Abstract
Cytochrome P450 enzymes activate oxygen at heme iron centers to oxidize relatively inert substrate carbon-hydrogen bonds. Cysteine thiolate coordination to iron is posited to increase the pKa of compound II, an iron(IV)hydroxide complex, correspondingly lowering the one-electron reduction potential of compound I, the active catalytic intermediate, and decreasing the driving force for deleterious autooxidation of tyrosine and tryptophan residues in the enzyme’s framework. Here we report the preparation of an iron(IV)hydroxide complex in a P450 enzyme (CYP158) in ≥ 90% yield. Using rapid mixing technologies in conjunction with Mössbauer, ultraviolet/visible, and X-ray absorption spectroscopies, we determine a pKa value for this compound of 11.9. Marcus theory analysis indicates that this elevated pKa results in a >10,000 fold reduction in the rate constant for oxidations of the protein framework, making these processes noncompetitive with substrate oxidation.
The cytochrome P450 class of thiolate-ligated heme proteins use dioxygen and the formal equivalents of molecular hydrogen (2H+ + 2e−) to oxidize a broad spectrum of biologically active molecules. P450s are potent catalysts. They have been likened to a biological blowtorch for their ability to oxidize chemically inert hydrocarbons. (1) Compound I, the active intermediate in P450 catalysis, is capable of cleaving unactivated C-H bonds with an observed rate constant of 1.1 × 107 M−1s−1. The oxidation of bound substrate can exceed 1000 s−1. (2) A major goal of bioinorganic chemistry has been to elucidate the factors that govern these remarkable transformations. (3–5)
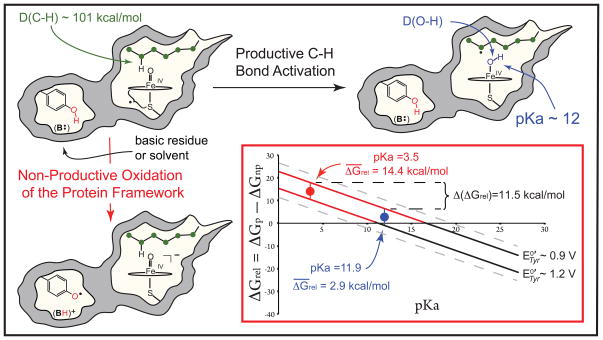
Experiments have shown that, during the course of productive C-H bond activation, compound I abstracts hydrogen from substrate to yield an iron(IV)hydroxide species (compound II) and a substrate radical (Fig. 1). (2, 6) An enigmatic aspect of P450 catalysis is the enzyme’s ability to perform this demanding oxidation ( ) (7, 8) without damage to its own relatively fragile protein framework: The oxidation of tyrosine and tryptophan residues found throughout the tertiary structures of these enzymes represents an energetically favorable “short circuit” for compound I’s oxidizing equivalents. Indeed, tyrosine oxidation ( ), which is known to dominate non-productive decay in P450s, requires ~13 kcal/mol less driving force than alkane oxidation. (9–11)
Fig. 1.
Productive and non-productive pathways for compound I decay. During productive C-H bond activation, compound I (a ferryl-radical species) abstracts hydrogen from substrate to yield compound II (an iron(IV)hydroxide complex) and a substrate radical. The non-productive pathway shown involves the oxidation of a tyrosine residue contained within the protein framework, with the phenolic proton being transferred to B:, a basic residue or solvent molecule, upon tyrosine oxidation. The inset shows the relative free energies (ΔGrel, Eq. 5) for the productive and non-productive pathways as a function of the compound II pKa for the range of aqueous tyrosine potentials. When ΔGrel is negative, the productive pathway is thermodynamically preferred. As a result of the ± 2 kcal/mol error in Eq. 2, a small range of tyrosine potentials could yield a given ΔGrel. The uncertainty in the solid lines as a result of this error is ± 0.1 V.
These unfavorable thermodynamics prompt a kinetic explanation of P450’s preference for C-H bond activation. However, the intramolecular oxidation of tyrosine via long-range proton coupled electron transfer (PCET) can be rapid, with rate constants approaching 106 s−1. (12–14) Given rate constants of this magnitude and the substantial difference in pathway energetics, what mechanism precludes non-productive oxidations of the protein framework and biases P450 towards substrate C-H bond activation?
We have argued that P450’s axial thiolate ligand promotes C-H bond activation through the generation of basic iron(IV)oxo (or ferryl) species. (15, 16) To understand this proposal, it is useful to consider the free energies of the productive and non-productive pathways illustrated in Fig. 1. The driving force for C-H bond activation, ΔGp, is given by the difference between the energies of the C-H bond broken, D(C-H), and the O-H bond made, D(O-H), Eq 1.
| (1) |
In heme systems, D(O-H) is determined by the one-electron reduction potential of compound I, and the pKa of compound II, making ΔGp a function of both of these thermodynamic parameters, Eq. 2. C is a constant that depends on the solvent and reference electrode. Its value is 57.6 for aqueous solution with E° vs. normal hydrogen electrode (NHE). (8)
| (2) |
In contrast, ΔGnp, the driving force for non-productive oxidations is determined by the one-electron reduction potential of compound I ( ) and the proton-coupled reduction potential of tyrosine ( , which includes the energetics of proton transfer to a basic residue or solvent, shown as B: in Fig. 1), Eq. 3.
| (3) |
This means that a change in the compound II pKa alters the energetics of the system. To illustrate this point, we construct the relative free energy, ΔGrel [Eqs. 4 and 5 and supplementary materials (SM) Eqs. S20–S25], which is the difference between the free energies of the productive and non-productive pathways. The sign of ΔGrel indicates the thermodynamically preferred process. When ΔGrel < 0, the productive pathway is preferred. Note that ΔGrel is independent of the reduction potential of compound I. For a given tyrosine potential, ΔGrel depends only upon the compound II pKa, with a unit-increase in the pKa shifting the energetics of the system 1.37 kcal/mol towards the productive pathway (Fig. 1).
| (4) |
| (5) |
For a given ΔGp, a change in the compound II pKa affects not only the energetics but also the kinetics of the system. The rate constant for non-productive oxidations can be linked directly to , which decreases (by 59 mV/unit) with increasing pKa. Thus, a highly elevated pKa can dramatically attenuate the rate of non-productive decay.
Based on these arguments, we propose that a critical role of thiolate ligation in P450 is to alter the free energy landscape, shifting the relative free energy for the productive and non-productive pathways to a regime where the rate constant for C-H bond activation dominates that for non-productive decay.
Results from theoretical investigations and reactivity studies with model systems lend support to this hypothesis, (16–20) but an experiment (or experimental parameter) that could provide a quantitative measure of the thiolate’s impact on P450 catalysis has proven elusive. In what follows, we report the identification of a P450 that can be prepared in the compound II form in high yield over a wide pH range. This discovery has allowed us to determine an iron(IV)hydroxide pKa precisely.
Determination of an iron(IV)hydroxide pKa
Iron(IV)oxo moieties are generally thought to be electrophilic in nature, and hence rarely protonated. Although examples of synthetic ferryl porphyrins are numerous, (21) a definitive characterization of a synthetic iron(IV)hydroxide has not been reported. The same is true for histidine ligated iron centers in peroxidase and globin enzymes. A battery of spectroscopic measurements have shown that the compound II forms of these enzymes have pKa values ≤ 3.5. They are best described as iron(IV)oxo species. (22) These results stand in stark contrast to those obtained for thiolate ligated heme systems, where only the iron(IV)hydroxide state has been observed. (16)
Although a number of spectroscopic investigations have confirmed the basic nature of thiolate ligated ferryls, (15, 16, 23–26) poor yields and the inability to prepare compound II at high pH have precluded efforts to pinpoint the iron(IV)hydroxide pKa. Recently, we identified a P450 (CYP158 with a large, solvent exposed, active-site) in which the compound II form of the enzyme can be prepared in high yield over a wide pH range.
CYP158 is one of 18 P450 enzymes found in the prototypic soil bacterium Streptomyces coelicolor. (27, 28) The enzyme is thought to play a role in the synthesis of secondary metabolites that afford S. coelicolor protection against the harmful effects of UV irradiation. During efforts to characterize high-valent intermediates in the catalytic cycle of CYP158, we found that compound II (CYP158-II) could be prepared by reacting ferric enzyme with meta-chloroperbenzoic acid (m-CPBA).
The use of m-CPBA, a two-electron oxidant, to prepare P450 compound II (which is only one oxidizing equivalent above ferric enzyme) can be understood in terms of the highly reactive nature of P450-I, which in the absence of substrate can strip an electron from the protein framework to form compound II and a protein-based radical (9–11): To avoid the uncoupling of redox equivalents during normal aerobic turnover, P450s rely on precise control of proton delivery. Two protons must be delivered to the distal oxygen of the reduced ferrous-oxy complex to generate compound I, while subsequent delivery of protons to the ferryl oxygen must be avoided to prevent its reduction to water (29, 30). In substrate free enzyme, solvent derived protons can readily access the iron(IV)oxo unit. The coupling of ferryl protonation to the reduction of compound I significantly increases the driving force for oxidation of the protein framework. Under turnover conditions with substrates that fit poorly in the active site, these processes are known to lead to oxidase chemistry. (31–33) In rapid mixing experiments with m-CPBA, they can present a major obstacle to the isolation of P450-I. (9–11) As we will show, CYP158 allows us to use this facet of P450 chemistry to our advantage.
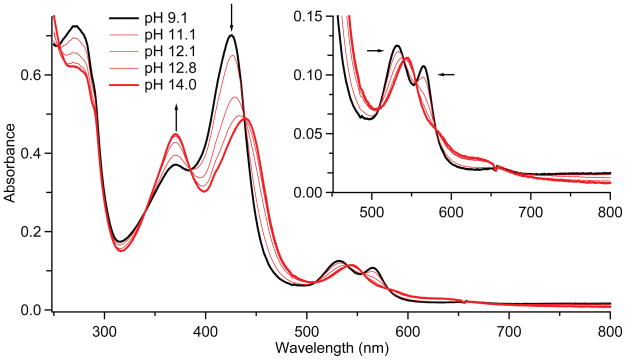
The reaction of CYP158 with m-CPBA generates compound II in good yield in the pH range from 7 to 10, with maximum formation (> 90%, fig. S1) occurring at pH 9. The intermediate is relatively stable at this pH, decaying at ~ 0.01s−1. The UV/Visible spectrum of CYP158-II at pH 9.0 is hyperporphyrin-like: it features a split Soret band with absorption maxima at 370 and 426 nm and Q-bands at 532 and 565 nm (Fig. 2). Mössbauer measurements confirm that this species is best described as an iron(IV)-hydroxide complex.
Fig. 2.
UV/Visible spectra of CYP158-II at varying pH. Samples were prepared in a double-mix stopped-flow experiment. 30μM ferric CYP158 (10 mM Tris-HCl, pH 9.0) was first mixed with 150 μM m-CPBA and held until maximum formation of CYP158-II was achieved (2.5 seconds). This solution was then mixed with a strongly buffered solution (containing 200 mM phosphate, 200 mM carbonate, pH adjusted with KOH) to obtain the desired final pH. Mixing was 1:1:1. The inset shows a magnification of the 450–800 nm region.
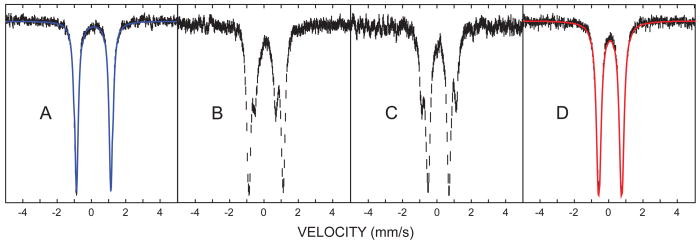
The 57Fe Mössbauer spectrum of CYP158-II at pH 9 consists of a single quadrupole doublet (Fig. 3A). Best fits of the data provide a quadrupole splitting of ΔEQ = 2.05 mm/s and an isomer shift of δ = 0.10 mm/s. These values are in good agreement with Mössbauer parameters reported for other thiolate ligated compound II species. (23, 26)
Fig. 3.
Mössbauer spectra of CYP158-II at varying pH. (A) pH 9.0 (B) pH 11.8 (C) 12.2 (D) 13.3. Samples were prepared in a double-mix freeze-quench experiment. 6 mM protein was mixed 2:1 with 60 mM m-CPBA to form CYP158-II at pH 9. This species was then mixed 3:1 with an arginine/NaOH buffer (pH 14). The strength of the arginine/NaOH buffer was varied (10mM to 108.16mM) to achieve the desired final pH. The reaction mixture was sprayed into liquid ethane 7 ms after the first mix (SM Materials and Methods). Fits for the intermediate pH samples (B and C) are shown in fig. S2.
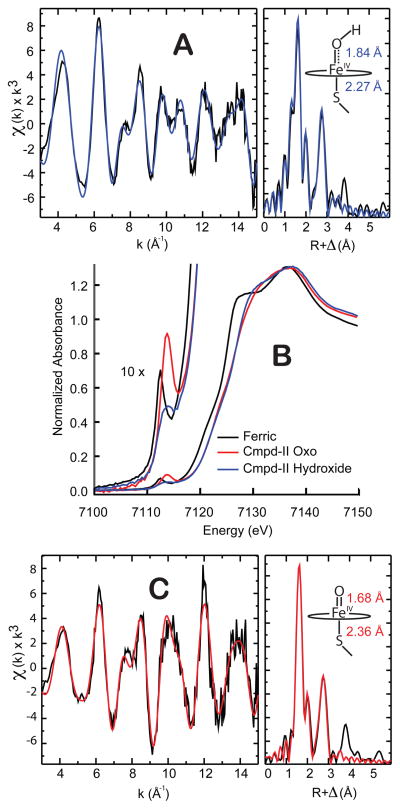
Given the high yield and stability of CYP158-II at pH 9, we wondered if the pH 9 intermediate could serve as platform from which CYP158-II could be prepared at higher pH via rapid sequential-mixing, allowing a well-bounded pKa measurement. Figures 2 and 3 show spectra of CYP158A-II samples prepared from pH 9.0 to pH 14.0. With increasing pH, the iron(IV)hydroxide complex converts cleanly to a new species. The UV/Visible spectrum of the high pH intermediate is also hyperporphyrin-like, with absorption maxima at 371 and 437 nm and a single Q-band centered at 545 nm. Mössbauer measurements at pH 13.3 again reveal the presence of a single iron(IV) species. This species has a reduced quadrupole splitting that is indicative of an iron(IV)oxo porphyrin (δ = 0.09 mm/s, ΔEQ=1.30 mm/s). (22)
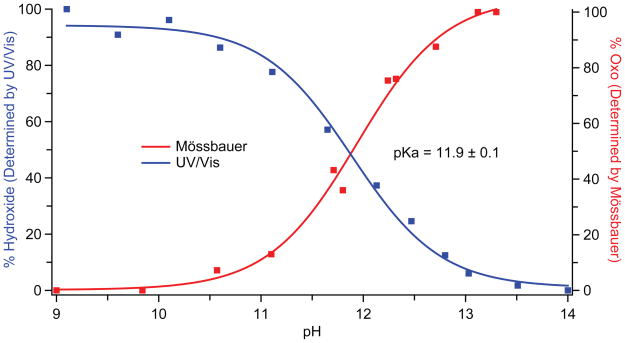
Fig. 2 reveals that the UV/Visible spectra obtained as a function of pH possess multiple isosbestic points, indicating that pH driven changes in the spectra result only from the hydroxo to oxo reaction. This result is consistent with our Mössbauer experiments (Fig. 3) as well as the observed decay rates, which indicate that there is minimal degradation (< 3% at pH 14, fig S3) during the time required for our measurements. Analyses of the data reveal that the iron(IV) species are in equilibrium: Plotting either the change in UV/Visible absorbance or the relative concentration of the oxo species as a function of pH results in the pH titration curves shown in Fig. 4. Fits of these curves provide an iron(IV)hydroxide pKa of 11.9. The reversibility of this transition was confirmed using triple-mix stopped-flow experiments, in which the oxo form (prepared at pH 13.5) was protonated to the hydroxo form (at pH 9) by an additional mixing stage with 1M low-pH Tris-HCl (fig. S4).
Fig. 4.
pH titration curves for CYP158-II. Data obtained from stopped-flow UV/Visible (blue) and Mössbauer spectroscopies (red) yield the same pKa value of 11.9. UV/Visible data points were obtained from the change in absorbance at 423 nm (Fig. 2). Mössbauer data points were obtained from the ratio of the areas of the iron(IV)oxo and iron(IV)hydroxide subspectra (fig S2).
Given the extreme pH at which this transition occurs, could a pH driven conformational change result in the loss of thiolate ligation and associated driving force for ferryl protonation? The observation of a split Soret band at high pH effectively rules this possibility out. A split Soret, or hyperporphyrin spectrum, is known to be a key indicator of thiolate ligation in heme proteins. The Soret band typically arises from porphyrin π –> π* electronic transitions, but in thiolate ligated systems this band is split through interactions with a sulfur –> porphyrin charge-transfer band. (34) The two Soret bands at 371 and 437 nm in the UV/Visible spectrum of the high pH intermediate are the hallmarks of this interaction. Their presence indicates the retention of thiolate ligation at high pH.
Iron K-edge x-ray absorption measurements (Fig. 5) further confirm the retention of thiolate ligation and support the assignment of the compound II pKa. Both the high and low pH forms of the intermediate have an absorption edge that lies ~ 1.5 eV above the ferric edge, consistent with the presence of an iron(IV) state. Fits of the extended x-ray absorption fine structure (EXAFS) data yield Fe-O/Fe-S distances of 1.68/2.36 Å at pH 13.3 and 1.84/2.27 Å at pH 9.0. The short Fe-O distance at pH 13.3 is indicative of an iron(IV)oxo species. With decreasing pH, the Fe-O bond length increases by 0.15 Å, consistent with protonation of the ferryl moiety. (15)
Fig. 5.
X-ray absorption spectroscopy. A) EXAFS of CYP158-II at pH 9.0. B) X-ray absorption edges. C) EXAFS of CYP158-II at pH 13.3. All EXAFS samples were analyzed by Mössbauer spectroscopy prior to data collection. See tables S1 and S2 for further details.
Examinination of the CYP158 crystal structure suggests a number of candidates for the location of the protein derived reducing equivalent which converts P450-I to P450-II in our rapid mixing experiments. (27, PDB accession code: 1S1F) Chief among them is Tyr-352, a solvent exposed residue, which is adjacent to the Cys-353 thiolate ligand (fig. S5). Incorporation of a phenylalanine at this position, through site-directed mutagenesis, results in protein that generates P450-I in high (~80%) yield (figs. S6, S7, and S8). Samples of the Y352F CYP158-I variant show the presence of residual compound II (~3%) as well as a protein-based radical (~5%). Both can be completely eliminated through the removal of a second tyrosine at position 318. The Y318F/Y352F variant shows increased CYP158-I production (~90% yield), and a compound I EPR spectrum that is devoid of protein radicals (figs. S6, S7, and S8).
Using insights gained from these site-directed mutagenesis studies, we sought to determine the compound II pKa in a second P450. The reaction of the thermophilic CYP119 with m-CPBA is known to produce P450-I in high (~75%) yield: Compound II does not accumulate. Incorporation of a tyrosine residue at the position corresponding to that of Tyr-352 in CYP158, results in an L316Y CYP119 variant that generates compound II in high yield. Similar pH jump experiments set the pKa of the CYP119-II variant at 12.2 (figs. S9–S12).
It is conceivable that an electron deficient residue adjacent to the proximal cysteine could diminish the electron donating ability of the thiolate ligand, thereby decreasing the iron(IV)hydroxide pKa. However the solvent exposed nature of the tyrosine residues under consideration renders this possibility unlikely. To alleviate any concerns that a neighboring Tyr-O• might suppress the compound II pKa from its natural value, DFT calculations were performed on an active-site model, using implicit solvation. These calculations, which examined the energy of ferryl protonation in the presence and absence of tyrosine oxidation, revealed that the Tyr-O• has little (if any) effect on the pKa of the iron(IV)hydroxide complex. When “solvated” in a dielectric continuum with ε = 4.0 (a value typically chosen for the protein matrix), (35) the Tyr-O• suppresses the iron(IV)hydroxide pKa by only 0.3 units. In water (ε = 80), the effect is less than 0.1 units (fig. S13).
The observation of a similar compound II pKa (~12) in both CYP119 and CYP158 is important. These P450s have significantly different active-site pockets and substrate preferences. (27, 28, 36, 37) Our results suggest that an elevated iron(IV)hydroxide pKa is a general feature of P450s and thiolate-ligated hemes.
Implications for Reactivity and the Role of Thiolate Ligation in P450 Catalysis
A central question in the field of P450 catalysis has been the thiolate’s role in hydrocarbon oxidations. A number of spectroscopic studies have revealed the impact of thiolate ligation on the electronic and geometric structures of the P450 active-site. These investigations have provided clear evidence for the strong electron-donating character of the axial thiolate ligand, (38, 39) but they have not provided a quantitative measure of the thiolate’s impact on reactivity. A compound II pKa of ~12 provides such a measure.
A pKa of ~12 is remarkable for an iron(IV)hydroxide, given the (typically) electrophilic nature of ferryl species. The compound II pKa values reported here are at least 8.5 units higher than the pKa values of histidine ligated hemes in peroxidase and globin enzymes. An increase of ≥ 8.5 units in the compound II pKa shifts the energetics of the system ≥ 11.5 kcal/mol towards the productive pathway (Eq. 5). This is illustrated in the inset of Fig. 1, where ΔGrel is shown for the full range of aqueous tyrosine reduction potentials, indicated by the dashed gray lines (E°Tyr-O/Tyr-O• = 0.7 V to E°Tyr-OH/Tyr-OH•+ = 1.4 V). The solid line at ~ 0.9 V in Fig. 1 separates the low- and mid-potential regions. It corresponds to the reduction potential of a solvent exposed tyrosine. The incorporation of a tyrosine residue into the protein framework is expected to place its reduction potential near or above this value. (40, 41) The line at separates mid- and high-potential regions: for tyrosines with reduction potentials above , ΔGrel changes sign as the iron(IV)hydroxide pKa increases from 3.5 to 12. That is, with thiolate ligation, the productive pathway becomes thermodynamically preferred.
This behavior is not observed in the important mid-potential region, which lies at the intersection of tyrosine potentials most likely to be found in P450s with those most readily oxidized by compound I. For these systems, with a compound II pKa of 3.5, non-productive tyrosine oxidation is favored over C-H bond activation by an average of 14 kcal/mol. As the pKa increases, the energetics of the system shift towards the productive pathway. With a pKa of 12, the preference for the non-productive pathway has been reduced to an average of only 3 kcal/mol.
We argue that this shift in the relative free energy places C-H bond activation in a regime of kinetic control. For a given ΔGp, a decrease of ≥ 11.5 kcal/mol in ΔGrel corresponds to a reduction of ≥ 0.5 V in . This drop in reduction potential dramatically attenuates the rate constants for non-productive oxidations, biasing the system towards C-H bond activation.
The impact of thiolate ligation on the non-productive rate constants can be illustrated through the application of Marcus theory. Although originally proposed to describe outer-sphere electron transfer reactions, Marcus theory has been shown to be applicable to a broad range of proton coupled electron transfer (PCET) processes, (42, 43) with the rate constant for PCET being given by Eq. 6,
| (6) |
where ΔG is the driving force for the reaction, γ is the intrinsic barrier, (44) and A is a function of the coupling between the donor and acceptor states.
Examinations of the intra- and intermolecular oxidations of tyrosine and substituted phenols have revealed that γ is large in these systems. Analyses of reactions employing oxidants with reduction potentials as high as 1.3V indicate that 2γ ≫ ΔG. (13, 45–47) Given PCET within this regime, Eq. 7 (derived in Eqs. S1–S19) allows a comparison of rate constants for non-productive oxidation by compound I species, k(np), as a function of compound II pKa values.
| (7) |
Implicit in the derivation of Eq. 7 is the assumption of an invariant driving force for productive substrate oxidations (i.e. ΔGp). Eq. 7 can be used to determine the magnitude by which thiolate ligation suppresses non-productive oxidations. Inserting ΔpKa ≥ 8.5 (the difference between histidine and thiolate ligated compound II pKa values) into Eq. 7 reveals that (relative to histidine ligation) thiolate ligation provides—ceteris paribus— a >10,000 fold reduction in the rate constant for non-productive oxidations. The ability of thiolate ligation to suppress these non-productive pathways is of critical importance given the apparent endergonic nature of C-H bond activation.
The O-H bond strength of P450-II is unknown, but theoretical methods predict that cleavage of an unactivated C-H bond by P450-I is ~6 kcal/mol endergonic. (48) To achieve similar energetics, a histidine ligated compound I would require a one-electron reduction potential of at least 1.4 V, yielding ΔGnp ~ 0.2–0.5 V for the tyrosine potentials displayed in Fig 1. Experiments on biological and synthetic model systems possessing similar driving forces have shown that the oxidation of tyrosines via long-range intramolecular PCET can be rapid, with experimentally determined rate constants ranging from 104 to 106 s−1. (12, 13, 49) The preceding analysis suggests that thiolate ligation lowers rate constants of even this magnitude to less than 50 s−1, making them non-competitive with C-H bond activation (< 5% uncoupling). Thus, we argue that thiolate ligation buys time for P450-I to accomplish endergonic oxidations, creating what is effectively a controlled burn of inert organic compounds.
Our analysis provides insight into the absence of histidine ligated P450-like enzymes in nature: the degree of uncoupling (via the non-productive oxidation of aromatic residues) in such a system would be unsustainable. Oxidations by the ferryl intermediates of non-heme enzymes such as AlkB, SyrB2, and TauD (50–53) and by the 5′-deoxyadenosyl 5′-radical of radical SAM enzymes (54–56) appear to be additional examples of this scheme in action. (57–60)
Supplementary Material
Acknowledgments
We thank M. Latimer and E. Nelson for onsite assistance at the synchrotron. This work was supported by the NIH (R01-GM101390). Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource, a Directorate of SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the U.S. Department of Energy Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393).
References
- 1.Schlichting I, et al. Science. 2000;287:1615. doi: 10.1126/science.287.5458.1615. [DOI] [PubMed] [Google Scholar]
- 2.Rittle J, Green MT. Science. 2010;330:933. doi: 10.1126/science.1193478. [DOI] [PubMed] [Google Scholar]
- 3.Sligar SG. Science. 2010;330:924. doi: 10.1126/science.1197881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bollinger JM, Jr, Broderick JB. Curr Opin Chem Biol. 2009;13:51. doi: 10.1016/j.cbpa.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 5.Groves JT. Proc Natl Acad Sci USA. 2003;100:3569. doi: 10.1073/pnas.0830019100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Groves JT, McClusky GA, White RE, Coon MJ. Biochem Biophys Res Commun. 1978;81:154. doi: 10.1016/0006-291x(78)91643-1. [DOI] [PubMed] [Google Scholar]
- 7.P450s are known to activate inert C-H bonds with bond dissociation energies (BDEs) on the order of 98–101 kcal/mol. In this work we consider a hypothetical C-H bond with a BDE that equals the average of this range, BDE = 99.5 kcal/mol. Assuming S0solv(CH) = S0solv(C•) for hydrocarbons adds 1.8 kcal/mol to the BDE, (8) for a bond dissociation free energy (BDFE) of ~ 101 kcal/mol. Using this value, we obtain the thermodynamic potential for hydrocarbon oxidation at pH 7. The oxidation of tyrosine at this pH requires a potential of . The difference in these potentials is equivalent to ~13 kcal/mol.
- 8.Warren JJ, Tronic TA, Mayer JM. Chem Rev. 2010;110:6961. doi: 10.1021/cr100085k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.In the absence of substrate, P450-I decays predominantly through the oxidation of tyrosine residues. (10,11) The oxidation of tryptophan is energetically feasible, but tyrosine with its (generally) lower potential is the preferred target. Tryptophan radicals have been observed following P450-I decay, but these species constituted only a minor component of the decay product. (10) The treatment of tyrosine oxidation presented here is an archetype for non-productive one-electron processes. It may be applied equally well to tryptophan oxidation.
- 10.Jung C, Schünemann V, Lendzian F. Biochem Biophys Res Commun. 2005;338:355. doi: 10.1016/j.bbrc.2005.08.166. [DOI] [PubMed] [Google Scholar]
- 11.Schunemann V, et al. J Biol Chem. 2004;279:10919. doi: 10.1074/jbc.M307884200. [DOI] [PubMed] [Google Scholar]
- 12.Irebo T, Johansson O, Hammarström L. J Am Chem Soc. 2008;130:9194. doi: 10.1021/ja802076v. [DOI] [PubMed] [Google Scholar]
- 13.Sjödin M, Styring S, Åkermark B, Sun L, Hammarström L. J Am Chem Soc. 2000;122:3932. [Google Scholar]
- 14.Ahlbrink R, et al. Biochemistry. 1998;37:1131. doi: 10.1021/bi9719152. [DOI] [PubMed] [Google Scholar]
- 15.Green MT, Dawson JH, Gray HB. Science. 2004;304:1653. doi: 10.1126/science.1096897. [DOI] [PubMed] [Google Scholar]
- 16.Green MT. Curr Opin Chem Biol. 2009;13:84. doi: 10.1016/j.cbpa.2009.02.028. [DOI] [PubMed] [Google Scholar]
- 17.Kang Y, et al. Chemistry - A European Journal. 2009;15:10039. [Google Scholar]
- 18.Parsell TH, Yang MY, Borovik AS. J Am Chem Soc. 2009;131:2762. doi: 10.1021/ja8100825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prokop KA, de Visser SP, Goldberg DP. Angew Chem Int Ed. 2010;49:5091. doi: 10.1002/anie.201001172. [DOI] [PubMed] [Google Scholar]
- 20.Sastri CV, et al. Proc Natl Acad Sci USA. 2007;104:19181. doi: 10.1073/pnas.0709471104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Terner J, et al. J Inorg Biochem. 2006;100:480. doi: 10.1016/j.jinorgbio.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 22.Behan RK, Green MT. J Inorg Biochem. 2006;100:448. doi: 10.1016/j.jinorgbio.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 23.Behan RK, Hoffart LM, Stone KL, Krebs C, Green MT. J Am Chem Soc. 2006;128:11471. doi: 10.1021/ja062428p. [DOI] [PubMed] [Google Scholar]
- 24.Green MT. J Am Chem Soc. 2006;128:1902. doi: 10.1021/ja054074s. [DOI] [PubMed] [Google Scholar]
- 25.Stone KL, Behan RK, Green MT. Proc Natl Acad Sci USA. 2006;103:12307. doi: 10.1073/pnas.0603159103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stone KL, Hoffart LM, Behan RK, Krebs C, Green MT. J Am Chem Soc. 2006;128:6147. doi: 10.1021/ja057876w. [DOI] [PubMed] [Google Scholar]
- 27.Zhao B, et al. J Biol Chem. 2005;280:11599. doi: 10.1074/jbc.M410933200. [DOI] [PubMed] [Google Scholar]
- 28.Zhao B, Guengerich FP, Voehler M, Waterman MR. J Biol Chem. 2005;280:42188. doi: 10.1074/jbc.M509220200. [DOI] [PubMed] [Google Scholar]
- 29.de Montellano PRO, editor. Cytochrome P450: Structure, Mechanism, and Biochemistry. 3. Kluwer Academic/Plenum Publishers; New York: 2004. [Google Scholar]
- 30.Kadkhodayan S, Coulter ED, Maryniak DM, Bryson TA, Dawson JH. J Biol Chem. 1995;270:28042. doi: 10.1074/jbc.270.47.28042. [DOI] [PubMed] [Google Scholar]
- 31.Gorsky LD, Koop DR, Coon MJ. J Biol Chem. 1984;259:6812. [PubMed] [Google Scholar]
- 32.Atkins WM, Sligar SG. J Am Chem Soc. 1987;109:3754. [Google Scholar]
- 33.Grinkova YV, Denisov IG, McLean MA, Sligar SG. Biochem Biophys Res Commun. 2013;430:1223. doi: 10.1016/j.bbrc.2012.12.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanson LK, et al. J Am Chem Soc. 1976;98:2672. doi: 10.1021/ja00425a050. [DOI] [PubMed] [Google Scholar]
- 35.Gilson MK, Honig BH. Biopolymers. 1986;25:2097. doi: 10.1002/bip.360251106. [DOI] [PubMed] [Google Scholar]
- 36.Yano JK, et al. J Biol Chem. 2000;275:31086. doi: 10.1074/jbc.M004281200. [DOI] [PubMed] [Google Scholar]
- 37.Park SY, et al. J Inorg Biochem. 2002;91:491. doi: 10.1016/s0162-0134(02)00446-4. [DOI] [PubMed] [Google Scholar]
- 38.Dawson and coworkers (46) have shown that exogenous thiol ligands bound trans to the proximal thiolate of P450 have enhanced basicity relative to analogous myoglobin adducts.
- 39.Sono M, Andersson LA, Dawson JH. J Biol Chem. 1982;257:8308. [PubMed] [Google Scholar]
- 40.Hay S, Westerlund K, Tommos C. Biochemistry. 2005;44:11891. doi: 10.1021/bi050901q. [DOI] [PubMed] [Google Scholar]
- 41.Tommos C, Valentine KG, Martinez-Rivera MC, Liang L, Moorman VR. Biochemistry. 2013;52:1409. doi: 10.1021/bi301613p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mayer JM. J Phys Chem Lett. 2011;2:1481. doi: 10.1021/jz200021y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mayer JM. Acc Chem Res. 2011;44:36. doi: 10.1021/ar100093z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Warren JJ, Winkler JR, Gray HB. FEBS Lett. 2012;586:596. doi: 10.1016/j.febslet.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang MT, Irebo T, Johansson O, Hammarström L. J Am Chem Soc. 2011;133:13224. doi: 10.1021/ja203483j. [DOI] [PubMed] [Google Scholar]
- 46.Markle TF, Rhile IJ, DiPasquale AG, Mayer JM. Proc Natl Acad Sci USA. 2008;105:8185. doi: 10.1073/pnas.0708967105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rhile IJ, et al. J Am Chem Soc. 2006;128:6075. doi: 10.1021/ja054167+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Visser SP, Kumar D, Cohen S, Shacham R, Shaik S. J Am Chem Soc. 2004;126:8362. doi: 10.1021/ja048528h. [DOI] [PubMed] [Google Scholar]
- 49.Immoos CE, et al. Inorg Chem. 2004;43:3593. doi: 10.1021/ic049741h. [DOI] [PubMed] [Google Scholar]
- 50.Matthews ML, et al. Biochemistry. 2009;48:4331. doi: 10.1021/bi900109z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Price JC, Barr EW, Tirupati B, Bollinger JM, Krebs C. Biochemistry. 2003;42:7497. doi: 10.1021/bi030011f. [DOI] [PubMed] [Google Scholar]
- 52.Krebs C, GalonićFujimori D, Walsh CT, Bollinger JM. Acc Chem Res. 2007;40:484. doi: 10.1021/ar700066p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simmons JM, Mueller TA, Hausinger RP. Dalton Transactions. 2008:5132. doi: 10.1039/b803512a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Booker SJ. Curr Opin Chem Biol. 2009;13:58. doi: 10.1016/j.cbpa.2009.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Challand MR, Driesener RC, Roach PL. Nat Prod Rep. 2011;28:1696. doi: 10.1039/c1np00036e. [DOI] [PubMed] [Google Scholar]
- 56.Duschene KS, Veneziano SE, Silver SC, Broderick JB. Curr Opin Chem Biol. 2009;13:74. doi: 10.1016/j.cbpa.2009.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davies DM, Jones P, Mantle D. Biochem J. 1976;157:247. doi: 10.1042/bj1570247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Annino G, Cassettari M, Martinelli M. IEEE Trans Microwave Theory Tech. 2009;57:775. [Google Scholar]
- 59.Reijerse E, Lendzian F, Isaacson R, Lubitz W. J Magn Reson. 2012;214:237. doi: 10.1016/j.jmr.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 60.Gaussian 09, Revision C.01. Gaussian Inc; Wallingford, CT: 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.