Abstract
Signaling pathways function as information-passing mechanisms of the cell. A number of extensively manually curated databases maintain the current knowledge-base for signaling pathways, inviting computational approaches for prediction and analysis. Such methods require an accurate and computable representation of signaling pathways. Pathways are often described as sets of proteins or as pairwise interactions between proteins. However, many signaling mechanisms cannot be described using these representations. In this opinion, we highlight an underutilized representation for signaling pathways: the hypergraph. We demonstrate the usefulness of hypergraphs in this context and discuss challenges and opportunities for the scientific community.
Keywords: signaling pathways, graphs, hypergraphs
Signaling Pathways and their Representations
Signaling pathways mediate a cell’s response to its environment, starting with the recognition of an external stimulus at receptors, proceeding through intra-cellular protein interactions and activation of transcription factors, and culminating in the perturbation of the expression of target genes. Due to their importance in cellular communication, signaling pathways are often perturbed in diseases. Numerous publicly-available and often manually-curated databases store information about signaling pathways [1–6]. Despite the growing knowledge of signaling pathways from experimental data, these databases face a number of obstacles when storing and conveying this information. Databases that constitute signaling pathways from manual curation of the literature produce high-quality interactions, but are time-consuming to construct, are often incomplete or outdated, and might be biased based on the curators’ expertise [7–10]. Databases that use automated methods for literature searching, such as predictive text mining, are relatively easy to maintain but tend to have many erroneous entries [11]. Different databases may represent the same biological event in different ways, making them difficult to standardize for computational use.
In this opinion, we lay out the common representations that have been used in computational analyses of signaling pathways. After examining the limitations of these representations, we encourage the use of hypergraphs as models that better capture the complex relationships in the underlying biological mechanisms. We describe three applications to motivate the need for more powerful representations of signaling pathways. Pathway enrichment assesses whether discovered proteins are significantly enriched for proteins/interactions in a pathway of interest. Pathway reconstruction aims to explicitly reconstruct and discover missing proteins and interactions in a pathway of interest. Finally, pathway crosstalk tries to capture how the stimulation of one pathway may result in alternate downstream responses.
Current Representations of Signaling Pathways
Signaling Pathways as Sets of Proteins
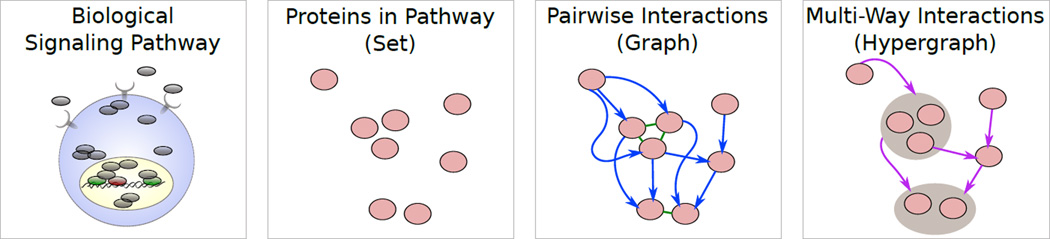
The simplest representation of a pathway is a list of its members, i.e., the set of proteins that are involved in the pathway (Figure 1). Catalogs such as the Gene Ontology [1] and the Molecular Signatures Database [12] provide signaling pathways in this format. With this representation, pathway enrichment identifies pathways whose members occur surprisingly often in a set of experimentally-identified proteins (e.g., from differential gene expression analysis) [13]. However, such set-based approaches ignore the relationships between proteins within a pathway, thereby providing no clues as to how interactions may alter gene expression [8]. These methods can correct and adjust for proteins shared among multiple pathways [14, 15], thus accounting for crosstalk to some extent. By definition, purely set-based methods can reconstruct only the proteins in a pathway and not its interactions [16].
Figure 1. Signaling Pathway Representations.
There are three main ways of representing signaling pathways. A signaling pathway may be simply represented as a set of proteins, with no additional information. Graphs encode pairwise interactions between proteins; these interactions may be undirected (green) or directed (blue). Hypergraphs, the focus of this article, encode multi-way interactions and reactions (see Box 1 for examples).
Signaling Pathways as Directed Graphs
Signaling pathways are also conceptualized as graphs, where nodes represent proteins and edges represent pairwise interactions between proteins (Figure 1 and Glossary). The edges are often directed in signaling pathways, such as when a kinase phosphorylates a substrate. Recent enrichment methods make use of pathway topology in their scoring metrics by taking the interaction among member proteins into account [8]. This class of approaches continues to be in active development. Pathway reconstruction algorithms on graphs typically use a large background interactome (such as a protein-protein interaction network) and identify pathways as subgraphs of proteins and interactions within the interactome. These approaches often try to find connections between signaling initiators (membrane receptors) and downstream regulators (transcription factors). Pathway reconstruction algorithms leverage many well-known concepts from graph theory [17–21]. Graph-based approaches to assess pathway crosstalk rely on the notion that two crosstalking pathways (each represented as a set of genes) will have statistically more interactions connecting their members than expected in a random network [22, 23]. However, these approaches fail to compute the specific paths of signaling interactions that contribute to crosstalk.
Graph representations of signaling pathways are an improvement from the “set of proteins” representation because they capture pairwise relationships between proteins. However, signaling pathways contain more complicated relationships that are problematic for graph representations. For example, graphs often represent a complex by connecting all its members, which can artificially increase the number of edges (Box 1, panel a) [24]. More importantly, graphs do not accurately represent several types of molecular reactions, including protein complex assembly and dis-assembly or regulation (e.g., activation and inhibition) (Box 1, panel b). Finally, graphs do not typically distinguish between inactive and active forms of a protein or complex (Box 1, panel c).
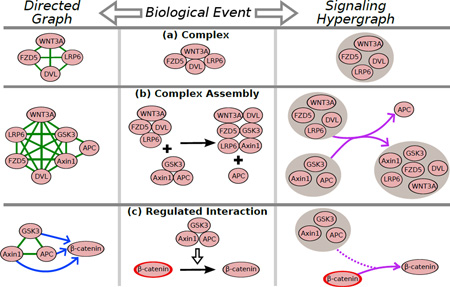
Box 1: A comparison of representations for events that occur in signaling pathways.
Protein Complexes (Figure I, panel a). A complex is a set of proteins that bind together to carry out a biological function. Upon stimulation of FZD5 by the WNT3A ligand, a four-protein complex (FZD5/WNT3A/LRP6/DVL) assembles at the plasma membrane. In a graph, complexes are commonly represented as cliques; as a result it is unclear whether the proteins form a complex or if they interact in pairs. A signaling hypergraph represents the complex as a single hypernode.
Complex Assembly (Figure I, panel b). Many biochemical reactions involve complex assembly and disassembly, e.g., the four-protein complex (FZD5/WNT3A/LRP6/DVL) sequesters GSK3 and Axin1, thereby disassembling the destruction complex (GSK3/Axin1/APC). A graph representation connects every pair of proteins within each complex; in addition to the ambiguity arising from complexes, the exact complex reconstitution is unclear. In a signaling hypergraph, complex rearrangement may be represented as a single directed hyperedge. Unlike the graph representation, the hyperedge representation clarifies the reactants, the products, and the “direction” of the reaction.
Regulation (Figure I, panel c). Many cellular reactions are regulated by proteins, small molecules, or complexes. For instance, phosphorylation (and inactivation) of β-catenin is regulated by the destruction complex (GSK3/Axin1/APC). A graph may represent this regulation using directed edges from every protein in the regulator (in this case, a complex) to β-catenin, creating the misleading appearance that each protein in the complex may independently regulate β-catenin. Moreover, graphs do not typically distinguish between inactive and active forms of a protein. Instead, hypergraphs can represent such a reaction as a regulated hyperedge, which indicates that the complex is required for the inactivation of β-catenin.
Other Representations of Signaling Pathways
While directed graphs have been useful for representing signaling pathways, their limitations are widely-recognized. A number of approaches have modified and extended graph representations. Compound graphs [25] and metagraphs [26] represent a complex as a single entity, and allow a nested structure. Factor graphs [27] and Petri nets [28] introduce different types of nodes into the directed graph in order to represent events involving sets of proteins. Multimodal networks [29] associate four entities with each edge: a head, a tail, a regulator, and a mode. The head, tail, and regulator can each be a set of proteins, and the mode specifies how the regulator controls the transition from head to tail, e.g., activation or repression.
These models of signaling pathways seek to address the shortcomings of directed graphs. However, each approach has drawbacks, including an inability to comprehensively model the complexity of signaling pathways, applicability to a limited range of computational problems, or under-utilization in systems biology. Nodes from compound graphs and metagraphs focus on protein complexes. Multimodal networks do not support the hierarchical structure of signaling networks. Factor graphs and Petri nets are not ideal for generalizations of common graph-theoretic operations such as paths, connectivity, and random walks. In the next section, we seek to unify these models under the umbrella of signaling hypergraphs.
Signaling Pathways as Hypergraphs
Hypergraphs are a generalization of graphs which are capable of representing relationships among two or more proteins (Figure 1). Typically, directed hypergraphs consist of a set of nodes and a set of directed hyperedges, where each hyperedge connects two sets of nodes (see Glossary). Directed hypergraphs are an attractive alternative to directed graphs for representing complex facets of cellular processes, especially for metabolic networks [29–33]. They have also been shown to be advantageous for signaling networks [30, 34, 35]; however, they still remain an under-utilized tool.
In our definition, a signaling hypergraph consists of a set of hypernodes, directed hyperedges, and regulated hyperedges. Each hypernode represents an individual protein or a set of proteins, each directed hyperedge connects one set of hypernodes to another, and each regulated hyperedge is a directed hyperedge with one or more hypernodes that act as regulators (see Glossary). In Box 1, we motivate this definition of signaling hypergraphs by describing three biological events in the canonical Wnt signaling pathway. These biological events (protein complexes, the assembly of protein complexes, and the regulation of proteins and complexes) commonly occur in signaling pathways. Each event may be represented as a graph consisting of multiple edges (Figure I left), or as a single hyperedge in a signaling hypergraph (Figure I right). Thus, each of these biological events has a more direct interpretation when represented as a signaling hypergraph rather than as a directed graph.
Figure I. Representations of three types of events in signaling pathways.
In each panel, the biological representation (middle) can be converted into a graph (left) or a hyper- graph (right). Green: undirected edge; blue: directed edge; gray circle: hypernode; purple: hyperedge; red outline: active form; dashed purple: regulatory component of hyperedge.
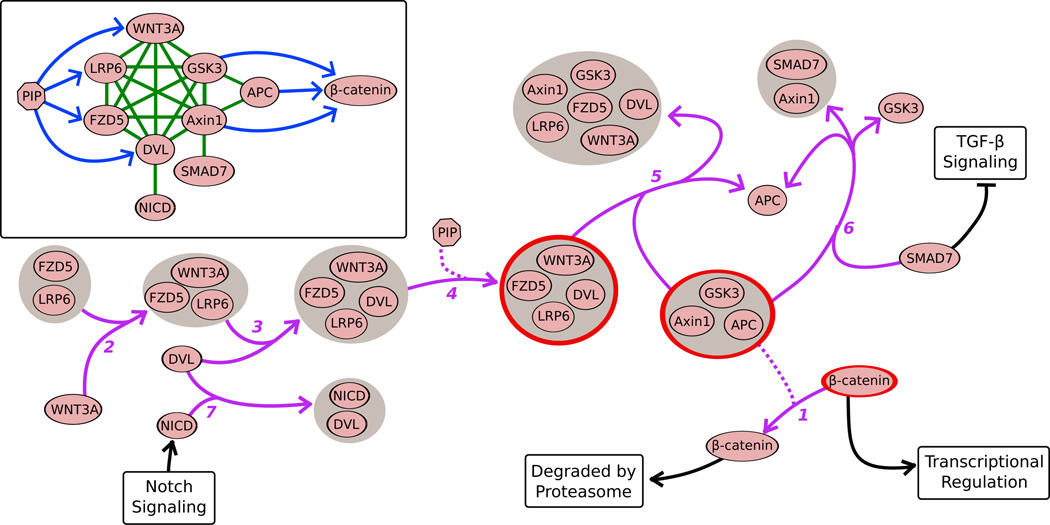
When these biological events are considered as components of the larger Wnt signaling pathway, it becomes clear that a typical graph-based representation simply does not have the power to accurately model sequences of signaling events (Figure 2). The first five hyperedges in Figure 2 represent the canonical Wnt signaling pathway (we discuss hyperedges 6 and 7 in the context of pathway crosstalk in the next section). The primary function of Wnt signaling is to control the activity status of β-catenin (hyperedge 1); active β-catenin regulates the transcription of target genes, and inactive β-catenin is degraded by the proteasome. The maintainance of active β-catenin starts with WNT3A binding to the FZD5/LRP6 complex (hyperedge 2). Next, DVL binds to the FZD5/LRP6/WNT3A complex to form FZD5/LRP6/WNT3A/DVL (hyperedge 3), which the small molecule PIP subsequently activates (hyperedge 4). Activated FZD5/LRP6/WNT3A/DVL interferes with the destruction complex (Axin1/APC/GSK3) by sequestering Axin1 and GSK3, and releasing APC (hyperedge 5). However, in the absence of Wnt signaling, the activated destruction complex persists, and marks β-catenin for degradation by phosphorylation (hyperedge 1).
Figure 2.
Events following Wnt signaling that lead to release of β-catenin. Black edges denote connections to other signaling pathways and biological processes. The inset shows a possible graph representation of these events. Green: undirected edge; blue: directed edge; purple: hyperedge; dashed purple: regulated component of hyperedge; gray circle: hypernode; red outline: active form.
These examples indicate that signaling hypergraphs offer a more accurate representation than directed graphs of the underlying biological events that occur in signaling pathways. Moving to a hypergraph-based representation brings with it several computational challenges and opportunities. From what datasets can we build signaling hypergraphs? Which algorithmic questions need to be solved for signaling hypergraphs? How exactly can problems in computational systems biology such as pathway enrichment, extension, and crosstalk benefit from this representation? We turn our attention to these questions below.
Building Signaling Hypergraphs
Although hypergraphs are not widely utilized for the analysis of signaling pathways, the increasing popularity of standardized data exchange formats such as BioPAX [36] and SBML [37] can accelerate their adoption. These file formats explicitly support reaction networks [2, 36, 37], which are in essence hypergraphs. As a result, we can directly build signaling hypergraphs from these representations. In addition, algorithms that operate on signaling hypergraphs can take advantage of the rich information embedded in BioPAX and similar formats.
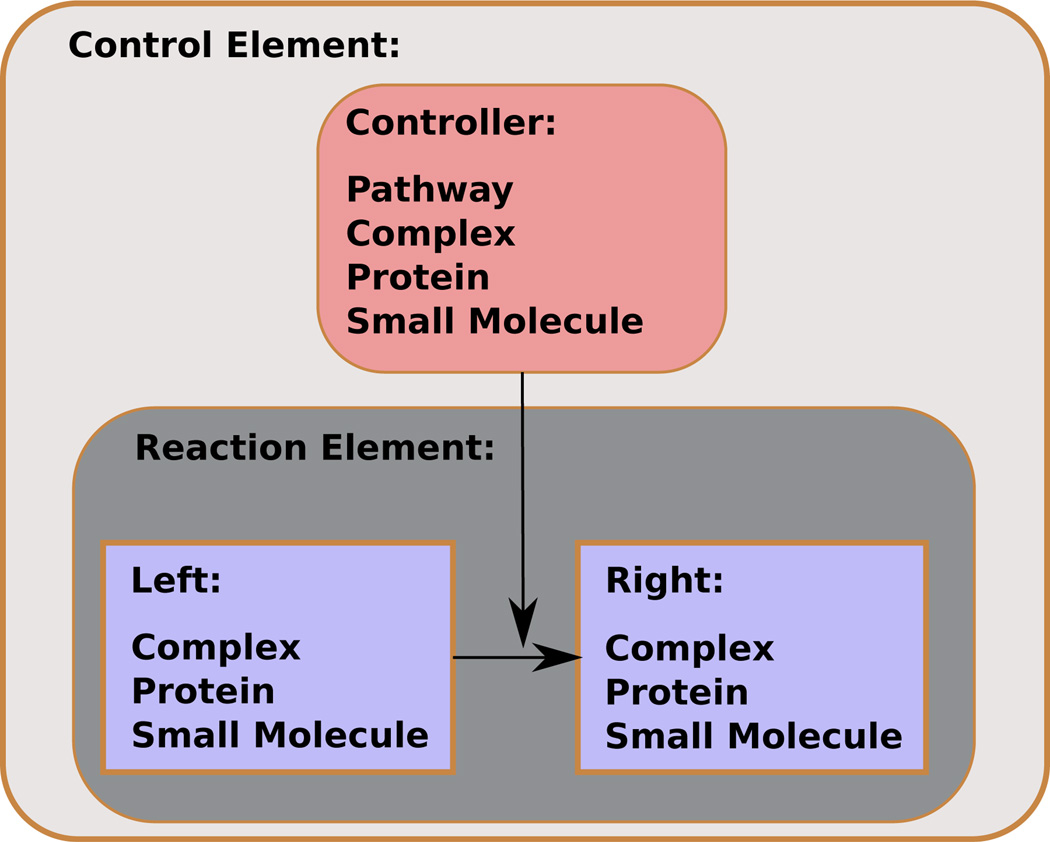
BioPAX is a format that aims to enable the integration and exchange of reactions and biological pathways. It has facilities for representing the diversity of interactions within a signaling pathway, including notions such as complex assembly, biochemical reactions and control mechanisms. In essence, BioPAX represents each of these notions as a hypergraph, although the notion of a hypergraph is not explicit in BioPAX. Figure 3 illustrates the scheme of a generic reaction in BioPAX. For example, a complex is simply a set of proteins that we call a hypernode. Complexes in BioPAX can be nested within each other, and be members of reactions. The representation of a reaction such as complex assembly explicitly specifies the reactants and products, each of which can be a set of complexes, proteins, or small molecules, called a directed hyperedge in our nomenclature. BioPAX takes regulation into account as well: a reaction may have a controller, which can be a complex, protein, small molecule, or even another pathway (regulated hyperedge). Thus, it should not be difficult to convert BioPAX-like formats into signaling hypergraphs. In addition to representing various elements in a signaling pathway, BioPAX makes explicit the notion that all members of a complex must be present (and bound to each other) before participating in or regulating a reaction. Since graphs model only pairwise representations, they cannot represent such requirements conveniently.
Figure 3. Example nested organization of BioPAX pathway elements.
This shows the nested hierarchy of the BioPAX file formats. Hypernodes may be complexes, proteins or small molecules. Reaction elements in BioPAX are equivalent to directed hyperedges, and controlled reactions to regulated hyperedges.
Applications of Signaling Hypergraphs
While hypergraphs have been an established area of mathematics since the 1960s [38], their application to systems biology may have been hampered by the lack of powerful algorithms. Since graphs are a \special case" of signaling hypergraphs (where each hypernode contains exactly one node and each hyperedge contains exactly two nodes), computational problems on hypergraphs are likely to be at least as hard as the corresponding problems for graphs [30]. In fact, many problems that can be solved in polynomial time in graphs become computationally difficult (𝒩𝒫-Complete) when posed on hypergraphs, e.g., computing shortest paths in directed hypergraphs [39]. Nevertheless, as we discuss below, hypergraphs are a promising representation for the pathway enrichment, reconstruction, and crosstalk applications. We motivate these applications with examples from the Wnt signaling pathway.
Pathway Enrichment
Using the signaling hypergraph in Figure 2, consider the scenario where only WNT3A and DVL are differentially expressed. Set-based methods may not identify Wnt signaling as significantly enriched because WNT3A and DVL are the only two proteins used to compute enrichment. A hypergraphical approach may observe that WNT3A or DVL participate in seven hypernodes and five hyperedges, and may conclude that Wnt signaling is perturbed. IfWNT3A and DVL were down-regulated, then the method may conclude that Wnt signaling would not occur, the destruction complex would maintain integrity, and the degradation of β-catenin would ensue. Signaling hypergraphs impose an added layer of complexity since they can represent the formation of a complex, both the active and inactive forms of a complex, and the regulation of reactions. We expect that ideas borrowed from factor graphs [40] and pathway enrichment techniques that consider-edge structure [8, 41] will be useful for developing hypergraph-based approaches for pathway enrichment.
Pathway Reconstruction
Existing methods have relied on well-known graph algorithms, but the corresponding theory for hypergraphs is considerably less well developed [42]. However, pathway reconstruction is an excellent application to motivate the development of new mathematical, statistical, and algorithmic research on hypergraphs. Suppose we are given the hypergraph representation in Figure 2 where the FZD5/LRP6 complex represents a signal initiator and the active form of β-catenin represents a transcriptional regulator. Hypergraph-based pathway reconstruction seeks to determine a series of hyperedges that links the FZD5/LRP6 complex with the active form of β-catenin, as shown by hyperedges 1–5 in Figure 2. By using the hyperedges and hypernodes as the units of connection in the pathway reconstruction problem, we can preserve the integrity of complexes and reactions, whereas graph-based methods will struggle to discern these entities.
Pathway Crosstalk
Crosstalking pathways are frequently identified by the activation of genes downstream of one pathway after a stimulus for another pathway. Extant methods for computationally estimating crosstalk use the intuition that it results from proteins shared by both pathways [15, 43]. Hypergraph-based approaches have the potential to unveil connected sequences of interactions that contribute to crosstalk between pathways. Figure 2 shows crosstalk of the Wnt signaling pathway with the TGF-β (hyperedge 6) and Notch pathways (hyperedge 7). SMAD7, a negative regulator of the TGF-β signaling pathway, mediates the crosstalk with the Wnt pathway. Specifically, SMAD7 catalyzes the dissociation of the destruction complex by binding with Axin1 and releasing APC and GSK3. This event results in increased transcriptional regulation by β-catenin as well as increased TGF-β signaling [44]. Crosstalk between the Notch and Wnt signaling pathways occurs through an interaction between NICD (NOTCH intracellular domain) and DVL [45]. Once DVL is bound to NICD, Wnt signaling cannot proceed, allowing the destruction complex to mark β-catenin for degradation. Thus, activation of Notch signaling down-regulates Wnt signaling. Computationally discovering these sequences of events that lead to crosstalk is an outstanding problem in both graph and hypergraph representations.
Outstanding Challenges
Several issues confront both graph- and hypergraph-based approaches for the analysis of signaling pathways. Current interaction datasets remain highly incomplete and/or contain many false positive interactions. The same pathway can have considerably different representation in different databases, sometimes resulting from the focus of the curator. Estimating the reliability of interactions (from the type of experiment or the curation method) continues to be an important challenge. While most signaling pathways in databases are organism specific, interactomes established by combining multiple databases are not tissue-specific.
Existing graph-based approaches to pathway reconstruction often analyze the entire interactome (i.e., all pairs of proteins known to interact). The interactome is often constructed from a combination of signaling pathway databases, more general protein-protein interaction databases, and high-throughput experiments. Hence, the quality of solutions from pathway reconstruction algorithms depend on the reliability of this interactome [43]. To use signaling hypergraphs for pathway reconstruction, we must establish a corresponding hypergraph interactome that represents the interactions among multiple proteins. As a start, one may construct a hypergraph interactome by beginning with a graph built from protein-protein interaction data and add hyperedges as they are represented in the signaling pathway databases (e.g. using BioPAX). Constructing a high-quality and high-coverage hypergraph interactome is an important research direction, since it will ultimately determine the quality of solutions from hypergraph-based algorithms.
For any representation, we can ask how the activity (or inactivity) of signaling pathway members affect the rest of the pathway. Boolean networks, for example, allow each node in a graph to take two values: 0 (inactive) or 1 (active). Logical models [35, 46] use Boolean gates to represent how a set of proteins may inuence another protein. Here, each gate is a special case of a directed hyperedge with many nodes in the tail (inputs) and one node in the head (output). Developing extensions of logical models that are appropriate for signaling hypergraphs is an important direction of research.
Signaling hypergraphs do not include stoichiometric or kinetic information; they focus more on capturing pathway structure. In contrast, dynamic models of signaling pathways capture both the structure and the stoichiometry of signaling pathways [46, 47]. However, these models tend to scale poorly since they require extensive prior experimental knowledge to fit parameters. Extensions of logical models on signaling hypergraphs that incorporate stochiometric information as well may provide a scalable alternative to dynamic models.
Concluding Remarks
The study of signaling pathways is a cornerstone of modern molecular and cellular biology. Unfortunately, their representation in common signaling pathway databases varies widely in terms of completeness, quality, and standardization. Furthermore, methods that computationally analyze such pathways may often ignore important characteristics of their structure. These gaps necessitate the development of a fresh representational approach. We have introduced signaling hypergraphs, an under-utilized representation in systems biology, to overcome these limitations.
Our goal is not to replace graph-based approaches in systems biology. Rather, our intent is to stimulate and encourage the use of hypergraph-based methods in contexts such as signaling pathways where their unique capabilities will have considerable impact. In the future, we hope that hypergraph-based analyses can encompass other types of cellular processes in conjunction with methods that infer hypergraphs directly from systems biology datasets [48–51].
We acknowledge that computational analysis of signaling hypergraphs will be challenging since the theory for hypergraphs is much less well developed than for graphs. Nonetheless, the development of pathway enrichment, pathway reconstruction, and pathway crosstalk algorithms on hypergraphs show promise of better representing the biological signaling pathway. We hope that our advocacy of signaling hypergraphs will stimulate new directions of mathematical, statistical, and algorithmic research. These results can power the increased use of signaling hypergraphs in computational systems biology.
Highlights.
We review common computational representations of signaling pathways.
These approaches cannot adequately model multi-way molecular reactions that are common in pathways.
We highlight hypergraphs as an under-utilized abstraction for representing such interactions.
We summarize challenges and opportunities presented by signaling hypergraphs
Glossary
Graph Notation: Nodes and Edges
*In a directed graph, an undirected edge between nodes u and υ is replaced by two directed edges (u, υ) and (υ, u).
- Node
an element (Protein; Compound)
- Undirected edge*
an unordered pair of nodes (Physical interaction between two proteins)
- Directed edge
an ordered pair of nodes (Kinase phosphorylates a substrate)
Signaling Hypergraph Notation: Hypernodes and Hyperedges
- Hypernode
a set of node(s) (Protein; Protein complex)
- Directed hyperedge
an ordered pair of sets of hypernodes (Complex assembly)
- Regulated hyperedge
a directed hyperedge regulated by a hypernode (Kinase phosphorylates a protein complex, thereby activating it)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Research. 2012;40(Database issue):D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B, Garapati P, Hemish J, Hermjakob H, Jassal B, Kanapin A, Lewis S, Mahajan S, May B, Schmidt E, Vastrik I, Wu G, Birney E, Stein L, D’Eustachio P. Reactome knowledge-base of human biological pathways and processes. Nucleic Acids Research. 2009;37(Database issue):D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kandasamy K, Mohan SS, Raju R, Keerthikumar S, Kumar GS, Venugopal AK, Telikicherla D, Navarro JD, Mathivanan S, Pecquet C, Gollapudi SK, Tattikota SG, Mohan S, Padhukasahasram H, Subbannayya Y, Goel R, Jacob HK, Zhong J, Sekhar R, Nanjappa V, Balakrishnan L, Subbaiah R, Ramachandra YL, Rahiman BA, Prasad TS, Lin JX, Houtman JC, Desiderio S, Renauld JC, Constantinescu SN, Ohara O, Hirano T, Kubo M, Singh S, Khatri P, Draghici S, Bader GD, Sander C, Leonard WJ, Pandey A. NetPath: a public resource of curated signal transduction pathways. Genome Biology. 2010;11(1):R3. doi: 10.1186/gb-2010-11-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paz A, Brownstein Z, Ber Y, Bialik S, David E, Sagir D, Ulitsky I, Elkon R, Kimchi A, Avraham KB, Shiloh Y, Shamir R. SPIKE: a database of highly curated human signaling pathways. Nucleic Acids Research. 2011;39(suppl 1):D793–D799. doi: 10.1093/nar/gkq1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breuer K, Foroushani AK, Laird MR, Chen C, Sribnaia A, Lo R, Winsor GL, Hancock RE, Brinkman FS, Lynn DJ. InnateDB: systems biology of innate immunity and beyond–recent updates and continuing curation. Nucleic Acids Research. 2013;41(Database issue):D1228–D1233. doi: 10.1093/nar/gks1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bauer-Mehren A, Furlong LI, Sanz F. Pathway databases and tools for their exploitation: benefits, current limitations and challenges. Molecular Systems Biology. 5(1) doi: 10.1038/msb.2009.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khatri P, Sirota M, Butte AJ. Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Computational Biology. 2012;8(2):e1002375. doi: 10.1371/journal.pcbi.1002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korcsmáros T, Farkas IJ, Szalay MS, Rovó P, Fazekas D, Spiró Z, Böde C, Lenti K, Vellai T, Csermely P. Uniformly curated signaling pathways reveal tissue-specific cross-talks and support drug target discovery. Bioinformatics. 2010;26(16):2042–2050. doi: 10.1093/bioinformatics/btq310. [DOI] [PubMed] [Google Scholar]
- 10.Fearnley LG, Davis MJ, Ragan MA, Nielsen LK. Extracting reaction networks from databases–opening Pandora’s box. Briefings in Bioinformatics. 2013 doi: 10.1093/bib/bbt058. bbt058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cusick ME, Yu H, Smolyar A, Venkatesan K, Carvunis AR, Simonis N, Rual JF, Borick H, Braun P, Dreze M, Vandenhaute J, Galli M, Yazaki J, Hill DE, Ecker JR, Roth FP, Vidal M. Literature-curated protein interaction datasets. Nature Methods. 2009;6(1):39–46. doi: 10.1038/nmeth.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bauer S, Gagneur J, Robinson PN. GOing Bayesian: model-based gene set analysis of genome-scale data. Nucleic Acids Research. 2010;38(11):3523–3532. doi: 10.1093/nar/gkq045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donato M, Xu Z, Tomoiaga A, Granneman JG, MacKenzie RG, Bao R, Than NG, Westfall PH, Romero R, Draghici S. Analysis and correction of crosstalk effects in pathway analysis. Genome Research. 2013;23(11):1885–1893. doi: 10.1101/gr.153551.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharan R, Ulitsky I, Shamir R. Network-based prediction of protein function. Mol Syst Biol. 2007;3:88. doi: 10.1038/msb4100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tuncbag N, Braunstein A, Pagnani A, Huang SS, Chayes J, Borgs C, Zecchina R, Fraenkel E. Simultaneous reconstruction of multiple signaling pathways via the prize-collecting Steiner forest problem. Journal of Computational Biology. 2013;20(2):124–136. doi: 10.1089/cmb.2012.0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mostafavi S, Ray D, Warde-Farley D, Grouios C, Morris Q. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biology. 2008;9(Suppl 1):S4. doi: 10.1186/gb-2008-9-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yosef N, Zalckvar E, Rubinstein AD, Homilius M, Atias N, Vardi L, Berman I, Zur H, Kimchi A, Ruppin E, Sharan R. ANAT: A tool for constructing and analyzing functional protein networks. Science Signaling. 2011;4(196) doi: 10.1126/scisignal.2001935. pl1+. [DOI] [PubMed] [Google Scholar]
- 20.Poirel CL, Rodrigues RR, Chen KC, Tyson JJ, Murali TM. Top-down network analysis to drive bottom-up modeling of physiological processes. Journal of Computational Biology. 2013;20(5):409–418. doi: 10.1089/cmb.2012.0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Navlakha S, Gitter A, Bar-Joseph Z. A network-based approach for predicting missing pathway interactions. PLoS Computational Biology. 2012;8(8):e1002640+. doi: 10.1371/journal.pcbi.1002640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dotan-Cohen D, Letovsky S, Melkman AA, Kasif S. Biological process linkage networks. PLoS One. 2009;4(4):e5313. doi: 10.1371/journal.pone.0005313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCormack T, Frings O, Alexeyenko A, Sonnhammer EL. Statistical Assessment of Crosstalk Enrichment between Gene Groups in Biological Networks. PloS one. 2013;8(1):e54945. doi: 10.1371/journal.pone.0054945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schelhorn S-E, Mestre J, Albrecht M, Zotenko E. Inferring physical protein contacts from large-scale purification data of protein complexes. Molecular & Cellular Proteomics. 2011;10(6) doi: 10.1074/mcp.M110.004929. M110.004929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fukuda K, Takagi T. Knowledge representation of signal transduction pathways. Bioinformatics. 2001;17(9):829–837. doi: 10.1093/bioinformatics/17.9.829. [DOI] [PubMed] [Google Scholar]
- 26.Hu Z, Mellor J, Wu J, Kanehisa M, Stuart JM, DeLisi C. Towards zoomable multidimensional maps of the cell. Nature Biotechnology. 2007;25(5):547–554. doi: 10.1038/nbt1304. [DOI] [PubMed] [Google Scholar]
- 27.Gat-Viks I, Shamir R. Refinement and expansion of signaling pathways: the osmotic response network in yeast. Genome Research. 2007;17(3):358–367. doi: 10.1101/gr.5750507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.David R, Alla H. Discrete, Continuous, and Hybrid Petri Nets. 2nd Edition. Springer Publishing Company, Incorporated; 2010. [Google Scholar]
- 29.Heath LS, Sioson AA. Semantics of multimodal network models. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 2009;6(2):271–280. doi: 10.1109/TCBB.2007.70242. [DOI] [PubMed] [Google Scholar]
- 30.Klamt S, Haus U-U, Theis F. Hypergraphs and cellular networks. PLoS Computational Biology. 2009;5(5):e1000385. doi: 10.1371/journal.pcbi.1000385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou W, Nakhleh L. Properties of metabolic graphs: biological organization or representation artifacts? BMC Bioinformatics. 2011;12(1):132. doi: 10.1186/1471-2105-12-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Christensen T, Oliveira A, Nielsen J. Reconstruction and logical modeling of glucose repression signaling pathways in Saccharomyces cerevisiae. BMC Systems Biology. 2009;3(1):7. doi: 10.1186/1752-0509-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramadan E, Perincheri S, Tuck D. Proceedings of the First ACM International Con3 ference on Bioinformatics and Computational Biology, BCB ’10. ACM: New York, NY, USA; 2010. A hyper-graph approach for analyzing transcriptional networks in breast cancer; pp. 556–562. [Google Scholar]
- 34.Klamt S, Rodriguez JS, Lindquist J, Simeoni L, Gilles E. A methodology for the structural and functional analysis of signaling and regulatory networks. BMC Bioinformatics. 2006;7(1):56+. doi: 10.1186/1471-2105-7-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Samaga R, Klamt S. Modeling approaches for qualitative and semi-quantitative analysis of cellular signaling networks. Cell Commun. Signal. 2013;11(1):43. doi: 10.1186/1478-811X-11-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Demir E, et al. The BioPAX community standard for pathway data sharing. Nature Biotechnology. 2010;28(9):935–942. doi: 10.1038/nbt.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hucka M, Finney A, Sauro H, Bolouri H, Doyle J, Kitano H, Arkin A, Bornstein B, Bray D, Cornish-Bowden A, Cuellar A, Dronov S, Gilles E, Ginkel M, Gor V, Goryanin I, Hedley W, Hodgman T, Hofmeyr J, Hunter P, Juty N, Kasberger J, Kremling A, Kummer U, Le Novere N, Loew L, Lucio D, Mendes P, Minch E, Mjolsness E, Nakayama Y, Nelson M, Nielsen P, Sakurada T, Schaff J, Shapiro B, Shimizu T, Spence H, Stelling J, Takahashi K, Tomita M, Wagner J, Wang J. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19(4):524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 38.Berge C. Hypergraphs, Volume 45: Combinatorics of Finite Sets (North-Holland Mathematical Library) 1st Edition. North Holland: 1989. [Google Scholar]
- 39.Gallo G, Longo G, Pallottino S, Nguyen S. Directed hypergraphs and applications. Discrete Applied Mathematics. 1993;42(23):177–201. [Google Scholar]
- 40.Vaske CJ, Benz SC, Sanborn JZ, Earl D, Szeto C, Zhu J, Haussler D, Stuart JM. Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics. 2010;26(12):i237–i245. doi: 10.1093/bioinformatics/btq182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gonçalves JP, Francisco AP, Mira NP, Teixeira MC, Sá-Correia I, Oliveira AL, Madeira SC. TFRank: network-based prioritization of regulatory associations underlying transcriptional responses. Bioinformatics. 2011;27(22):3149–3157. doi: 10.1093/bioinformatics/btr546. [DOI] [PubMed] [Google Scholar]
- 42.Ausiello G, Franciosa PG, Frigioni D. Theoretical Computer Science, Vol. 2202 of Lecture Notes in Computer Science. Springer Berlin Heidelberg; 2001. Directed hypergraphs: Problems, algorithmic results, and a novel decremental approach; pp. 312–328. [Google Scholar]
- 43.Kirouac DC, Saez-Rodriguez J, Swantek J, Burke JM, Lauffenburger DA, Sorger PK. Creating and analyzing pathway and protein interaction compendia for modelling signal transduction networks. BMC Systems Biology. 2012;6:29. doi: 10.1186/1752-0509-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo X, Wang X-F. Signaling cross-talk between TGF-β/BMP and other pathways. Cell Research. 2009;19(1):71–88. doi: 10.1038/cr.2008.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ranganathan P, Weaver KL, Capobianco AJ. Notch signalling in solid tumours: a little bit of everything but not all the time. Nature Reviews Cancer. 2011;11(5):338–351. doi: 10.1038/nrc3035. [DOI] [PubMed] [Google Scholar]
- 46.Karlebach G, Shamir R. Modelling and analysis of gene regulatory networks. Nature Reviews Molecular Cell Biology. 2008;9(10):770–780. doi: 10.1038/nrm2503. [DOI] [PubMed] [Google Scholar]
- 47.Aldridge BB, Burke JM, Lauffenburger DA, Sorger PK. Physicochemical modelling of cell signalling pathways. Nat. Cell Biol. 2006;8(11):1195–1203. doi: 10.1038/ncb1497. [DOI] [PubMed] [Google Scholar]
- 48.Tian Z, Hwang T, Kuang R. A hypergraph-based learning algorithm for classifying gene expression and arrayCGH data with prior knowledge. Bioinformatics. 2009;25(21):2831–2838. doi: 10.1093/bioinformatics/btp467. [DOI] [PubMed] [Google Scholar]
- 49.Battle A, Jonikas MC, Walter P, Weissman JS, Koller D. Automated identification of pathways from quantitative genetic interaction data. Molecular Systems Biology. 2010;6(1):379. doi: 10.1038/msb.2010.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sumazin P, Yang X, Chiu H-SS, Chung W-JJ, Iyer A, Llobet-Navas D, Rajbhandari P, Bansal M, Guarnieri P, Silva J, Califano A. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011;147(2):370–381. doi: 10.1016/j.cell.2011.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rahman A, Poirel CL, Badger DG, Murali TM. Reverse engineering molecular hypergraphs. Proceedings of the ACM Conference on Bioinformatics, Computational Biology, and Biomedicine. 2012:68–75. doi: 10.1109/TCBB.2013.71. [DOI] [PMC free article] [PubMed] [Google Scholar]