Abstract
Clinical and field-portable diagnostic devices require the detection of atto- to zeptomoles of biological molecules rapidly, easily and at low cost, with stringent requirements in terms of robustness and reliability. Though a number of creative approaches to this difficult problem have been reported1–9, numerous unmet needs remain in the marketplace, particularly in resource-poor settings10–12. Using rational materials design, we investigated harnessing the amplification inherent in a radical chain polymerization reaction to detect molecular recognition. Polymerization-based amplification is shown to yield a macroscopically observable polymer, easily visible to the unaided eye, as a result of as few as ~1,000 recognition events (10 zeptomoles). Design and synthesis of a dual-functional macromolecule that is capable both of selective recognition and of initiating a polymerization reaction was central to obtaining high sensitivity and eliminating the need for any detection equipment. Herein, we detail the design criteria that were used and compare our findings with those obtained using enzymatic amplification. Most excitingly, this new approach is general in that it is readily adaptable to facile detection at very low levels of specific biological interactions of any kind.
The extensive molecular level understanding of pathogens and of disease states that has emerged in recent years enables the diagnosis of disease on the basis of the detection of nucleic acids, proteins, and other biological molecules in patient samples. Molecular diagnostics are exceptionally valuable when they provide rapid, reliable answers at reduced cost compared with traditional laboratory diagnosis using culture, polymerase chain reaction and histology. Although these traditional methods are the gold standard, they are frequently expensive, time consuming, skilled-labour intensive, and simply not possible in numerous settings. Immunochromatography is the main alternative technology that is currently robust and cost-effective enough to enjoy widespread use outside of the clinical setting. Home pregnancy tests are one prominent example, and they make use of antibodies conjugated to either enzyme or colloid labels that effect a colour change if the hormone hCG is present at a sufficient level (40 pM, or 2.4 × 1011 molecules in 10 ml). Enzyme-linked immunosorbent assays (ELISAs) carried out in microtitre plates with fluorescent or chemiluminescent readouts are perhaps the next most widely used, relatively simple diagnostics. This type of ELISA is more costly and requires hours rather than minutes, skilled labour and detection instrumentation, but these tradeoffs are accompanied by gains in sensitivity. A number of groups have investigated using gold nanoparticle labels followed by reductive silver staining as an advantageous alternative to enzymatic amplification1,2,4. The present work has a shared goal of developing a non-enzymatic, material-based amplification strategy that improves on the sensitivity of ELISAs and eliminates as many of the aforementioned drawbacks as possible. Here, we have successfully used polymer chemistry in the place of nanoparticles and silver staining. Inspired by the biological amplification accomplished by enzymes, we have fabricated intelligent macroinitiators capable of both selective binding and subsequent polymerization of organic monomers as a facile chemical analogue to enzymatic amplification.
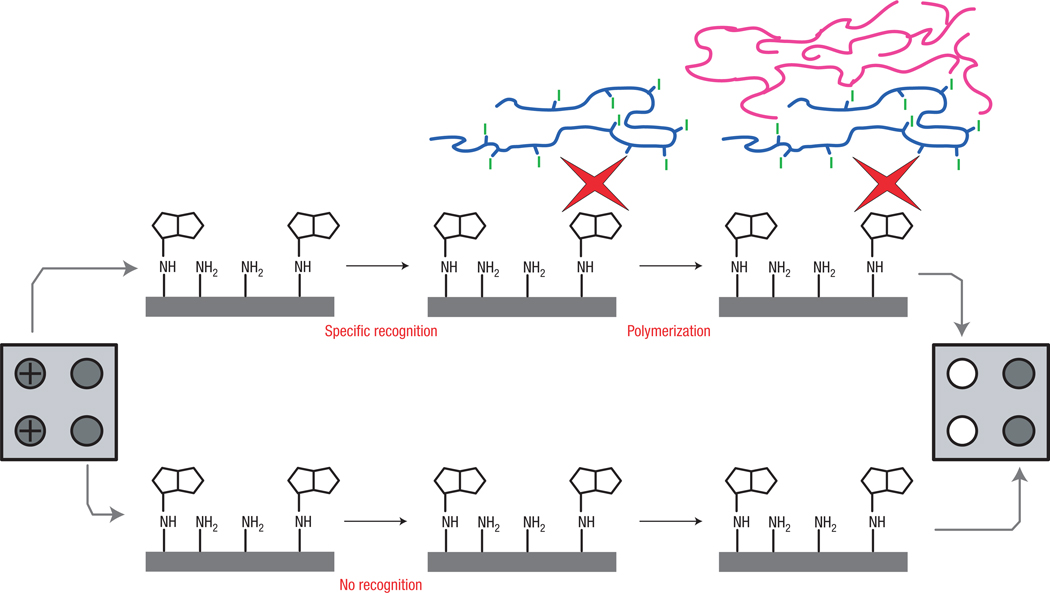
In photoinitiated free-radical polymerization13, carbon-based radicals derived from organic initiator molecules react with the carbon–carbon double bonds of acrylate monomers, and polymers are formed via a chain-growth mechanism. The concept of amplification is inherent in chain-growth polymerization reactions owing to the extremely large number of propagation steps that result from a single initiation event. The scope of this study is to explore the possibility of coupling a polymerization reaction to a biochemical binding event, and to determine how many binding events are required to result in readily detectable polymer formation. Figure 1 conceptually describes the photopolymerization of acrylate monomers as a means of signal amplification following a molecular-recognition event. This generalized exploration uses biotinylated oligonucleotides covalently bound to a surface, and the recognition event occurs between biotin and avidin14, though any specific biological interaction such as hybridization or antigen–antibody binding could be detected in an analogous manner. Though often thought of as a model system, biotin–avidin detection reagents enjoy widespread practical use in applications ranging from ELISAs to gene expression arrays. Assays using surfaces enable simple multiplexing because a single fluid sample can be interrogated for many biomolecules of interest simultaneously using an array of appropriate complementary molecules. For easy, relevant comparison to current technology used in commercial molecular diagnostic devices, thin-film biosensor surfaces15–17 (Inverness Medical-Biostar) were used, and side-by-side tests of the enzymatic amplification strategy used in their rapid, point-of-care tests and polymerization-based amplification were carried out.
Figure 1. Conceptual depiction of detection using polymerization-based signal amplification.
A surface that may contain biotin-labelled biomolecules of interest is placed in contact with a dual-functional macrophotoinitiator, a water-soluble macromolecule that contains one or two pendant Neutravidins (red shapes) and ~100 pendant photoinitiator moieties. The Neutravidin-functionalized macroinitiator binds only to the spots that contain biotin (illustrated with a ‘+’ in the first column) and does not bind elsewhere (negative spots illustrated schematically in the second column). The entire surface is then contacted with a monomer solution and irradiated to initiate polymerization. Recognition of biotin by avidin is demonstrated by the formation of a solid polymer only on areas of the surface where molecular recognition and subsequent polymerization occurred. Numerous other biological molecules can be detected with a similar approach by incorporating appropriately designed macroinitiators.
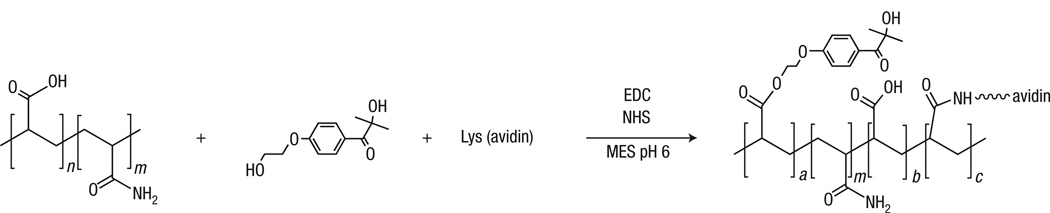
Design and synthesis of a molecule that is capable both of molecular recognition and of initiating a polymerization reaction are prerequisites for implementation of the proposed signal amplification method. Though polymers that are capable of molecular recognition18–20 and macroinitiators21–23 have each been reported in the literature, dual functional polymeric materials that are simultaneously capable of both functions, highly efficient initiation and biological recognition, have not been reported. Dual-functional macrophotoinitiators were synthesized by coupling water-soluble photoinitiators and Neutravidin to a fraction of the carboxylate residues of a high-molecular-weight copolymer of acrylic acid and acrylamide (Fig. 2) using aqueous carbodiimide coupling chemistry24. Ultraviolet absorbance measurements showed that each macroinitiator contained an average of 140 initiators per chain, and 4′-hydroxyazobenzene-2-carboxylic acid (HABA) assays25 revealed one or two pendant Neutravidins per chain with retention of biotin-binding capability.
Figure 2. Design and synthesis of molecules to enable polymerization following a binding event.
Polymeric materials that are capable both of selective binding and of initiating a radical polymerization reaction subsequent to binding were synthesized according to this reaction. Initiators and proteins were linked to the –COOH groups of a high-molecular-weight copolymer of acrylic acid and acrylamide through ester linkages (initiators) and amide linkages (proteins). Though the acrylamide subunits were not involved in the conjugation reaction, their presence was important in the surface bound detection results shown in Figs 3 and 4 (macroinitiators made with an acrylic-acid starting material in the place of poly(acrylic acid-co-acrylamide) do not function similarly).
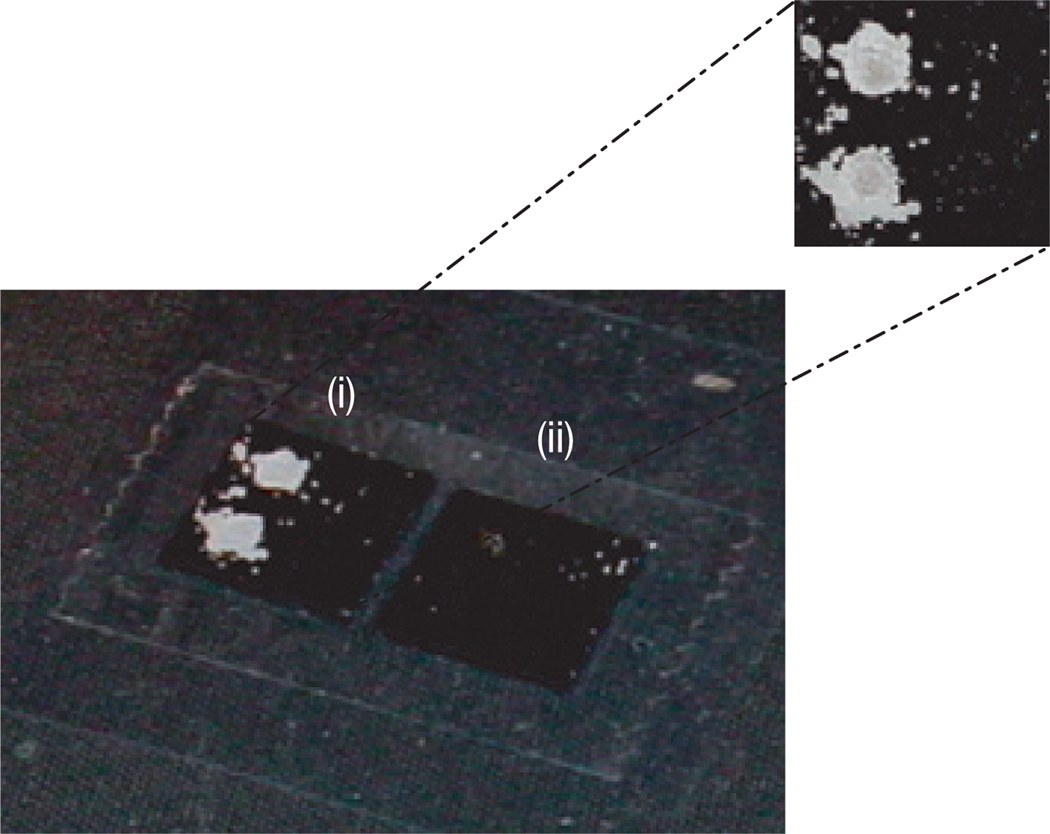
The ability of the macrophotoinitiator to recognize biotin-labelled oligonucleotides and to initiate polymerization of water-soluble monomers was tested on thin-film biosensors15–17. These surfaces, because of a specifically designed optical interference layer in the substrate15, were ideally suited to testing and optimizing polymerization conditions because films as thin as 5 nm result in an easily observable colour change of the surface from gold to blue15. Further, the colour of the film is a direct measure of the thickness of the film, and hence a quantitative measure of the amount of polymer that is formed. Figure 3 shows that, following a 10 min dose of 5 mW cm−2, 365 nm light, polymer grew only from the two spots containing biotinylated oligonucleotides (5 and 0.5 fmol) and did not grow from the two spots containing unlabelled oligonucleotides. The polymer spots rapidly exceeded 100 nm in thickness, below which the surfaces would yield quantitative information on the amount of polymer. Thus, the unique optical properties of the surfaces were not necessary for detecting a positive response, as all polymer films beyond 100 nm look white. The special optical properties of the surfaces were, however, useful for assessing the occurrence of bulk polymerization or non-specific polymerization, as even small amounts of polymer would cause a colour change on the surface. Colour changes were not observed, indicating the lack of both false negatives and bulk polymerization. A negative control chip was subjected to identical conditions except for a lack of exposure to the macrophotoinitiator. No polymerization resulted (at or above the visible limit of 5 nm), ruling out concerns that a molecule other than the macroinitiator initiated polymerization.
Figure 3. Polymerization for signal amplification following a binding event.
Digital-camera image of 2 × 2 oligonucleotide arrays spotted on thin-film biosensors15–17. The spots in the first column contain biotinylated oligonucleotides (5 fmol in the upper spot, and 0.5 fmol in the lower spot), whereas the spots in the second column contain unlabelled oligonucleotides (5 fmol and 0.5 fmol).
(i) Polymeric amplification only in spots containing biotinylated oligonucleotides.
(ii) Negative control (same surface, monomer and light, but no macrophotoinitiator).
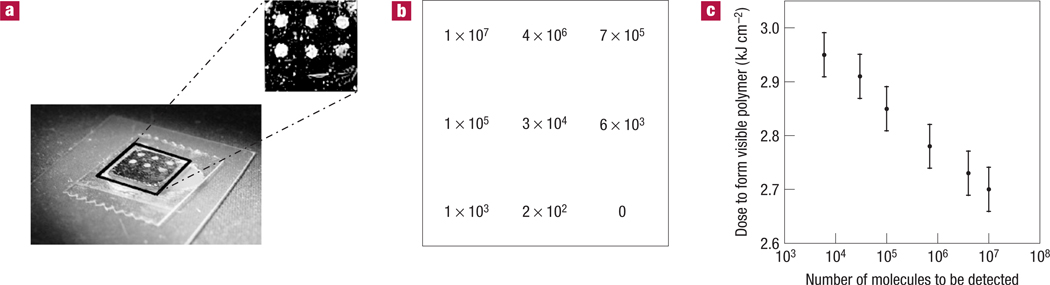
To determine a detection limit for surface-bound oligonucleotides, a dilution chip was prepared with spots containing from femtomoles to zeptomoles of biotinylated oligonucleotides. A negative control spot containing only unlabelled oligonucleotides was included on the chip. Figure 4 shows the results obtained using photopolymerization for signal amplification. The most dilute spot that is detected through the formation of a visible amount of polymer contains of the order of 1,000 biotinylated oligonucleotides, orders of magnitude fewer than the numbers that are detectable using enzymatic amplification methods. The picture shown in Fig. 4a is representative of 20 trials; no false positives or false negatives (above the limit of detection) occurred.
Figure 4. Quantification of the number of binding events necessary for a macroscopic, visible response using a 3 × 3 dilution array of biotinylated oligonucleotides.
a, Polymer spots were visible over the first six spots in the array following a 10 min incubation with avidin-functionalized macroinitiator and a 10 min dose of 5 mW cm−2, 365 nm light. Using enzymatic amplification (horseradish peroxidase/3,3′,5,5′-tetramethylbenzidine–dextran), only the first two spots were visible. b, The maximum number of biotinylated oligonucleotides present in each spot. The bottom right spot contained only unlabelled oligonucleotides and served as a negative control. c, The minimum radiation dose that was delivered to each spot before observation of polymer formation, where the dose is a product of the light intensity and exposure time. The error bars reflect variations in the intensity output from the lamp as measured with a radiometer.
Further, with constant light-intensity exposure, spots containing varying concentrations of the analyte do not appear simultaneously. Rather, as the surface-localized polymerization reactions progress, spots containing higher concentrations of biotinylated oligonucleotides become visible before spots with lower concentrations of biotinylated oligonucleotides become visible. Figure 4c shows the dose of light that was necessary to see each spot. This response of lower concentrations polymerizing with larger irradiation doses was observed in a differentiable manner across more than three orders of magnitude in analyte concentration. This outcome provides a facile means for converting the qualitative detection scheme that is the focus of this manuscript into a technique that is readily able to quantify the amount of an analyte present at these levels. Simply by changing the exposure time systematically across an array of sample spots, for example by the movement of an opaque film across the surface, it would be possible to quantify an analyte, even at these extremely low levels. Thus, a simple change in exposure time across an array would enable quantitative analysis of the target-molecule concentration.
The macroscopic, visible response generated by this small number of possible recognition events is remarkable and, to our knowledge, not possible using any other technique. We hypothesize that the large degree of polymerization is a result of high radical initiation rates occurring only at the desired surface whereas radicals that propagate into the bulk do not encounter radicals that lead to termination. The large degree of polymerization is further enhanced by the formation of a cross-linked, hydrogel polymer. Here, the monomer formulation was optimized to contain a cross-linking agent that hinders radical termination and facilitates the formation of extremely large amounts of polymer from each radical generated. The relationship between the colour of the spot and the thickness of the thin film within the spot has been reported previously: white spots correspond to film thicknesses of at least 100 nm (ref. 16). Using this thickness as a lower limit for the thicknesses of the polymer spots in Fig. 4, combined with knowledge of the number of possible binding events and the density of the polymer, we calculate a minimum amplification factor of 1011 monomers polymerized per binding event in the most dilute spot in Fig. 4. The density of biotinylated biomolecules in the last visible spot is ~0.005 µm−2; this density is below the limit of detection of even the newest high-end fluorescence scanners, instruments that cost tens of thousands of dollars.
False positives are a substantial concern with any signal amplification approach to detection. Though we did not encounter problems with false positives in this study, the result shown in Fig. 4c provides evidence that, should false positives arise with more complex samples or recognition pairs that have less specificity than avidin and biotin, it would be possible to shift the threshold of the positive response to exclude non-specific interactions by selecting an appropriate dose of light. Specificity of detection reagents is a limitation in many diagnostic assays, including ELISAs. Usually, it is not the enzymatic amplification step that limits sensitivity in these assays, but rather the antibody specificity. In this study, we compared the ‘top’ enzymatic amplification step used in many ELISA sandwich assays with polymerization-based amplification. This comparison, in which polymerization provided an improvement in sensitivity by orders of magnitude, is fair but perhaps not relevant until the specificity of detection reagents is improved. The exciting and rapidly progressing discoveries toward this end26–30 promise to make our findings more practically relevant in the coming years.
In its first incarnation, photopolymerization as a method of signal amplification has both strengths and weaknesses. It provides a limited amount of quantitative information about the number of binding events, but it does provide a rapid, highly sensitive, inexpensive, reliable yes/no response above a tunable threshold, which is a primary need in the case of initial screens for deadly pathogens or infectious agents that should not be present at any level in a healthy individual. A few examples of such applications that are given high priority by the World Health Organisation are tuberculosis, leprosy, syphilis, African trypanosomiasis, viral hemorrhagic fevers, gonorrhoea and chlamydia10. It is a method that is easily multiplexed; many biomolecules of interest can be screened for simultaneously in an array format. Inhibitors of polymerization must be excluded during the polymerization step; though this is manageable, future identification of more robust chemical reactions and/or strategies to combat inhibitors that do not diminish sensitivity will improve the method.
By detecting biotinylated oligonucleotides bound to surfaces, we set aside the separate problem of sample capture (for which there is a vast body of scientific literature) and focused on the signal-amplification problem. The high affinity of avidin for biotin certainly makes it a more forgiving recognition event to detect than hybridized nucleic acids or antigen–antibody interactions. However, we have already successfully used this polymerization-based amplification to detect nucleic-acid hybridization and antigen–antibody interactions. Detection in these real-world systems will be the focus of follow-up publications.
Though not ready for field application at this early stage, the new approach we have described improves on the detection limit usually associated with rapid techniques to a degree that could greatly impact near-patient testing even in remote settings by enabling the testing of small quantities of blood directly, such as from a finger prick. Identifying those at high risk for common diseases such as severe malaria or tuberculosis with a simple, rapid, and easy to use blood test requiring little or no instrumentation would help to better focus limited health-care resources in the developing world.
METHODS
MACROPHOTOINITIATOR SYNTHESIS AND CHARACTERIZATION
A 1 mg ml−1solution of N-hydroxysuccinimide (NHS) (Aldrich) in 0.1 M 2-morpholinoethanesulfonic acid (MES) 0.5 M NaCl buffer, pH 6, and a 1 mg ml−1solution of the commercial initiator Irgacure 2959 (Ciba) in distilled water, were prepared and placed on a vortexer for 10 min until fully dissolved. 0.8 mg of poly(acrylic acid co-acrylamide) (200,000 MW, Aldrich) and 1 mg of the coupling agent 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC) (Aldrich) were weighed out during this time. 260 µl of the NHS solution was pipetted into the tube containing 0.8 mg of poly(acrylic acid co-acrylamide). 1 ml of MES buffer was quickly added to the tube containing 1 mg EDC, and 440 µl of this solution was quickly added to the tube containing NHS and poly(acrylic acid co-acrylamide). This solution was placed on a shaker on a low setting for 15 min to allow time for activation of the carboxylic acids of poly(acrylic acid co-acrylamide) by EDC and NHS. 685 µl of the initiator solution and 50 µl of a 10 mg ml−1 Neutravidin solution (Pierce) were added to the activated poly(acrylic acid co-acrylamide) solution, and the reaction was allowed to proceed on a shaker on a low setting for one hour and 45 min. At this time, the high-molecular-weight product was separated from unreacted smaller molecules using a 100,000-molecular-weight cut-off filter (Millipore) and a centrifuge. The spin rates and times recommended by Millipore were used. Purified products were brought up to 500 µl total volume with MES buffer, and ultraviolet spectra were collected. A calibration curve of initiator absorbance as a function of concentration was made and used to determine the average number of initiator substituents per macrophotoinitiator. HABA assays were carried out as described in ref. 25 to determine the average number of Neutravidin substituents per macrophotoinitiator, and to verify retention of biotin binding capability. In our hands, this reaction was very sensitive to any deviation from the above procedures. The non-standard stoichiometry of reactants described above was reached by trial and error. Initial stoichiometry was 1 × poly(acrylic acid co-acrylamide): 1,000 × photoinitiator: 1,000 × EDC : 1,000 × NHS: 1 × Neutravidin; however, using this stoichiometry the resulting product was a cross-linked hydrogel that was not useful. Nuclear magnetic resonance was also used to verify the product of the above reaction. Representative nuclear magnetic resonance, HABA and ultraviolet–visible spectra are available as Supplementary Information.
ARRAYS
Optical thin-film biosensor surfaces were prepared and oligonucleotides were covalently coupled to the surfaces through hydrazone linkages according to previously published procedures17. To make the dilution chip shown in Fig. 4, spotting solutions were delivered to the surface in 60 nl droplets using a robotic microarrayer (custom instrument, Biodot); the resulting spots measured approximately 600 µm in diameter. Biotinylated oligonucleotides were diluted into spotting solutions containing unlabelled oligonucleotides so that the total oligonucleotide concentration remained constant across the array.
ENZYMATIC AMPLIFICATION
Detection of biotinylated oligonucleotides using enzymatic methods was accomplished by incubating thin-film biosensors with 50 µl of a 1 mg ml−1 solution of antibiotin–horseradish peroxidase conjugate (Jackson ImmunoResearch Laboratories) in a buffer consisting of 5 × standard sodium citrate (SSC), 0.1% SDS and 0.5% BlockAid (Invitrogen-Molecular Probes) for 10 min. Following thorough rinsing with 0.1 × SSC buffer, thin-film biosensors were incubated with 60 µl of a 3,3′,5,5′-tetramethylbenzidine–dextran solution (BioFX Laboratories) for 15 min. After rinsing with distilled water, thin-film biosensors were visually inspected for a colour change from gold to blue as described in ref. 16.
POLYMERIZATION-BASED AMPLIFICATION
Detection of biotinylated oligonucleotides using photopolymerization was accomplished by incubating thin-film biosensors with 50 µl of a 1.6 mg ml−1 solution of a macrophotoinitiator in a buffer consisting of 5 × SSC, 0.1% SDS, and 0.5% BlockAid (Invitrogen-Molecular Probes) for 10 min. No steps were taken to protect the macroinitiator from ambient light; it is stable under ambient light conditions. Immediately following rinsing with 0.1 × SSC buffer, while the surfaces were still wet, 50 µl of an argon-purged monomer solution (97% by weight hydroxyethylacrylate and 3% by weight ethyleneglycol dimethacrylate cross-linker, each triply distilled to remove inhibitors) was pipetted over the entire array. Polymerization from spots containing biotin and bound macrophotoinitiator was accomplished using a 10 min dose of 5 mW cm−2 ultraviolet light centred around 365 nm from a Blak-Ray B Series-100A lamp. The percentage of the cross-linker in the monomer formulation and the dose of radiation were optimized by systematic variation of each.
Polymerized arrays were photographed with a digital camera without any rinsing or other post-polymerization treatment.
Supplementary Material
Acknowledgments
H.D.S. acknowledges support from the National Human Genome Research Institute (NSRA F32-HG003100) and the Burroughs Wellcome Fund (Career Award at the Scientific Interface). R.R.H., L.M.J., K.L.R. and C.N.B. acknowledge support from NSF SGER 0442047 and NIH R41 AI060057. Thin-film biosensors, buffers and enzymatic detection reagents were generously provided by Inverness Medical-Biostar.
Footnotes
Supplementary Information accompanies this paper on www.nature.com/naturematerials.
Author contributions
K.L.R., J.W.B. and C.N.B. came up with the concept, C.N.B., H.D.S. and R.J. designed the experiments, H.D.S., R.R.H. and L.M.J. performed the experiments and H.D.S., C.N.B. and R.J. wrote the paper.
References
- 1.Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science. 1997;277:1078–1081. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- 2.Nam J-M, Thaxton CS, Mirkin CA. Nanoparticle-based bio-bar codes for the ultrasensitive detection of proteins. Science. 2003;301:1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 3.Jenison R, Yang S, Haeberli A, Polisky B. Interference-based detection of nucleic acid targets on optically coated silicon. Nature Biotechnol. 2001;19:62–65. doi: 10.1038/83530. [DOI] [PubMed] [Google Scholar]
- 4.Sia SK, Linder V, Parviz BA, Siegel A, Whitesides GM. An integrated approach to a portable and low-cost immunoassay for resource-poor settings. Angew. Chem. Int. Edn. 2004;43:498–502. doi: 10.1002/anie.200353016. [DOI] [PubMed] [Google Scholar]
- 5.Lee HJ, Li Y, Wark AW, Corn RM. Enzymatically amplified SPR imaging detection of DNA by exonuclease III digestion of DNA microarrays. Anal. Chem. 2005;77:5096–5100. doi: 10.1021/ac050815w. [DOI] [PubMed] [Google Scholar]
- 6.Cooper MA. Label-free screening of bio-molecular interactions. Anal. Bioanal. Chem. 2003;377:834–842. doi: 10.1007/s00216-003-2111-y. [DOI] [PubMed] [Google Scholar]
- 7.Fan C, Plaxco KW, Heeger AJ. Biosensors based on binding-modulated donor–acceptor distances. Trends Biotechnol. 2005;23:186–192. doi: 10.1016/j.tibtech.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 8.Cunin F, et al. Biomolecular screening with encoded porous-silicon photonic crystals. Nature Mater. 2002;1:39–41. doi: 10.1038/nmat702. [DOI] [PubMed] [Google Scholar]
- 9.Asher SA, et al. Photonic crystal carbohydrate sensors: low ionic strength sugar sensing. J. Am. Chem. Soc. 2003;125:3322–3329. doi: 10.1021/ja021037h. [DOI] [PubMed] [Google Scholar]
- 10.Mabey D, Peeling RW, Ustianowski A, Perkins MD. Diagnostics for the developing world. Nature Rev. Microbiol. 2004;2:231–240. doi: 10.1038/nrmicro841. [DOI] [PubMed] [Google Scholar]
- 11.Bell J. Predicting disease using genomics. Nature. 2004;429:453–456. doi: 10.1038/nature02624. [DOI] [PubMed] [Google Scholar]
- 12.Darr AS, et al. Top ten biotechnologies for improving health in developing countries. Nature Genet. 2002;32:229–232. doi: 10.1038/ng1002-229. [DOI] [PubMed] [Google Scholar]
- 13.Kloosterboer JG. Network formation by chain crosslinking photopolymerization and its applications in electronics. Adv. Polym. Sci. 1988;84:1–61. [Google Scholar]
- 14.Wilcheck M, Bayer EA, Livnah O. Essentials of biorecognition: The (strept)avidin–biotin system as a model for protein–protein and protein–ligand interaction. Immunol. Lett. 2006;103:27–32. doi: 10.1016/j.imlet.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 15.Jenison R, La H, Haeberli A, Ostroff R, Polisky B. Silicon-based biosensors for rapid detection of protein or nucleic acid targets. Clin. Chem. 2001;47:1894–1900. [PubMed] [Google Scholar]
- 16.Jenison R, Yang S, Haeberli A, Polisky B. Interference-based detection of nucleic acid targets on optically coated silicon. Nature Biotechnol. 2001;19:62–65. doi: 10.1038/83530. [DOI] [PubMed] [Google Scholar]
- 17.Zhong X, et al. Single-nucleotide polymorphism genotyping on optical thin-film biosensor chips. Proc. Natl Acad. Sci. USA. 2003;100:11559–11564. doi: 10.1073/pnas.1934783100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mosbach K, Ramstrom O. The emerging technique of molecular imprinting and its future impact on biotechnology. Nature Biotechnol. 1996;14:163–170. [Google Scholar]
- 19.Peppas NA, Hilt JZ, Khademhosseini A, Langer R. Hydrogels in biology and medicine: from molecular principles to bionanotechnology. Adv. Mater. 2006;18:1345–1360. [Google Scholar]
- 20.O’Connor NA, Paisner DA, Huryn D, Shea KJ. Screening of 5-HT1A receptor antagonists using molecularly imprinted polymers. J. Am. Chem. Soc. 2007;129:1680–1689. doi: 10.1021/ja067276c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hawker CJ, Bosman AW, Harth E. New polymer synthesis by nitroxide mediated living polymerizations. Chem. Rev. 2001;101:3661–3688. doi: 10.1021/cr990119u. [DOI] [PubMed] [Google Scholar]
- 22.Coessens V, Pintauer T, Matyjaszewski K. Functional polymers by atom transfer radical polymerization. Prog. Polym. Sci. 2001;26:337–377. [Google Scholar]
- 23.Guacher G, et al. Block copolymer micelles: Preparation, characterization, and application in drug delivery. J. Control. Rel. 2005;109:169–188. doi: 10.1016/j.jconrel.2005.09.034. [DOI] [PubMed] [Google Scholar]
- 24.Staros JV, Wright RW, Swingle DM. Enhancement by N-hydroxysulfosuccinimide of water-soluble carbodiimide-mediated coupling reactions. Anal. Biochem. 1986;156:220–222. doi: 10.1016/0003-2697(86)90176-4. [DOI] [PubMed] [Google Scholar]
- 25.Green NM. A spectrophotometric assay for avidin and biotin based on binding of dyes by avidin. Biochem. J. 1965;94:23c–24c. doi: 10.1042/bj0940023c. [DOI] [PubMed] [Google Scholar]
- 26.Jayasena SD. Aptamers: An emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999;45:1628–1650. [PubMed] [Google Scholar]
- 27.Binz HK, Amstutz P, Pluckthun A. Engineering novel binding proteins from nonimmunoglobulin domains. Nature Biotechnol. 2005;23:1257–1268. doi: 10.1038/nbt1127. [DOI] [PubMed] [Google Scholar]
- 28.Brandt O, Hoheisel JD. Peptide nucleic acids on microarray and other biosensors. Trends Biotechnol. 2004;22:617–622. doi: 10.1016/j.tibtech.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 29.Liu H, et al. A four-base paired genetic helix with expanded size. Science. 2003;302:868–871. doi: 10.1126/science.1088334. [DOI] [PubMed] [Google Scholar]
- 30.Boder ET, Midelfort KS, Wittrup KD. Directed evolution of antibody fragments with monovalent femtomolar antigen binding affinity. Proc. Natl Acad. Sci. 2000;97:10701–10705. doi: 10.1073/pnas.170297297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.