Abstract
There are three predominant forms of co-translational mRNA surveillance: nonsense-mediated decay (NMD), no-go decay (NGD) and non-stop decay (NSD). While discussion of these pathways often focuses on mRNA fate, there is growing consensus that there are other important outcomes of these processes that must be simultaneously considered. Here, we seek to highlight similarities between NMD, NGD and NSD and their likely origins on the ribosome during translation.
In eukaryotes, grossly aberrant mRNAs, such as those lacking a 5′ cap or 3′ poly(A) tail, are unlikely to effectively engage in the translational cycle. However, mRNAs containing more subtle errors cannot be as easily discriminated against. Some of these mRNAs, if translated, can produce aberrant protein products that are detrimental to the cell. To minimize these errors, cells have evolved mechanisms to monitor mRNAs as they are translated and to degrade troublesome transcripts – these processes are broadly referred to as “mRNA surveillance”. Most of the recognition events occur on the ribosome, thus directly implicating translation in these processes. It follows as no surprise that increasing evidence shows that the effects of these surveillance pathways are not restricted to the mRNA, but rather have broad consequences for the translational output of a cell. Studies on mRNA surveillance have traditionally focused on mRNA fate and many excellent reviews cover this area of interest (e.g. ref. 1,2). In this review, we focus on exploring mRNA surveillance from the perspective of its origins on the ribosome. We hope that this approach provides a new perspective from which to consider mRNA surveillance and will lead to new and unanticipated insights that inform future experiments.
mRNA surveillance: what defines a substrate?
There are three classically identified mRNA surveillance pathways in eukaryotes: nonsense-mediated decay (NMD), non-stop decay (NSD), and no-go decay (NGD). Historically, the hallmark activity of each process is the selective degradation of a class of aberrant mRNAs: NMD specifically targets mRNAs containing a premature termination codon (PTC), NSD targets mRNAs lacking a termination codon and NGD targets mRNAs containing a range of potential stall-inducing sequences. In this section, we discuss in more specific terms our current understanding of the molecular features that define these three classes of targeted mRNA.
NMD
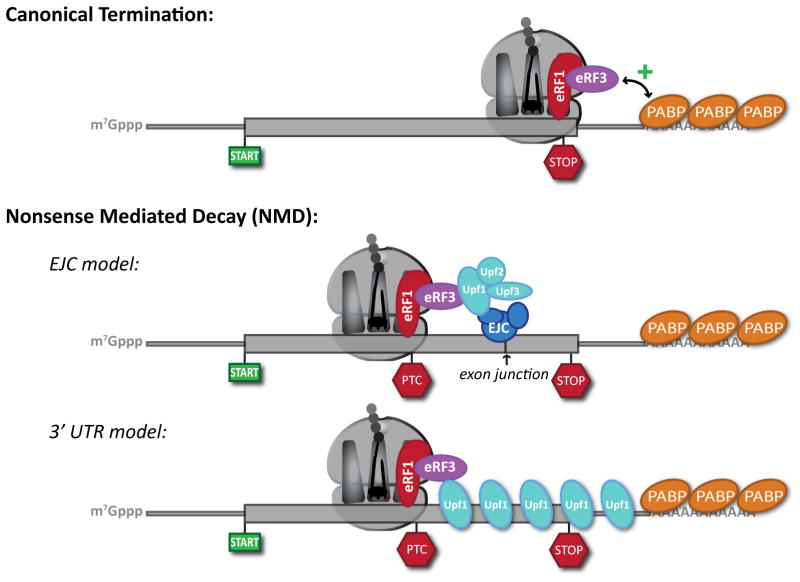
All stop codons must initially be recognized by the canonical translation termination factors eRF1 and eRF3 (Fig. 1a). What then distinguishes a “premature” stop codon from an authentic one? In higher eukaryotes, premature termination codons are generally thought to be recognized by their proximity to protein complexes (called exon-junction complexes, or EJCs) deposited near exon junctions during pre-mRNA splicing in the nucleus3,4. As authentic stop codons are typically located in the 3′ exon of spliced mRNAs, the presence of an EJC downstream of a stop codon immediately marks an mRNA as suspect (Fig 1b). Given that translating ribosomes likely displace such bound protein complexes, EJCs effectively define mRNA status during an initial, or “pioneer,” round of translation5. We note however that NMD does not strictly depend on the presence of an EJC even in higher eukaryotes6.
Figure 1. Recognition of NMD surveillance targets.
(a) Canonical termination. Capped and polyadenylated messages are translated through the open reading frame until recognition of a stop codon by the eukaryotic release factors, eRF1 and eRF3. Close proximity of authentic stop codons with the poly(A) tail is proposed to facilitate interactions between eRF3 and poly(A)-binding proteins (PABP) that positively contribute to peptide release. (b) Nonsense-mediated decay (NMD). In the case of a premature stop codon (PTC), lack of proximity is proposed to disrupt interaction between eRF3 and PABP. Canonical termination is further modified by the presence of NMD factors. In the EJC model of higher eukaryotes, this results from encountering a stop codon upstream of an exon-junction complex (EJC). In this model, communication between the termination factors and the EJC is effectively bridged by Upf1 in coordination with Upf2 and Upf3. In the 3′ UTR model, a PTC effectively extends the de facto 3′ untranslated region (UTR) of the message. This provides a larger binding platform for Upf1, which drives the termination event towards NMD rather than classical termination. Thick line, open reading frame; thin line, 5′ and 3′ UTR.
Broad applicability of this model is further compromised by the fact that there are few introns in some organisms, including the model yeast S. cerevisiae, and yet NMD in these organisms is robust. In these organisms, and perhaps elsewhere, NMD is proposed to be induced by recognition of a stop codon upstream of an extended 3′ untranslated region (UTR)7–9 (Fig 1b). Whether this feature defines a PTC because of the increased binding of Upf1 to the extended 3′ UTR8 or because of increased separation of positive termination effectors, such as poly(A)-binding protein (PABP), from the site of termination6 is not clear. And, while the presence of a poly(A) tail and PABP are non-essential for recognition of NMD substrates in yeast10, such studies still leave room for models where Upf1 is a positive effector of NMD that competes with PABP as a positive effector of normal termination11. Regardless of the trigger, a unifying theme is that RNA elements downstream of the PTC must interface with the translational apparatus to define stop codons as authentic or premature.
NGD and NSD: are all stalls equal?
NGD, as its name suggests, is a blanket term for a process that targets mRNAs with sequence features that cause translating ribosomes to “not go” or stall (typically at sense codons). The most effective NGD-targeting sequences to be studied are those induced by inhibitory mRNA structures such as stable stem-loops, pseudoknots, GC-rich sequences or damaged RNA bases12,13 (Fig. 2a, top). It is suspected that more subtle perturbations in the mRNA, such as strings of certain codons14 or certain peptide sequences15, may also stimulate an NGD response (Fig 2b). Evidence suggests that, at least in the case of peptide-mediated arrest, these stalls are dependent on the conserved ribosomal protein RACK115, though a complete analysis of the role of RACK1 in NGD has yet to be presented. As we will discuss below, such mRNA stalling features typically result in endonucleolytic cleavage of the mRNA, which in turn likely identifies these ribosomes complexes as prime targets for surveillance.
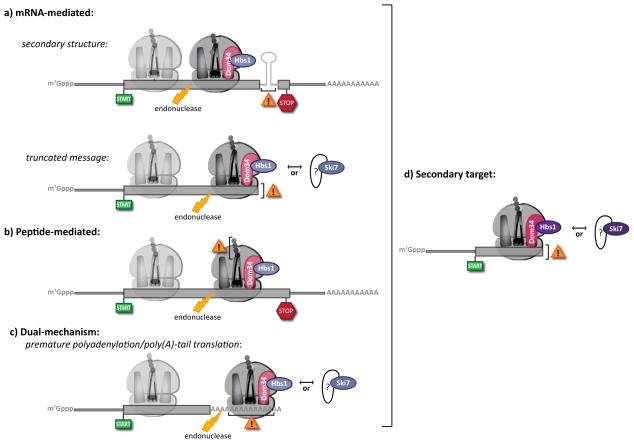
Figure 2. Recognition and initiation of NGD and NSD complexes.
No-go decay (NGD) and non-stop decay (NSD) both involve the recognition of stalled ribosome complexes. These stalls can arise through multiple mechanisms. (a) mRNA-mediated targets. Inhibitory mRNA secondary structures stall ribosomes at internal loci (top), while truncated mRNAs result in terminal stalls (bottom). While classically distinguished as NGD and NSD targets, respectively, increasing evidence suggests that such distinctions belie common mechanistic features. (b) Peptide-mediated targets. Inhibitory peptide sequences lead to internally stalled ribosomes, classically defined as NGD substrates. (c) Dual-mechanism targets. Translation of the poly(A) tail, originally considered to mimic a truncated message and invoke NSD, likely induces ribosome stalling prior to its arrival at the end of the message. As such, the distinction between NGD and NSD under these conditions is ambiguous in the absence of further experimentation. In all cases, (a), (b) and (c), endonucleolytic cleavage occurs upstream of the stalled ribosome, potentially stimulated by Dom34 and Hbs1. This tentative role for Dom34 and Hbs1 prior to cleavage is indicated by the increased transparency of these factors in (a–c). Following cleavage, the trailing ribosome (shown transparently) elongates to the point of cleavage, generating an ideal target for Dom34/Hbs1 (or Ski7) recognition (d). At present, no Dom34-like factor has been identified that interacts with Ski7.
As a technical note, while the term “stalling” is broadly used in the literature, kinetically distinguishing between transient pauses and stable stalls is difficult with currently available techniques (e.g. toeprinting and ribosomal profiling). So, for example, while these techniques identify high ribosome density at certain proline-rich sequence motifs16,17, the duration of such a pause in the cell remains to be determined. To maintain consistency with the literature, translational pauses that are sufficient to induce NGD will be referred to as stalls throughout this review. Implicit in this characterization, however, is the understanding that in most cases the kinetic features of these ‘stalling’ determinants remain to be thoroughly evaluated.
NSD was similarly discovered as a mechanism to resolve ribosome complexes stalled on defective mRNA. As the name implies, nonstop decay is broadly interpreted as a process for eliminating mRNAs that lack a stop codon18,19. mRNAs not carrying an in-frame stop signal can be of two types: the first class includes truncated mRNAs (Fig 2a, bottom) where the ribosome simply runs to the end of the template; the second class includes mRNAs lacking a stop codon but with a poly(A) tail (Fig 2c). In the latter case, it was initially assumed that ribosomes would translate through the poly(A) tail, reach the 3′ end of the mRNA and stall at the end of the template. If this were true, NGD and NSD substrates would differ based on whether the inducing stall occurs mid-message (NGD) or at the end of the message (NSD).
Recently, however, these distinctions between NGD and NSD have become blurred. It is now thought that translation of poly(A) sequences into poly-lysine can cause ribosome stalling through interactions between the positively charged peptide and the overwhelmingly negatively charged exit channel of the ribosome20,21. The potency of these stalls increases with length, but can be observed after incorporating as few as 6 lysine residues (corresponding to the translation of as few as 18 adenosine nucleotides)20. Since the typical length of a poly(A) tail is ~70 nucleotides in yeast and ~200 in human cells22, a ribosome that translates into the poly(A) region likely will stall long before reaching the 3′ end of the mRNA. Therefore, poly(A) read-through, which was previously referred to as an end-of-message stall, or NSD, may also involve peptide-mediated internal stalling, reminiscent of NGD. Regardless of how they are classified, what ultimately appears to unify all NGD and NSD substrates is the formation, following endonucleolytic cleavage, of a secondary stall formed by the upstream translating ribosome reaching the cleavage site (Fig 2d). This secondary stall is a clear target for Dom34:Hbs1 or, in yeast, Ski7.
Independent of the cause, all stalls require similar resolution of the ribosome complex. In the end, ‘unnatural’ stalls and stochastic translational pausing must be distinguished from one another. While the mechanism of this discrimination is unknown, it seems likely that the kinetics of these events play a critical role such that the surveillance machinery efficiently recognizes only sufficiently long-lasting stalls. Such models have been well supported in other systems; for example, in protein quality control, specifically during protein folding, cells rely on the length of time a misfolded species exists to distinguish between transient folding intermediates and terminally misfolded protein products23.
Ribosome recognition by key mRNA surveillance factors
If a stalled ribosome complex is a substrate for surveillance, what are the specialized cellular factors that recognize these ribosome complexes and target them for resolution?
NMD
Some of the key factors involved in NMD – the UPF (for ‘UPstream Frameshifting’) genes Upf1, Upf2 and Upf3 - were identified in early genetic screens in yeast24–27. Each of the three factors is highly conserved in eukaryotes and implicated in NMD in a broad range of organisms28. Upf1 is an enzyme containing both ATPase and helicase activities29; inhibition of either of these activities impedes NMD30. Upf1 interacts with both eRF1 and eRF3 and is likely present during initial recognition of a premature stop codon31,32 (Fig 1b). Upf1 also interacts directly with Upf2 and Upf333. Upf2 and Upf3 modulate Upf1 activity and are thought to function as protein scaffolds34–36; any direct catalytic function for Upf2 and Upf3 is unknown. Further studies in higher eukaryotes have implicated numerous other critical and conserved factors involved in NMD37. While several of these factors will be discussed in this perspective, in particular those that directly engage the translational machinery, for a more extensive review of the role of these other factors in NMD we direct the reader to ref. 28.
While some is known about the core Upf factors and what they can do in isolation, less is known about how these factors help to identify premature stop codons or begin to specify the downstream events of NMD. As described in the previous section, during NMD, something about the mRNA sequence downstream of a stop codon informs the ribosome termination complex that termination is occurring too early. The Upfs are proposed, for example, to bridge the interaction between the ribosome and this downstream mRNA signal35,38. In the EJC model, this signal includes the EJC proteins, MAGOH, Y14, and eIF4AIII, which directly interact with the Upfs and, in turn, the terminating ribosome (Fig. 1b)35. Another model (the 3′ UTR model) suggests that Upf1 directly interacts with (and even coats) the 3′ UTR of an mRNA; as such, a PTC will be associated with a longer 3′ UTR sequence than an authentic termination codon, and thus with a larger target for Upf1 binding8. An additional consequence of a long 3′ UTR is that the poly(A) tail is less proximal to the stop codon. Some studies have argued that proximity to the poly(A) tail lowers the likelihood of a stop codon being recognized as premature6 while other studies argue that the polyA tail plays little if any role in NMD10. Finally, still other models argue that Upf1 is directly associated with the small ribosomal subunit, evaluating encounters with termination codons as they appear in the decoding center39.
In all of the models, Upf1 is involved as a critical effector molecule, though little is known about the actual mechanism of PTC recognition or the effects of Upf recognition on downstream ribosome function. It will be important to make these connections in moving forward.
NGD and NSD
Two protein factors, Dom34 (or Pelota) and Hbs1, were originally implicated in NGD through genetic approaches showing that the decay of mRNAs on which ribosomes stalled depended on their presence12. Strikingly, Dom34 and Hbs1 are structurally related to the canonical termination factors eRF1 and eRF340–45, immediately suggesting that NGD, like NMD, involves a modified termination event. Indeed, Dom34 and Hbs1 interact directly with the A site of the ribosome, like the canonical termination factors, but promote an event akin to ribosome recycling46,47. That Dom34 and Hbs1 function directly on the ribosome suggests that the effects of these surveillance pathways may have broad consequences for ribosome function and translational output.
Also characteristic of the initiation of NGD is an early endonucleolytic cleavage event. It is not fully understood how the endonucleolytic cleavage is triggered, but cleavage increases in the presence of Dom34, suggesting that Dom34 could play a stimulatory role12. While Dom34 was originally proposed to act directly as the endonuclease44, many biochemical and genetic studies have since argued against such an activity15,41,48.
Critical insight into the specificity of recognition by the Dom34:Hbs1 complex was recently obtained in in vitro studies showing that these factors operate most efficiently on ribosome complexes with very little mRNA sequence extending 3′ of the site of stalling49 (Fig 2d); subsequent studies further established that this limitation is imposed by Hbs1 and not by Dom3450. Consistent with this idea, structural studies position the N-terminus of Hbs1 at the mRNA entry channel46, poised to monitor mRNA length. In this way, mRNA length detection by Hbs1 drives the events of NGD specifically on ribosome complexes stalled proximal to the 3′ end of an mRNA50. The central importance of the endonucleolytic cleavage event becomes clear in the light of this biochemical result; cleavage of the mRNA generates a strong inducing signal for Dom34:Hbs1 activity. As a caveat, it is important to reiterate that genetic experiments suggest a role for Dom34 binding prior to cleavage. The interplay between Dom34 and the unknown endonuclease, and which signal ultimately initiates NGD, is poorly understood. It is interesting to speculate that recognition of stalled ribosomes in bacterial systems as mediated by tmRNA:SmpB and/or YaeJ relies on similar clues. Recent X-ray structures of bacterial ribosomes bound to these different “rescue” factors reveal specific protein moieties located near the decoding center where mRNA length could be directly monitored51,52.
Ski7 is a factor that has specifically been implicated in recognition of nonstop-stalled ribosome complexes during NSD19 (Fig. 2). Ski7 is a translational GTPase, closely related to the NGD factor Hbs1 (and thus to eRF3), having arisen through the duplication of a common ancestral gene53. Ski7 is known to interact with the exosome, placing it at the interface of ribosome recognition and mRNA degradation. Yet Ski7 is rare, found only in a small subset of yeasts40; organisms lacking this gene likely rely on the related Hbs1 to function in both NGD and NSD. Consistent with this prediction, Hbs1 from a yeast lacking Ski7 (S. kluyveri) can complement both Hbs1 and Ski7 deletions in S. cerevisiae53. On a mechanistic level, many questions remain concerning the initial recognition of NSD-targeted ribosome complexes. For example, no binding partner, such as Dom34 for Hbs1 or eRF1 for eRF3, has been identified for Ski7 in yeast. Moreover, the interaction of Ski7 with the ribosome remains wholly uncharacterized. As Dom34 and Hbs1 are seen to be active on ribosomes stalled at the 3′ end of messages, Ski7 may play a redundant role with Dom34/Hbs1 as suggested by recent studies from Inada’s group54. That said, if NGD and NSD are essentially equivalent processes, it is unclear why Ski7 would be preserved in yeast. Appreciation for significant overlap between NGD and NSD is newly developing and will require further experiments to deconvolute.
What are the consequences of aberrant translation?
Despite their several differences, what ultimately unites all three surveillance pathways – NMD, NSD and NGD – is the presence in the cell of a problematic ribosome complex that must be resolved on multiple levels (Fig. 3). First, the unproductive mRNA must be eliminated. Second, the incomplete protein product may have dominant or toxic effects, and so again elimination makes sense. Third, and perhaps most significantly, ribosomes are energetically costly to replace and so the cell will ideally seek to recover stalled subunits for subsequent rounds of translation. Each of these events is discussed individually below.
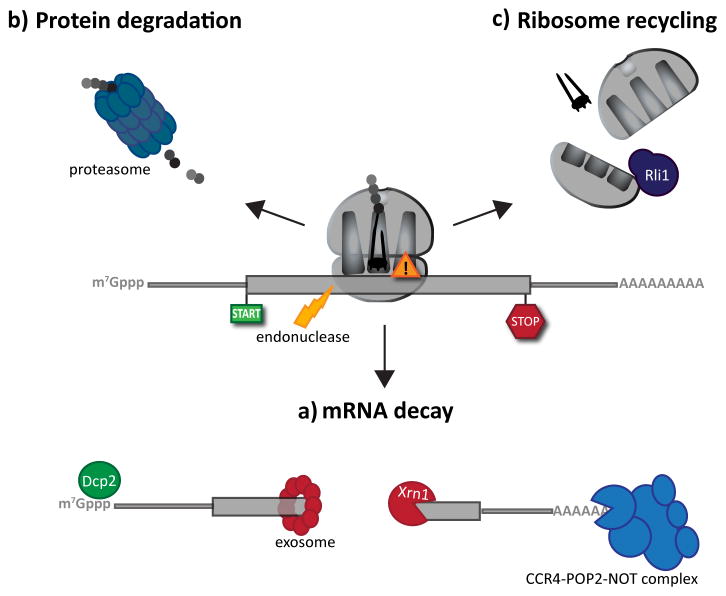
Figure 3. mRNA surveillance pathway outcomes.
Following the recognition of NMD, NGD or NSD ribosome complexes, at least three discrete salvage pathways are invoked: mRNA decay, protein degradation and ribosome recycling. (a) mRNA decay. Endonucleolytic cleavage subverts the need for deadenylation, by the CCR4-POP2-NOT complex, and decapping, by Dcp2, prior to mRNA decay. Rapid mRNA degradation then proceeds through canonical means, including 5′-3′ degradation by Xrn1 and 3′-5′ degradation by the exosome. (b) Protein degradation. Targeted degradation of aberrant peptides occurs via the ubiquitin-proteasome system. Several E3 ligases have been implicated in this process, but the molecular features of substrate recognition remain to be determined. (c) Ribosome recycling. Dom34:Hbs1 are known to exploit the canonical recycling activities of Rli1 to effect ribosome recycling during NGD and NSD. Recycling of ribosome complexes during NMD are less well-characterized, but may involve Upf1.
mRNA decay
The hallmark of mRNA surveillance pathways has long been the selective degradation of aberrant mRNAs. Canonical mRNA degradation occurs in both the 5′-3′ direction, by the exonuclease Xrn1, and the 3′-5′ direction, by the exosome, Ski7 and the Ski complex55,56 (Box 1). In NMD, targeted mRNAs undergo accelerated decay from both directions57,58. In vivo, Upf1 associates with multiple factors implicated in mRNA degradation, which suggests plausible mechanisms for this acceleration59,60. For example, the tethering of human Smg7, a Upf1-interacting protein, to the 3′ UTR of a reporter gene bypasses the requirement for Upf1 function in NMD in mammalian cells61; as such, these studies argue that Smg7 may be directly involved in recruitment of mRNA decay components that act downstream of Upf1. Additionally, endonucleolytic cleavage of PTC-containing mRNAs has been observed in several higher eukaryotes62,63. In both Drosophila and humans, this endonucleolytic cleavage event is catalyzed by the PilT N-terminus (PIN) domain of Smg6, an NMD factor conserved in metazoans62,64. However, no PIN domain containing proteins have been implicated in NMD in yeast nor has endonucleolytic activity been observed in this organism during NMD.
Box 1. Classical mRNA degradation mechanisms in yeast.
Turnover of stable messenger RNA occurs through two general mechanisms: 5′-3′ and 3′-5′ degradation (for reviews, see refs 92,93). In both cases, decay initiates via deadenylation56 catalyzed by the CCR4-POP2-NOT complex94,95. Substantial deadenylation (leaving behind fewer than ~10 adenosines) is required for mRNA degradation to further progress. After this, 5′-3′ degradation is thought to be the primary direction of mRNA degradation in yeast56. This process begins with removal of the mRNA cap structure by the decapping enzyme Dcp296. Removal of the 5′ cap sensitizes mRNA to degradation by the 5′ exonuclease Xrn197. Recent evidence suggests that 5′-3′ degradation by Xrn1 occurs co-translationally98, allowing ribosomes to complete a round of translation while the trailing mRNA is degraded. Prior to this recent report, Xrn1-mediated degradation had been observed only on non-translating mRNPs that appear to accumulate in discrete cytoplasmic foci, called P bodies (reviewed in ref. 99).
As mentioned above, an alternative (3′-5′) degradation pathway also exists. This process also follows deadenylation and is catalyzed by a multifactor ring complex termed the exosome55. The core exoribonuclease activity of the yeast exosome resides in one subunit, Dis3100. The remaining subunits, while catalytically inactive, form a pore-like structure through which the RNA is threaded101. While the exosome has both nuclear and cytoplasmic RNA processing functions, the cytoplasmic activities seem to be primarily responsible for 3′-5′ degradation of bulk mRNA102. The cytoplasmic exosome further requires Ski7 and the Ski complex – composed of Ski2, Ski3, and Ski8 – which tether the exosome to mRNA targets55,103,104. We recall that an additional role for Ski7 in NSD has also been proposed, as discussed in the main text. The differential roles of Ski7 in NSD and basal exosome function are of ongoing interest.
The rate of decay is ultimately influenced by multiple features of an mRNA. The most well characterized features are protein binding sites including, for example, AU-rich elements (AREs) or PUF protein binding sites. These features are typically found in the 3′ UTR and can have either positive or negative effects on mRNA half-life. The complex determinants (including the binding sites for various trans acting factors) that control half-life for the majority of mRNAs are poorly understood, and likely vary by organism. There is currently no generalized, predictive model for mRNA half-life.
NSD-targeted messages have recently been shown to be endonucleolytically cleaved upstream of stalled ribosomes52,63. The catalytic subunit of the exosome, Rrp44p (alternatively called Dis3p), can promote both endonucleolytic and exonucleolytic activities, both of which appear to be involved in degradation of nonstop messages65,66. In yeast, recruitment of the exosome to these messages is promoted by Ski719. A favored model is that Ski7 fulfills a bridging function: the C-terminal domain of Ski7, which resembles a translational GTPase, binds to NSD-targeted ribosome complexes while the N-terminal domain recruits the exosome19.
NGD-targeted mRNAs are also generally subject to endonucleolytic cleavage, as discussed above (Fig 2). Following endonucleolytic cleavage, the 3′ and 5′ mRNA fragments are subsequently degraded by Xrn1 and the exosome, respectively12 (Fig 3a). Endonucleolytic cleavage during NGD occurs upstream of the stalling site in the mRNA and results in a 5′ mRNA fragment lacking a poly(A) tail40,52. This fragment, if translated by another ribosome, results in another stalled complex – a conspicuous target for additional rounds of mRNA surveillance (Fig 2d). If secondary stalls induce additional cleavages, multiple cleavage events should occur with ribosome-sized spacing upstream of the initial stall site. In fact, several groups have confirmed such a prediction and reported regularly spaced cleavage events positioned just upstream of mRNA stall sequences of interest52,65.
Endonucleolytic cleavage has been implicated in all three mRNA surveillance pathways though the cellular factor responsible for the cleavage and the actual inducing stimuli are incompletely defined (with the exception of the cleavage factor, Smg6, involved in NMD in higher eukaryotes). That said, endonucleolytic cleavage is a potent mechanism for triggering mRNA decay. A single endonucleolytic event circumvents the need for the normal initial steps in mRNA decay, decapping and deadenylation, which are typically slow and tightly regulated (Box 1). As such, cleavage is likely to be an irreversible process that commits a stalled ribosome (and its mRNA) to the surveillance pathway. The extent to which there is overlap between NMD-, NGD- and NSD-based endonucleolytic cleavage mechanisms is yet to be resolved. Deciphering how cleavage occurs, including how complexes are selected for cleavage and the identification of the endonuclease(s) involved in NGD and NSD, will greatly advance our understanding of these processes in vivo.
Degrading the peptide
The partial peptide derived from the stall-inducing mRNA is not likely to play a positive physiological role in the cell and so these peptides are typically targeted for degradation. Various studies have identified NMD-, NSD- and NGD-derived protein products as readily processed substrates for the proteasome20,67,68. These data suggest that quality control pathways can accelerate the degradation of stalled or incomplete protein products. In the case of both NGD and NSD, the terminated protein product likely originates from peptidyl-tRNA that is directly produced by the actions of Dom34 and Hbs154.
Two E3 ligases, Not4 and Ltn1 (also known as either YMR247C or Rkr1 in yeast), have been shown to target NSD protein products for polyubiquitylation and subsequent degradation68–70. It is unknown whether these same E3 ligases are similarly involved in the destabilization of NGD- or NMD-derived proteins. One intriguing alternative candidate for this role is the N-terminus of Upf1, which itself has an E3 ligase motif that is known to contribute to NMD36. Additional work will be required to fully elucidate the mechanisms by which these different classes of stalled peptides are recognized, as well as the extent to which these degradative events occur co-translationally as a function of mRNA surveillance or post-translationally through more canonical pathways.
Recovering the ribosomes
Ribosomes are large cellular machines that are energetically costly to synthesize and thus are worth preserving if their malfunction is not the source of the problem. To recover ribosomes, some form of ribosome recycling must take place to allow for the dissociated ribosomal subunits to engage in re-initiation.
For NGD and NSD, as anticipated from similarities to the translation termination factors eRF1 and eRF3, Dom34 and Hbs1 were shown to directly bind to the A site of the ribosome, in a codon-independent manner, and to dissociate ribosome complexes47. In vitro, this subunit splitting activity is further stimulated by Rli149,50, an essential ATPase known to be required for canonical ribosome dissociation71,72. Structural data suggest that Rli1 forces Dom34 – or in the case of canonical recycling, eRF1 – through the ribosomal subunit interface, disrupting critical intersubunit bridges in the process and leading directly to subunit dissociation73. In vivo, genetic studies demonstrate that Dom34, Hbs1 and, presumably, Rli1 are required for subunit dissociation during both NSD and NGD54; because Rli1 is an essential gene, it is has been difficult to establish its specific roles in vivo. The splitting of ribosome complexes by Dom34 facilitates subsequent rounds of translation initiation49,74. As for NMD, given the essential role of Upf1, a known ATPase, in the process, and the typical energetic demands of a ribosome splitting reaction, Upf1 is a viable candidate for filling this role. While there are some data consistent with such a model in yeast 7,39, it is also possible that the NMD factors simply serve to recruit more canonical recycling factors in the cell such as Rli1, and even Dom34/Hbs1.
Eliminating ribosomes when they are faulty
An interesting twist in this survey of mRNA surveillance is the apparently related process of nonfunctional ribosome decay (NRD). Dom34, the eRF1 homolog that is intimately involved in NGD and NSD, is critical for the rapid turnover of demonstrably faulty small ribosomal subunits75. This was clearly demonstrated when small ribosomal subunits carrying debilitating mutations in key residues involved in tRNA selection were specifically targeted for rapid turnover76. Given the dependence of this process on Dom3475, NRD likely initiates like NGD with a stalled ribosome complex, except that the stall-inducing signal in this case is located within the ribosome rather than within the mRNA. While it seems unlikely that Dom34 specifically targets ribosomes for degradation, ribosomes that are repeatedly dissociated by Dom34 will be repeatedly exposed to the degradative machinery, thus resulting in an acceleration in the rate of their degradation. These data very clearly define a specific role for the surveillance machinery in ribosome fate.
Broad surveillance mechanism or specific gene regulator?
Some of the most interesting questions surrounding mRNA surveillance pathways revolve around their effects at an organismal level. To what extent are these surveillance mechanisms quality control pathways and to what extent do cells exploit these pathways to selectively modulate broader translational events? We know that the ubiquitin-proteasome system, for example, is involved in both basal and selective protein turnover.
NMD
There are numerous studies establishing that NMD modulates the stability of a variety of specific transcripts including alternatively spliced messages, messages containing upstream ORFs, and transcripts that derive from transposons, pseudogenes, or out-of-frame gene rearrangements (as in T cell receptor and immunoglobulin genes)37. In all, NMD regulates a high number and broad range of transcripts in vivo with estimates indicating as many as 10% of all eukaryotic genes77–80. While each of the above mRNAs could contain a premature stop codon, there is evidence to suggest this is not always the case9. The mechanism by which non-PTC containing genes might be targeted by the NMD machinery is unclear though it is certainly possible that additional factors might be involved.
There has long been interest in NMD because of its strong connection to human disease; indeed, some 30% of inherited genetic disorders are thought to involve gene mutations which result in premature stop codons81. The ability to modulate NMD and selectively increase read-through of these stop codons82,83 has shown promise as a therapeutic strategy for diseases such as cystic fibrosis84,85. Further insights into NMD and stop codon read-through should aid in identifying additional drug targets and advancing these therapies.
NSD/NGD
While genome-wide efforts at characterizing NSD and NGD targets have not been published, genome-wide analysis has revealed that alternative polyadenylation sites are common in both higher and lower eukaryotes18. Premature polyadenylation occurring within coding sequences is likely to elicit NGD/NSD in response to translation of poly-lysine tracts. NGD is also involved in responding to chemically damaged mRNAs, as depurinated mRNA appears to stall translation, leading to mRNA degradation in a Dom34-dependent manner13. Oxidative mRNA damage is similarly likely to cause ribosome stalling86; a role for Dom34 in responding to oxidative mRNA damage has not directly been explored although deletion of DOM34 sensitizes yeast to a variety of oxidative stressors87. Intriguingly, oxidative mRNA damage may be clinically relevant as it is involved in the early pathogenesis of many neurological diseases such as Alzheimer’s and amyotrophic lateral sclerosis (ALS)88,89.
Another intriguing case of potential NGD targeting is found in work done on the CGS1 coding sequence in Arabidopsis. A region within CGS1, MTO1, arrests translation and induces mRNA degradation in the presence of S-adenosyl-L-methionine90. Ribosomal stalling in this context is peptide-mediated, caused by compaction of the nascent chain in the exit tunnel, and results in subsequent mRNA cleavage91 – both characteristic of NGD. At present, involvement of Dom34 and Hbs1 in this process is merely speculative.
Conclusions
In this review we hope to have emphasized the interconnectedness of translation and mRNA surveillance, as well as the multifaceted response of the cell to translation of aberrant transcripts. NMD, NGD and NSD effect strikingly similar fates for aberrant mRNAs, the ribosomes translating them and the altered protein products they encode. Indeed, broadly overlapping strategies are similarly used to resolve translational stalls in bacteria, indicative of the broad utility of such a three-pronged approach (Box 2). The term ‘mRNA surveillance’ inadvertently downplays the capacity of surveillance systems to broadly address translation of aberrant mRNAs, and it is not clear that mRNA degradation is even the most critical outcome. The majority of factors implicated in these processes either interact directly with known translation factors or are themselves translation factor homologs. We suggest that it will ultimately be informative to consider these surveillance events from the perspective of their origins on the ribosome.
Box 2. tmRNA: universality of surveillance outcomes.
Bacterial mRNAs are significantly less stable than their eukaryotic counterparts. Furthermore, the coupling of transcription and translation in bacteria limits the opportunity to assess mRNA quality prior to translation. As such, one might anticipate there would be a larger number of aberrant mRNAs actively being translated and so the need for surveillance systems to deal with such transcripts seems great.
Not surprisingly, parallel systems do appear to be found in bacteria. Stalled bacterial ribosome complexes result in 1) the degradation (with possible endonucleolytic cleavage) of the stalled mRNA105,106, 2) the tagging of the peptide for proteolysis107, and 3) the rescue of ribosomes through canonical recycling processes108. Indeed, these effects are strikingly reminiscent of the outcomes observed during eukaryotic ribosome rescue. However, none of the factors required for these activities in eukaryotes are conserved in bacteria. Instead, there seem to be several different systems in place to deal with ‘stalled’ ribosome complexes, some better characterized than others.
The best characterized of these ribosome rescue events depends on an intriguing functional RNA referred to as tmRNA (for a review, see ref. 109). tmRNA is an RNA that contains regions resembling a charged alanyl-tRNA (the t for transfer) and a short open reading frame ending in a stop codon (the m for mRNA). tmRNA requires two protein factors, SmpB and EFTu, for its function. Like Dom34:Hbs1, tmRNA:SmpB:EFTu:GTP binds to the A site of ribosome complexes, and is most efficient on ribosome complexes carrying only a short 3′ mRNA extension110; a recent structure of ribosome-bound tmRNA quaternary complex reveals how mRNA length may be directly monitored52. Upon tmRNA binding, the alanine residue is directly incorporated into the growing peptide chain, and the ORF-containing region of the tmRNA substitutes for the problematic stalled mRNA sequence. Translation resumes on the tmRNA ORF, resulting in a hybrid protein product that is ultimately tagged for degradation by bacterial proteases, such as the ClpXP system. tmRNA-rescued ribosomes are competent for subsequent recycling through canonical means since translational termination is effectively routine.
Other less well-characterized ribosome rescue events may depend on release factor homologs (typically missing the codon recognition domain) that survey the cell for stalled ribosomal complexes. While the molecular requirements of such rescue events have not been characterized, a recent X-ray structure of YaeJ bound to the ribosome reveals the presence of a protein domain that appears to engage, and likely monitor, the mRNA channel51. Thus, while the factors involved and the molecular mechanisms of ribosome rescue in bacteria and eukaryotes are quite distinct, the outcomes of the pathways are markedly conserved.
The molecular mechanisms by which these surveillance pathways are initiated on the ribosome remain a significant question in the field. Also of considerable interest are questions concerning the cell’s ability to exploit these surveillance processes to selectively regulate translation of non-aberrant transcripts. As our molecular understanding of these processes grows, and our ability to analyze translation in vivo increases, our ability to interpret biological relevance should similarly expand.
References
- 1.Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes & development. 2007;21:1833–56. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- 2.Wilson MA, Meaux S, van Hoof A. Diverse aberrancies target yeast mRNAs to cytoplasmic mRNA surveillance pathways. Biochimica et biophysica acta. 2008;1779:550–7. doi: 10.1016/j.bbagrm.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. The EMBO journal. 2000;19:6860–9. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maquat LE, Li X. Mammalian heat shock p70 and histone H4 transcripts, which derive from naturally intronless genes, are immune to nonsense-mediated decay. RNA. 2001;7:445–56. doi: 10.1017/s1355838201002229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maquat LE, Tarn WY, Isken O. The pioneer round of translation: features and functions. Cell. 2010;142:368–74. doi: 10.1016/j.cell.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh G, Rebbapragada I, Lykke-Andersen J. A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol. 2008;6:e111. doi: 10.1371/journal.pbio.0060111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amrani N, et al. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 2004;432:112–8. doi: 10.1038/nature03060. [DOI] [PubMed] [Google Scholar]
- 8.Hogg JR, Goff SP. Upf1 senses 3′UTR length to potentiate mRNA decay. Cell. 2010;143:379–89. doi: 10.1016/j.cell.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muhlrad D, Parker R. Aberrant mRNAs with extended 3′ UTRs are substrates for rapid degradation by mRNA surveillance. RNA. 1999;5:1299–307. doi: 10.1017/s1355838299990829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meaux S, van Hoof A, Baker KE. Nonsense-mediated mRNA decay in yeast does not require PAB1 or a poly(A) tail. Mol Cell. 2008;29:134–40. doi: 10.1016/j.molcel.2007.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kervestin S, Li C, Buckingham R, Jacobson A. Testing the faux-UTR model for NMD: Analysis of Upf1p and Pab1p competition for binding to eRF3/Sup35p. Biochimie. 2012 doi: 10.1016/j.biochi.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–4. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gandhi R, Manzoor M, Hudak KA. Depurination of Brome mosaic virus RNA3 in vivo results in translation-dependent accelerated degradation of the viral RNA. The Journal of biological chemistry. 2008;283:32218–28. doi: 10.1074/jbc.M803785200. [DOI] [PubMed] [Google Scholar]
- 14.Letzring DP, Dean KM, Grayhack EJ. Control of translation efficiency in yeast by codon-anticodon interactions. RNA. 2010;16:2516–28. doi: 10.1261/rna.2411710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuroha K, et al. Receptor for activated C kinase 1 stimulates nascent polypeptide-dependent translation arrest. EMBO reports. 2010;11:956–61. doi: 10.1038/embor.2010.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanner DR, Cariello DA, Woolstenhulme CJ, Broadbent MA, Buskirk AR. Genetic identification of nascent peptides that induce ribosome stalling. J Biol Chem. 2009;284:34809–18. doi: 10.1074/jbc.M109.039040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frischmeyer PA, et al. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science. 2002;295:2258–61. doi: 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- 19.van Hoof A, Frischmeyer PA, Dietz HC, Parker R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002;295:2262–4. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- 20.Ito-Harashima S, Kuroha K, Tatematsu T, Inada T. Translation of the poly(A) tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast. Genes & development. 2007;21:519–24. doi: 10.1101/gad.1490207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu J, Deutsch C. Electrostatics in the ribosomal tunnel modulate chain elongation rates. Journal of molecular biology. 2008;384:73–86. doi: 10.1016/j.jmb.2008.08.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eckmann CR, Rammelt C, Wahle E. Control of poly(A) tail length. Wiley interdisciplinary reviews RNA. 2011;2:348–61. doi: 10.1002/wrna.56. [DOI] [PubMed] [Google Scholar]
- 23.Smith MH, Ploegh HL, Weissman JS. Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science. 2011;334:1086–90. doi: 10.1126/science.1209235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cui Y, Hagan KW, Zhang S, Peltz SW. Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes & development. 1995;9:423–36. doi: 10.1101/gad.9.4.423. [DOI] [PubMed] [Google Scholar]
- 25.Leeds P, Peltz SW, Jacobson A, Culbertson MR. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes & development. 1991;5:2303–14. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- 26.Leeds P, Wood JM, Lee BS, Culbertson MR. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Molecular and cellular biology. 1992;12:2165–77. doi: 10.1128/mcb.12.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peltz SW, Brown AH, Jacobson A. mRNA destabilization triggered by premature translational termination depends on at least three cis-acting sequence elements and one trans-acting factor. Genes & development. 1993;7:1737–54. doi: 10.1101/gad.7.9.1737. [DOI] [PubMed] [Google Scholar]
- 28.Conti E, Izaurralde E. Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Current opinion in cell biology. 2005;17:316–25. doi: 10.1016/j.ceb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 29.Czaplinski K, Weng Y, Hagan KW, Peltz SW. Purification and characterization of the Upf1 protein: a factor involved in translation and mRNA degradation. RNA. 1995;1:610–23. [PMC free article] [PubMed] [Google Scholar]
- 30.Weng Y, Czaplinski K, Peltz SW. Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Molecular and cellular biology. 1996;16:5477–90. doi: 10.1128/mcb.16.10.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Czaplinski K, et al. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes & development. 1998;12:1665–77. doi: 10.1101/gad.12.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kashima I, et al. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes & development. 2006;20:355–67. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He F, Brown AH, Jacobson A. Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Molecular and cellular biology. 1997;17:1580–94. doi: 10.1128/mcb.17.3.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chakrabarti S, et al. Molecular mechanisms for the RNA-dependent ATPase activity of Upf1 and its regulation by Upf2. Molecular cell. 2011;41:693–703. doi: 10.1016/j.molcel.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 35.Chamieh H, Ballut L, Bonneau F, Le Hir H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nature structural & molecular biology. 2008;15:85–93. doi: 10.1038/nsmb1330. [DOI] [PubMed] [Google Scholar]
- 36.Takahashi S, et al. Upf1 potentially serves as a RING-related E3 ubiquitin ligase via its association with Upf3 in yeast. RNA. 2008;14:1950–8. doi: 10.1261/rna.536308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annual review of biochemistry. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 38.Lykke-Andersen J, Shu MD, Steitz JA. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 2000;103:1121–31. doi: 10.1016/s0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- 39.Ghosh S, Ganesan R, Amrani N, Jacobson A. Translational competence of ribosomes released from a premature termination codon is modulated by NMD factors. RNA. 2010;16:1832–47. doi: 10.1261/rna.1987710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Atkinson GC, Baldauf SL, Hauryliuk V. Evolution of nonstop, no-go and nonsense-mediated mRNA decay and their termination factor-derived components. BMC evolutionary biology. 2008;8:290. doi: 10.1186/1471-2148-8-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen L, et al. Structure of the Dom34-Hbs1 complex and implications for no-go decay. Nature structural & molecular biology. 2010;17:1233–40. doi: 10.1038/nsmb.1922. [DOI] [PubMed] [Google Scholar]
- 42.Graille M, Chaillet M, van Tilbeurgh H. Structure of yeast Dom34: a protein related to translation termination factor Erf1 and involved in No-Go decay. The Journal of biological chemistry. 2008;283:7145–54. doi: 10.1074/jbc.M708224200. [DOI] [PubMed] [Google Scholar]
- 43.Kobayashi K, et al. Structural basis for mRNA surveillance by archaeal Pelota and GTP-bound EF1alpha complex. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:17575–9. doi: 10.1073/pnas.1009598107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee HH, et al. Structural and functional insights into Dom34, a key component of no-go mRNA decay. Molecular cell. 2007;27:938–50. doi: 10.1016/j.molcel.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 45.van den Elzen AM, et al. Dissection of Dom34-Hbs1 reveals independent functions in two RNA quality control pathways. Nature structural & molecular biology. 2010;17:1446–52. doi: 10.1038/nsmb.1963. [DOI] [PubMed] [Google Scholar]
- 46.Becker T, et al. Structure of the no-go mRNA decay complex Dom34-Hbs1 bound to a stalled 80S ribosome. Nature structural & molecular biology. 2011;18:715–20. doi: 10.1038/nsmb.2057. [DOI] [PubMed] [Google Scholar]
- 47.Shoemaker CJ, Eyler DE, Green R. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science. 2010;330:369–72. doi: 10.1126/science.1192430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Passos DO, et al. Analysis of Dom34 and its function in no-go decay. Molecular biology of the cell. 2009;20:3025–32. doi: 10.1091/mbc.E09-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pisareva VP, Skabkin MA, Hellen CU, Pestova TV, Pisarev AV. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. The EMBO journal. 2011;30:1804–17. doi: 10.1038/emboj.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shoemaker CJ, Green R. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proceedings of the National Academy of Sciences of the United States of America. 2011 doi: 10.1073/pnas.1113956108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gagnon MG, Seetharaman SV, Bulkley D, Steitz TA. Structural basis for the rescue of stalled ribosomes: structure of YaeJ bound to the ribosome. Science. 2012;335:1370–2. doi: 10.1126/science.1217443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Neubauer C, Gillet R, Kelley AC, Ramakrishnan V. Decoding in the absence of a codon by tmRNA and SmpB in the ribosome. Science. 2012;335:1366–9. doi: 10.1126/science.1217039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Hoof A. Conserved functions of yeast genes support the duplication, degeneration and complementation model for gene duplication. Genetics. 2005;171:1455–61. doi: 10.1534/genetics.105.044057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsuboi TKK, Kudo K, Makino S, Inoue E, Kashima I, Inada T. Dom34:Hbs1 Plays a General Role in Quality Control Systems by Dissociation of a Stalled Ribosome at the 3′ End of Aberrant mRNA. Mol Cell. 2012 doi: 10.1016/j.molcel.2012.03.013. in press. [DOI] [PubMed] [Google Scholar]
- 55.Anderson JS, Parker RP. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. The EMBO journal. 1998;17:1497–506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Muhlrad D, Decker CJ, Parker R. Turnover mechanisms of the stable yeast PGK1 mRNA. Molecular and cellular biology. 1995;15:2145–56. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mitchell P, Tollervey D. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′-->5′ degradation. Molecular cell. 2003;11:1405–13. doi: 10.1016/s1097-2765(03)00190-4. [DOI] [PubMed] [Google Scholar]
- 58.Muhlrad D, Parker R. Premature translational termination triggers mRNA decapping. Nature. 1994;370:578–81. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- 59.Lejeune F, Li X, Maquat LE. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Molecular cell. 2003;12:675–87. doi: 10.1016/s1097-2765(03)00349-6. [DOI] [PubMed] [Google Scholar]
- 60.Lykke-Andersen J. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Molecular and cellular biology. 2002;22:8114–21. doi: 10.1128/MCB.22.23.8114-8121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Unterholzner L, Izaurralde E. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol Cell. 2004;16:587–96. doi: 10.1016/j.molcel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 62.Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen TH. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nature structural & molecular biology. 2009;16:49–55. doi: 10.1038/nsmb.1530. [DOI] [PubMed] [Google Scholar]
- 63.Gatfield D, Izaurralde E. Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature. 2004;429:575–8. doi: 10.1038/nature02559. [DOI] [PubMed] [Google Scholar]
- 64.Huntzinger E, Kashima I, Fauser M, Sauliere J, Izaurralde E. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA. 2008;14:2609–17. doi: 10.1261/rna.1386208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schaeffer D, et al. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nature structural & molecular biology. 2009;16:56–62. doi: 10.1038/nsmb.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schaeffer D, van Hoof A. Different nuclease requirements for exosome-mediated degradation of normal and nonstop mRNAs. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:2366–71. doi: 10.1073/pnas.1013180108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kuroha K, Tatematsu T, Inada T. Upf1 stimulates degradation of the product derived from aberrant messenger RNA containing a specific nonsense mutation by the proteasome. EMBO reports. 2009;10:1265–71. doi: 10.1038/embor.2009.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wilson MA, Meaux S, van Hoof A. A genomic screen in yeast reveals novel aspects of nonstop mRNA metabolism. Genetics. 2007;177:773–84. doi: 10.1534/genetics.107.073205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bengtson MH, Joazeiro CA. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature. 2010;467:470–3. doi: 10.1038/nature09371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dimitrova LN, Kuroha K, Tatematsu T, Inada T. Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. The Journal of biological chemistry. 2009;284:10343–52. doi: 10.1074/jbc.M808840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barthelme D, et al. Ribosome recycling depends on a mechanistic link between the FeS cluster domain and a conformational switch of the twin-ATPase ABCE1. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3228–33. doi: 10.1073/pnas.1015953108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pisarev AV, et al. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Molecular cell. 2010;37:196–210. doi: 10.1016/j.molcel.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Becker T, et al. Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature. 2012 doi: 10.1038/nature10829. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bhattacharya A, McIntosh KB, Willis IM, Warner JR. Why Dom34 stimulates growth of cells with defects of 40S ribosomal subunit biosynthesis. Molecular and cellular biology. 2010;30:5562–71. doi: 10.1128/MCB.00618-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cole SE, LaRiviere FJ, Merrikh CN, Moore MJ. A convergence of rRNA and mRNA quality control pathways revealed by mechanistic analysis of nonfunctional rRNA decay. Molecular cell. 2009;34:440–50. doi: 10.1016/j.molcel.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.LaRiviere FJ, Cole SE, Ferullo DJ, Moore MJ. A late-acting quality control process for mature eukaryotic rRNAs. Molecular cell. 2006;24:619–26. doi: 10.1016/j.molcel.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 77.He F, et al. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Molecular cell. 2003;12:1439–52. doi: 10.1016/s1097-2765(03)00446-5. [DOI] [PubMed] [Google Scholar]
- 78.Lelivelt MJ, Culbertson MR. Yeast Upf proteins required for RNA surveillance affect global expression of the yeast transcriptome. Mol Cell Biol. 1999;19:6710–9. doi: 10.1128/mcb.19.10.6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nature genetics. 2004;36:1073–8. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- 80.Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E. Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA. 2005;11:1530–44. doi: 10.1261/rna.2160905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Frischmeyer PA, Dietz HC. Nonsense-mediated mRNA decay in health and disease. Human molecular genetics. 1999;8:1893–900. doi: 10.1093/hmg/8.10.1893. [DOI] [PubMed] [Google Scholar]
- 82.Karijolich J, Yu YT. Converting nonsense codons into sense codons by targeted pseudouridylation. Nature. 2011;474:395–8. doi: 10.1038/nature10165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kerem E, et al. Effectiveness of PTC124 treatment of cystic fibrosis caused by nonsense mutations: a prospective phase II trial. Lancet. 2008;372:719–27. doi: 10.1016/S0140-6736(08)61168-X. [DOI] [PubMed] [Google Scholar]
- 84.Peltz SW, et al. Nonsense suppression activity of PTC124 (ataluren) Proc Natl Acad Sci U S A. 106:E64. doi: 10.1073/pnas.0901936106. author reply E65 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Holbrook JA, Neu-Yilik G, Hentze MW, Kulozik AE. Nonsense-mediated decay approaches the clinic. Nat Genet. 2004;36:801–8. doi: 10.1038/ng1403. [DOI] [PubMed] [Google Scholar]
- 86.Shan X, Chang Y, Lin CL. Messenger RNA oxidation is an early event preceding cell death and causes reduced protein expression. The FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2007;21:2753–64. doi: 10.1096/fj.07-8200com. [DOI] [PubMed] [Google Scholar]
- 87.Thorpe GW, Fong CS, Alic N, Higgins VJ, Dawes IW. Cells have distinct mechanisms to maintain protection against different reactive oxygen species: oxidative-stress-response genes. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:6564–9. doi: 10.1073/pnas.0305888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chang Y, et al. Messenger RNA oxidation occurs early in disease pathogenesis and promotes motor neuron degeneration in ALS. PloS one. 2008;3:e2849. doi: 10.1371/journal.pone.0002849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nunomura A, et al. Oxidative damage is the earliest event in Alzheimer disease. Journal of neuropathology and experimental neurology. 2001;60:759–67. doi: 10.1093/jnen/60.8.759. [DOI] [PubMed] [Google Scholar]
- 90.Onouchi H, et al. Nascent peptide-mediated translation elongation arrest coupled with mRNA degradation in the CGS1 gene of Arabidopsis. Genes & development. 2005;19:1799–810. doi: 10.1101/gad.1317105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Onoue N, et al. S-adenosyl-L-methionine induces compaction of nascent peptide chain inside the ribosomal exit tunnel upon translation arrest in the Arabidopsis CGS1 gene. The Journal of biological chemistry. 2011;286:14903–12. doi: 10.1074/jbc.M110.211656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Meyer S, Temme C, Wahle E. Messenger RNA turnover in eukaryotes: pathways and enzymes. Critical reviews in biochemistry and molecular biology. 2004;39:197–216. doi: 10.1080/10409230490513991. [DOI] [PubMed] [Google Scholar]
- 93.Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nature structural & molecular biology. 2004;11:121–7. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 94.Chen J, Chiang YC, Denis CL. CCR4, a 3′-5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. The EMBO journal. 2002;21:1414–26. doi: 10.1093/emboj/21.6.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tucker M, Staples RR, Valencia-Sanchez MA, Muhlrad D, Parker R. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. The EMBO journal. 2002;21:1427–36. doi: 10.1093/emboj/21.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Coller J, Parker R. Eukaryotic mRNA decapping. Annual review of biochemistry. 2004;73:861–90. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- 97.Hsu CL, Stevens A. Yeast cells lacking 5′-->3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Molecular and cellular biology. 1993;13:4826–35. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hu W, Sweet TJ, Chamnongpol S, Baker KE, Coller J. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature. 2009;461:225–9. doi: 10.1038/nature08265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Molecular cell. 2007;25:635–46. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 100.Dziembowski A, Lorentzen E, Conti E, Seraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nature structural & molecular biology. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- 101.Bonneau F, Basquin J, Ebert J, Lorentzen E, Conti E. The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell. 2009;139:547–59. doi: 10.1016/j.cell.2009.08.042. [DOI] [PubMed] [Google Scholar]
- 102.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′-->5′ exoribonucleases. Cell. 1997;91:457–66. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 103.Araki Y, et al. Ski7p G protein interacts with the exosome and the Ski complex for 3′-to-5′ mRNA decay in yeast. The EMBO journal. 2001;20:4684–93. doi: 10.1093/emboj/20.17.4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Brown JT, Bai X, Johnson AW. The yeast antiviral proteins Ski2p, Ski3p, and Ski8p exist as a complex in vivo. RNA. 2000;6:449–57. doi: 10.1017/s1355838200991787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hayes CS, Sauer RT. Cleavage of the A site mRNA codon during ribosome pausing provides a mechanism for translational quality control. Molecular cell. 2003;12:903–11. doi: 10.1016/s1097-2765(03)00385-x. [DOI] [PubMed] [Google Scholar]
- 106.Sunohara T, Jojima K, Yamamoto Y, Inada T, Aiba H. Nascent-peptide-mediated ribosome stalling at a stop codon induces mRNA cleavage resulting in nonstop mRNA that is recognized by tmRNA. RNA. 2004;10:378–86. doi: 10.1261/rna.5169404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Keiler KC, Waller PR, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–3. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 108.Huang C, Wolfgang MC, Withey J, Koomey M, Friedman DI. Charged tmRNA but not tmRNA-mediated proteolysis is essential for Neisseria gonorrhoeae viability. The EMBO journal. 2000;19:1098–107. doi: 10.1093/emboj/19.5.1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Moore SD, Sauer RT. The tmRNA system for translational surveillance and ribosome rescue. Annual review of biochemistry. 2007;76:101–24. doi: 10.1146/annurev.biochem.75.103004.142733. [DOI] [PubMed] [Google Scholar]
- 110.Ivanova N, Pavlov MY, Felden B, Ehrenberg M. Ribosome rescue by tmRNA requires truncated mRNAs. Journal of molecular biology. 2004;338:33–41. doi: 10.1016/j.jmb.2004.02.043. [DOI] [PubMed] [Google Scholar]