAT10, URM1, Atg12, Atg8, FUB1, and HUB1 [4]. Among these ubiquitin-like proteins, NEDD8 has the highest identity (58%) to ubiquitin [5] (Figure 1A). NEDD8 was first cloned from mouse brain in 1993 and is ubiquitously expressed in various tissues and cell types [5], with highest expression in cardiac and skeletal muscles [6]. NEDD8 is evolutionarily highly conserved (83% identity between human and Arabidopsis and 100% identity among rat, mouse and human) (Figure 1B), suggesting its conserved function in eukaryotic cells. Like ubiquitination, protein modification by NEDD8, termed neddylation, requires NEDD8 specific E1 activating enzyme, E2 conjugating enzyme and E3 ligases. Neddylation can be reversed by a set of NEDD8 specific proteases (deneddylase) via a process called deneddylation. With quickly expanding knowledge on the functional consequence of NEDD8 modification on target proteins, neddylation has emerged as a critical regulatory process controlling ubiquitination, protein transcription, signaling transduction, mitochondria turnover, autophagy, cell death, etc. Moreover, dysregulation of neddylation has been linked to a broad spectrum of pathological conditions ranging from tumorigenesis [7], neurodegeneration [8], inflammation [9-11], immunodeficiency [12], and heart failure [13,14]. In this review, we summarize the current understanding on the biology of neddylation and deneddylation, the NEDD8 proteome and involved biological processes, and highlight the influence of neddylation in cardiac function under physiological and pathological conditions.
Figure 1.

NEDD8 is a ubiquitin-like protein highly conserved among species. A. The amino acid sequences of human ubiquitin and NEDD8 are aligned. B. The amino acid sequences of NEDD8 precursor from various species are aligned. The conserved amino acids and the amino acids with identical properties, as well as those with weakly similar properties, are represented in light blue, dark red and yellow, respectively. Arrow points to the site cleaved by NEDD8 protease. The exposed C-terminal glycine is to be fused with the lysine residue of the substrate. A. thaliana, Arabidopsis thaliana; S. cerevisiae, Saccharomyces cerevisiae; C. elegans, Caenorhabditis elegans; D. melanogaster, Drosophila melanogaster; R. norvegicus, Rattus norvegicus; M. musculus, Mus musculus; H. sapiens, Homo sapiens.
The biochemistry of the NEDD8 pathway
NEDD8 maturation
NEDD8, in yeast or plants also known as RUB1 (related to ubiquitin), is first produced as a non-conjugatable 81-amino-acid precursor in mammals [5]. Maturation of NEDD8 requires cleavage of its C-terminal additional five amino acids to expose glycine 76, which forms an isopeptide bond with the lysine residue on the substrate. The hydrolysis is catalyzed by NEDD8 proteases NEDP1 (NEDD8-specific protease 1, also known as DEN1 and SENP8) and UCH-LC3 [15,16]. NEDP1 is a NEDD8 specific protease, while UCH-L3 is also able to process ubiquitin precursors. UCH-L3 knockout mice are viable and neddylated proteins were accumulated in UCH-L3 null cells [17], suggesting that UCH-LC3 is not essential for NEDD8 processing in vivo. It remained to be understood why NEDD8 and other ubiquitin-like proteins are not directly produced as a mature form. One possibility is that this strategy prevents unprocessed precursors from entering into the conjugation system, offering additional regulatory mechanism for the conjugation of ubiquitin and ubiquitin-like proteins to target substrates.
Neddylation
Similar to ubiquitination, conjugation of mature NEDD8 to target proteins is executed by an E1-E2-E3 multienzyme cascade (Figure 2). Neddylation begins with the activation of mature NEDD8 by the E1 NEDD8 activating enzyme (NAE), a heterodimer of NAE1 (also known as APP-BP1) and UBA3, through forming a NAE-S~NEDD8 thioester-bond in an ATP-dependent manner [18]. Activated NEDD8 is then transferred to the E2 NEDD8 conjugating enzyme Ubc12, creating another thioester bond [18]. An E3 NEDD8 ligase transiently interacts with NEDD8-charged E2 and subsequently transfers NEDD8 to the substrate through forming an isopeptide bond between the C-terminal glycine of NEDD8 and a lysine residue on the substrate. The discharged E2 is then released from the E3, allowing another charged E2 to associate with the E3 to initiate next round of NEDD8 transfer.
Figure 2.
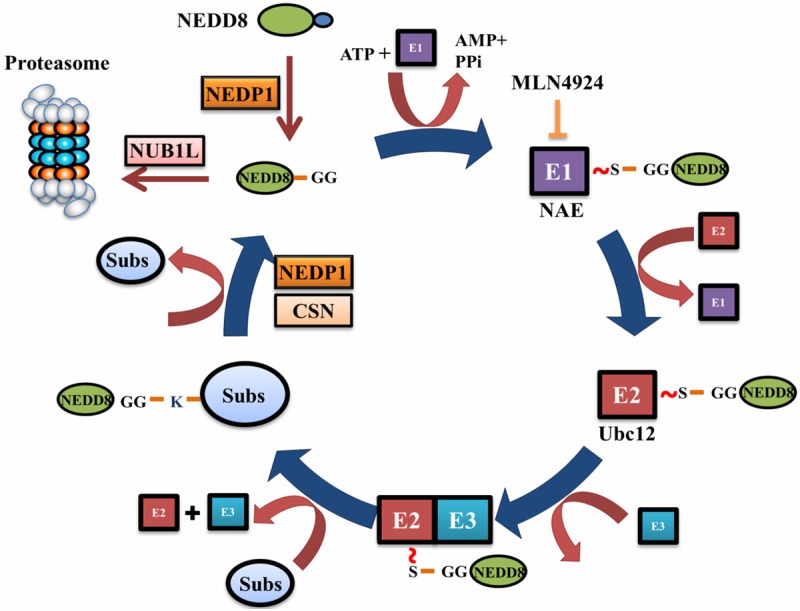
Protein modification by NEDD8. NEDD8 is translated into a precursor form that has to be processed by NEDD8 protease such as NEDP1 before NEDD8 can be attached to target proteins. Mature NEDD8 is conjugated to target proteins in an ATP-dependent manner by a serial reactions catalyzed by E1 (NAE), E2 (Ubc12) and E3 sequentially. With the help of these enzymes, the C-terminal glycine of NEDD8 forms an isopeptide bond with the lysine of target protein. An adenosine sulfamate analogue MLN4924 binds to the ATP-binding site in NAE and forms an irreversible MLN4924-NAE adduct, thus inhibiting the activation of neddylation. Deneddylases such as CSN and NEDP1 deconjugate NEDD8 from the neddylated proteins, freeing the substrate and NEDD8. NUB1L can specifically bind to and direct NEDD8 to the proteasome for degradation.
The structure of the neddylation enzymes is hierarchical. One or two E1s and E2s face towards a presumably huge and yet unknown number of E3s, which targets numerous substrates. NAE is the only dedicated NEDD8 E1. An adenosine sulfamate derivative MLN4924 can irreversibly form a NEDD8-MLN4924 adduct at the ATP-binding site of NEDD8, which prevents the binding of NAE to NEDD8, therefore specifically and potently inhibiting the activation of neddylation [19]. Besides NAE, the ubiquitin E1 activating enzyme UBE1 can also function as an atypical NEDD8 E1 to mediate the attachment of NEDD8 to the ubiquitin chain [20]. So far, only two E2s are identified, Ubc12 and UBE2F. In vivo, Ubc12 and UBE2F showed distinct patterns of cullin neddylation: Ubc12 activates neddylation of Cullins1-4 while UBE2F preferably activates neddylation of cullin 5 [21]. In yeast, DCN1 (defective in cullin neddylation 1 protein) is a bona fide E3 NEDD8 ligase by serving as a scaffold for the interaction of its cullin substrates and the NEDD8 E2 Ubc12 [22,23]. In mammalian cells, there are 5 DCN1-like proteins (DCNL), of which DCNL1 and DCNL3 facilitates cullin 1 and cullin 3 neddylation respectively [24,25]. The precise in vivo contributions of each DCNL protein to cullin neddylation remain to be defined. Other ubiquitin ligases such as MDM2, c-Cbl, parkin and IAP, can also function as the E3 NEDD8 ligases to promote the neddylation of various cellular proteins [26-30]. It is still mysterious how these ligases distinguish signals for neddylation from ubiquitination on the same substrate.
Proteins can be modified by one or multiple NEDD8 in trans, or by a chain of NEDD8 through extending on its own lysine residue. At least four of the nine lysines of NEDD8 (Lys 11, 22, 48, and 60) can be used for NEDD8 chain extension [31]. The NEDD8 chain is first built up on the E2 Ubc12 and subsequently transferred to the substrate [32]. Supporting the existence of polyneddylation, high-molecular-weight NEDD8-immunoreactive species were often observed when affinity purification of NEDD8 targets was carried out from cells expressing tagged NEDD8 [29,33-35]. It is well-accepted that the length and linkage types of the ubiquitin chain on a given substrate dictate the fate of the ubiquitinated proteins. In contrast, the biological significance of polyneddylation remains to be elucidated.
Deneddylation
Neddylation is dynamically regulated and counter balanced by the actions of deneddylases, termed deneddylation. A number of deneddylases are capable of removing NEDD8 from the modified proteins, including the COP9 signalosome (CSN), NEDP1, USP21, Ataxin-3, UCH-L1, and UCH-L3 [7]. Among these enzymes, only CSN and NEDP1 are NEDD8-specific, whereas the rest are also known to regulate deubiquitination [36,37].
CSN, a zinc metalloprotease containing 8 subunits from CSN1 to CSN8, is the best characterized deneddylase [38]. CSN was originally discovered in Arabidopsis and subsequently found to be evolutionarily conserved from plants to mammals. CSN is repeatedly demonstrated to remove NEDD8 from cullin proteins [38]. Loss of CSN function in mice accumulates neddylated cullins and various neddylated proteins with unknown identities, suggesting that CSN is also able to remove NEDD8 from non-cullin proteins [13,14]. The isopeptidase activity of CSN is conferred by the metalloenzyme-containing CSN5, but requires all 8 subunits to form a holo-complex. Loss of any CSN subunits disrupts the complex formation and impairs the deneddylation activity [39-43].
The cysteine protease NEDP1 is another specific deneddylase with no activity on processing ubiquitin and SUMO. Mutation of its cysteine to alanine suffices to abolish the protease activity [44]. Besides processing NEDD8 precursor, NEDP1 also removes NEDD8 from NEDD8 conjugates. Although NEDP1 was shown to efficiently deconjugate NEDD8 from neddylated cullin 1 and cullin 3 in vitro, deficiency of NEDP1 in Drosophila does not accumulate neddylated cullin 1 and cullin 3 [44-46]. Surprisingly, silencing of NEDP1 blunts stress-induced neddylation of cullin 1 and 2 in cultured mammalian cell lines, suggesting that NEDP1 may be necessary for cullin neddylation [9,47]. Probing its deneddylation activity in a loss-of-function mouse model, which so far is still lacking, will help explain the discrepancy. Despite its inconclusive role in cullin neddylation, NEDP1 appears to control the deneddylation of many non-cullin proteins such as p53, MDM2, Tap73, BCA3, and E2F1 [28,35,48-50]. Interestingly, CSN interacts with NEDP1 in Aspergillus nidulans and human cells and may target NEDP1 for proteasomal degradation [51]. Whether these two deneddylases coordinately regulate deneddylation in mammal remains to be determined.
NUB1L (NEDD8 ultimate buster 1 long), a negative regulator of neddylation
NUB1L is an interferon (IFN)-inducible protein that negatively regulates neddylation [52-56]. Overexpression of NUB1L in culture cells decreases the levels of NEDD8 and neddylated proteins, which is abrogated by proteasome inhibition [56,57]. It was first postulated that NUB1L directs neddylated proteins to the proteasome for degradation. This notion is now challenged by the observation that proteasome inhibition induces atypical neddylation in UBE1-dependent manner [20,58,59]. Therefore, the recovery of neddylated proteins by proteasome inhibition in NUB1L-overexpressed cells could be due to activation of atypical neddylation, rather than blockade of proteasomal degradation of neddylated proteins. Indeed, NUB1L specifically interacts with NEDD8, but not ubiquitin, and facilitates the transfer of NEDD8 to the proteasome for degradation through coordinating with P97-UFD1-NPL4 complex [60,61]. Thus, NUB1L suppresses neddylation by promoting the proteasomal degradation of NEDD8.
NUB1L is highly conserved among species [62]. Human NUB1L gene encodes two functionally nondifferentiable transcript variants, with the short form absent in other species. Structurally, NUB1L possesses a ubiquitin-like (UBL) domain, three ubiquitin-associated (UBA) domains and interacts with the 19S proteasome subunit Rpn10 and Rpn1 via its UBL domain [62,63]. Despite the similarity of NEDD8 and ubiquitin in sequence and structure, NUB1L binds to NEDD8 through its C-terminal sequence but not UBA domains [57]. Additionally NUB1L recognizes and promotes the degradation of another ubiquitin-like protein FAT10, whose substrates and function remain poorly defined [64].
Neddylated proteins and their biological functions
Neddylation targets
In contrast to a wide spectrum of proteins subjected to ubiquitin modification, the identities of the NEDD8 targets are poorly understood. Identification of NEDD8 targets is challenging owing to the relative low steady-state abundance of endogenous neddylated proteins and the antagonization of neddylation by deneddylases. In general, a bona fide NEDD8 target should meet at least a few of the criteria below: 1) the target protein is covalently modified by NEDD8 in vivo at its one or multiple lysine residues. This can be revealed by mass spectrometry analysis and/or mutagenesis analysis; 2) the neddylation should occur at endogenous condition in vivo and can be reconstituted in vitro; and 3) the neddylation relies on specific neddylation enzymes and/or is countered by deneddylases in vivo. Modulation of these enzyme activities, either by genetic or pharmacological means, should alter the neddylation status.
The NEDD8 proteome is quickly expanding. The first- and best-studied NEDD8 targets are cullin proteins that scaffold the assembly of cullin-RING ubiquitin ligases (CRLs) [65]. Besides cullins, an increasing list of cellular proteins are recently validated as NEDD8 substrates (Table 1) [26,27,30,33-35,49,66-72]. Importantly, many of these proteins such as Mdm2, p53, HIF1α, parkin, PINK1, and TGF-β receptor II (TGF-βRII) are known to regulate various cellular pathways important to cardiomyocyte survival and function, urging the needs to determine the impact of their neddylation in cardiomyocytes. The NEDD8 proteome is further enlarged by the finding that NEDD8 can be added into the existing ubiquitin chain [20], suggesting that any ubiquitinated proteins could be NEDD8 targets. By affinity purification of tagged-NEDD8 modified proteins followed by mass spectrometry analysis, several proteomics studies have identified a vast number of potential NEDD8 targets that regulate diverse cellular pathways involving DNA repair, replication, transcription and chromatin remodeling [31,73]. It should be pointed out that the di-glycine addition to the substrate upon trypsin digestion is common to NEDD8, ubiquitin and ISG15 modifications in mass spectrometry analysis. Other peptidase such as Lys-C has been suggested to distinguish NEDD8 from other ubiquitin-like protein modification by mass spectrum [20].
Table 1.
Neddylation targets
| Neddylated protein | E1/E2/E3 | NEDD8 Protease | Modified Sites | Consequence of Neddylation | Ref |
|---|---|---|---|---|---|
| Regulation of UPS | |||||
| Cullins | NAE/Ubc12/DCN1 | CSN | C-terminal lysine | Induces the assembly of CRLs | [65,74] |
| Parkin | NAE/-/- | - | K76 | Increases ubiquitin ligase activity | [33,34] |
| Mdm2 | -/Ubc12/Mdm2 | NEDP1 | - | Increases the stability | [27,50] |
| Smurf1 | -/Ubc12/Smurf1 | - | Multiple lysines | Increases ubiquitin ligase activity | [76] |
| XIAP | -/Ubc12/IAP | NEDP1 | - | ? | [30,126] |
| Ubiquitin | UBE1/-/- | CSN? | K29 | Terminate ubiquitin chain extension? | [20,79] |
| Regulation of transcription factors | |||||
| E2F1 | NAE/-/- | NEDP1 | Multiple lysines | Decreases the stability and transcriptional activity | [83,84] |
| HuR | -/-/Mdm2 | NEDP1 | K283, 313, 326 | Enables nuclear localization and stabilization, increases transcriptional activity | [26] |
| HIF1α | NAE/-/- | - | - | Increases protein stability and activity | [69] |
| AICD/APP | - | - | Multiple lysines | Inhibits the transcriptional activity | [127] |
| RCAN1 | - | - | K96, 104, 107 | Increases protein stability | [78] |
| BCA3 | - | NEDP1 | Multiple lysines | Increases its binding to p65; recruits SIRT1 | [49] |
| BRAP2 | - | - | K432 | Promotes NF-κB nuclear translocation and transcription activity | [128] |
| P73 | SMC/-/Mdm2 | NEDP1 | K321,327, 331 | Promotes cytoplasmic localization; inhibits transcription activity | [28] |
| Regulation of p53 signaling | |||||
| P53 | NAE/-/Mdm2 (FBOX11) | NEDP1 | K370, 372, 373 | Inhibit the transcriptional activity | [27,50,90] |
| L11 | - | NEDP1 | - | Increases stability; enhances P53 activity | [67] |
| S14 | -/-/HDM2 | NEDP1 | - | Increases stability; enhances P53 activity | [92] |
| Regulation of receptor-mediated signaling | |||||
| EGFR | NAE/Ubc12/c-Cbl | - | Multiple lysines | Reduces its stability | [72] |
| TGFβRII | NAE/Ubc12/c-Cbl | - | K556, 567 | Stabilizes TGFβRII, promotes TGFβ signaling | [66] |
| Regulation of cell death effectors | |||||
| Caspase7? | NAE/Ubc12/IAP1 | NEDP1 | K142 and others | Inhibits caspase activity | [30,126] |
| RIPK1 | -/-/cIAP1 | - | - | - | [30] |
| Caspase1? | NAE/-/- | - | - | Activates catalytic activity | [129] |
| Others | |||||
| pVHL | - | - | K159 | Inhibits its binding to cul2; Promotes fibronectin matrix assembly | [70,71] |
| PINK1 | NAE/-/- | - | - | Stabilizes its 55kD cleaved form | [33] |
| SHC | NAE/Ubc12/- | - | K3 | Forms ZAP70-SHC-Grb2 complex; activates Erk | [11] |
| Histone H4 | -/Ubc12/RNF111 | - | N-terminal lysines | Alters the chromatin orientation for DNA repair | [130] |
Like any other post-translational modification, neddylation influences protein function by changing the target protein’s conformation, half-life, subcellular distribution or DNA/protein binding partners. Based on the identified NEDD8 targets, we highlight a selection of studies from several cellular pathways, which are known to play crucial roles in cardiac physiology and pathology, to demonstrate the diverse functions of neddylation.
Regulation of the ubiquitin proteasome system (UPS)
Majority of the intracellular proteins are degraded through the UPS, which consists of two steps: ubiquitination of the target protein and the degradation by the proteasome. Ubiquitin ligases specifically recognize the target proteins and catalyze the ubiquitination, thereby controlling the specificity of UPS-mediated proteolysis. In recent years, there has been growing appreciation of the crosstalk between ubiquitination and neddylation. The NEDD8 pathway appears to impact UPS function at least through regulating ubiquitin ligase activity, antagonizing ubiquitination or controlling ubiquitin chain extension.
First, neddylation fine-tunes the activity of CRLs, the largest known class of ubiquitin ligases [65,74]. CRLs are typically composed of one of seven cullin family members (Cul-1, -2, -3, -4A, -4B, -5, -7), an E2 interacting RING finger protein such as Rbx1 or Rbx2, a F-box interacting adaptor protein such as Skp1, and a substrate-recognizing F-box protein such as Skp2. By pairing individual cullin with different F-box proteins, CRLs control the degradation of a vast number of cellular proteins and, not surprisingly, involve in many aspects of biological processes. In general, neddylation of cullin at their conserved C-terminal lysine residue induces conformational change of cullin, which dislodges the CRL inhibitory protein CAND1 from cullin and facilitates simultaneous binding of Rbx1 and Skp1 to cullin. Furthermore, Rbx1 recruits charged ubiquitin E2 while Skp1 associates with Skp2, which further recruits the substrate to be ubiquitinated. Therefore, neddylation of cullins triggers the assembly of functional CRLs, brings the ubiquitin charged E2 and the substrate to the proximity, and promotes the transfer of ubiquitin to the substrates. After completion of ubiquitination, CSN enables deneddylation of cullins, leading to the disassembly of CRL and release of the ubiquitin E2 and the substrate for the next round of ubiquitination. Although neddylation is required for the activation of CRLs, CSN-mediated deneddylation of cullins is also essential to optimal CRL activity. Defective CSN function causes sustained cullin neddylation, leading to autoubiquitination of CRL components and subsequently self-destruction [38,75]. Thus, dynamical cycling of neddylation and deneddylation represents an important mechanisms by which CRL activity is regulated (Figure 3).
Figure 3.
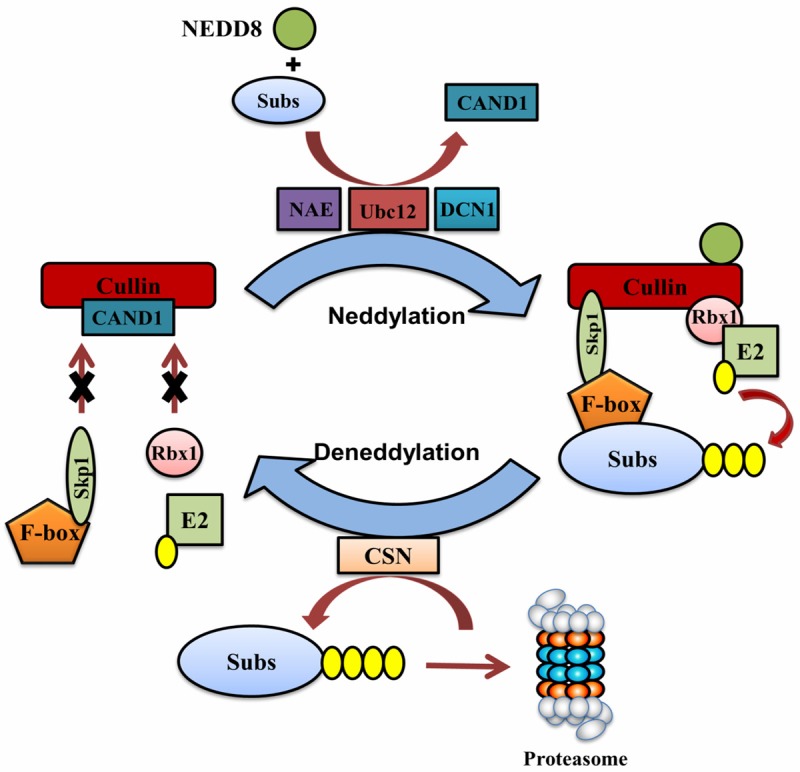
Regulation of cullin-RING ubiquitin ligase (CRL) activity by neddylation and deneddylation. CRL consists of a scaffold protein cullin, a RING protein Rbx1 that recruits ubiquitin E2, an adaptor protein Skp1 that interacts with a F-box protein, and a substrate (Subs)-recognizing F-box protein. NAE-Ubc12-DCN1-mediated neddylation of cullin changes the conformation of cullin, which prevents the binding of a CRL inhibitory protein CAND1 to cullin but allows the recruitment of Skp1 and Rbx1. The assembly of a functional CRL brings ubiquitin charged E2 and the substrate to the proximity, allowing the transfer of ubiquitin to the substrate. After ubiquitination, the deneddylase CSN removes NEDD8 from cullin, leading to the disassembly of CRL. The released cullin is then ready to recruit another ubiquitin charged E2 for next round of ubiquitination. Dynamic cycling of neddylation and deneddylation is essential for optimal CRL activity.
Besides cullins, non-cullin ubiquitin E3 ligases such as Mdm2, parkin and Smurf1 can also be modified by NEDD8 and such modification affects their ubiquitin ligase activities [27,33,34,76]. For instance, Smurf1 is a HECT ubiquitin ligase that plays a crucial role in multiple processes including cell cycle progression, cell proliferation, differentiation, maintenance of genomic stability and metastasis [77]. Smurf1 catalyzes its own neddylation on multiple lysine sites and requires an active site at cysteine 426 for the neddylation. Smurf1 neddylation is repressed by neddylation inhibitor MLN4924 or silencing of Uba3 and Ubc12 respectively. Neddylation of Smurf1 enhances its association with ubiquitin E2 and augments the ubiquitin ligase activity of Smurf1 [76]. In human colorectal cancers, the elevated expression of Smurf1, NEDD8, NAE1 and Ubc12 correlates with cancer progression.
Third, neddylation of a number of cellular proteins such as PINK1, HuR and RCAN1 were shown to antagonize ubiquitination and increase their stabilities [33,66,78]. For example, HuR is an oncogenic RNA-binding protein that regulates cell proliferation and survival. HuR is modified by NEDD8 at its lysine 283, 313 and 326, which is mediated by NEDD8 E3 ligase Mdm2. Neddylation of HuR suppresses its ubiquitination, increases its stability and promotes its nuclear localization [26]. It remains unclear whether NEDD8 competes with ubiquitin for a common lysine target to stabilize the modified protein.
Finally, proteins can be simultaneously modified by both NEDD8 and ubiquitin upon proteasome inhibition and other stresses, in a form of mixed ubiquitin-NEDD8 chain or “in trans” on different sites on the target [20,79]. In yeast and human cells, NEDD8 forms NEDD8-ubiquitin heterodimer with ubiquitin and can be extended on existing ubiquitin chain, possibly acting as a chain terminator [79]. In vitro, ubiquitin shuttle proteins and receptors recognize NEDD8-ubiquitin heterodimer with equivalent binding affinity to ubiquitin-ubiquitin homodimer [79]. Proteomic studies revealed the interaction of neddylated proteins with proteasome components [31]. Given the sequence and structure similarity between ubiquitin and NEDD8, it is possible that upon (proteotoxic) stress, NEDD8 functions as a substitute for ubiquitin and caps the ubiquitin chain, therefore preventing excessive ubiquitin chain extension and depletion of ubiquitin pool, which otherwise would be catastrophic to the cells. On the other hand, the heterologous ubiquitin-NEDD8 chains can be cleaved by CSN and 26S proteasome in vitro [79], suggesting that the proteins with mixed ubiquitin-NEDD8 chain may be still degraded by the proteasome, maybe at a less efficient rate compared to those modified with homologous ubiquitin chain. The functional consequence of such mixed ubiquitin-NEDD8 chain is to be determined.
Regulation of protein transcription
A number of transcription factors are found to be modified by NEDD8, which leads to either activation or suppression of their transcription activity. E2F1 is an important transcription factor that lies downstream of retinoblastoma protein, promotes apoptosis and suppresses cell proliferation [80]. E2F1 deficiency has been shown to suppress cardiomyocyte cell death and protect the heart against ischemia-reperfusion injury in mice [81,82]. Neddylation of E2F1 decreases its stability and inhibits the interaction with its transcription cofactor MCPH1, leading to reduced transcription activity. Moreover, overexpression of NEDP1 reduces neddylated E2F1 and promotes its transcription activity, while silencing of NEDP1 does the opposite [83,84]. Another example is HIF1α, a transcription factor that functions as a master regulator of oxygen homeostasis. HIF1α are protective to the pathogenesis of ischemic heart disease and pressure-overload heart disease [85]. Neddylation of HIF1α stabilizes and activates HIF1α at baseline and upon hypoxia, which is attenuated by silencing of NAE1 [69].
Neddylation can also indirectly regulate gene expression by modulation of the regulator of transcription factor. Calcineurin-NFAT signaling promotes the nuclear translocation of transcription factor NFAT and is necessary and sufficient to induce pathological cardiac hypertrophy [86]. RCAN1 is a feedback inhibitor of calcineurin by directly binding to the catalytic subunit of calcineurin and inhibiting its activity [87]. Neddylation of RCAN1 increases its stability by inhibiting ubiquitination and promotes its nuclear translocation and binding to calcineurin, leading to suppression of NFAT activity in HEK293 cells [78].
Regulation of p53 signaling
The tumor suppressor p53 plays an important role in apoptosis, autophagy and angiogenesis in the heart and has been linked to a number of cardiac disorders including cardiac hypertrophy, dilated cardiomyopathy, diabetic cardiomyopathy and cardiac ischemia reperfusion injury [88]. Mounting evidence suggests that neddylation regulates p53 signaling at different levels. First, p53 is the direct neddylation target. Neddylation of p53 prevents the nuclear translocation of p53 and inhibits its transcriptional activity. In contrast, neddylation-resistant mutants retain the transcriptional activity [27]. Consistently, overexpression or downregulation of NAE1 inhibits or enhances p53 activity respectively [89]. Second, FBXO11 and Mdm2 promote p53 neddylation by acting as E3 NEDD8 ligases [27,90]. The stability of Mdm2 and FBXO11 are directly or indirectly controlled by neddylation. Neddylation of Mdm2 increases its stability and NEDP1-induced deneddylation results in destabilization of Mdm2 [50]. FBXO11 is an F-box protein and a component of the Skp1-Cullin1- F-box ubiquitin ligase. As mentioned above, the stability of F-box protein is often affected by neddylation of cullins [91]. Third, ribosome proteins activate p53 upon nucleolar stress by binding to and inhibiting Mdm2. Ribosome protein L11 and S14 are identified as NEDD8 targets [68,92]. Neddylation of L11 and S14 controls their stability and subcellular localization, leading to p53 activation [67,68,92].
Regulation of receptor internalization
The epidermal growth factor receptor (EGFR), a receptor tyrosine kinase, is crucial to cardiac development and pathological hypertrophy [93,94]. EGFR can be directly modified by multiple NEDD8 molecules. Neddylation of EGFR is stimulated by EGF and enhances its ubiquitination, resulting in downregulation of EGFR. These effects are attenuated by either overexpression of dominant-negative mutant of Ubc12 or silencing of NEDD8 [72]. Therefore, neddylation of EGFR blunts its downstream signaling. In contrast, neddylation of TGFβreceptor II (TGFβ-RII) has positive effect on its downstream signaling [29]. TGFβRII is essential to TGFβ-mediated signaling and is critical to pathological cardiac remodeling [95]. Neddylation of TGFβRII occurs at its lysine556 and lysine567 at plasma membrane, and requires NEDD8 E2 Ubc12 and NEDD8 E3 c-Cbl, a ligase with dual function for both ubiquitination and neddylation. Neddylation of TGFβRII inhibits ubiquitination and subsequent degradation of TGFβRII by regulating receptor compartmentalization. Furthermore, a c-Cbl mutant identified from leukemia patients has defective neddylation activity, which may account for the low TGFβ responsiveness in human leukemia cells [29].
Regulation of mitochondria turnover
Mitochondria are cellular fuel plants critical for matching the energy demands of eukaryotic cells and precise control of mitochondria quality has to be achieved to prevent cellular damage. Defective mitochondria turnover has been linked to many forms of cardiac pathological conditions [96]. Although direct evidence is lacking, a couple of recent studies have suggested the potential roles of neddylation in mitochondria turnover [17,18]. Parkin and PINK1 are known to be important to mitochondria turnover and are implicated in Parkinson’s disease. It is generally believed that upon mitochondrial depolarization, the mitochondrial localized protein kinase PINK1 phosphorylates parkin, which leads to the translocation of parkin from cytoplasm to depolarized mitochondria, whereby parkin functions as a ubiquitin ligase to promote the ubiquitination of mitochondrial proteins and thus autophagic degradation of damaged mitochondria [97]. Both parkin and PINK1 can be neddylated [33,34]. Neddylation of parkin elevates its ubiquitin ligase activity, which results from enhanced association of parkin with ubiquitin E2 enzyme and the substrates. Neddylation of PINK1 leads to stabilization of a PINK1 fragment with unknown functional significance [33]. Moreover, expression of NAE1 partially rescues abnormalities induced by PINK1 knockdown in Drosophila, while neurotoxin MPP(+) suppresses the neddylation of parkin and PINK1 in HEK293 cells [33,34], suggesting that neddylation of parkin could be protective against stress. Further studies to determine the consequence of neddylation of parkin and PINK1 on mitochondria turnover are clearly warranted.
Neddylation and deneddylation are essential for embryonic development
Emerging evidence has revealed a critical role of the intact NEDD8 pathway in embryonic development. Disruption of either neddylation or deneddylation caused lethality at an early embryonic development stage in most model organisms (Table 2). Given the enormous capacity of neddylation in regulating CRL activity and that CRLs mediate the degradation of most cell cycle and survival regulators, it is anticipated that the observed phenotypes are more or less associated with aberrant cell cycle progression and prevalent cell death. Indeed, the Uba3-deficient mouse embryos displayed defective blastocyst formation and hatching at as early as embryonic day (E)3.5 and eventually died at the periimplantation stage [98]. Massive apoptosis and accumulation of cyclin E and CDK are evident in Uba3-deficient embryos at E6.5. Furthermore, suppression of deneddylation by germline deletion of any CSN subunit (CSN2, CSN3, CSN5, CSN6 and CSN8) in mice all results in early embryonic lethality [39-43]. CSN5 null embryos survived to the blastocyst stage with relatively smaller size, and died quickly after implantation without undergoing gastrulation. Increase of p27, p53, cyclin E and massive apoptosis were observed in the CSN5 null embryos, suggesting that defective cell cycle progression and apoptosis are at least in part attributable to the development arrest [41]. Interestingly, an attempt to generate transgenic mice overexpressing the E3 NEDD8 ligase SCRRO (DCNL1) has not been successful [99], raising the possibility that enhancement of neddylation may not be desired to embryonic development. In mice, cardiogenesis starts at around E7.5 and the development continues up to postnatal day 7 (P7) [100]. The early embryonic lethality of these knockout mice precludes the examination of the specific role of neddylation in cardiac development. Conditional deletion of neddylation or deneddylation enzymes in cardiac progenitor cells will be helpful to address this question.
Table 2.
Roles of components of the NEDD8 pathway in embryonic development
| Components | Phenotypes | Mechanisms | Species | Ref |
|---|---|---|---|---|
| NEDD8/Rub1 | ||||
| Rub1 & 2 mutant | Embryonic lethal or retarded growth | - | Arabidopsis | [131] |
| NEDD8 mutant | Embryonic lethal | Defects in cell proliferation and survival | Drosophila | [132] |
| NEDD8 KD | Embryonic lethal | - | C. elegans | [133] |
| NEDD8 conjugation enzymes | ||||
| ULA1* KD | Embryonic lethal | - | C. elegans | [133] |
| Uba3 KO | Embryonic lethal | Apoptosis and cell cycle arrest | Mouse | [98] |
| Ubc12 KD | Embryonic lethal | - | C. elegans | [133] |
| SCCRO KO | Viable | Compensation by other paralogues? | Mouse | [134] |
| SCCRO O/E | Embryonic lethal | - | Mouse | [99] |
| Deneddylase | ||||
| NEDP1 & CSN double mutant | Impaired multicellular development | - | A.nidulans | [51] |
| CSN4 & 5 mutant | Larval lethal | Defective oogenesis, hypersensitive to DNA damage | Drosophila | [135] |
| CSN8 mutant | Larval lethal | Defective oogenesis | Drosophila | [136] |
| CSN2 KO | Embryonic lethal | Cell cycle arrest | Mouse | [39] |
| CSN3 KO | Embryonic lethal | - | Mouse | [40] |
| CSN5 KO | Embryonic lethal | Impaired proliferation, apoptosis | Mouse | [41] |
| CSN6 KO | Embryonic lethal | Mouse | [42] | |
| CSN8 KO | Embryonic lethal | - | Mouse | [43] |
Abbreviations used in the table: KO, knockout; KD, knockdown; O/E overexpression;
Uba3 homologous in C. elegans;
C. elegans, Caenorhabditis elegans; A. nidulans, Aspergillus nidulans.
Impact of neddylation and deneddylation in cardiac disease
The importance of the neddylation in health and disease has begun to be understood, especially in the field of cancer. Neddylation is hyperactivated in several cancer cell lines [76]. Expression of NEDD8 E3 ligase SCCRO is oncogenic in mice [99]. The deneddylase CSN has also been closely linked to tumorigenesis [75]. Moreover, the specific and potent NAE inhibitor MLN4924 was shown to effectively suppress tumor growth in animal models and in clinical trials [7,101,102]. Targeting neddylation is now emerging as a novel anticancer therapeutic strategy.
Dysfunction of neddylation is also implicated in neurodegenerative disorders [8]. Accumulation of NEDD8 was frequently found in neuronal and glial inclusions from a number of neurodegenerative diseases [103]. NUB1L, the negative regulator of neddylation, promotes the degradation of mutant Huntington and synphilin-1 [55,104], prevents tau aggregation [105], and incorporates into the inclusion bodies in diseased brains [106-108], implying its potential role in protein quality control.
Our knowledge with respect to the importance of neddylation in other tissues, including the heart, is lagging behind. This is largely due to the lack of animal models in which neddylation/deneddylation is genetically manipulated in a tissue-specific manner. To this end, we have recently created a mouse model in which the eighth subunit of CSN (Csn8) gene is specifically deleted in cardiomyocytes [13,14,109]. Using this mouse model, we have obtained the first line of evidence for the impact of neddylation in cardiac homeostasis.
Deneddylase CSN is essential for the integrity of the structure and function of postnatal and adult heart
CSN8 is the smallest and least conserved CSN subunit. Using Cre-loxP strategy, we were able to specifically ablate Csn8 gene in mouse cardiomyocytes at both perinatal and adult stage. Loss of CSN8 in neonatal mouse hearts disrupts the formation of CSN complex and accumulates neddylated cullins and non-cullin proteins, confirming an essential role of CSN in deneddylation [13]. The CSN8-deficient hearts are indistinguishable from the littermate controls at 1 week of age but develop hypertrophy by 2 weeks of age, which quickly progresses into dilated cardiomyopathy with significantly reduced contractility and relaxation at 3 weeks of age. The neonatal CSN8-deficient mice eventually die of heart failure at around 4 weeks of age [13]. Similarly, loss of CSN8 in adult mouse hearts also greatly impairs CSN deneddylation activity and causes rapid heart failure and premature death [14]. These findings provide compelling evidence that CSN-mediated deneddylation is indispensable for the maintenance of cardiac structure and function.
CSN controls UPS-mediated proteolysis in the heart
Insufficient proteasomal proteolysis is being recognized as an important pathogenic factor to various forms of cardiac disease [110,111]. Although CSN has been long viewed as a UPS regulator due to its ability to control CRL activity and its structural similarity to the 19S proteasome lid [112], the in vivo experimental evidence is still lacking. By probing the UPS function with a reporter mouse, it was found that loss of CSN8 accumulated the UPS surrogate substrate in the hearts, suggesting severe impairment of UPS function. Consistently, massive ubiquitinated and oxidized proteins as well as protein aggregates were evident in CSN8-deficient mouse hearts [13,14]. Several chaperone proteins such as Hsp25, Hsp90 and α-Crystallin B (CryAB) were upregulated in CSN8-deficient hearts, presumably due to adaptive response to proteotoxic stress [13]. Together, these data indicate that CSN-mediated deneddylation is critical for proteasomal proteolysis in the heart and that the compromised UPS function can be attributed to the severe cardiac phenotype of CSN8-deficient mice.
Persistent neddylation of cullins led to destabilization of CRL components and accumulation of their substrates in non-cardiac cells [38,91]. In the heart, loss of CSN8 reduces the expression of several F-box proteins but surprisingly does not affect the abundances of their substrates, suggesting that CRL dysfunction does not likely account for the impairment of UPS function. Instead, CSN deficiency increases proteasome abundance and proteasomal peptidase activities [13,14]. Based on the accumulation of ubiquitinated proteins and contrasting elevated proteasomal peptidase activities, it is speculated that CSN dysfunction may uncouple the ubiquitination with the subsequent degradation of misfolded proteins in cardiomyocytes.
The mammalian 19S proteasome has an intrinsic “deubiquitinase” (DUB) subunit RPN11 to couple ubiquitination and proteasome degradation of the substrate. RPN11 removes ubiquitin from the substrate before it is translocated to the 20S proteasome to be hydrolyzed [113]. The release of ubiquitin prevents it from degradation, minimizing the fluctuations in ubiquitin pools. Thus, RPN11 is required for economic and efficient degradation of the substrates. It is recently found that NEDD8 can incorporate into and cap the ubiquitin chain, particularly under stress conditions [20,58]. Interestingly, CSN interacts with proteasome in vivo and cleave off NEDD8 from the ubiquitin chain in vitro [79]. Therefore, it is possible that CSN may facilitate proteasomal degradation of misfolded proteins by decapping NEDD8 from the mixed NEDD8-ubiquitin chain, in a way similar to processing of ubiquitin chain by RPN11.
CSN regulates autophagic proteolysis in the heart
Autophagy is a metabolic process with bulk protein degradation capacity [114]. It begins with membrane nucleation, which further elongates and sequestrates cytoplasmic contents including protein aggregates, damaged organelles and invading pathogens, to form autophagosome with characteristic double membranes. The autophagsome is then fused with lysosome to form autolysosome, whereby the sequestered contents are degraded by lysosomal enzymes. It becomes apparent that both excessive activation and suppression of autophagy contribute to the development of cardiac diseases [114,115].
Neddylation appears to regulate autophagy in the heart. CSN8 deficiency increases the abundance of autophagosomes, which is due to blockade of autophagsome clearance but not activation of autophagy [14,109]. The compromised autophagosome degradation is at least in part caused by a defect in the fusion of autophagsome with lysosome. Downregulation of Rab7, a critical protein for autophagosome maturation, likely accounts for the defective autophagosome maturation in CSN8-deficient heart [109]. These findings indicate that CSN-mediated deneddylation is crucial for autophagic degradation and that impaired autophagic function likely contributes to cardiac dysfunction in CSN8-deficient mice.
Additionally, neddylation also seems to control the initiation of autophagy, which is known to be repressed by activation of mTOR signaling. In cancer cells, administration of NAE inhibitor MLN4924 inactivates CRL and accumulates a mTOR inhibitory protein Deptor, leading to suppression of mTOR activity and activation of autophagy [116]. Consistently, silencing of a CRL component ROC1 also accumulates Deptor, inhibited mTOR activity and induced autophagy [117].
Neddylation and necrosis in the heart
Necrosis is a major form of cell death that is morphologically distinct from apoptosis [118]. Recognition of necrosis as a regulatory process has reignited cardiac researchers’ enthusiasm to understand how necrosis is regulated in the heart [119]. In CSN8-deficient mouse hearts, massive necrotic but not apoptotic cardiomyocytes were observed, suggesting that necrosis may be an underlying mechanism of cardiac dysfunction [13,14,109]. The necrotic cardiomyocytes were accumulated with autophagic vesicles in CSN8-deficient hearts, raising a possibility that the defective autophagy is causative to necrotic cell death [109]. Indeed, inhibition of autophagic degradation is associated with necrosis and cardiac disease [120,121]. In contrast to the absence of prevalent apoptosis in the heart, loss of CSN8 in hepatocytes induces massive apoptosis and impaired liver function [122,123], suggesting that CSN8-deficiency induced cell death may be cell-type specific.
Necrosis can occur in a highly regulated and genetically controlled manner. Recent studies have identified RIPK1-RIPK3 signaling as an underlying mechanism of TNFα-induced regulated necrosis [118]. Pharmacological or genetic inhibition of RIPK1-RIPK3 signaling has been shown to protect the heart against cardiac insults [124,125]. Interestingly, RIPK1 seems to be modified by NEDD8, which is mediated by IAP, a novel NEDD8 ligase [30]. It will be interesting to investigate whether CSN regulates necrosis through modulating RIPK1-RIK3 pathway in the heart.
Concluding remarks and future perspectives
Over the past decades, we have achieved a better understanding of the functional significance of neddylation and its association with diseases. Existing evidence strongly indicate that neddylation impacts profoundly on many basic cellular processes and that fine-tuning of neddylation is critical for the maintenance of cardiac structure and function. With assuming equal prominence to the ubiquitination and other ubiquitin-like protein modification, it is anticipated that virtually no complex cellular process will be untouched by neddylation. Therefore, a deeper understanding of the role of neddylation in cardiac development and pathological remodeling is needed for the development of novel therapeutic strategy to treat cardiac disease. In our opinion, future studies may be extended and directed to the following areas, using a combination of biochemical, proteomics, pharmacological and genetic approaches.
First of all, the association of dysregulation of neddylation with cardiac diseases remains to be further established. So far, no clinical evidence is available regarding the association of mutations within the NEDD8 networks with any inherited cardiomyopathies. Also, the functional status of neddylation/deneddylation in different forms of cardiac disorders remains to be defined, both in patients and in animal models.
Second, novel gain-of-function and loss-of-function mouse models awaits to be established to specifically target the major components of the NEDD8 pathway in the heart, including NAE, Ubc12, UBE2F, DCNL1/2/3/4/5, NEDP1 and NUB1L. Creation and characterization of these mice will not only provide us the entire picture of physiological functions of neddylation/deneddylation in the heart, but also differentiate distinct versus redundant functions between family members. These mice will also be valuable tools to dissect the impact of neddylation in cardiac pathological remodeling.
Third, efforts need to be directed to investigate cellular mechanisms by which neddylation controls the integrity of cardiomyocyte structure and function. Despite the observations on the involvement of neddylation in the UPS, autophagy and necrosis, the exact molecular mechanisms are to be elucidated. Considering the wide spectrum of NEDD8 targets in cardiomyocytes, novel processes regulated by neddylation remain to be discovered.
Finally, our understanding of protein neddylation will be strongly improved by systematic, unbiased and proteome-wide studies to identify NEDD8 targets in the heart, at both baseline and in response to physiological/pathological stimuli. This is relevant for better understanding of deregulation of neddylation in cardiac diseases and for the development of drugs that target components of neddylation signaling to battle cardiac diseases.
Acknowledgements
Our research was supported by NIH grant R01HL124248 (to H.S.), R01HL124251 (to I.K.), and American Heart Association grant 11SDG6960011 (to H.S.), 14SDG18970040 (to I.K.). We apologize to those whose work could not be cited due to space restrictions.
Disclosure of conflict of interest
None.
References
- 1.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 2.Grabbe C, Husnjak K, Dikic I. The spatial and temporal organization of ubiquitin networks. Nat Rev Mol Cell Biol. 2011;12:295–307. doi: 10.1038/nrm3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Popovic D, Vucic D, Dikic I. Ubiquitination in disease pathogenesis and treatment. Nat Med. 2014;20:1242–1253. doi: 10.1038/nm.3739. [DOI] [PubMed] [Google Scholar]
- 4.Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat Rev Mol Cell Biol. 2009;10:319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar S, Yoshida Y, Noda M. Cloning of a cDNA which encodes a novel ubiquitin-like protein. Biochem Biophys Res Commun. 1993;195:393–399. doi: 10.1006/bbrc.1993.2056. [DOI] [PubMed] [Google Scholar]
- 6.Kamitani T, Kito K, Nguyen HP, Yeh ET. Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J Biol Chem. 1997;272:28557–28562. doi: 10.1074/jbc.272.45.28557. [DOI] [PubMed] [Google Scholar]
- 7.Watson IR, Irwin MS, Ohh M. NEDD8 pathways in cancer, Sine Quibus Non. Cancer Cell. 2011;19:168–176. doi: 10.1016/j.ccr.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y, Neve RL, Liu H. Neddylation dysfunction in Alzheimer’s disease. J Cell Mol Med. 2012;16:2583–2591. doi: 10.1111/j.1582-4934.2012.01604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehrentraut SF, Kominsky DJ, Glover LE, Campbell EL, Kelly CJ, Bowers BE, Bayless AJ, Colgan SP. Central Role for Endothelial Human Deneddylase-1/SENP8 in Fine-Tuning the Vascular Inflammatory Response. J Immunol. 2013;190:392–400. doi: 10.4049/jimmunol.1202041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mathewson N, Toubai T, Kapeles S, Sun Y, Oravecz-Wilson K, Tamaki H, Wang Y, Hou G, Reddy P. Neddylation plays an important role in the regulation of murine and human dendritic cell function. Blood. 2013;122:2062–2073. doi: 10.1182/blood-2013-02-486373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin HS, Liao L, Park Y, Liu YC. Neddylation pathway regulates T-cell function by targeting an adaptor protein Shc and a protein kinase Erk signaling. Proc Natl Acad Sci U S A. 2013;110:624–629. doi: 10.1073/pnas.1213819110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stanley DJ, Bartholomeeusen K, Crosby DC, Kim DY, Kwon E, Yen L, Cartozo NC, Li M, Jager S, Mason-Herr J, Hayashi F, Yokoyama S, Krogan NJ, Harris RS, Peterlin BM, Gross JD. Inhibition of a NEDD8 Cascade Restores Restriction of HIV by APOBEC3G. PLoS Pathog. 2012;8:e1003085. doi: 10.1371/journal.ppat.1003085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su H, Li J, Menon S, Liu J, Kumarapeli AR, Wei N, Wang X. Perturbation of cullin deneddylation via conditional Csn8 ablation impairs the ubiquitin-proteasome system and causes cardiomyocyte necrosis and dilated cardiomyopathy in mice. Circ Res. 2011;108:40–50. doi: 10.1161/CIRCRESAHA.110.230607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su H, Li J, Osinska H, Li F, Robbins J, Liu J, Wei N, Wang X. The COP9 Signalosome Is Required for Autophagy, Proteasome-Mediated Proteolysis, and Cardiomyocyte Survival in Adult Mice. Circ Heart Fail. 2013;6:1049–57. doi: 10.1161/CIRCHEARTFAILURE.113.000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gan-Erdene T, Nagamalleswari K, Yin L, Wu K, Pan ZQ, Wilkinson KD. Identification and characterization of DEN1, a deneddylase of the ULP family. J Biol Chem. 2003;278:28892–28900. doi: 10.1074/jbc.M302890200. [DOI] [PubMed] [Google Scholar]
- 16.Frickel EM, Quesada V, Muething L, Gubbels MJ, Spooner E, Ploegh H, Artavanis-Tsakonas K. Apicomplexan UCHL3 retains dual specificity for ubiquitin and Nedd8 throughout evolution. Cell Microbiol. 2007;9:1601–1610. doi: 10.1111/j.1462-5822.2007.00896.x. [DOI] [PubMed] [Google Scholar]
- 17.Kwon J, Wang YL, Setsuie R, Sekiguchi S, Sato Y, Sakurai M, Noda M, Aoki S, Yoshikawa Y, Wada K. Two closely related ubiquitin C-terminal hydrolase isozymes function as reciprocal modulators of germ cell apoptosis in cryptorchid testis. Am J Pathol. 2004;165:1367–1374. doi: 10.1016/S0002-9440(10)63394-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gong L, Yeh ET. Identification of the activating and conjugating enzymes of the NEDD8 conjugation pathway. J Biol Chem. 1999;274:12036–12042. doi: 10.1074/jbc.274.17.12036. [DOI] [PubMed] [Google Scholar]
- 19.Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, Cullis CA, Doucette A, Garnsey JJ, Gaulin JL, Gershman RE, Lublinsky AR, McDonald A, Mizutani H, Narayanan U, Olhava EJ, Peluso S, Rezaei M, Sintchak MD, Talreja T, Thomas MP, Traore T, Vyskocil S, Weatherhead GS, Yu J, Zhang J, Dick LR, Claiborne CF, Rolfe M, Bolen JB, Langston SP. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 20.Leidecker O, Matic I, Mahata B, Pion E, Xirodimas DP. The ubiquitin E1 enzyme Ube1 mediates NEDD8 activation under diverse stress conditions. Cell Cycle. 2012;11:1142–1150. doi: 10.4161/cc.11.6.19559. [DOI] [PubMed] [Google Scholar]
- 21.Huang DT, Ayrault O, Hunt HW, Taherbhoy AM, Duda DM, Scott DC, Borg LA, Neale G, Murray PJ, Roussel MF, Schulman BA. E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol Cell. 2009;33:483–495. doi: 10.1016/j.molcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurz T, Chou YC, Willems AR, Meyer-Schaller N, Hecht ML, Tyers M, Peter M, Sicheri F. Dcn1 functions as a scaffold-type E3 ligase for cullin neddylation. Mol Cell. 2008;29:23–35. doi: 10.1016/j.molcel.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 23.Kurz T, Ozlu N, Rudolf F, O’Rourke SM, Luke B, Hofmann K, Hyman AA, Bowerman B, Peter M. The conserved protein DCN-1/Dcn1p is required for cullin neddylation in C. elegans and S. cerevisiae. Nature. 2005;435:1257–1261. doi: 10.1038/nature03662. [DOI] [PubMed] [Google Scholar]
- 24.Meyer-Schaller N, Chou YC, Sumara I, Martin DD, Kurz T, Katheder N, Hofmann K, Berthiaume LG, Sicheri F, Peter M. The human Dcn1-like protein DCNL3 promotes Cul3 neddylation at membranes. Proc Natl Acad Sci U S A. 2009;106:12365–12370. doi: 10.1073/pnas.0812528106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang G, Kaufman AJ, Ramanathan Y, Singh B. SCCRO (DCUN1D1) promotes nuclear translocation and assembly of the neddylation E3 complex. J Biol Chem. 2011;286:10297–10304. doi: 10.1074/jbc.M110.203729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Embade N, Fernandez-Ramos D, Varela-Rey M, Beraza N, Sini M, Gutierrez de Juan V, Woodhoo A, Martinez-Lopez N, Rodriguez-Iruretagoyena B, Bustamante FJ, de la Hoz AB, Carracedo A, Xirodimas DP, Rodriguez MS, Lu SC, Mato JM, Martinez-Chantar ML. Murine double minute 2 regulates Hu antigen R stability in human liver and colon cancer through NEDDylation. Hepatology. 2012;55:1237–1248. doi: 10.1002/hep.24795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xirodimas DP, Saville MK, Bourdon JC, Hay RT, Lane DP. Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell. 2004;118:83–97. doi: 10.1016/j.cell.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 28.Watson IR, Blanch A, Lin DC, Ohh M, Irwin MS. Mdm2-mediated NEDD8 modification of TAp73 regulates its transactivation function. J Biol Chem. 2006;281:34096–34103. doi: 10.1074/jbc.M603654200. [DOI] [PubMed] [Google Scholar]
- 29.Zuo W, Huang F, Chiang YJ, Li M, Du J, Ding Y, Zhang T, Lee HW, Jeong LS, Chen Y, Deng H, Feng XH, Luo S, Gao C, Chen YG. c-Cbl-mediated neddylation antagonizes ubiquitination and degradation of the TGF-beta type II receptor. Mol Cell. 2013;49:499–510. doi: 10.1016/j.molcel.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 30.Broemer M, Tenev T, Rigbolt KT, Hempel S, Blagoev B, Silke J, Ditzel M, Meier P. Systematic in vivo RNAi analysis identifies IAPs as NEDD8-E3 ligases. Mol Cell. 2010;40:810–822. doi: 10.1016/j.molcel.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 31.Jones J, Wu K, Yang Y, Guerrero C, Nillegoda N, Pan ZQ, Huang L. A targeted proteomic analysis of the ubiquitin-like modifier nedd8 and associated proteins. J Proteome Res. 2008;7:1274–1287. doi: 10.1021/pr700749v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohki Y, Funatsu N, Konishi N, Chiba T. The mechanism of poly-NEDD8 chain formation in vitro. Biochem Biophys Res Commun. 2009;381:443–447. doi: 10.1016/j.bbrc.2009.02.090. [DOI] [PubMed] [Google Scholar]
- 33.Choo YS, Vogler G, Wang D, Kalvakuri S, Iliuk A, Tao WA, Bodmer R, Zhang Z. Regulation of parkin and PINK1 by neddylation. Hum Mol Genet. 2012;21:2514–2523. doi: 10.1093/hmg/dds070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Um JW, Han KA, Im E, Oh Y, Lee K, Chung KC. Neddylation positively regulates the ubiquitin E3 ligase activity of parkin. J Neurosci Res. 2012;90:1030–1042. doi: 10.1002/jnr.22828. [DOI] [PubMed] [Google Scholar]
- 35.Aoki I, Higuchi M, Gotoh Y. NEDDylation controls the target specificity of E2F1 and apoptosis induction. Oncogene. 2012;32:3954–64. doi: 10.1038/onc.2012.428. [DOI] [PubMed] [Google Scholar]
- 36.Xirodimas DP. Novel substrates and functions for the ubiquitin-like molecule NEDD8. Biochem Soc Trans. 2008;36:802–806. doi: 10.1042/BST0360802. [DOI] [PubMed] [Google Scholar]
- 37.Rabut G, Peter M. Function and regulation of protein neddylation. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 2008;9:969–976. doi: 10.1038/embor.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei N, Serino G, Deng XW. The COP9 signalosome: more than a protease. Trends Biochem Sci. 2008;33:592–600. doi: 10.1016/j.tibs.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 39.Lykke-Andersen K, Schaefer L, Menon S, Deng XW, Miller JB, Wei N. Disruption of the COP9 signalosome Csn2 subunit in mice causes deficient cell proliferation, accumulation of p53 and cyclin E, and early embryonic death. Mol Cell Biol. 2003;23:6790–6797. doi: 10.1128/MCB.23.19.6790-6797.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan J, Walz K, Nakamura H, Carattini-Rivera S, Zhao Q, Vogel H, Wei N, Justice MJ, Bradley A, Lupski JR. COP9 signalosome subunit 3 is essential for maintenance of cell proliferation in the mouse embryonic epiblast. Mol Cell Biol. 2003;23:6798–6808. doi: 10.1128/MCB.23.19.6798-6808.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomoda K, Yoneda-Kato N, Fukumoto A, Yamanaka S, Kato JY. Multiple functions of Jab1 are required for early embryonic development and growth potential in mice. J Biol Chem. 2004;279:43013–43018. doi: 10.1074/jbc.M406559200. [DOI] [PubMed] [Google Scholar]
- 42.Zhao R, Yeung SC, Chen J, Iwakuma T, Su CH, Chen B, Qu C, Zhang F, Chen YT, Lin YL, Lee DF, Jin F, Zhu R, Shaikenov T, Sarbassov D, Sahin A, Wang H, Lai CC, Tsai FJ, Lozano G, Lee MH. Subunit 6 of the COP9 signalosome promotes tumorigenesis in mice through stabilization of MDM2 and is upregulated in human cancers. J Clin Invest. 2011;121:851–865. doi: 10.1172/JCI44111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Menon S, Chi H, Zhang H, Deng XW, Flavell RA, Wei N. COP9 signalosome subunit 8 is essential for peripheral T cell homeostasis and antigen receptor-induced entry into the cell cycle from quiescence. Nat Immunol. 2007;8:1236–1245. doi: 10.1038/ni1514. [DOI] [PubMed] [Google Scholar]
- 44.Mendoza HM, Shen LN, Botting C, Lewis A, Chen J, Ink B, Hay RT. NEDP1, a highly conserved cysteine protease that deNEDDylates Cullins. J Biol Chem. 2003;278:25637–25643. doi: 10.1074/jbc.M212948200. [DOI] [PubMed] [Google Scholar]
- 45.Chan Y, Yoon J, Wu JT, Kim HJ, Pan KT, Yim J, Chien CT. DEN1 deneddylates non-cullin proteins in vivo. J Cell Sci. 2008;121:3218–3223. doi: 10.1242/jcs.030445. [DOI] [PubMed] [Google Scholar]
- 46.Wu K, Yamoah K, Dolios G, Gan-Erdene T, Tan P, Chen A, Lee CG, Wei N, Wilkinson KD, Wang R, Pan ZQ. DEN1 is a dual function protease capable of processing the C terminus of Nedd8 and deconjugating hyper-neddylated CUL1. J Biol Chem. 2003;278:28882–28891. doi: 10.1074/jbc.M302888200. [DOI] [PubMed] [Google Scholar]
- 47.Curtis VF, Ehrentraut SF, Campbell EL, Glover LE, Bayless A, Kelly CJ, Kominsky DJ, Colgan SP. Stabilization of HIF through inhibition of Cullin-2 neddylation is protective in mucosal inflammatory responses. FASEB J. 2014 doi: 10.1096/fj.14-259663. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen LN, Liu H, Dong C, Xirodimas D, Naismith JH, Hay RT. Structural basis of NEDD8 ubiquitin discrimination by the deNEDDylating enzyme NEDP1. EMBO J. 2005;24:1341–1351. doi: 10.1038/sj.emboj.7600628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao F, Cheng J, Shi T, Yeh ET. Neddylation of a breast cancer-associated protein recruits a class III histone deacetylase that represses NFkappaB-dependent transcription. Nat Cell Biol. 2006;8:1171–1177. doi: 10.1038/ncb1483. [DOI] [PubMed] [Google Scholar]
- 50.Watson IR, Li BK, Roche O, Blanch A, Ohh M, Irwin MS. Chemotherapy induces NEDP1-mediated destabilization of MDM2. Oncogene. 2010;29:297–304. doi: 10.1038/onc.2009.314. [DOI] [PubMed] [Google Scholar]
- 51.Christmann M, Schmaler T, Gordon C, Huang X, Bayram O, Schinke J, Stumpf S, Dubiel W, Braus GH. Control of multicellular development by the physically interacting deneddylases DEN1/DenA and COP9 signalosome. PLoS Genet. 2013;9:e1003275. doi: 10.1371/journal.pgen.1003275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tanaka T, Nakatani T, Kamitani T. Inhibition of NEDD8-conjugation pathway by novel molecules: potential approaches to anticancer therapy. Mol Oncol. 2012;6:267–275. doi: 10.1016/j.molonc.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Su V, Lau AF. Ubiquitin-like and ubiquitin-associated domain proteins: significance in proteasomal degradation. Cell Mol Life Sci. 2009;66:2819–2833. doi: 10.1007/s00018-009-0048-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hosono T, Tanaka T, Tanji K, Nakatani T, Kamitani T. NUB1, an interferon-inducible protein, mediates anti-proliferative actions and apoptosis in renal cell carcinoma cells through cell-cycle regulation. Br J Cancer. 2010;102:873–882. doi: 10.1038/sj.bjc.6605574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu B, Al-Ramahi I, Valencia A, Wang Q, Berenshteyn F, Yang H, Gallego-Flores T, Ichcho S, Lacoste A, Hild M, Difiglia M, Botas J, Palacino J. Identification of NUB1 as a suppressor of mutant Huntington toxicity via enhanced protein clearance. Nat Neurosci. 2013;16:562–570. doi: 10.1038/nn.3367. [DOI] [PubMed] [Google Scholar]
- 56.Kito K, Yeh ET, Kamitani T. NUB1, a NEDD8-interacting protein, is induced by interferon and down-regulates the NEDD8 expression. J Biol Chem. 2001;276:20603–20609. doi: 10.1074/jbc.M100920200. [DOI] [PubMed] [Google Scholar]
- 57.Kamitani T, Kito K, Fukuda-Kamitani T, Yeh ET. Targeting of NEDD8 and its conjugates for proteasomal degradation by NUB1. J Biol Chem. 2001;276:46655–46660. doi: 10.1074/jbc.M108636200. [DOI] [PubMed] [Google Scholar]
- 58.Hjerpe R, Thomas Y, Chen J, Zemla A, Curran S, Shpiro N, Dick LR, Kurz T. Changes in the ratio of free NEDD8 to ubiquitin triggers NEDDylation by ubiquitin enzymes. Biochem J. 2012;441:927–936. doi: 10.1042/BJ20111671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hjerpe R, Thomas Y, Kurz T. NEDD8 overexpression results in neddylation of ubiquitin substrates by the ubiquitin pathway. J Mol Biol. 2012;421:27–29. doi: 10.1016/j.jmb.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 60.Liu S, Yang H, Zhao J, Zhang YH, Song AX, Hu HY. NEDD8 ultimate buster-1 long (NUB1L) protein promotes transfer of NEDD8 to proteasome for degradation through the P97UFD1/NPL4 complex. J Biol Chem. 2013;288:31339–49. doi: 10.1074/jbc.M113.484816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu S, Fu QS, Zhao J, Hu HY. Structural and mechanistic insights into the arginine/lysine-rich peptide motifs that interact with P97/VCP. Biochim Biophys Acta. 2013;1834:2672–2678. doi: 10.1016/j.bbapap.2013.09.021. [DOI] [PubMed] [Google Scholar]
- 62.Tanaka T, Kawashima H, Yeh ET, Kamitani T. Regulation of the NEDD8 conjugation system by a splicing variant, NUB1L. J Biol Chem. 2003;278:32905–32913. doi: 10.1074/jbc.M212057200. [DOI] [PubMed] [Google Scholar]
- 63.Tanji K, Tanaka T, Kamitani T. Interaction of NUB1 with the proteasome subunit S5a. Biochem Biophys Res Commun. 2005;337:116–120. doi: 10.1016/j.bbrc.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 64.Schmidtke G, Aichem A, Groettrup M. FAT10ylation as a signal for proteasomal degradation. Biochimica et Biophysica Acta. 2013;1843:97–102. doi: 10.1016/j.bbamcr.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 65.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 66.Zuo W, Huang F, Chiang YJ, Li M, Du J, Ding Y, Zhang T, Lee HW, Jeong LS, Chen Y, Deng H, Feng XH, Luo S, Gao C, Chen YG. c-Cbl-Mediated Neddylation Antagonizes Ubiquitination and Degradation of the TGF-beta Type II Receptor. Mol Cell. 2013;49:499–510. doi: 10.1016/j.molcel.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 67.Mahata B, Sundqvist A, Xirodimas DP. Recruitment of RPL11 at promoter sites of p53-regulated genes upon nucleolar stress through NEDD8 and in an Mdm2-dependent manner. Oncogene. 2012;31:3060–3071. doi: 10.1038/onc.2011.482. [DOI] [PubMed] [Google Scholar]
- 68.Sundqvist A, Liu G, Mirsaliotis A, Xirodimas DP. Regulation of nucleolar signalling to p53 through NEDDylation of L11. EMBO Rep. 2009;10:1132–1139. doi: 10.1038/embor.2009.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ryu JH, Li SH, Park HS, Park JW, Lee B, Chun YS. Hypoxia-inducible factor alpha subunit stabilization by NEDD8 conjugation is reactive oxygen species-dependent. J Biol Chem. 2011;286:6963–6970. doi: 10.1074/jbc.M110.188706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Russell RC, Ohh M. NEDD8 acts as a ‘molecular switch’ defining the functional selectivity of VHL. EMBO Rep. 2008;9:486–491. doi: 10.1038/embor.2008.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stickle NH, Chung J, Klco JM, Hill RP, Kaelin WG Jr, Ohh M. pVHL modification by NEDD8 is required for fibronectin matrix assembly and suppression of tumor development. Mol Cell Biol. 2004;24:3251–3261. doi: 10.1128/MCB.24.8.3251-3261.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oved S, Mosesson Y, Zwang Y, Santonico E, Shtiegman K, Marmor MD, Kochupurakkal BS, Katz M, Lavi S, Cesareni G, Yarden Y. Conjugation to Nedd8 instigates ubiquitylation and down-regulation of activated receptor tyrosine kinases. J Biol Chem. 2006;281:21640–21651. doi: 10.1074/jbc.M513034200. [DOI] [PubMed] [Google Scholar]
- 73.Xirodimas DP, Sundqvist A, Nakamura A, Shen L, Botting C, Hay RT. Ribosomal proteins are targets for the NEDD8 pathway. EMBO Rep. 2008;9:280–286. doi: 10.1038/embor.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lipkowitz S, Weissman AM. RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat Rev Cancer. 2011;11:629–643. doi: 10.1038/nrc3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee MH, Zhao R, Phan L, Yeung SC. Roles of COP9 signalosome in cancer. Cell Cycle. 2011;10:3057–3066. doi: 10.4161/cc.10.18.17320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xie P, Zhang M, He S, Lu K, Chen Y, Xing G, Lu Y, Liu P, Li Y, Wang S, Chai N, Wu J, Deng H, Wang HR, Cao Y, Zhao F, Cui Y, Wang J, He F, Zhang L. The covalent modifier Nedd8 is critical for the activation of Smurf1 ubiquitin ligase in tumorigenesis. Nat Commun. 2014;5:3733. doi: 10.1038/ncomms4733. [DOI] [PubMed] [Google Scholar]
- 77.David D, Nair SA, Pillai MR. Smurf E3 ubiquitin ligases at the cross roads of oncogenesis and tumor suppression. Biochim Biophys Acta. 2013;1835:119–128. doi: 10.1016/j.bbcan.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 78.Noh EH, Hwang HS, Min B, Im E, Chung KC. Covalent NEDD8 conjugation increases RCAN1 protein stability and potentiates its inhibitory action on calcineurin. PLoS One. 2012;7:e48315. doi: 10.1371/journal.pone.0048315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Singh RK, Zerath S, Kleifeld O, Scheffner M, Glickman MH, Fushman D. Recognition and cleavage of related to ubiquitin 1 (Rub1) and Rub1-ubiquitin chains by components of the ubiquitin-proteasome system. Mol Cell Proteomics. 2012;11:1595–1611. doi: 10.1074/mcp.M112.022467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Meng P, Ghosh R. Transcription addiction: can we garner the Yin and Yang functions of E2F1 for cancer therapy? Cell Death Dis. 2014;5:e1360. doi: 10.1038/cddis.2014.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Angelis E, Zhao P, Zhang R, Goldhaber JI, Maclellan WR. The role of E2F-1 and downstream target genes in mediating ischemia/reperfusion injury in vivo. J Mol Cell Cardiol. 2011;51:919–926. doi: 10.1016/j.yjmcc.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang K, An T, Zhou LY, Liu CY, Zhang XJ, Feng C, Li PF. E2F1-regulated miR-30b suppresses Cyclophilin D and protects heart from ischemia/reperfusion injury and necrotic cell death. Cell Death Differ. 2014 doi: 10.1038/cdd.2014.165. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Loftus SJ, Liu G, Carr SM, Munro S, La Thangue NB. NEDDylation regulates E2F-1-dependent transcription. EMBO Rep. 2012;13:811–818. doi: 10.1038/embor.2012.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Aoki I, Higuchi M, Gotoh Y. NEDDylation controls the target specificity of E2F1 and apoptosis induction. Oncogene. 2013;32:3954–3964. doi: 10.1038/onc.2012.428. [DOI] [PubMed] [Google Scholar]
- 85.Semenza GL. Hypoxia-inducible factor 1 and cardiovascular disease. Annu Rev Physiol. 2014;76:39–56. doi: 10.1146/annurev-physiol-021113-170322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Molkentin JD. Calcineurin-NFAT signaling regulates the cardiac hypertrophic response in coordination with the MAPKs. Cardiovasc Res. 2004;63:467–475. doi: 10.1016/j.cardiores.2004.01.021. [DOI] [PubMed] [Google Scholar]
- 87.Rothermel BA, Vega RB, Williams RS. The role of modulatory calcineurin-interacting proteins in calcineurin signaling. Trends Cardiovasc Med. 2003;13:15–21. doi: 10.1016/s1050-1738(02)00188-3. [DOI] [PubMed] [Google Scholar]
- 88.Portbury AL, Ronnebaum SM, Zungu M, Patterson C, Willis MS. Back to your heart: ubiquitin proteasome system-regulated signal transduction. J Mol Cell Cardiol. 2012;52:526–537. doi: 10.1016/j.yjmcc.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Guihard S, Ramolu L, Macabre C, Wasylyk B, Noel G, Abecassis J, Jung AC. The NEDD8 conjugation pathway regulates p53 transcriptional activity and head and neck cancer cell sensitivity to ionizing radiation. Int J Oncol. 2012;41:1531–1540. doi: 10.3892/ijo.2012.1584. [DOI] [PubMed] [Google Scholar]
- 90.Abida WM, Nikolaev A, Zhao W, Zhang W, Gu W. FBXO11 promotes the Neddylation of p53 and inhibits its transcriptional activity. J Biol Chem. 2007;282:1797–1804. doi: 10.1074/jbc.M609001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cope GA, Deshaies RJ. Targeted silencing of Jab1/Csn5 in human cells downregulates SCF activity through reduction of F-box protein levels. BMC Biochem. 2006;7:1. doi: 10.1186/1471-2091-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang J, Bai D, Ma X, Guan J, Zheng X. hCINAP is a novel regulator of ribosomal protein-HDM2-p53 pathway by controlling NEDDylation of ribosomal protein S14. Oncogene. 2012;33:246–54. doi: 10.1038/onc.2012.560. [DOI] [PubMed] [Google Scholar]
- 93.Lee KF, Simon H, Chen H, Bates B, Hung MC, Hauser C. Requirement for neuregulin receptor erbB2 in neural and cardiac development. Nature. 1995;378:394–398. doi: 10.1038/378394a0. [DOI] [PubMed] [Google Scholar]
- 94.Schreier B, Rabe S, Schneider B, Bretschneider M, Rupp S, Ruhs S, Neumann J, Rueckschloss U, Sibilia M, Gotthardt M, Grossmann C, Gekle M. Loss of epidermal growth factor receptor in vascular smooth muscle cells and cardiomyocytes causes arterial hypotension and cardiac hypertrophy. Hypertension. 2013;61:333–340. doi: 10.1161/HYPERTENSIONAHA.112.196543. [DOI] [PubMed] [Google Scholar]
- 95.Koitabashi N, Danner T, Zaiman AL, Pinto YM, Rowell J, Mankowski J, Zhang D, Nakamura T, Takimoto E, Kass DA. Pivotal role of cardiomyocyte TGF-beta signaling in the murine pathological response to sustained pressure overload. J Clin Invest. 2011;121:2301–2312. doi: 10.1172/JCI44824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Andres AM, Stotland A, Queliconi BB, Gottlieb RA. A time to reap, a time to sow: Mitophagy and biogenesis in cardiac pathophysiology. J Mol Cell Cardiol. 2015;78C:62–72. doi: 10.1016/j.yjmcc.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ni HM, Williams JA, Ding WX. Mitochondrial dynamics and mitochondrial quality control. Redox Biol. 2014;4C:6–13. doi: 10.1016/j.redox.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tateishi K, Omata M, Tanaka K, Chiba T. The NEDD8 system is essential for cell cycle progression and morphogenetic pathway in mice. J Cell Biol. 2001;155:571–579. doi: 10.1083/jcb.200104035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Broderick SR, Golas BJ, Pham D, Towe CW, Talbot SG, Kaufman A, Bains S, Huryn LA, Yonekawa Y, Carlson D, Hambardzumyan D, Ramanathan Y, Singh B. SCCRO promotes glioma formation and malignant progression in mice. Neoplasia. 2010;12:476–484. doi: 10.1593/neo.10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xin M, Olson EN, Bassel-Duby R. Mending broken hearts: cardiac development as a basis for adult heart regeneration and repair. Nat Rev Mol Cell Biol. 2013;14:529–541. doi: 10.1038/nrm3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Duncan K, Schafer G, Vava A, Parker MI, Zerbini LF. Targeting neddylation in cancer therapy. Future Oncol. 2012;8:1461–1470. doi: 10.2217/fon.12.131. [DOI] [PubMed] [Google Scholar]
- 102.Nawrocki ST, Griffin P, Kelly KR, Carew JS. MLN4924: a novel first-in-class inhibitor of NEDD8-activating enzyme for cancer therapy. Expert Opin Investig Drugs. 2012;21:1563–1573. doi: 10.1517/13543784.2012.707192. [DOI] [PubMed] [Google Scholar]
- 103.Mori F, Nishie M, Piao YS, Kito K, Kamitani T, Takahashi H, Wakabayashi K. Accumulation of NEDD8 in neuronal and glial inclusions of neurodegenerative disorders. Neuropathol Appl Neurobiol. 2005;31:53–61. doi: 10.1111/j.1365-2990.2004.00603.x. [DOI] [PubMed] [Google Scholar]
- 104.Tanji K, Tanaka T, Mori F, Kito K, Takahashi H, Wakabayashi K, Kamitani T. NUB1 suppresses the formation of Lewy body-like inclusions by proteasomal degradation of synphilin-1. Am J Pathol. 2006;169:553–565. doi: 10.2353/ajpath.2006.051067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Richet E, Pooler AM, Rodriguez T, Novoselov SS, Schmidtke G, Groettrup M, Hanger DP, Cheetham ME, van der Spuy J. NUB1 modulation of GSK3beta reduces tau aggregation. Hum Mol Genet. 2012;21:5254–5267. doi: 10.1093/hmg/dds376. [DOI] [PubMed] [Google Scholar]
- 106.Tanji K, Mori F, Kakita A, Zhang H, Kito K, Kamitani T, Takahashi H, Wakabayashi K. Immunohistochemical localization of NUB1, a synphilin-1-binding protein, in neurodegenerative disorders. Acta Neuropathol. 2007;114:365–371. doi: 10.1007/s00401-007-0238-1. [DOI] [PubMed] [Google Scholar]
- 107.Tanji K, Mori F, Kito K, Kakita A, Mimura J, Itoh K, Takahashi H, Kamitani T, Wakabayashi K. Synphilin-1-binding protein NUB1 is colocalized with nonfibrillar, proteinase K-resistant alphasynuclein in presynapses in Lewy body disease. J Neuropathol Exp Neurol. 2011;70:879–889. doi: 10.1097/NEN.0b013e3182303745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Odagiri S, Mori F, Tanji K, Kuroda N, Wakabayashi K. Immunohistochemical study of microscopic globular bodies of normal human brain. Biomed Res. 2011;32:337–342. doi: 10.2220/biomedres.32.337. [DOI] [PubMed] [Google Scholar]
- 109.Su H, Li F, Ranek MJ, Wei N, Wang X. COP9 signalosome regulates autophagosome maturation. Circulation. 2011;124:2117–2128. doi: 10.1161/CIRCULATIONAHA.111.048934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Su H, Wang X. The ubiquitin-proteasome system in cardiac proteinopathy: a quality control perspective. Cardiovasc Res. 2010;85:253–262. doi: 10.1093/cvr/cvp287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang X, Robbins J. Proteasomal and lysosomal protein degradation and heart disease. J Mol Cell Cardiol. 2014;71:16–24. doi: 10.1016/j.yjmcc.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li L, Deng XW. The COP9 signalosome: an alternative lid for the 26S proteasome? Trends Cell Biol. 2003;13:507–509. doi: 10.1016/j.tcb.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 113.Verma R, Aravind L, Oania R, McDonald WH, Yates JR 3rd, Koonin EV, Deshaies RJ. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 114.Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov. 2012;11:709–730. doi: 10.1038/nrd3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nemchenko A, Chiong M, Turer A, Lavandero S, Hill JA. Autophagy as a therapeutic target in cardiovascular disease. J Mol Cell Cardiol. 2011;51:584–593. doi: 10.1016/j.yjmcc.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Luo Z, Yu G, Lee HW, Li L, Wang L, Yang D, Pan Y, Ding C, Qian J, Wu L, Chu Y, Yi J, Wang X, Sun Y, Jeong LS, Liu J, Jia L. The Nedd8-activating enzyme inhibitor MLN4924 induces autophagy and apoptosis to suppress liver cancer cell growth. Cancer Res. 2012;72:3360–3371. doi: 10.1158/0008-5472.CAN-12-0388. [DOI] [PubMed] [Google Scholar]
- 117.Yang D, Li L, Liu H, Wu L, Luo Z, Li H, Zheng S, Gao H, Chu Y, Sun Y, Liu J, Jia L. Induction of autophagy and senescence by knockdown of ROC1 E3 ubiquitin ligase to suppress the growth of liver cancer cells. Cell Death Differ. 2013;20:235–247. doi: 10.1038/cdd.2012.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Vanden Berghe T, Linkermann A, Jouan-Lanhouet S, Walczak H, Vandenabeele P. Regulated necrosis: the expanding network of non-apoptotic cell death pathways. Nat Rev Mol Cell Biol. 2014;15:135–147. doi: 10.1038/nrm3737. [DOI] [PubMed] [Google Scholar]
- 119.Kung G, Konstantinidis K, Kitsis RN. Programmed necrosis, not apoptosis, in the heart. Circ Res. 2011;108:1017–1036. doi: 10.1161/CIRCRESAHA.110.225730. [DOI] [PubMed] [Google Scholar]
- 120.Kostin S, Pool L, Elsasser A, Hein S, Drexler HC, Arnon E, Hayakawa Y, Zimmermann R, Bauer E, Klovekorn WP, Schaper J. Myocytes die by multiple mechanisms in failing human hearts. Circ Res. 2003;92:715–724. doi: 10.1161/01.RES.0000067471.95890.5C. [DOI] [PubMed] [Google Scholar]
- 121.Miyata S, Takemura G, Kawase Y, Li Y, Okada H, Maruyama R, Ushikoshi H, Esaki M, Kanamori H, Li L, Misao Y, Tezuka A, Toyo-Oka T, Minatoguchi S, Fujiwara T, Fujiwara H. Autophagic cardiomyocyte death in cardiomyopathic hamsters and its prevention by granulocyte colony-stimulating factor. Am J Pathol. 2006;168:386–397. doi: 10.2353/ajpath.2006.050137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lei D, Li F, Su H, Liu J, Wei N, Wang X. Hepatic Deficiency of COP9 Signalosome Subunit 8 Induces Ubiquitin-Proteasome System Impairment and Bim-Mediated Apoptosis in Murine Livers. PLoS One. 2013;8:e67793. doi: 10.1371/journal.pone.0067793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lei D, Li F, Su H, Tian Z, Ye B, Wei N, Wang X. COP9 signalosome subunit 8 is required for postnatal hepatocyte survival and effective proliferation. Cell Death Differ. 2011;18:259–270. doi: 10.1038/cdd.2010.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Oerlemans MI, Liu J, Arslan F, den Ouden K, van Middelaar BJ, Doevendans PA, Sluijter JP. Inhibition of RIP1-dependent necrosis prevents adverse cardiac remodeling after myocardial ischemia-reperfusion in vivo. Basic Res Cardiol. 2012;107:270. doi: 10.1007/s00395-012-0270-8. [DOI] [PubMed] [Google Scholar]
- 125.Luedde M, Lutz M, Carter N, Sosna J, Jacoby C, Vucur M, Gautheron J, Roderburg C, Borg N, Reisinger F, Hippe HJ, Linkermann A, Wolf MJ, Rose-John S, Lullmann-Rauch R, Adam D, Flogel U, Heikenwalder M, Luedde T, Frey N. RIP3, a kinase promoting necroptotic cell death, mediates adverse remodelling after myocardial infarction. Cardiovasc Res. 2014;103:206–216. doi: 10.1093/cvr/cvu146. [DOI] [PubMed] [Google Scholar]
- 126.Nagano T, Hashimoto T, Nakashima A, Kikkawa U, Kamada S. X-linked inhibitor of apoptosis protein mediates neddylation by itself but does not function as a NEDD8-E3 ligase for caspase-7. FEBS Lett. 2012;586:1612–1616. doi: 10.1016/j.febslet.2012.04.056. [DOI] [PubMed] [Google Scholar]
- 127.Lee MR, Lee D, Shin SK, Kim YH, Choi CY. Inhibition of APP intracellular domain (AICD) transcriptional activity via covalent conjugation with Nedd8. Biochem Biophys Res Commun. 2008;366:976–981. doi: 10.1016/j.bbrc.2007.12.066. [DOI] [PubMed] [Google Scholar]
- 128.Takashima O, Tsuruta F, Kigoshi Y, Nakamura S, Kim J, Katoh MC, Fukuda T, Irie K, Chiba T. Brap2 regulates temporal control of NF-kappaB localization mediated by inflammatory response. PLoS One. 2013;8:e58911. doi: 10.1371/journal.pone.0058911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Segovia JA, Tsai SY, Chang TH, Shil NK, Weintraub ST, Short JD, Bose S. Nedd8 Regulates Inflammasome-Dependent Caspase-1 Activation. Mol Cell Biol. 2014 doi: 10.1128/MCB.00775-14. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ma T, Chen Y, Zhang F, Yang CY, Wang S, Yu X. RNF111-dependent neddylation activates DNA damage-induced ubiquitination. Mol Cell. 2013;49:897–907. doi: 10.1016/j.molcel.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bostick M, Lochhead SR, Honda A, Palmer S, Callis J. Related to ubiquitin 1 and 2 are redundant and essential and regulate vegetative growth, auxin signaling, and ethylene production in Arabidopsis. Plant Cell. 2004;16:2418–2432. doi: 10.1105/tpc.104.024943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ou CY, Lin YF, Chen YJ, Chien CT. Distinct protein degradation mechanisms mediated by Cul1 and Cul3 controlling Ci stability in Drosophila eye development. Genes Dev. 2002;16:2403–2414. doi: 10.1101/gad.1011402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jones D, Candido EP. The NED-8 conjugating system in Caenorhabditis elegans is required for embryogenesis and terminal differentiation of the hypodermis. Dev Biol. 2000;226:152–165. doi: 10.1006/dbio.2000.9847. [DOI] [PubMed] [Google Scholar]
- 134.Huang G, Stock C, Bommelje CC, Weeda VB, Shah K, Bains S, Buss E, Shaha M, Rechler W, Ramanathan Y, Singh B. SCCRO3 (DCU- N1D3) antagonizes the neddylation and oncogenic activity of SCCRO (DCUN1D1) J Biol Chem. 2014;289:34728–42. doi: 10.1074/jbc.M114.585505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Oron E, Mannervik M, Rencus S, Harari-Steinberg O, Neuman-Silberberg S, Segal D, Chamovitz DA. COP9 signalosome subunits 4 and 5 regulate multiple pleiotropic pathways in Drosophila melanogaster. Development. 2002;129:4399–4409. doi: 10.1242/dev.129.19.4399. [DOI] [PubMed] [Google Scholar]
- 136.Oren-Giladi P, Krieger O, Edgar BA, Chamovitz DA, Segal D. Cop9 signalosome subunit 8 (CSN8) is essential for Drosophila development. Genes Cells. 2008;13:221–231. doi: 10.1111/j.1365-2443.2008.01164.x. [DOI] [PubMed] [Google Scholar]


