Abstract
Borrelia burgdorferi sensu lato (B. burgdorferi s.l.), the group of bacterial species represented by Lyme Disease pathogens, has one of the most complex and variable genomic architectures among prokaryotes. Showing frequent recombination within and limited gene flow among geographic populations, the B. burgdorferi s.l. genomes provides an excellent window into the processes of bacterial evolution at both within- and between-population levels. Comparative analyses of B. burgdorferi s.l. genomes revealed a highly dynamic plasmid composition but a conservative gene repertoire. Gene duplication and loss as well as sequence variations at loci encoding surface-localized lipoproteins (e.g., the PF54 genes) are strongly associated with adaptive differences between species. There are a great many conserved intergenic spacer sequences that are candidates for cis-regulatory elements and non-coding RNAs. Recombination among coexisting strains occurs at a rate approximately three times the mutation rate. The coexistence of a large number of genomic groups within local B. burgdorferi s.l. populations may be driven by immune-mediated diversifying selection targeting major antigen loci as well as by adaptation to multiple host species. Questions remain regarding the ecological causes (e.g., climate change, host movements, or new adaptations) of the ongoing range expansion of B. burgdorferi s.l. and on the genomic variations associated with its ecological and clinical variability. Anticipating an explosive growth of the number of B. burgdorferi s.l. genomes sampled from both within and among species, we propose genome-based methods to test adaptive mechanisms and to identify molecular bases of phenotypic variations. Genome sequencing is also necessary to monitor the ongoing genetic admixture of previously isolated species and populations in North America and elsewhere.
Keywords: Lyme disease, phylogenomics, comparative genomics, phylogenetic footprinting, population genomics, recombination, gene conversion, genome-wide association study, pangenome, frequency-dependent selection, multiple-niche polymorphisms
1. Introduction
Borrelia burgdorferi is among the first bacterial species with a genome completely sequenced (Fraser et al., 1997). The first B. burgdorferi genome, from the type strain B31 and consisting of a 910-kilobase long linear chromosome and additional 610 kilobases of twenty-one linear and circular plasmids, was not closed until three years later (Casjens et al., 2000). It remains true today that members of B. burgdorferi sensu lato (B. burgdorferi s.l.), a term referring to the bacterial species complex represented by pathogens of Lyme disease, have one of the most complex prokaryotic genomes. Comparative studies of fully sequenced B. burgdorferi s.l. genomes began about a decade ago when draft genomes of additional four strains were published (Glöckner et al., 2006, 2004; Qiu et al., 2004). At present, the number of completed and draft B. burgdorferi s.l. genomes is close to thirty based on a search in the PATRIC database of bacterial genomes (Wattam et al., 2013) and the online GOLD registry of genome-sequencing projects (Pagani et al., 2012) (Table 1). While this number is poised for rapid growth with the increasing availability of high-throughput DNA sequencing technologies, it is expected that due to its genome complexity most current and future B. burgdorferi s.l. genomes will remain as draft genome assemblies, partial genome sequences, or both, rather than fully assembled genomes.
Table 1.
Completed and draft genomes of B. burgdorferisensu latoa
| Strain | Species | Geographic Origin |
Biological Origin |
Sequencing Status |
Genome Report |
|---|---|---|---|---|---|
| B31 | Bb sensu stricto | New York, US | I. scapularis | Complete | (Casjens et al., 2000 Fraser et al., 1997) |
| 64b | Bb sensu stricto | New York, US | Human | Draft | (Schutzer et al., 2011) |
| ZS7 | Bb sensu stricto | Germany | I. ricinus | Draft | (Schutzer et al., 2011) |
| JD1 | Bb sensu stricto | Massachusetts, US | I. scapularis | Complete | (Schutzer et al., 2011) |
| CA-11.2A | Bb sensu stricto | California, US | I. pacificus | Draft | (Schutzer et al., 2011) |
| CA382 | Bb sensu stricto | California, US | N.A. | Complete (chromosome only) | Unpublished |
| CA8 | Bb sensu stricto | California, US | N.A. | Draft | Unpublished |
| N40 | Bb sensu stricto | New York, US | I. scapularis | Complete | (Schutzer et al., 2011) |
| 72a | Bb sensu stricto | New York, US | Human | Draft | (Schutzer et al., 2011) |
| 156a | Bb sensu stricto. | New York, US | Human | Draft | (Schutzer et al., 2011) |
| WI91-23 | Bb sensu stricto | Wisconsin, US | Bird | Draft | (Schutzer et al., 2011) |
| 118a | Bb sensu stricto | New York, US | Human | Draft | (Schutzer et al., 2011) |
| 297 | Bb sensu stricto | Connecticut, US | Human | Complete (plasmids only) | (Schutzer et al., 2011) |
| 29805 | Bb sensu stricto | Connecticut, US | I. scapularis | Draft | (Schutzer et al., 2011) |
| Bol26 | Bb sensu stricto | Italy | I. ricinus | Draft | (Schutzer et al., 2011) |
| 94a | Bb sensu stricto | New York, US | Human | Draft | (Schutzer et al., 2011) |
| SV1 | B. finlandensis | Finland | I. ricinus | Draft | (Casjens et al., 2011a) |
| DN127 | B. bissettii | California, US | I. pacificus | Draft | (Schutzer et al., 2012) |
| PKo | B. afzelii | Germany | Human | Draft | (Casjens et al., 2011b Glöckner et al., 2006) |
| ACA-1 | B. afzelii | Sweden | Human | Draft | (Casjens et al., 2011b) |
| PBi | B. bavariensis | Germany | Human | Complete (chromosome, cp26, and lp54 only) | (Glöckner et al., 2004) |
| PBr | B. garinii | Denmark | Human | Draft | (Casjens et al., 2011b) |
| Far04 | B. garinii | Denmark | Bird | Draft | (Casjens et al., 2011b) |
| VS116 | B. valaisiana | Switzerland | I. ricinus | Draft | (Schutzer et al., 2012) |
| A14S | B. spielmani | The Netherlands | I. ricinus | Draft | (Schutzer et al., 2012) |
| BgVir | B. garinii | Russia | I. persulcatus | Draft | (Brenner et al., 2012) |
| NMJW1 | B. garinii | China | I. persulcatus | Complete | (B. Jiang et al., 2012) |
| HLJ01 | B. afzelii | China | Human | Complete | (B.-G. Jiang et al., 2012) |
Compiled based on a search of the PATRIC database (Wattam et al., 2013) and the GOLD registry of genome-sequencing projects (Pagani et al., 2012) in January of 2014.
Despite the permanently draft and incomplete status of many current and future B. burgdorferi s.l. genomes, comparative analysis of B. burgdorferi s.l. genomes is a highly valuable approach for uncovering the genetic basis of phenotypic variations of this globally distributed pathogen (Norris and Lin, 2011). For example, genome comparisons should implicate genetic variations contributing to human virulence, considering that B. burgdorferi s.l. strains apparently differ in human pathogenicity, clinical manifestations, and the ability for systemic dissemination (Hanincova et al., 2013; Stanek et al., 2012). Besides clinical applications, comparing the genomes of B. burgdorferi s.l. species helps identify genes associated with ecological traits such as host specificity by comparing the genomes from species differing in preferred reservoir hosts (Kurtenbach et al., 2006; Margos et al., 2011; Vollmer et al., 2013). A major challenge for identifying phenotype-associated genetic variations is that many, if not the majority of, genomic differences between B. burgdorferi s.l. species are either selectively neutral or unrelated to the traits of interest. Theories and methods for genome-wide association studies (Box 1) of B. burgdorferi s.l. (or any bacterial species) are relatively under-developed in comparison with those for humans and other eukaryotes. Clinical manifestations of B. burgdorferi s.l. appear to be associated more with multi-locus sequence types (MLSTs) than with alleles at antigen loci like ospC (Dykhuizen et al., 2008; Hanincova et al., 2013; Seinost et al., 1999). The genome-wide identities of B. burgdorferi s.l. genes causing their ecological and disease phenotypes (e.g., host specificity, diverse human symptoms, and different levels of invasiveness) remain to be identified.
Box 1. A glossary of evolutionary genomics.
Polymorphisms – Sequence variations within a population or species, e.g., single-nucleotide polymorphisms (SNPs). The amount of sequence polymorphisms at a locus is commonly measured by the average number of differences per nucleotide site (π).
Fixed differences – Nucleotide differences between two species that are constant within each species. The amount of sequence divergence between two species is commonly measured by the number of fixed differences per nucleotide site (K).
Synonymous substitutions – Nucleotide substitutions in a protein-coding sequence that do not cause amino-acid changes. Commonly used notations for levels of synonymous polymorphism and synonymous divergence are, respectively, πS and KS.
Nonsynonymous substitution – Nucleotide substitutions in a protein-coding sequence that causes amino-acid replacements. Commonly used notations for levels of nonsynonymous polymorphism and nonsynonymous divergence are, respectively, πA and KA.
The dN/dS ratio test of natural selection – Neutrally evolving protein-coding gene is expected to show the same rates of synonymous and nonsynonymous substitutions. In other words, if amino-acid substitutions are neutral, one would expect πA/πS =1 for within-species comparisons or KA/KS=1 for between-species comparisons. More generally, these two neutral expectations may be referred to as dN/dS =1. Most protein genes are under purifying (negative) natural selection for amino-acid substitutions, showing dN/dS <1. If a gene (or a part of it) shows dN/dS >1, the gene may be subject to positive natural selection (Hurst, 2002).
The MK test of adaption – Another expectation for a neutrally evolving protein gene is similar levels of selective constraints within and between species, i.e., πA/πS= KA/KS. The MK test tests for accelerated amino-acid replacements during speciation, i.e. KA/KS > πA/πS as evidence of adaptive evolution (McDonald and Kreitman, 1991).
Phylogenomics – Phylogeny-based prediction of gene functions. Orthologs, homologs due to speciation, are more likely to share the same molecular functions than paralogs, homologs due to gene duplication (Eisen, 1998). Orthologs are identified by comparing and reconcile a species phylogeny with a gene tree. At present, phylogenomics refer to any phylogeny-based inference of gene functions, including identification of genes and codon sites influenced by positive natural selection (Kumar et al., 2012). In contrast to population genomics (see below), phylogenomics generally compare genomes from different species.
Population genomics – Comparative study of genomes sampled from within a natural population (Guttman and Stavrinides, 2010). It is the application of classic population genetics to genome sequences for understanding population processes including changes of effective population size, rates of mutation and recombination, amount of migration, and selective forces.
Pangenomics – In comparing genomes from a bacterial group (e.g., a species or genus), each genome can be decomposed into a “core genome” – the set of genes present in every genome and an “accessory genome” – the set of genes that is uniquely present in this genome (Tettelin et al., 2005). All genes present in all genomes constitute the “pan-genome” of the bacterial group.
Phylogenetic footprinting – A method for identifying gene-regulatory sequences (e.g., promoters and non-coding RNAs) in non-coding parts of a genome based on evolutionary conservation (Eddy, 2005). The basic assumption is that functional intergenic elements are under purifying selection and show lower nucleotide substitution rates than neutrally evolving sequences such as synonymous sites in a protein-coding gene.
Phylogeography – The study of history, size, and genetic structure of geographic populations of a species using molecular markers (Avise, 2000). Understanding phylogeographical process of a species is challenging because it requires simultaneous considerations of phylogenetic, demographic, and selective processes.
Genome-wide association study (GWAS) – Identification of genetic loci contributing to phenotypic variations by comparing genome-wide markers individually in two phenotypically distinct random population samples. This approach is different from the traditional pedigree-based approach of identifying Quantitative-Trait Loci (QTLs) and is believed to be more powerful in identifying genetic basis of a complex trait (Risch and Merikangas, 1996).
Gene conversion – A type of homologous DNA recombination that results in an allelic replacement rather than an exchange of DNA arms (“crossing over”). Gene conversion is the predominant form of recombination in bacteria. It causes a loosening of linkage disequilibrium (LD) at short genomic scales without significantly affecting LD at long-distance scales (Wiuf and Hein, 2000).
Evolutionary biology provides a unified theoretical framework as well as a rich set of technical tools to meet these challenges of genome comparison (Felsenstein, 2004; Hartl and Clark, 2007; Harvey and Pagel, 1998). For example, phylogenomics (Box 1) is an evolution-informed approach to annotate functions of unknown genes based on identification of gene orthology and signatures of adaptive evolution (Eisen, 1998; Kumar et al., 2012). Also based on evolutionary principles, phylogenetic footprinting (Box 1) is a computational approach for ab initio identification of gene-regulatory elements in non-coding parts of a genome (Brohée et al., 2011; Degnan et al., 2011; Katara et al., 2012). By generating and testing specific expectations on the level and pattern of sequence variability, evolutionary analyses are powerful to distinguish biologically important genome variations from a noisy background of stochastic neutral or nearly neutral variations. For example, genome comparisons confirmed the evolution-derived expectation that genes encoding major surface-exposed antigens in B. burgdorferi s.l., including the outer-surface protein C gene (ospC), the decorin-binding protein A gene (dbpA), and the Vmp-like sequence (vls) locus, exhibit the highest levels of non-synonymous nucleotide variability genome-wide, consistent with their roles in host invasion, escape from host adaptive immunity, or both (Glöckner et al., 2006; Graves et al., 2013; Mongodin et al., 2013; Qiu et al., 2004).
For Borrelia, a robust phylogeny of 23 sequenced B. burgdorferi s.l. genomes has been reconstructed using genome-wide nucleotide variations (Mongodin et al., 2013). An estimate of the rate of homologous recombination among co-existing B. burgdorferi strains has been obtained and a model of sympatric genome diversification driven by negative frequency-dependent selection has been proposed (Haven et al., 2011). To understand climatic and ecological mechanisms causing the on-going global endemics of Lyme disease, intensive efforts have been made to infer the geographic origin, size, and migratory history of natural populations of B. burgdorferi s.l. using multilocus sequence typing (Brisson et al., 2010; Hoen et al., 2009; Margos et al., 2012, 2008; Qiu, 2008; Rudenko et al., 2013). We share the optimism expressed by the other authors that the use of genome-wide sequence variations will lead to more precise and accurate estimates of these critical population parameters (Brisson et al., 2012; Margos et al., 2011; Ogden et al., 2013).
Evolutionary analysis of B. burgdorferi s.l. genomes has grown from a comparison of two or three genomes a decade ago (Glöckner et al., 2006, 2004; Qiu et al., 2004) to a comparison of up to twenty-three genomes from eight species in recent studies (Casjens et al., 2012; Haven et al., 2011; Mongodin et al., 2013). This review is intended, first, to summarize results made in the first decade of comparative genomic studies of B. burgdorferi s.l. We take the liberty to update and re-interpret some of these results with new analyses. For narrative convenience, this review is organized into various aspects of evolutionary genomics although they are inherently inseparable. These aspects include (i) genome sampling and genome phylogeny, which form the basis of all subsequent analyses, (ii) pan-genomics (Box 1), which aims to identify lineage-specific genes and to estimate rates of gene acquisition during genome diversification, (iii) phylogenomics (Box 1), which identifies host-interacting, virulence-associated genes through analysis of sequence evolution rates, (iv) phylogenetic footprinting (Box 1), which searches for conserved gene-regulatory sequences in intergenic spacers (IGSs), (v) population genomics (Box 1), which investigates evolutionary mechanisms such as mutation, recombination, and natural selection, and (vi) genome-based biogeography, which tests biogeographic hypotheses using genome sequences.
The second goal of this review is to provide an outlook on the future of evolutionary genomics of B. burgdorferi s.l. There is a consensus among ecologists and evolutionary geneticists of Lyme disease that genetic and ecological causes of the ongoing range expansion of B. burgdorferi s.l. leave detectable footprints in the pathogen genomes, and, hence, genome variability is a key to revealing these causes through hypothesis testing (Brisson et al., 2012; Margos et al., 2011; Ogden et al., 2013; Ostfeld, 2010). There is a similar optimism that comparative genomics is now in a position to reveal genetic variations associated with clinical variability among B. burgdorferi s.l. strains (Norris and Lin, 2011). We propose studies with which these promises of evolutionary genomics of B. burgdorferi s.l. may be delivered in the near future.
2. Genome sampling and genome phylogeny
A phylogeny-motivated genome-sampling strategy
Strategy for sampling bacterial isolates for whole-genome sequencing is a highly consequential decision that has to be made early on during a genome-sequencing project. Clinically motivated studies tend to sequence genomes differing in pathogenicity, disease manifestation, or degree of virulence. Ecologically motivated studies may choose to sequence strains differing in host/vector specificity, geographic distribution, or natural abundance. B. burgdorferi s.s., B. garinii, and B. afzelii, the three B. burgdorferi s.l. species causing the majority of Lyme diseases cases worldwide, are over-represented in currently available genomes (Table 1). There also have been considerations of host and geographic diversity, such as the inclusion of WI91-23, a Midwestern US strain and a bird isolate; CA-11.2A, a Western US strain; and BOL26 and ZS7, two European B. burgdorferi s.s. strains. Main motivations behind the selection of strains during the beginning years of B. burgdorferi s.l. genome sequencing, however, were evolutionary and guided by the multilocus phylogeny of a large panel of isolates (Mongodin et al., 2013). Specifically, a multitude of phylogeny-based considerations were built into the genome-sampling strategy, each with an eye for a post-genomic comparative analysis. First, to maximally cover the entire phylogenetic diversity within the species complex, eight formally named species are represented, even though some species (e.g., B. bissettii) have never been isolated from human patients. Second, to best resolve genomic changes specific to the highly pathogenic species B. burgdorferi s.s., the European strain SV1 (later designated as the type strain of a proposed species “ B. finlandensis”), which represents the closest known outgroup of B. burgdorferi s.s., was chosen (Casjens et al., 2011a). Third, to reveal mechanisms driving the con-specific genome diversification among coexisting strains, one species (B. burgdorferi s.s.) was sampled in great depth to include fourteen ospC-defined genomic lineages, twelve of which are sympatric in the Northeast United States. Fourth, to resolve the most recent evolutionary changes associated with the emergence of new clonal groups, three pairs of phylogenetic sister-group strains were chosen including the 297/156a pair, the 118a/72a pair, and the Bol26/ZS7 pair.
A rooted genome phylogeny
An organismal phylogeny serves as an overall framework for comparative analyses (Harvey and Pagel, 1998) such as the pan-genome analysis, reconstruction of gene gains and losses, tests of natural selection, and inference of horizontal genetic exchanges (Sections 3, 4, and 6). Obtaining a statistically robust organismal phylogeny using genome-wide variability is one goal of whole-genome sequencing. Organismal phylogeny based on sequences at single or multiple loci often contain poorly resolved branches as a result of insufficient number of variable sites, re-assortment of polymorphisms due to recombination, a history of rapid evolutionary diversification, or any combinations of these effects (Morlon et al., 2012). The use of whole-genome single-nucleotide variations indeed improved statistical confidence of both the between-species and within-species phylogenies, judging from their consistency with features less prone to genetic exchange such as chromosomal re-arrangements, large genomic indels, and gene duplications and losses (Section 4.1) (Mongodin et al., 2013).
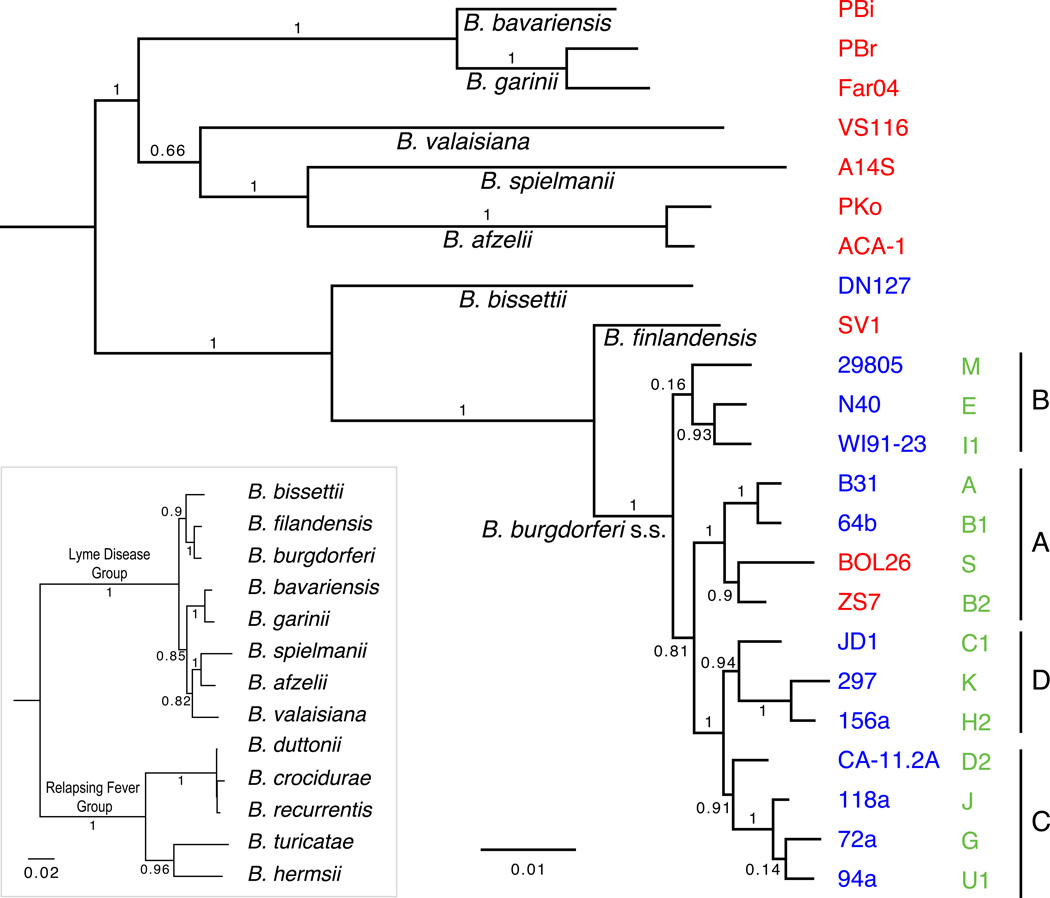
Here, a newly rooted tree of 23 sequenced B. burgdorferi s.l. genomes is derived from an alignment of cp26 plasmid sequences from a region that is not heavily influenced by recombination (Figure 1, main panel). The root position, which is arbitrarily put at the midpoint of the tree, is supported by an independent phylogenetic analysis based on a concatenated alignment of 24 conserved protein sequences including those from five relapsing-fever Borrelia strains (Figure 1, inset). Both trees place the common ancestor of the B. burgdorferi s.l. species complex on the branch between B. bissettii (a North American species) and a large Eurasian clade. This root position is consistent with that from previous multilocus sequence analyses, which similarly suggested that the deepest and most ancestral cladogenesis in B. burgdorferi s.l. is between a Eurasian clade and a North American/New World clade, although these studies did not use relapsing-fever Borrelia as the outgroup (Margos et al., 2011, 2010; Rudenko et al., 2009). This biogeography hypothesis, however, is now challenged with the discovery of a New World species from Chile “ B. chilensis”, which appears to be the most basal B. burgdorferi s.l. species known so far (Ivanova et al., 2013) (Section 7).
Figure 1. Genome sampling and phylogeny: Borrelia phylogeny.
(Main Panel) A phylogeny of 23 sequenced Lyme Group (LD) Borrelia genomes. The tree is based on an alignment of a 6.3-kb region on the cp26 plasmid, encompassing genes b14, b16 oppAIV, b17 (guaA), and, b18 (guaB). Since it does not include ospC, this region is relatively free from recombination and therefore more informative for phylogenetic inference. The cp26 sequences were aligned by using MUGSY (version 1.2.1) (Angiuoli and Salzberg, 2011) in a LINUX environment, the alignment slice was extracted by using customized Perl scripts, and the tree was inferred with FastTree (version 2.1.7) (Enright et al., 2002). The tree is rooted at the midpoint, which is consistent with the root suggested by a tree including Relapsing-Fever Borrelia as outgroups (Inset). The latter tree is based on protein sequences at 24 single-copy conserved loci, including infB (encoding an initiation factor), lepA (encoding a GTP-binding protein), pheS (encoding phenylalanyl-tRNA synthetase subunit alpha), rplB/C/D/E/F/K/N/O/P (encoding 50S ribosomal proteins), and rpsB/C/E/G/H/I/J/K/L/M/Q/S (encoding 30S ribosomal proteins) (Wu and Eisen, 2008) (Norris and Tyagi, personal communications). The protein sequences were aligned by using MUSCLE (version 3.8.31) (Edgar, 2004), the resulting alignments were concatenated by using a customized Perl script, and the tree was inferred with FastTree (version 2.1.7) (Enright et al., 2002). The B. burgdorferi s.l. strains were chosen for whole-genome sequencing to allow for at least three levels of phylogenetic comparisons: (i) between eight B. burgdorferi s.l. species, with the goal of identifying species-specific variations; (ii) between fourteen clonal groups within a single species (B. burgdorferi sensu stricto), with the goal of identifying strain-specific variations; and (iii) between members of sister-group genomes, e.g., within SNP groups A–D (Mongodin et al., 2013), with the goal of identifying most recent genomic changes.
Challenges and opportunities
The process of genome divergence in bacteria is more accurately described with a reticulate network than with a bifurcating phylogeny. Both the horizontal gene transfer between distantly related species and the frequent recombination within populations contribute to the reticulation. Nonetheless, empirical and simulation work supports the use of phylogenetic trees as a useful approximation of population and speciation processes in bacteria (Didelot et al., 2010; Haven et al., 2011; Touchon et al., 2009; Wu et al., 2013). For Borrelia, potentials of the phylogenetic-motivated genome-sample strategy have just begun to be realized. Further efforts await to fully implement the post-genomic analyses envisioned by the original genome-sampling strategy including, in particular, a genome-wide catalog of adaptive changes specific to the three most pathogenic species B. burgdorferi s.s., B. garinii, and B. afzelii. Many more B. burgdorferi s.l. species and genomic groups have since been discovered. The total number of B. burgdorferi s.l. species is at least twenty including the eighteen that have been previously reviewed and the more recently proposed species “ B. finlandensis” and “B. chilensis” (Casjens et al., 2011a; Ivanova et al., 2013; Margos et al., 2011). From this latest catalog of species, it is clear that the New World species are under-represented in the currently available genomes. Within the species of B. burgdorferi s.s., strains specific to Europe and Midwest US are also under-represented in sequenced genomes.
3. Pan-genomics: a critical assessment
A highly stable gene repertoire
The terms “pan-genome” and “core-genome” refer to the total and shared gene repertoire among genomes of a bacterial species, respectively. Pan-genome analysis is based on the assumption that individual genomes of a bacterial species consist of an indispensable, species-specific set of “core” genes and a dispensable, strain-specific set of “accessory” genes (Medini et al., 2005; Tettelin et al., 2008). Using this “bag-of-genes” model of bacterial genome composition, estimates on the gain and loss of genes were obtained using mathematic functions with no obvious biological interpretations, such as the exponential decay function used for modeling the decrease of core genome size with increasing number of genomes used in comparison (Tettelin et al., 2005). Later pan-genomics studies take a more evolutionary approach by estimating rates of gene gains and losses in a bacterial species using coalescent- or phylogeny-based models (Donati et al., 2010; Touchon et al., 2009). Both the exponential-decay and phylogenetic models have been applied to the estimation of the pan-genome sizes of a single B. burgdorferi s.l. species and the species complex as a whole (Mongodin et al., 2013). The phylogenetic distance explains the majority of pan-genome size variations, with an R2 (coefficient of determination) value of approximately 0.6 and 0.9 for intra-specific and inter-specific pan-genome sizes, respectively (Mongodin et al., 2013). The more modest R2 for the intra-specific pan-genome size is likely due to that fact that recombination is more frequent than spontaneous point mutations in driving intra-specific genome evolution in B. burgdorferi s.l. (Haven et al., 2011), so that intra-specific phylogenetic distances were greatly inflated. The authors conclude that the gene repertoire of B. burgdorferi s.l. is highly stable and no significant number of novel genes are expected to be found with the sequencing of additional genomes (Casjens et al., 2012; Mongodin et al., 2013).
A lack of resolution
While informative for inferring functional differences between genomes based on their total gene repertoire, pan-genome analysis is broad-brushed and does not easily lend itself to the identification and validation of gains and losses of specific genes. Nor would a pan-genome analysis, at least in its original design, reveal lineage-specific genomic changes since phylogenetic information among the genomes is under-utilized. More of a concern is the fact that results of a pan-genome analysis vary depending on the cutoff values chosen for determining whether two genes are homologous or not, based largely on the e-value and length coverage of BLAST (Camacho et al., 2009) hits. In fact, we suspect that the observed strong linear dependency of the B. burgdorferi s.l. pan-genome sizes on the phylogenetic distance (Mongodin et al., 2013) is an inevitable consequence of the fact that genome sequences diverge at a roughly constant rate, a fact evidenced by the general success of sequence-evolution models in phylogenetic reconstruction (Felsenstein, 2004). In other words, the pan-genome gene counts may be in essence a summary statistic of a vast number of BLAST searches. The rate of “gene gains” may be misleading since it could take an arbitrarily high or low value by choosing more or less stringent BLAST cutoff values. In particular, the pan-genome framework is not well suited for identifying lineage-specific gene gains or losses in B. burgdorferi s.l. genomes, since B. burgdorferi s.l. appears to have a “closed” genome with little, if any, gene acquisition from distantly related species through horizontal gene transfers (Casjens et al., 2012; Mongodin et al., 2013). Genome variations in B. burgdorferi s.l.are due almost exclusively to duplications and losses of homologous genes and sequence variations among orthologous genes (Mongodin et al., 2013).
Challenges and opportunities
Although B. burgdorferi s.l. genomes have little horizontally acquired accessory genes, sequencing more genomes is necessary for, e.g., obtaining a more complete phylogeny (Section 2), clinical and ecological association studies (Section 4), and identifying gene-regulatory elements using phylogenetic footprinting (Section 5). As such, we would like to caution here that conclusions on genome stability should by no means be interpreted as if there is no need for sequencing more B. burgdorferi s.l. genomes.
4. Phylogenomics
More informative studies for identifying strain-specific genomic changes are those comparing the genomes of closely related strains (Casjens et al., 2012; Qiu et al., 2004; Wywial et al., 2009). A key advantage of such studies is that they are based on carefully identified orthologous (or partially orthologous) plasmids (Casjens et al., 2012). Similarly, gene orthology is carefully identified by a combination of reciprocal BLAST searches, genome synteny, and reconciliation between gene and a strain trees (Qiu et al., 2004; Wywial et al., 2009). Indeed, distinguishing between orthologous and paralogous copies of a homologous gene family is the key insight of the original phylogenomics approach for predicting gene functions (Eisen, 1998).
Based on orthologous genes, genetic exchanges among coexisting strains were discovered based on incongruent gene trees among genomic loci (Qiu et al., 2004). Orthology analysis helps to infer functions of PF54 genes contributing to host resistance (Wywial et al., 2009). Upon the identification of orthologous plasmids and ortholgous plasmid segments in four completed B. burgdorferi s.s. genomes, a detailed catalog of genome-evolution events has been meticulously compiled and documented, revealing numerous cases of genome re-arrangement, lineage-specific gene decay (pseudogenization), and plasmid duplications and losses (Casjens et al., 2012).
4.1 Gene duplications and losses
As an update from an earlier study (Wywial et al., 2009) we report here an analysis of gene gains and losses on the lp54 plasmids using the expanded dataset of 23 sequenced genomes (Figure 2). Among all replicons in the B31 genome, the lp54 plasmid is over-represented in genes differentially expressed between the tick and mammalian environments (Caimano et al., 2007).
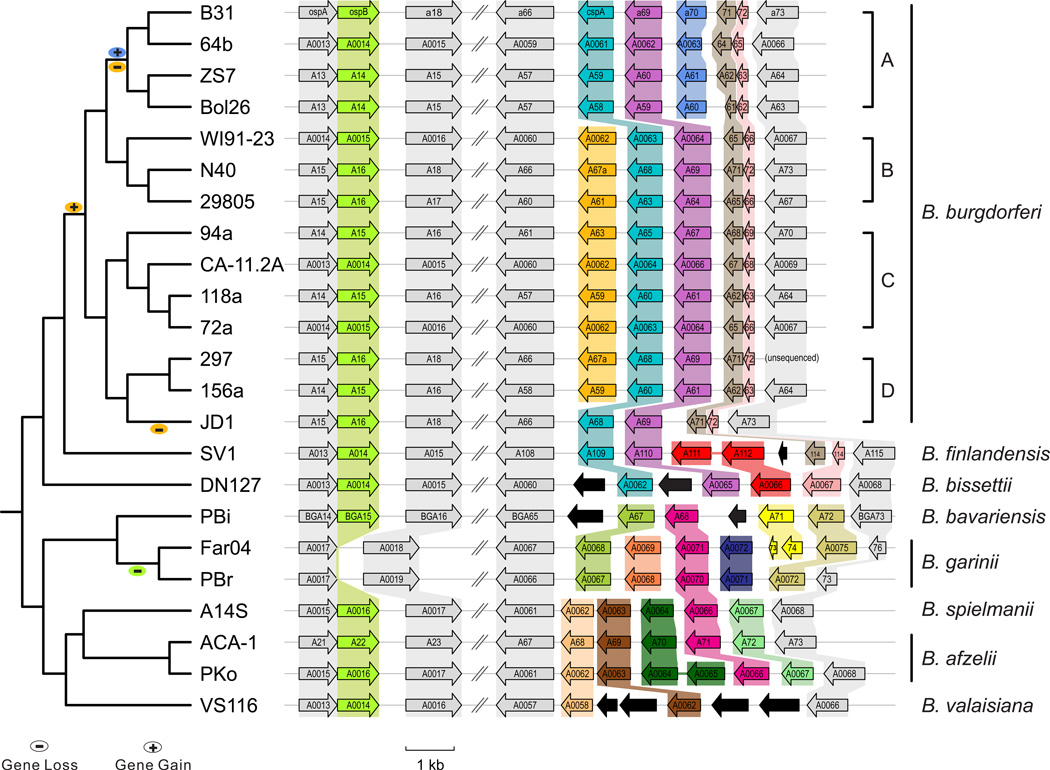
Figure 2. Phylogenomics: Gene duplications and losses on lp54.
Parsimony analysis based on the chromosomal SNP tree (left panel, topological diagram) suggests a loss of ospB (dark green) in B. garinii, a bird-adapted species. The PF54 lipoprotein gene array, which consists of paralogs of the host-resistant gene bba68/cspA, evolves rapidly and in a species- and lineage-specific manner. Orthologs, each set of which is shaded in the same color, were determined by reconciliation of a tree of all PF54 homologs with the genome-based phylogeny, as described previously (Wywial et al., 2009). N40_a67a (yellow) orthologs appear to have lost in SNP Group A and in JD1 independently. The B31_a70 (blue, encoding a plasminogen contributing to disabling host complement system) (Koenigs et al., 2013) ortholog appears to be a recently duplicated copy of its neighboring gene a71 and is present only in SNP Group A, which is associated with disseminative Lyme disease (Dykhuizen et al., 2008 Hanincova et al., 2013). ORFs in black have no apparent orthologs in sequenced genomes. Far04_A74* is a merged version of Far04_A73 and Far04_A74 in GenBank. Orthology among PF54 genes provides an independent line of corroborating evidence for the SNP-based phylogeny (Figure 1).
Loss of ospB in B. garinii may be a host adaptation
Parsimony analysis suggests that ospB has been lost in the European B. garinii lineages (represented here by PBr and Far04). Sequence alignments further revealed that the loss of ospB in B. garinii was not due to pseudogenization but to a deletion of sequences encompassing ospB and its upstream 9-base intergenic sequences, while leaving its downstream flanking gene a18 (a PF62 plasmid-partitioning gene) and its associated promoter intact (Figure 2). Although ospA and ospB share a common promoter, the level of ospB transcripts is much higher than those of ospA in experimentally infected mice (Liang et al., 2004). The loss of ospB, which appears to function predominantly during infections within a mammalian host, may therefore be due to the host specificity of B. garinii, a bird-adapted species (Vollmer et al., 2013).
Gain of a70 in high-virulence strains
The SNP Group A of B. burgdorferi s.s. belongs to the ribotype RST1, a group of strains associated with disseminative Lyme borreliosis (Dykhuizen et al., 2008; Hanincova et al., 2013). The current analysis suggests that a67a (in orange, Figure 2) was present in the common ancestor of B. burgdorferi s.s. and subsequently lost independently in SNP Group A and in JD1. An alternative hypothesis, which posits two independent gains at SNP Groups B and C/D and one loss at JD1, is less parsimonious. In addition, a70 (in blue, Figure 2) was apparently a recently gained PF54 gene in Group A. Phylogenetic analysis showed that a70 is result of a gene duplication of a71. A70 is a surface-localized plasminogen-recruiting protein that contribute to both the dissemination of the pathogen through host tissues and its escape from host immunity (Koenigs et al., 2013). It would be interesting to test experimentally if the evolutionary loss of a67a and, especially, the gain of a70 in B. burgdorferi s.s. SNP Group A contribute directly to this group’s ability to cause disseminative infections in humans.
Fast evolution of PF54 gene array associated with speciation
The PF54 gene array is the most variable region on the lp54 plasmid and consists of paralogs of a68/cspA, one of the three classes of complement regulator acquiring surface proteins (CRASPs) involved in neutralizing host innate defenses (Gilmore et al., 2008; Hallström et al., 2013a; Hammerschmidt et al., 2014; Koenigs et al., 2013). Besides a64, a65, a66 and a73, which are present in all strains, the PF54 gene cluster vary in strain-specific (e.g., a67a and a70 within B. burgdorferi s.s.) and species-specific manners (Figure 2). Broadly, three major types of PF54 gene arrays are distinguishable based on gene orthology, each of which is phylogenetically consistent and associated with a group of species. One type is associated with the species group consisting of B. bissettii (DN127), B. “finlandensis” (SV1), and B. burgdorferi s.s., a second type with the species group consisting of B. bavariensis (PBi) and B. garinii (PBr and Far04), and the third type with the species group consisting of B. spielmanii (A14S), B. afzelii (ACA1 and PKo), and B. valaisiana (VS116) (Figure 2). In addition to the lineage-specific gains and losses of paralogous members, the PF54 genes show accelerated amino-acid replacements during species divergence (Section 4.2). In sum, three lines of evidence support the notion that rapid evolution of the PF54 gene array may reflect adaptation to different host species and may contribute to differential clinical manifestations in humans among the three common pathogenic species. Such evidence includes (i) strain- and species-specific gene duplications and losses, (ii) accelerated amino-acid sequence variations between (but not within) species, and (iii) host-resistance functions of PF54 members like CspA and A70 (Caesar et al., 2013; Hallström et al., 2013a; Koenigs et al., 2013).
Challenges and opportunities
Currently, phylogenomic analysis of gene gains and losses is attainable only for the three conserved replicons including the main chromosome and the lp54 and cp26 plasmids. Systematic and programmatic phylogenomic analysis is challenging for other replicons, many of which house other important antigen genes including the vls locus (Casjens et al., 2012; Glöckner et al., 2006; Graves et al., 2013). Synteny among these replicons is difficult to assess due to limited orthology, non-universal presence in strains, and incomplete genome assemblies.
4.2 Genes under positive natural selection
Current phylogenomics approaches to predicting functional importance of a gene include analysis of rates of synonymous (dS) and non-synonymous (dN) nucleotide substitutions (Kumar et al., 2012). This approach is based on the expected parity between the rate of amino acid-changing and the amount of synonymous nucleotide substitutions for selectively neutral variations, and a greater rate of non-synonymous than that of synonymous substitutions for beneficial amino-acid replacements (Hurst, 2002).
Targets of within-species balancing selection: ospC, dbpA, a07, and b08
The dN/dS ratio test can be used for analysis of within-species synonymous and non-synonymous nucleotide polymorphisms as well. To distinguish between the within- and the between-species dN/dS analyses, we use π, the conventional notation for the level of within-species nucleotide polymorphisms, for within-species analyses. An elevated πA/πS ratio is a reliable indication of genes under diversifying selection, with the well-known example of ospC in B. burgdorferi s.s. (Wang et al., 1999). A number of lipoprotein genes with high πA/πS ratios have been identified from a whole-genome comparison of three B. burgdorferi s.s. strains (Qiu et al., 2004). The ospC and dbpA are apparently subject to strong balancing selection, consistent with their molecular roles in immune escape, host invasion, or both (Schmit et al., 2011; Tilly et al., 2013). It turns out that the high πA/bπS ratios of three PF54 genes (cspA, a69, and a70) found in the earlier study (Qiu et al., 2004) was due to incorrectly identified orthologs. With more accurate ortholog identification as a result of using more genomes, it is now clear that sequences of PF54 genes vary little within B. burgdorferi s.s. but greatly between species (Wywial et al., 2009). The variable region of the PF54 gene array experiences frequent gene duplications and losses so that many genes do not have convincing orthologs beyond closely related species groups (Section 4.1, Figure 2). For example, a70 orthologs exist only in genomes of the B. burgdorferi s.s. SNP Group A (Figure 2).
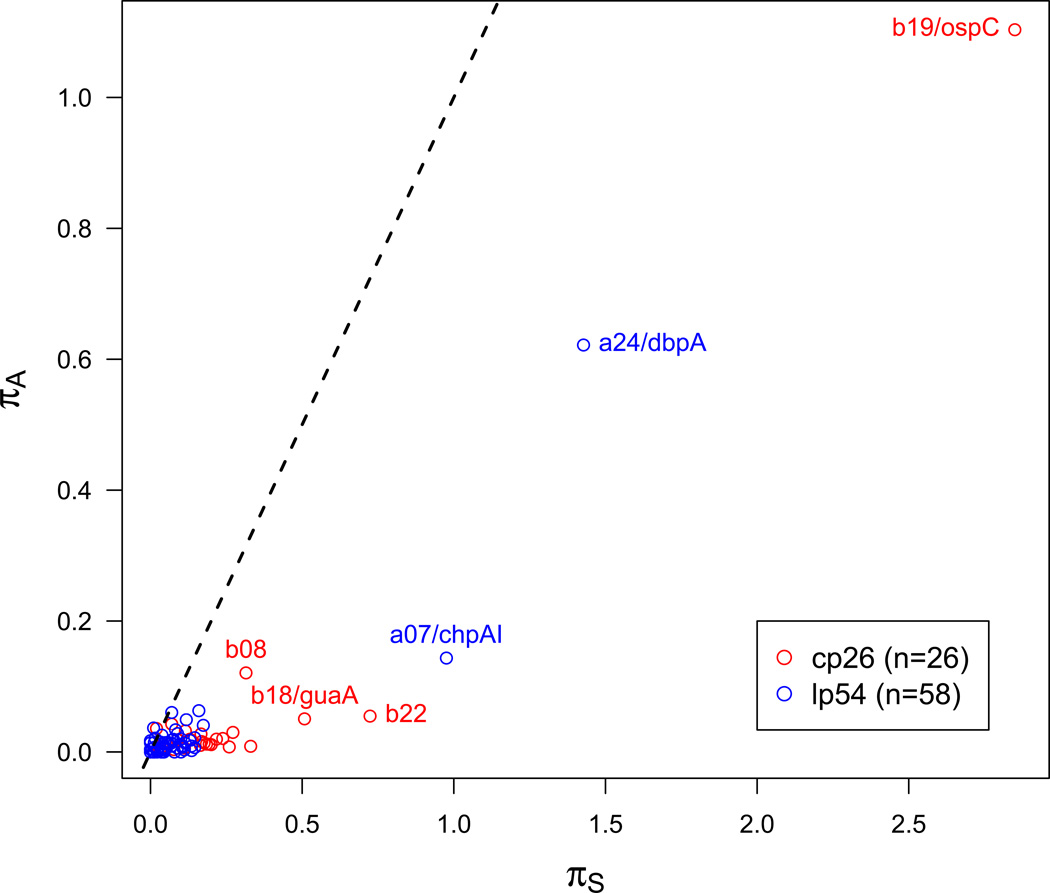
Here, we performed the πA/πS analysis for 58 genes on the lp54 plasmid and 26 genes on the cp26 plasmid from 14 B. burgdorferi s.s. genomes. As in a previous study (Qiu et al., 2004), ospC and dbpA show the highest πA and πS values (Figure 3D). To a lesser but significant degree, b08 and a07 also display high πA values, suggesting the roles of their gene products in interacting with the host (Figure 3D). B08 is predicted to be a lipoprotein and has not been functionally characterized. A07 is predicted to be the lipoprotein ChpAI and has been characterized as an essential protein for Borrelia transmission from the tick to the mammalian host (Xu et al., 2010a). The a07 gene in strain B31 is one of the approximately 150 genes differentially expressed between the tick and mammalian environments (Caimano et al., 2007). Through phylogenetic footprinting, we identified conserved sequence motifs in its promoter region that appear to be binding sites to RpoS, a key transcription factor (Section 5). The existence of RpoS binding sites implies that a07 expression is directly under control of the Rrp-RpoN-RpoS pathway, the main transcriptional control mechanism regulating the phase transitions between the tick and mammalian environments in B. burgdorferi s.l. (Samuels, 2011; Tilly et al., 2013).
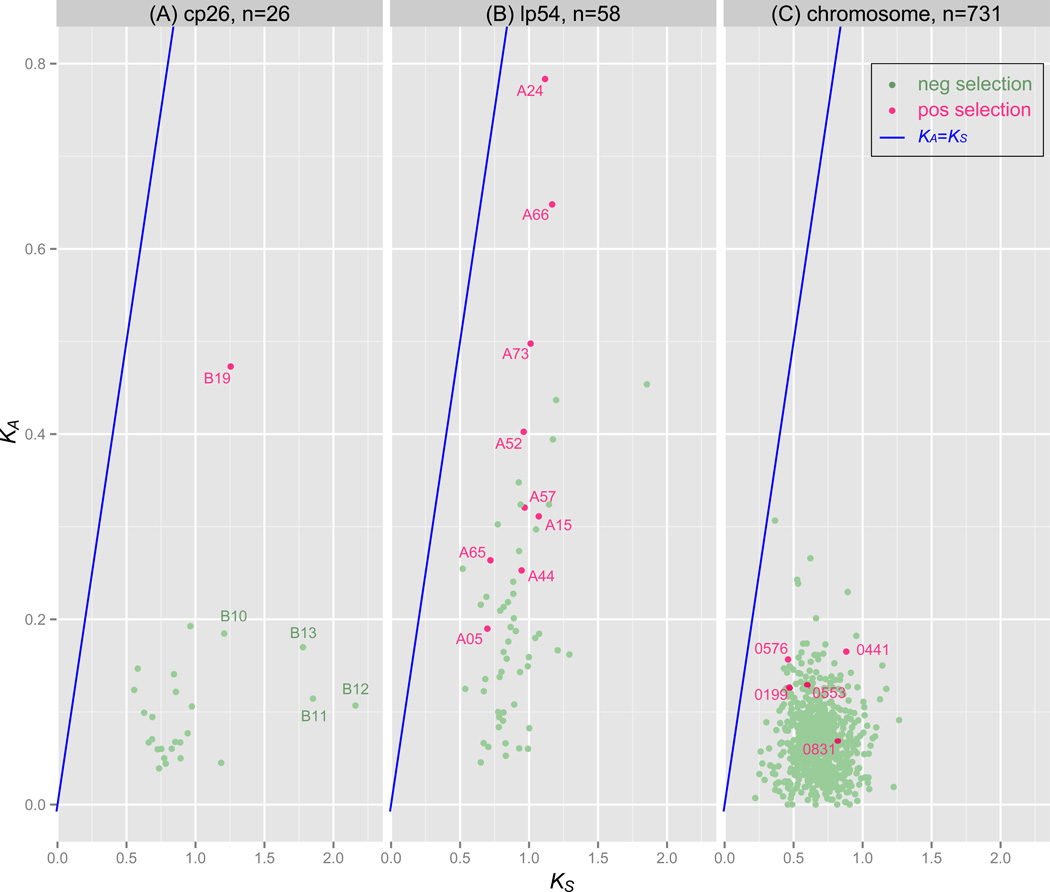
Figure 3. Phylogenomics: positively selected genes.
The between-species synonymous (KS) and non-synonymous (Ka) nucleotide substitution rates of genes on the cp26 plasmid (A), the lp54 plasmid (B), and the chromosome (C). In each panel, the blue line indicates the neutral expectation (Ks=Ka). ORFs containing codon sites under positive selection are colored in red and their molecular functions are discussed in Section 4.2. Other genes (colored in green) are predominantly under purifying selection. The lp54 plasmid contains a large proportion of ORFs evolving adaptively between B. burgdorferi s.l. species. The majority of ORFs on cp26 are under strong purifying selection especially the plasmid-partitioning genes b10, b11, b12, and b13, justifying the use of these genes as plasmid markers (Casjens et al., 2012). The high KS values of ORFs on cp26 reflect high effective recombination rates caused by strong balancing selection at ospC. Also shown are within-species synonymous (πS) and non-synonymous (πa) polymorphisms of genes on the cp26 and lp54 plasmids (D). The dashed line indicates the neutral expectation (πA= πS). Two lipoprotein genes on lp54 (a07/chpA1 and a24/dbpA) show highly elevated πS an indication of the roles of their gene products in interacting with the host. Similar inference could be made for two lipoprotein genes on cp26 (b08 and b19/ospC), which also show elevated πA values. Two genes flanking ospC (b18/guaA and b22) show high πS values apparently due to high effective recombination rates at ospC, but are under purifying selection since their πA values are close to normal. All substitution rates were obtained by using the CODEML program of the PAML software package (version 4.4c) (Yang, 2007) and a phylogeny based on genome-wide SNPs (Mongodin et al., 2013). Plots were made using R (R Core Team, 2013; Wickham, 2009). Fifty-eight genes on lp54 are a01, a03–05, a08–16, a18–21, a23-a25, a30-a34, a36-a57, a59-a62, a64–66, a73, a74, and a76. Twenty-six genes on cp26 are b01–14, b16-b19, and b22–29.
Plasmid lp54 is enriched in genes associated with host specificity
More frequently, the dN/dS ratio test is used for analysis of between-species variations, in which case it is also called the KA/KS analysis. By obtaining maximum likelihood estimates based on a gene tree, the KA/KS analysis is powerful enough to identify adaptive amino-acid changes at individual codon sites and at a particular tree branch (Yang, 2007). First, the PAML analyses of orthologs in eight B. burgdorferi s.l. species suggest that genes on the cp26 plasmid are under strong purifying selection except for b19/ospC (Figure 3A). The relatively high KS values of cp26 genes are apparently a result of high effective recombination rates near the ospC locus (Section 6.1).
Second, a relatively large proportion of genes on the lp54 plasmid show high KA values (Figure 3B). Nine genes contain codon sites with a KA/KS ratio significantly (p< 0.01) greater than one, including a05, a15/ospA, a24/dbpA, a44, a52, a57, a65, a66, and a73/p35 (Figure 3B). Among these genes, a05 codes for the S1 antigen (Xu et al., 2010b), a44 is uncharacterized, a52 is predicted to code for a membrane protein, and a57 encodes a surface lipoprotein and a virulence factor implicated in arthritis (Yang et al., 2013). The a65, a66, and a73 are PF54 genes present in all genomes. In the strain B31, genes bba64, bba65, bba66, and bba73 encodes surface-localized lipoproteins that are required for persistent infection in mice (Gilmore et al., 2008). Genes in the variable part of the PF54 gene array are present in a limited number of strains (Figure 2) and were excluded from this KA/KS analysis. However, an earlier KA/KS study showed evidence for adaptive sequence variations between species at a68/cspA and a69 (Wywial et al., 2009). Summarizing these results, we conclude that the lp54 plasmid is highly enriched in genes (~20% of the total) evolving adaptively during B. burgdorferi s.l. speciation. Since many of these positively selected genes encode outer surface proteins and are differentially expressed between tick and mammalian environments (Caimano et al., 2007), they are strong candidates for genes associated with vector/host specificity among B. burgdorferi s.l. species. The hypothetical role of vector/host specificity these lp54 genes play is supported by the additional evidence that, except for a24/dbpA, their sequences are conserved within species despite large variations between species. The high variability of lp54 genes (e.g., cspA and a70) between species is consistent with their molecular functions of providing resistance against host innate immunity (Hallström et al., 2013b; Koenigs et al., 2013, p. 70), while their conservation within species suggest that they do not interact with host adaptive immunity as directly or strongly as genes like ospC, dbpA, and vlsE.
Third, genes on the main chromosome in general show both low KA/KS ratios (Figure 3C), indicating that the main chromosome lacks in genes subject to positive selection. Five genes contain codon sites significantly under positive selection including 0199 (encoding a putative membrane protein), 0441 (encoding rnpA, a ribonuclease P component), 0553 (uncharacterized), 0576 (uncharacterized), and 0831 (encoding a xylose operon regulatory protein).
An earlier study identified the following, mostly uncharacterized genes having elevated KA/KS ratios between species: a33, a53, a54, and a65 on the lp54 plasmid and bb0102 and bb0404 on the main chromosome (Mongodin et al., 2013). The overlapping but different gene lists from the two KA/KS studies have to do with the fact that the earlier study calculated the KA/KS ratio between a gene and its B31 ortholog and did not use a tree-based approach as here. In both studies, the four genes responsible for the partitioning of the cp26 plasmid, b10, b11, b12, and b13, consistently stand out as the most conserved genes on these two plasmids (Figure 3A), justifying them as optimal markers for plasmid identification (Casjens et al., 2012).
Challenges and opportunities
Phylogenomic analysis of the amount of synonymous (dN) and non-synonymous (dS) nucleotide substitutions is a general analytical framework for screening genes under positive or negative natural selection. It is in fact the main tool for distinguishing ecologically and functionally important sequence variations from selectively neutral ones in the genomes of a species including bacterial species (Su et al., 2013; Vos et al., 2013). To identify functionally important sequence variability associated with the emergence of the pathogenic species B. burgdorferi s.s., statistical tests can be designed and computational algorithms be developed to search for fixed non-synonymous nucleotide differences between B. burgdorferi s.s. and its closest outgroup species “ B. finlandensis”. Similarly intriguing is the prospect of scanning for non-synonymous differences between pairs of sister-group genomes as a way to identify selectively driven variations associated with the most recent divergence between con-specific strains. The same statistical design can be applied to the identification virulence factors by comparing the genomes of strains that vary in human pathogenicity and invasiveness.
5. Phylogenetic footprinting
Phylogenetic footprinting is a computational approach for ab initio prediction of gene-regulatory elements in intergenic spacer (IGS) regions by identifying excessively conserved sequences (Brohée et al., 2011; Katara et al., 2012). The basic assumption of phylogenetic footprinting is that cis-regulatory sequences and non-coding RNAs (ncRNAs) are subject to purifying selection and therefore evolve at a lower rate than less functional, more neutrally evolving intergenic sequences (Eddy, 2005). It is especially effective for identifying regulatory elements in non-model bacterial species (Degnan et al., 2011).
Previous work: three ncRNAs and two transcriptional terminators
Five conserved IGS sequences with potential for forming stable RNA secondary structures have been identified based on a comparison of three B. burgdorferi s.l. genomes (Delihas, 2009). Sequence #1 is a 60-base long sequence found in a variety of linear plasmids including lp25, lp28, and lp60-2 in B. afzelii and lp17, lp38, lp28s, and lp36 in B. burgdorferi s.s. There is no apparent consistency in the functions of its flanking genes and it is likely that it encodes a conserved ncRNA. Sequence #2 is a 34-base long sequence located consistently downstream of the PF60 lipoprotein gene family, which includes BB_h32, BB_i34, and BB_e31 in the B31 genome. It is predicted to be an intrinsic transcriptional terminator. Sequence #3 is apparently a 70-base long Rho-independent transcription terminator for PF54 genes including cspA. Sequence #4 is a 122-base long sequence that may represent another ncRNA. Sequence #5 is a 150-base long sequence that contains a perfect inverse repeat (IR). Unlike the Sequences #1 to #4 in the above, it is present as a single copy in each of the three genomes and 95% of its sequence is identical among the three species. It is located downstream of the plasmid-partitioning cluster (consisting of a18, a19, a20 and a21) on the lp54 plasmid and predicted to be another conserved ncRNA.
Extensive conservation of IGS sequences and six putative ncRNAs
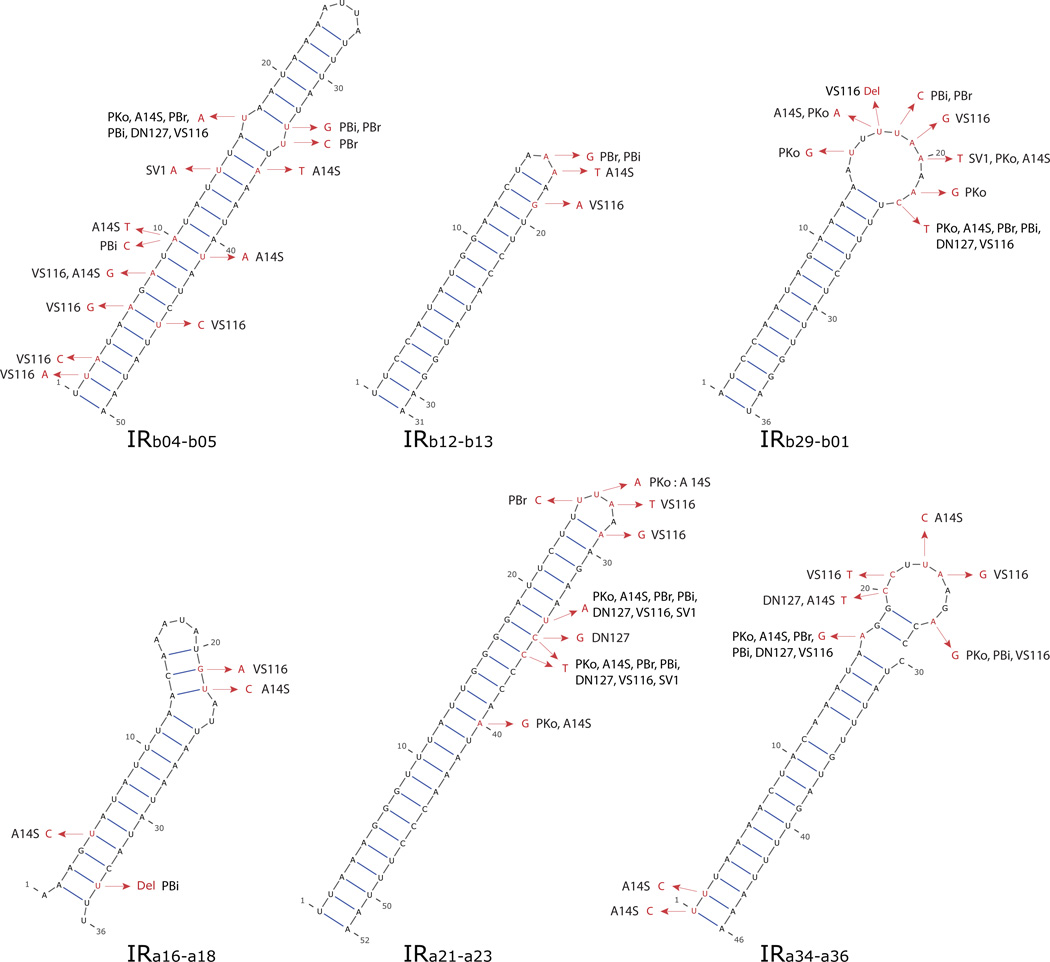
With the availability of 23 genome sequences, we have initiated a systematic genome-wide search for gene-regulatory elements using phylogenetic footprinting. While the project is still ongoing, the protocol and some preliminary findings are described in the following. Approximately 800 sets of orthologous IGS sequences have been identified and extracted based on previously identified orthologous ORFs on the main chromosome and the lp54 and cp26 plasmids (Haven et al., 2011). For each set of orthologous ORFs, a consensus start-codon position was determined based on the majority start-codon positions among the eight B. burgdorferi s.l. species. The 5’ sequence of an ORF was extended or shortened if it did not conform to the majority consensus. This re-adjustment of start-codon positions is necessary because of large discrepancies in predicted start-codon positions among orthologous ORFs. After obtaining consensus start-codon positions, the orthologous IGS sequences were aligned using MUSCLE (Edgar, 2004). The between-species variability of IGS sequences is much more informative than the within-species variability and was thus used to assess the level of evolutionary conservation of nucleotide sequences at each IGS locus. IGSs contain numerous conserved sequence motifs, over 30% of which are perfectly conserved between the eight species. While these conserved IGS sequence motifs include well-known regulatory elements such as ribosomal-binding sites and RpoS-binding sites, functions of most of these conserved elements are unknown. We used all-against-all BLAST+ (Camacho et al., 2009) to exhaustively search for shared sequence motifs among all IGS sequences in the B31 genome. Shared motifs with less than 90% average sequence identify among the eight species were excluded. This protocol is capable of identifying self-similar inverted repeats (IRs) in addition to similar sequence motifs across all IGSs in the genome. Predicted RNA secondary structures of six longest conserved IRs on lp54 and cp26 plasmid are shown in Figure 4. Among these, the inverted repeat IRa21-a23 is the same as the conserved motif Sequence #5 discovered by (Delihas, 2009) (see above), providing a validation between this study and the previous one.
Figure 4. Phylogenetic footprinting: Putative non-coding RNAs.
Predicted secondary structures of six longest putative ncRNAs on cp26 and lp54 plasmids. Each putative ncRNA contains a highly conserved inverted repeat (IR) sequence. These conserved IRs were identified by running all-against-all BLAST (Camacho et al., 2009) among B31 IGS sequences. Only IRs that share 90% or more sequence identity among the eight B. burgdorferi s.l. species (represented by strains B31, SV1, DN127, PBi, PBr, A14S, PKo, and VS116) were retained. Each RNA structure was predicted by using the B31 sequence with the program RNAalifold (Bernhart et al., 2008) and plotted by using VARNA (Darty et al., 2009). Base substitutions (in red) represent variations between species. These predicted structures are supported by patterns of sequence variability, which is enriched within the loop regions and compensatory in some stem regions. The IRa21-a23 has previously been described (Delihas, 2009) and the other five putative ncRNA are new findings.
Challenges and opportunities
The statistical power of detecting conserved gene-regulatory sequence motifs using phylogenetic footprinting increases with the number of genome sequences (Eddy, 2005). Comparing more genomes, especially those from different B. burgdorferi s.l. species, would greatly reduce the number of falsely predicted regulatory elements, which may appear to be conserved among sampled genomes purely by chance. A challenge is to identify transcription factor-binding sequences shared among co-regulated genes. For example, the RpoS-binding motif consists of two disjoint parts intercepted by a region that vary in both sequences and lengths (Caimano et al., 2007). Prediction of RpoS-binding site is made harder by the fact that it varies among co-regulated genes. Nevertheless, phylogenetic footprinting is a highly effective computational approach to reveal genome-wide candidate cis-and trans-regulatory elements in a non-model organism like B. burgdorferi s.l. In combination with other motif-finding algorithms, phylogenetic footprinting has the potential to computationally reconstruct genome-wide regulatory networks (Brohée et al., 2011).
6. Population genomics
Theory and applications
Unlike phylogenomics and phylogenetic footprinting, both of which are motivated by the goal of characterizing gene and genome functions, population genomics concerns itself primarily with an understanding of evolutionary and ecological forces operating in the natural populations of a species (Ellegren, 2014; Whitaker and Banfield, 2006). By sampling multiple genomes within and between populations, theories of population genetics can be used to reveal key evolutionary and ecological parameters and processes in a microbial species, such as its population size based on the standing genetic variability, relative rates of mutation and recombination inferred from single-nucleotide polymorphisms (SNPs), history of divergence and migration reconstructed from the amount of genetic differentiation between populations, and forces of natural selection manifested in differential patterns of sequence variability among genomic loci (DeLong, 2004; Guttman and Stavrinides, 2010; Whitaker and Banfield, 2006). Population genomics is therefore more than a tool for gene and genome annotation but in fact the foundation for all evolutionary analyses of genomes.
The ideal dataset most amenable to population genomic analysis is a random, unbiased sample of genomes from a natural population, which in classic population genetics is defined as a group of individuals sharing a common history of genetic drift, subject to the same selective forces, and capable of DNA exchange with one another (Hartl and Clark, 2007). Without these assumptions, it would be harder to generate precise, theory-based expectations on the amount and pattern of genetic variability at a genetic locus. For free-living bacteria species, such idealized samples rarely exist due to a complex natural history that includes frequent population admixture and fluctuating populations sizes. In a well-known pan-genome study of twenty pathogenic and commensal strains of Escherichia coli, for example, isolates were sampled from worldwide archives with diverse geographic, temporal, and ecological backgrounds (Touchon et al., 2009). While the analyses were powerful enough to reconstruct the history and mechanisms of genome evolution in E. coli, the study leaves many population-level questions unanswered including its population size, population differentiation, and selective forces driving the genome differentiation.
An ideal population-genomic dataset
The twelve genomes of B. burgdorferi s.s. from the United States (Table 1) represent a nearly ideal dataset amenable for population genomic analyses. First, ten of the twelve genomes are samples from the Northeastern US population, which is separated from other US populations with a well-defined boundary (Margos et al., 2012). While all North American B. burgdorferi s.s. populations share a common ancestral population, the far Western, upper Midwestern, Northeastern, and perhaps Southeastern populations have diverged significantly (Brisson et al., 2010; Hoen et al., 2009; Margos et al., 2012; Qiu, 2008; Rudenko et al., 2013). Although migrations occur among these North American populations as well as between the North American and European populations, the migratory events may be rare and relatively recent (see Section 7). Second, strains coexisting within the Northeastern US population recombine frequently. In fact, recombination is the main mechanism driving genome differentiation within local B. burgdorferi s.s. populations (Haven et al., 2011; Qiu et al., 2004). Third, coexisting strains from the Northeastern US share a common enzootic transmission cycle and are therefore likely to subject to similar selective forces (Brisson et al., 2012; Diuk-Wasser et al., 2012; Kurtenbach et al., 2006; Margos et al., 2011; Vuong et al., 2013). In sum, with genetic isolation, frequent recombination, and shared ecological conditions, the ten B. burgdorferi s.s. genomes from the Northeastern US represent a rare, if not unique, whole-genome sample of a natural bacterial population in a strict population-genetic sense. Although not a random sample and representing only about a half of known genomic groups existing in the Northeastern US, these genomes nonetheless offer an excellent opportunity to look into the processes of bacterial genome evolution at both within- and between-population levels. Indeed, evolutionary analyses of these twelve genomes have revealed some key mechanisms of genome diversification within natural B. burgdorferi s.l. populations, including recombination and selective forces.
6.1 Recombination and its implications to associate study
Recombination is an essential facilitator of adaptation, without which species would quickly go extinct due to accumulation of deleterious mutations (“Mueller’s Ratchet”) as well as an inability to fix beneficial ones (“Hill-Robinson Effect”) (Barton, 1995; Hill and Robertson, 1966; Muller, 1964). Although reproducing asexually, bacteria species are no exception to a dependency on homologous recombination for long-term sustainability (Didelot and Maiden, 2010; Dykhuizen and Green, 1991; Fraser et al., 2007). The ability to recombine is apparently itself a nearly universal adaptation maintained by strong natural selection despite a substantial fitness cost to the individuals (Barton, 2009).
Population structure: clonality with frequent recombination
Initial study of natural populations of B. burgdorferi s.l. revealed a clonal genetic structure, in which gene trees inferred from sequences at multiple loci are highly congruent (Dykhuizen et al., 1993). Interpretation of such multilocus clonality as a result of low recombination in B. burgdorferi s.l. (Dykhuizen and Baranton, 2001), however, proved to be premature when evidence emerged for extensive recombination among coexisting strains including incongruent gene trees between loci and clusters of SNPs in genome sequences (Qiu et al., 2004). Using the technique of sister-group comparisons (Guttman and Dykhuizen, 1994), the rate of recombination among coexisting strains in the Northeastern US populations of B. burgdorferi s.s. has been estimated to be approximately three times the mutation rate (Haven et al., 2011; Qiu et al., 2004). In other words, sequence differences between a randomly selected pair of coexisting strains consist of on average 75% of pre-existing nucleotide variations and only 25% of de novo mutations.
High effective (but not intrinsic) recombination rates at ospC
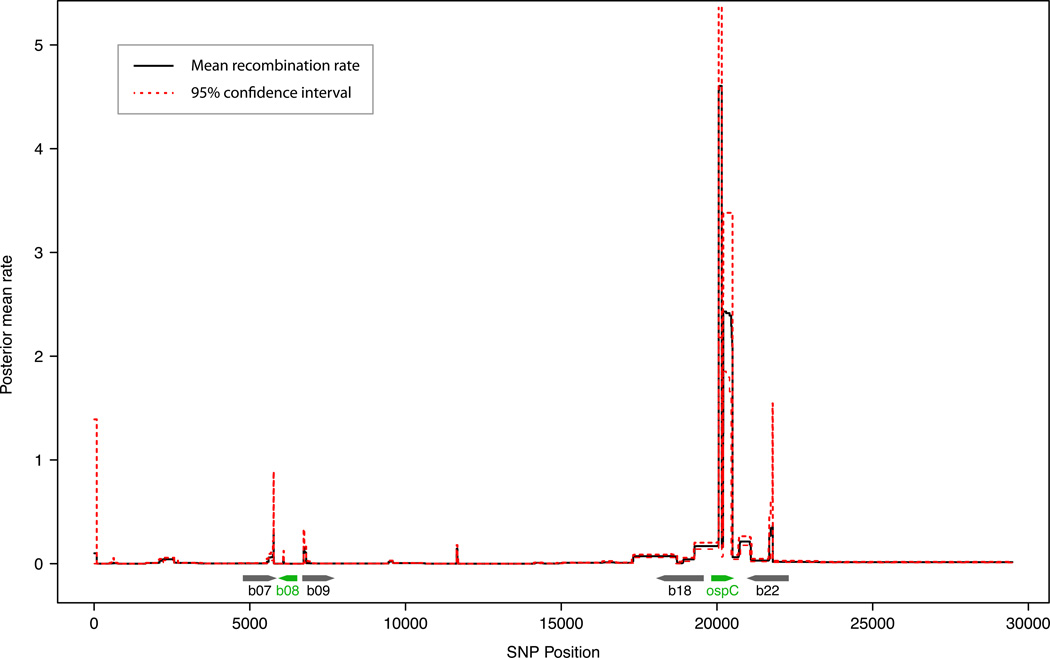
The recombination rate obtained from sister-group analysis may be considered as an estimate of the intrinsic or neutral recombination rate – denoted here as r0– since the majority of nucleotide differences between sister-group are synonymous. We formulate the neutral recombination rate (r0) in analogy to the neutral substitution rate (µ0), which is equal to the mutation rate at a neutrally evolving locus, a key prediction of the Neutral Theory of molecular evolution (Kimura, 1984). The actual or observed recombination rate (r) at a particular locus, however, may deviate significantly from the intrinsic recombination rate as a result of natural selection. For example, at a locus subject to strong negative (i.e., purifying) selection, most spontaneous mutations would be removed while SNPs introduced by recombination from existing strains is likely to be retained, resulting in a greatly reduced overall level of observed sequence variations. In contrast, at a locus subject to strong positive (i.e., adaptive or diversifying) selection, both the observed mutation and recombination rates are expected to be greatly elevated due to selection for sequence variability. Computer programs for estimating recombination rates based on genome-wide SNPs typically calculate observed, realized, or effective recombination rates (r) rather than the intrinsic, neutral recombination rate (r0). For example, we used the computer program LDhat (McVean et al., 2002) to estimate recombination rates along the cp26 plasmid using the twelve B. burgdorferi s.s. genomes from the US. While one may call the ospC locus as a recombination “hot spot” based on the extremely high recombination rate observed at that locus (Figure 5), one should be careful in concluding the existence of an intrinsically recombination-prone mechanism at this locus. As we explain below, the high observed recombination rate at ospC is more parsimoniously explained as a consequence of diversifying selection targeting at this locus, which otherwise has a normal intrinsic recombination rate (r0).
Figure 5. Population genomics: Selection-driven recombination hotspots.
Recombination rates along the cp26 plasmid. Two recombination hotspots are observed, one centered on b08 (encoding an uncharacterized lipoprotein) and the other on ospC. As explained in the text (Section 6.1), these recombination hotspots are most likely due to preferential retainment of amino-acid variations by strong balancing selection and not necessarily due to intrinsic pro-recombination mechanisms. As such, genome-wide recombination analysis of within-population samples helps identify loci under diversifying selection. Recombination rates for all pairs of 951 SNPs on the cp26 plasmids among twelve US B. burgdorferi s.s. genomes (Table 1) were estimated by using LDhat (version 2.2) (McVean et al., 2002). The cp26 sequences were co-linearized according to the B31 sequence (all starting at b01 and ending at b29) and then aligned by using MUGSY (version 1.2.1) (Angiuoli and Salzberg, 2011) in a LINUX environment. The longest alignment was extracted by using a MUGSY-supplied Perl script. SNPs were identified by using the CONVERT program of LDhat. Pair-wise recombination rates were estimated by using the PAIRWISE program of LDhat, which were subsequently plotted in the R statistical computing environment (R Core Team, 2013) with the “summarise.pairwise” R function included in the LDhat package. Recombination hotspots were estimated by using the INTERVAL program of LDhat and plotted with the supplied R function “summarise.interval”.
Robust within-population phylogeny despite recombination
Recombination in B. burgdorferi s.l. predominantly takes the form of gene conversion, which is the replacement of a short (typically less than 1 kilobases) DNA segment in a recipient genome by a homologous fragment from a donor genome (Dykhuizen and Baranton, 2001; Haven et al., 2011). Contrary to the crossing-over recombination, gene conversion tends to reduce local linkage disequilibrium (LD) and does not cause LD decay over genomic distances commonly seen in Eukaryotes (Wiehe et al., 2000; Wiuf and Hein, 2000). The weak local LD in conjunction with strong genome-wide LD in bacterial species had been captured by the concept of “clonal frames” (Desjardins et al., 1995; Milkman and Bridges, 1990). One consequence of the bacterial clonal frame is that phylogeny of bacterial strains are more reliably inferred by using genome-wide SNPs than using sequences at a few loci. This may be true because the use of SNPs from the whole genome maximizes the number of phylogenetically informative SNPs, leading to increased statistical support for individual branches. Indeed, statistically robust within-species phylogenies have successfully been obtained using genome-wide SNPs for B. burgdorferi s.s. and E. coli despite high rates of localized recombination in both species (Mongodin et al., 2013; Touchon et al., 2009).
Challenges and opportunities
The special mode of recombination in bacteria calls for the development of novel analytical methods distinct from those developed for eukaryotes (Ansari and Didelot, 2014; Didelot et al., 2010). Specifically, genome-wide association studies in bacteria should consider and take advantage of two uniquely bacterial patterns of linkage disequilibrium. First, the genealogical process of a sample of bacterial genomes can be approximated by a single coalescent tree due to a genome-wide clonal frame. Such a tree, which can be inferred by using a large number of SNPs, in turn helps identification of recombination events at individual loci. Second, linkage disequilibrium (LD) among neighboring SNPs is loosened by frequent gene conversion. It has been shown analytically that the sample size required for detecting disease-causing genes linked with a marker gene increases with the rate of local recombination rate (Pritchard and Przeworski, 2001). This theoretical result suggests that, while it is important to discover that Borrelia invasiveness is associated strongly with MLST haplotypes (Hanincova et al., 2013), it is essential to identify virulence-causing genes by testing genome-wide SNPs individually and by using a large number of genome sequences. At the cusp of the coming wave of genomes sampled from within populations, we envision a whole-genome linkage map for local B. burgdorferi s.l. populations. Under a framework an intra-specific phylogeny, such a genetic map would categorize each SNP in each genome as either a novel mutations, a part of laterally transferred DNA due to gene conversion, or a randomly sorted ancestral polymorphism. Genes associated with ecological and clinical variations could then be identified as the loci where genetic changes are correlated with phenotypic changes.
6.2 Natural selection
Predicting functional importance of genomic variations takes more than a description of observable variations and requires an understanding of causative population processes. Standing genomic variations among coexisting strains are a result of complex population processes including both stochastic mechanisms, such as random genetic drift and fluctuating population sizes, and more deterministic forces such as natural selection. Among these, natural selection is the key for identifying functionally important genomic variations. Distinguishing between selectively maintained genomic variations and stochastic neutral variations is the main challenge in all genomic analyses of natural populations (Ellegren, 2014; Guttman and Stavrinides, 2010; Li et al., 2008; Whitaker and Banfield, 2006).
Two selective hypotheses
Natural selection in the form of host specialization is apparently the main force maintaining a diverse array of B. burgdorferi s.l. species in regions where they overlap geographically, whether in Europe or in North America (Kurtenbach et al., 2006; Margos et al., 2011). Large adaptive genomic differences at loci encoding host-resistance lipoproteins, e.g., the PF54 gene array (Figure 2; Section 4), are strong candidates as being associated with host adaption. More controversial is the selective forces causing and maintaining the high levels of sympatric diversity of genomic groups within a single population such as B. burgdorferi s.s. in the Northeast US (Brisson et al., 2012). Two main selective hypotheses have been formulated including the multiple-niche polymorphism (MNP, Box 1) hypothesis (Brisson and Dykhuizen, 2004) and the negative frequency-dependent selection (NFD, Box 1) model (Haven et al., 2011). Consistent with empirical evidence including host preferences and clinical variability among the genomic groups, the MNP model proposes that coexisting B. burgdorferi s.s. strains are maintained by fine-grained niche partitioning such as vector, host, and tissue specializations (Brisson and Dykhuizen, 2004; Brisson et al., 2012). Consistent with observations such as the coexistence of a large number of genomic groups within a single local population and the strong linkage between local genomic lineages with major antigen loci such as ospC, the NFD model proposes that diversifying selection driven by escape from host immunity is the main selective force within natural B. burgdorferi s.s. populations (Haven et al., 2011). Since NFD does not assume host-adaptive differences among coexisting strains while MNP does, testing of these two hypotheses is at the heart of investigations into the degree of ecological specialization and clinical variability among coexisting B. burgdorferi s.l. strains.
Tests with genome sequences
Evolutionary analyses of whole-genome sequences offer a way for testing these two seemingly competing hypotheses. The NFD model predicts that genetic variations at antigen loci should be adaptive and have high dN/dS values, while those at housekeeping loci should be under purifying selection and have low dN/dS values (Haven et al., 2011). In contrast, the MNP model would predict that at least a portion of non-synonymous substitutions at housekeeping loci are fixed adaptive differences between the ospC-marked genomic groups. This prediction is based on the assumption that, although host-adaptation is primarily associated with a few surface antigen loci like ospC in an MVP model, secondary host-adaptation mutations are expected to occur and be fixed at housekeeping loci over time. Measuring the portion of fixed non-synonymous substitutions between genomic groups at housekeeping loci is therefore a way of testing the two hypotheses. Certainly, there are numerous fixed nucleotide differences among the strains, without which there would not be any phylogenetic signal. It is however not yet known what proportion of the fixed nucleotide differences at housekeeping loci among coexisting strains is synonymous or nonsynonymous. This analysis lends itself to a McDonald-Kreitman (MK) test (McDonald and Kreitman, 1991) with the neutral expectation, using the notation of (Stoletzki and Eyre-Walker, 2011), that the ratio of between- to within-strain non-synonymous substitutions (Dn/Pn) is the same as the ratio of the between- to within-strain synonymous substitutions (Ds/Ps). Assessed by the Fisher’s Exact test, a positive test would support the MNP model if the null hypothesis is rejected on evidence of an elevated level of fixed nonsynonymous differences (Dn) between strains at housekeeping loci.
Challenges and opportunities
Since the NFD model is a simple and parsimonious explanation of the coexistence of a large number of intra-specific genomic groups, it could be considered a null selective hypothesis. Certainly, there is room for reconciliation and integration between the NFD and MNP models. One possibility is the synergistic accumulation of host-adapting and immune-escape variations. It is conceivable, for example, that strains diverge initially at major antigen loci driven by NFD and, subsequently, mutations for host preferences become fixed during the prolonged period of coexistence of multiple genomic lineages. Feasibility of such a mixed model may be evaluated using computer simulations (Haven et al., 2011). A mixed model should be able to reconcile the observation of a large number of coexisting strains and the empirical evidence of ecological and clinical variations among these strains. Questions may rise on the relative importance of these two selective forces. Again, the MK test described above is a means to quantitatively estimate the relative strength of the two selective forces. A strongly positive MK test result would suggest substantial host adaptation between the coexisting strains, while a weak or insignificant MK test result would suggest that the within-species strain diversity in local B. burgdorferi s.l. populations is maintained predominantly by immune escape.
7. Genomic phylogeography
Biogeography of B. burgdorferi s.l. is not only important for reconstructing its history of worldwide diversification and migration, but also for predicting future risks of Lyme disease by understanding evolutionary and ecological mechanisms underlining its accelerating global range expansion (Kurtenbach et al., 2006; Margos et al., 2011; Ogden et al., 2013). One consensus opinion emerged is the realization and hope that B. burgdorferi s.l. genome variability holds signatures of biogeographic processes, making evolutionary-genomic analysis a key to testing ecological hypotheses, including climate changes leading to shifts in tick seasonality, migration of hosts, and habitat degradation (Diuk-Wasser et al., 2012; Gatewood et al., 2009; Ogden et al., 2013).
Unresolved global phylogeography
Multilocus and genome-based phylogenies of B. burgdorferi s.l. species, rooted by relapsing-fever Borrelia, support two large species assemblages of B. burgdorferi s.l., one found in Eurasia and the other in the New World (Figure 1). This geographic pattern seems to suggest that the most ancestral divergence of B. burgdorferi s.l. probably coincide with and may be caused by the breakage of the two continents, a process that started 150 million year ago. This biogeographic history, however, is inconsistent with the contemporary geographic distribution of B. burgdorferi s.s., which is widely distributed in both continents and its closest outgroup species (e.g., “B. finlandensis”) is found only in Europe (Margos et al., 2008; Qiu, 2008). The ancestral Eurasia-New World divergence is also not consistent with the geographic range of the newly proposed species “B. chilensis”, a New World species but appearing to be the most basal B. burgdorferi s.l. species known so far (Ivanova et al., 2013). Clearly, a coherent worldwide phylogeographic history of B. burgdorferi s.l. is yet to emerge, which can be better resolved by sequencing more genomes, especially those of recently discovered New World species. The use of genome-wide nucleotide variations or concatenated alignments of conserved protein sequences (Wu et al., 2013) improves statistical confidence in phylogenetic reconstruction of the B. burgdorferi s.l. species complex and will allow better resolution on its biogeographic histories and mechanisms at both global and regional levels. Any inconsistency of geographic origins with a species phylogeny would suggest ancestral or recent migration events, since random sorting of ancestral polymorphisms is less of a problem in genome-based phylogenetic reconstruction.
Cross-species introgression in Northeast US
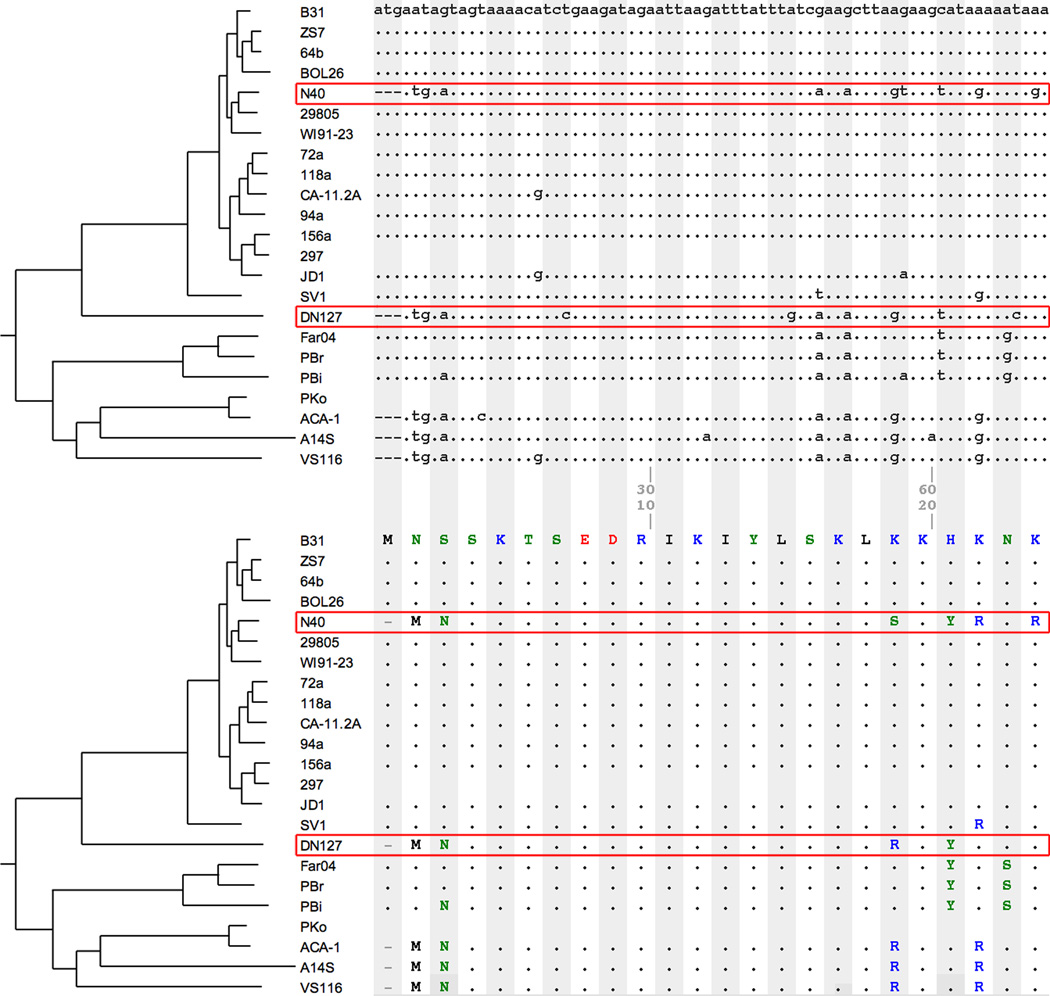
Infected ticks from Eurasia frequently contain a mixture of B. burgdorferi s.l. species while coexistence of species is less common in the New World (Margos et al., 2011). Genome analysis, however, has revealed not only coexistence but also genomic admixture (Box 1) between species in North America. For example, we identified that an approximately 2.2-kilobase long DNA fragment from a DN127-like genome has been transferred and incorporated into the N40 genome (Figure 6). B. bissettii-like strains found in Northeastern US have been renamed as members of a newly proposed species B. kurtenbachii, with strain 23015 as the type strain (Margos et al., 2010). Since B. bissettii is rarely found in the Northeast US or isolated from I. scapularis ticks, it is likely that the donor of this transferred DNA fragment is a member of B. kurtenbachii. The cluster of SNPs at this region has been identified previously based on a comparison of three genomes, but the identity of the donor genome was unknown at the time for lack of a large panel of reference genomes (Qiu et al., 2004). While this chromosomal region contains housekeeping genes and these SNPs are not obviously adaptive, this unambiguous case of cross-species recombination offers a glimpse on the genetic consequences of B. burgdorferi s.l. expansion in this region and elsewhere.
Figure 6. Genomic phylogeography: A cross-species hybridization.
(Top Panel) A codon alignment of the 5’ sequences at the main chromosomal locus 0082 (encoding an uncharacterized conserved protein). B31 sequence is used as the reference and an “.” in other sequences represents the same nucleotide as in the B31 sequence at the same aligned site. Neighboring codons are shaded in alternating colors. The corresponding amino-acid alignment (Bottom Panel) shows coding consequences (synonymous or non-synonymous) of individual nucleotide variations. The N40 sequence at this region displays a large phylogenetic inconsistency: it is more similar to the sequences of non-B. burgdorferi s.s. orthologs than to its con-specific strains. Since all outgroups of B. burgdorferi s.s. used here are distributed exclusively in Europe or Western US, it is likely that the donor is a DN127-like species in Eastern US, such as B. kurtenbachii (Margos et al., 2010). Further analysis identified the two ends of this single cross-species gene-conversion event and showed that it affects a 2.2-kilobase region from the 3’ end of the 0081 locus (encoding a permease) to the 5’ end of the 0084 locus (encoding an aminotransferase). While this gene-conversion event is not obviously adaptive, this analysis shows that (i) there is cross-species hybridization in Northeast USA, (ii) the hybridization may be recent (see Section 7), and (iii) evolutionary genomic analysis is powerful to reveal population history. In this case, signatures of recombination are discovered based on phylogenetically mismatched SNPs (“homoplasies”). The graph was prepared by using BorreliaBase.org, a phylogeny-centered browser designed for evolution-informed comparative analysis of Borrelia genomes (Di et al., 2014). Future studies will screen for genome-wide signatures of species hybridization and population admixture.
First, it suggests that there is little intrinsic barrier to recombination between B. burgdorferi s.l. species. The boundary between B. burgdorferi s.l. species is therefore delineated more by geographic isolation than by ecological specialization. Second, it implies that B. kurtenbachii and B. burgdorferi s.s. share tick vector, host species, or both, although it is not clear if such a niche sharing between the two species is accidental or common. Third, since this recombination event involves housekeeping loci it is a reflection of intrinsic and neutral rate of gene flow. If cross-species hybridization is common and historically ongoing, one would expect extensive genomic signatures of gene flow at housekeeping loci. Although we have not yet performed genome-wide search for such signatures of cross-species or cross-population gene flow, our preliminary assessment is that such neutral gene flows are not common. We therefore speculate that the cross-species contact and genetic admixture in the Northeast US is either historical but sporadic, or new and increasing as a result of range expansion of B. burgdorferi s.l.
Recent secondary contacts between species and populations
Genetic variations at non-housekeeping genes provide further evidence for increasing contacts between previously isolated B. burgdorferi s.l. species and populations. In Europe, the B. burgdorferi s.s. strain BOL26 has been converted at ospC and the two neighboring loci by a 1.9-kilobase DNA fragment from a coexisting B. afzelii strain (Haven et al., 2011). Cross-continent and cross-population gene flows involving ospC appear to be common in latest multilocus surveys of US local populations (Hanincova et al., 2013; Rudenko et al., 2013). A genome-wide identification of DNA signatures of cross-species DNA exchange would lead to more precise estimates on the age and frequency of such events. If the species admixture is relatively recent, the level of genome-wide cross-species gene exchange would be low. Much higher and more extensive levels of genomic admixture are expected if species hybridization is ancient. Methods have been developed to estimate the age of species hybridization in humans and other eukaryotes based on genome-wide heterogeneity using ad hoc speciation models, which are then evaluated with approximate Bayesian computation (ABC) (Prüfer et al., 2014; Roux et al., 2013; Vernot and Akey, 2014). Presumably, similar approaches can be applied to estimating the age of hybridizations between B. burgdorferi s.l. species in Eurasia and the New World.
Population admixture: sharing of recent ancestors or accelerating migration?
At a more regional level, multilocus sequence analysis (MLSA, Box 1) revealed a history of a shared ancestral population and migration among regional populations of B. burgdorferi s.s. in the US (Brisson et al., 2010; Hoen et al., 2009; Margos et al., 2012; Qiu, 2008). It remains unclear the age of population boundaries, the level of migrations, or the effective sizes of ancestral and contemporary populations. Answers to these questions help predict the future Lyme disease risks by identifying possible ecological causes of range expansion of B. burgdorferi s.s. By sequencing multiple genomes from within local populations, it will be possible to obtain more precise estimates on these population parameters using approaches such as isolation-migration modeling (Pickrell and Pritchard, 2012; Wang and Hey, 2010) and multilocus coalescence (Liu et al., 2009). An estimate of genetic differentiation among regional populations is not informative by itself since it could either be an estimate of the age of divergence from a common ancestral population or an estimate of ongoing migration rates. The isolation-with-migration model is designed to disentangle multiple causative mechanisms of recently diverged populations (Pickrell and Pritchard, 2012; Wang and Hey, 2010). The multilocus coalescent model maximally uses phylogenetic information at individual loci, an approach that is alternative to and more rigorous than the commonly used method of inference of multilocus phylogeny using concatenated alignments (Edwards, 2009; Liu et al., 2009). So far, these newly developed, genome-inspired biogeographic methods have not yet been applied to the study of B. burgdorferi s.l. populations.
Challenges and opportunities
Genome-wide extent and age distribution of DNA fragments introduced by cross-species recombination in Europe and North America remain to be determined. So are the extent and age of population admixture occurring within the continents. Genome admixture of B. burgdorferi s.l. species and populations may be a relatively recent phenomenon due to an increase of long-distance dispersal and regional range expansion. Population growth of a pathogen has at least two evolutionary consequences that are disturbing from the perspective of public health. First, theory (Takahata, 1993) and simulations (Mongodin et al., 2013) predicts a burst of pathogen diversity with an increase in population size under strong balancing selection. Second, population expansion leads to increased hybridization of previously isolated populations in a highly recombinogenic species such as B. burgdorferi s.l. Both of these evolutionary consequences facilitate pathogen adaptation and increase its public-health risks, since the resulting new genotypes may be ecologically more invasive and clinically more virulent.
8. Concluding remarks
The first decade of evolutionary genomics of B. burgdorferi s.l. covered much of the worldwide phylogenetic, geographic, clinical, and host/vector diversity known at the time. A key decision was also made to sequence multiple genomes from a single population of a single species. Without these between- and with-species genome sequences, it would not have been possible to obtain a robust strain phylogeny, define the gene repertoire, identify adaptively evolving lipoporteins, reveal conserved regulatory elements, discover genome-wide recombination, or detect recent cross-species genetic introgression. With the incoming wave of genome sequences from more species and strains worldwide, evolutionary genomics of B. burgdorferi s.l. will be at the forefront of investigating the ecological and evolutionary mechanisms and monitoring the trend of the global expansion of the Lyme disease endemic. In addition to field and experimental studies of Lyme disease ecology, future evolutionary studies of B. burgdorferi s.l. genomes may include, for example, reconstructing genome-wide linkage maps based on SNPs using population genomics, identifying adaptive genetic variations associated with a specific species, strain, or clinical isolate using phylogenomics, and testing ecological hypotheses by estimating population size, migration rate, strain frequencies, and genetic admixture using genome-level phylogeography. The main challenge will shift increasingly from the unavailability of genome sequences to a lack of theoretical models and informatics tools. Especially under-developed are models and tools that would allow for distinctly bacterial mechanisms of localized recombination, mixed selective forces including immune escape and host specialization, and enzootic transmission. In sum, comparative studies of worldwide Lyme disease endemics have made B. burgdorferi s.l. a model system for predicting risks of vector-borne diseases based on evolutionary and ecological principles (Keesing et al., 2010; Kurtenbach et al., 2006; Wood and Lafferty, 2013). With a theory-amenable genetic structure of worldwide distribution, well-defined population boundaries, and high diversity within populations, we are hopeful that B. burgdorferi s.l. will become a model system for understanding mechanisms of genome evolution in bacteria as well.
Highlights.
Genomes of Lyme disease pathogens vary in plasmid composition but has a conserved gene repertoire
Host-resistant PF54 genes contribute to host adaptation and clinical variations
Antigen genes including ospC, dbpA, vls, b08 and a07 are under diversifying selection
Borrelia range expansion leads to genome hybridization and new virulent strains
Patterns of genome evolution reflect mechanisms of Borrelia virulence and expansion
Acknowledgments
We thank two anonymous reviewers for careful, extensive, and constructive critiques of our draft. We thank Dr Lia Di of Hunter College for preparing figures, Dr Steve Norris and Surabhi Tyagi of University of Texas Health Sciences Center for providing protein sequences for the inference of Borrelia phylogeny, and Dr Sherwood Casjens of University of Utah School of Medicine for analyzing the PF54 gene evolution. This work was supported by the Public Health Service Awards AI107955 from the National Institute of Allergy and Infectious Diseases (NIAID) and MD007599 (to Hunter College) from the National Institute on Minority Health and Health Disparities (NIMHD) of the National Institutes of Health (NIH). The content of this manuscript is solely the responsibility of the authors and do not necessarily represent the official views of NIAID, NIMHD, or NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Angiuoli SV, Salzberg SL. Mugsy: fast multiple alignment of closely related whole genomes. Bioinforma. Oxf. Engl. 2011;27:334–342. doi: 10.1093/bioinformatics/btq665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari MA, Didelot X. Inference of the properties of the recombination process from whole bacterial genomes. Genetics. 2014;196:253–265. doi: 10.1534/genetics.113.157172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avise JC. Phylogeography: The History and Formation of Species. Harvard University Press; 2000. [Google Scholar]
- Barton NH. Linkage and the limits to natural selection. Genetics. 1995;140:821–841. doi: 10.1093/genetics/140.2.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton NH. Why sex and recombination? Cold Spring Harb. Symp. Quant. Biol. 2009;74:187–195. doi: 10.1101/sqb.2009.74.030. [DOI] [PubMed] [Google Scholar]
- Bernhart SH, Hofacker IL, Will S, Gruber AR, Stadler PF. RNAalifold: improved consensus structure prediction for RNA alignments. BMC Bioinformatics. 2008;9:474. doi: 10.1186/1471-2105-9-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner EV, Kurilshikov AM, Stronin OV, Fomenko NV. Whole-Genome Sequencing of Borrelia garinii BgVir, Isolated from Taiga Ticks (Ixodes persulcatus) J. Bacteriol. 2012;194:5713. doi: 10.1128/JB.01360-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisson D, Drecktrah D, Eggers CH, Samuels DS. Genetics of Borrelia burgdorferi. Annu. Rev. Genet. 2012;46:515–536. doi: 10.1146/annurev-genet-011112-112140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisson D, Dykhuizen DE. ospC Diversity in Borrelia burgdorferi Different Hosts Are Different Niches. Genetics. 2004;168:713–722. doi: 10.1534/genetics.104.028738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisson D, Vandermause MF, Meece JK, Reed KD, Dykhuizen DE. Evolution of Northeastern and Midwestern Borrelia burgdorferi, United States. Emerg. Infect. Dis. 2010;16:911–917. doi: 10.3201/eid1606.090329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohée S, Janky R, Abdel-Sater F, Vanderstocken G, André B, Helden Jvan. Unraveling networks of co-regulated genes on the sole basis of genome sequences. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caesar JJE, Wallich R, Kraiczy P, Zipfel PF, Lea SM. Further structural insights into the binding of complement factor H by complement regulator-acquiring surface protein 1 (CspA) of Borrelia burgdorferi. Acta Crystallograph. Sect. F Struct. Biol. Cryst. Commun. 2013;69:629–633. doi: 10.1107/S1744309113012748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caimano MJ, Iyer R, Eggers CH, Gonzalez C, Morton EA, Gilbert MA, Schwartz I, Radolf JD. Analysis of the RpoS regulon in Borrelia burgdorferi in response to mammalian host signals provides insight into RpoS function during the enzootic cycle. Mol. Microbiol. 2007;65:1193–1217. doi: 10.1111/j.1365-2958.2007.05860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S, Palmer N, Van Vugt R, Mun Huang W, Stevenson B, Rosa P, Lathigra R, Sutton G, Peterson J, Dodson RJ, Haft D, Hickey E, Gwinn M, White OM, Fraser C. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 2000;35:490–516. doi: 10.1046/j.1365-2958.2000.01698.x. [DOI] [PubMed] [Google Scholar]
- Casjens SR, Fraser-Liggett CM, Mongodin EF, Qiu W-G, Dunn JJ, Luft BJ, Schutzer SE. Whole Genome Sequence of an Unusual Borrelia burgdorferi Sensu Lato Isolate. J. Bacteriol. 2011a;193:1489–1490. doi: 10.1128/JB.01521-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens SR, Mongodin EF, Qiu W-G, Dunn JJ, Luft BJ, Fraser-Liggett CM, Schutzer SE. Whole-Genome Sequences of Two Borrelia afzelii and Two Borrelia garinii Lyme Disease Agent Isolates. J. Bacteriol. 2011b;193:6995–6996. doi: 10.1128/JB.05951-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens SR, Mongodin EF, Qiu W-G, Luft BJ, Schutzer SE, Gilcrease EB, Huang WM, Vujadinovic M, Aron JK, Vargas LC, Freeman S, Radune D, Weidman JF, Dimitrov GI, Khouri HM, Sosa JE, Halpin RA, Dunn JJ, Fraser CM. Genome Stability of Lyme Disease Spirochetes: Comparative Genomics of Borrelia burgdorferi Plasmids. PLoS ONE. 2012;7:e33280. doi: 10.1371/journal.pone.0033280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darty K, Denise A, Ponty Y. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinforma. Oxf. Engl. 2009;25:1974–1975. doi: 10.1093/bioinformatics/btp250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan PH, Ochman H, Moran NA. Sequence Conservation and Functional Constraint on Intergenic Spacers in Reduced Genomes of the Obligate Symbiont Buchnera. PLoS Genet. 2011;7:e1002252. doi: 10.1371/journal.pgen.1002252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delihas N. Intergenic regions of Borrelia plasmids contain phylogenetically conserved RNA secondary structure motifs. BMC Genomics. 2009;10:101. doi: 10.1186/1471-2164-10-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong EF. Microbial population genomics and ecology: the road ahead. Environ. Microbiol. 2004;6:875–878. doi: 10.1111/j.1462-2920.2004.00668.x. [DOI] [PubMed] [Google Scholar]
- Desjardins P, Picard B, Kaltenböck B, Elion J, Denamur E. Sex in Escherichia coli does not disrupt the clonal structure of the population: evidence from random amplified polymorphic DNA and restriction-fragment-length polymorphism. J. Mol. Evol. 1995;41:440–448. doi: 10.1007/BF00160315. [DOI] [PubMed] [Google Scholar]
- Di L, Pagan PE, Martin CI. BorreliaBase: a phylogeny-centered browser of Borrelia genomes. (Submitted) 2014 doi: 10.1186/1471-2105-15-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X, Lawson D, Darling A, Falush D. Inference of homologous recombination in bacteria using whole-genome sequences. Genetics. 2010;186:1435–1449. doi: 10.1534/genetics.110.120121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X, Maiden MCJ. Impact of recombination on bacterial evolution. Trends Microbiol. 2010;18:315–322. doi: 10.1016/j.tim.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diuk-Wasser MA, Hoen AG, Cislo P, Brinkerhoff R, Hamer SA, Rowland M, Cortinas R, Vourc’h G, Melton F, Hickling GJ, Tsao JI, Bunikis J, Barbour AG, Kitron U, Piesman J, Fish D. Human risk of infection with Borrelia burgdorferi, the Lyme disease agent, in eastern United States. Am. J. Trop. Med. Hyg. 2012;86:320–327. doi: 10.4269/ajtmh.2012.11-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati C, Hiller NL, Tettelin H, Muzzi A, Croucher NJ, Angiuoli SV, Oggioni M, Hotopp JCD, Hu FZ, Riley DR, Covacci A, Mitchell TJ, Bentley SD, Kilian M, Ehrlich GD, Rappuoli R, Moxon ER, Masignani V. Structure and dynamics of the pan-genome of Streptococcus pneumoniae and closely related species. Genome Biol. 2010;11:R107. doi: 10.1186/gb-2010-11-10-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykhuizen DE, Baranton G. The implications of a low rate of horizontal transfer in Borrelia. Trends Microbiol. 2001;9:344–350. doi: 10.1016/s0966-842x(01)02066-2. [DOI] [PubMed] [Google Scholar]
- Dykhuizen DE, Brisson D, Sandigursky S, Wormser GP, Nowakowski J, Nadelman RB, Schwartz I. The propensity of different Borrelia burgdorferi sensu stricto genotypes to cause disseminated infections in humans. Am. J. Trop. Med. Hyg. 2008;78:806–810. [PMC free article] [PubMed] [Google Scholar]
- Dykhuizen DE, Green L. Recombination in Escherichia coli and the definition of biological species. J. Bacteriol. 1991;173:7257–7268. doi: 10.1128/jb.173.22.7257-7268.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykhuizen DE, Polin DS, Dunn JJ, Wilske B, Preac-Mursic V, Dattwyler RJ, Luft BJ. Borrelia burgdorferi is clonal: implications for taxonomy and vaccine development. Proc. Natl. Acad. Sci. U. S. A. 1993;90:10163–10167. doi: 10.1073/pnas.90.21.10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. A model of the statistical power of comparative genome sequence analysis. PLoS Biol. 2005;3:e10. doi: 10.1371/journal.pbio.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SV. Is a new and general theory of molecular systematics emerging? Evol. Int. J. Org. Evol. 2009;63:1–19. doi: 10.1111/j.1558-5646.2008.00549.x. [DOI] [PubMed] [Google Scholar]
- Eisen JA. Phylogenomics: Improving Functional Predictions for Uncharacterized Genes by Evolutionary Analysis. Genome Res. 1998;8:163–167. doi: 10.1101/gr.8.3.163. [DOI] [PubMed] [Google Scholar]
- Ellegren H. Genome sequencing and population genomics in non-model organisms. Trends Ecol. Evol. 2014;29:51–63. doi: 10.1016/j.tree.2013.09.008. [DOI] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Inferring Phylogenies. Sinauer Associates, Incorporated. 2004 [Google Scholar]
- Fraser C, Hanage WP, Spratt BG. Recombination and the nature of bacterial speciation. Science. 2007;315:476–480. doi: 10.1126/science.1127573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, Gwinn M, Dougherty B, Tomb J-F, Fleischmann RD, Richardson D, Peterson J, Kerlavage AR, Quackenbush J, Salzberg S, Hanson M, Vugt Rvan, Palmer N, Adams MD, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fujii C, Cotton MD, Horst K, Roberts K, Hatch B, Smith HO, Venter JC. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- Gatewood AG, Liebman KA, Vourc’h G, Bunikis J, Hamer SA, Cortinas R, Melton F, Cislo P, Kitron U, Tsao J, Barbour AG, Fish D, Diuk-Wasser MA. Climate and Tick Seasonality Are Predictors of Borrelia burgdorferi Genotype Distribution. Appl. Environ. Microbiol. 2009;75:2476–2483. doi: 10.1128/AEM.02633-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore RD, Jr., Howison RR, Schmit VL, Carroll JA. Borrelia burgdorferi expression of the bba64, bba65, bba66, and bba73 genes in tissues during persistent infection in mice. Microb. Pathog. 2008;45:355–360. doi: 10.1016/j.micpath.2008.08.006. [DOI] [PubMed] [Google Scholar]
- Glöckner G, Lehmann R, Romualdi A, Pradella S, Schulte-Spechtel U, Schilhabel M, Wilske B, Sühnel J, Platzer M. Comparative analysis of the Borrelia garinii genome. Nucleic Acids Res. 2004;32:6038–6046. doi: 10.1093/nar/gkh953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glöckner G, Schulte-Spechtel U, Schilhabel M, Felder M, Sühnel J, Wilske B, Platzer M. Comparative genome analysis: selection pressure on the Borrelia vls cassettes is essential for infectivity. BMC Genomics. 2006;7:211. doi: 10.1186/1471-2164-7-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves CJ, Ros VID, Stevenson B, Sniegowski PD, Brisson D. Natural Selection Promotes Antigenic Evolvability. PLoS Pathog. 2013;9:e1003766. doi: 10.1371/journal.ppat.1003766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman DS, Dykhuizen DE. Clonal divergence in Escherichia coli as a result of recombination, not mutation. Science. 1994;266:1380–1383. doi: 10.1126/science.7973728. [DOI] [PubMed] [Google Scholar]
- Guttman DS, Stavrinides J. Population Genomics of Bacteria. In: Robinson DA, Falush D, Feil EJ, editors. Bacterial Population Genetics in Infectious Disease. Hoboken, New Jersey: John Wiley & Sons, Inc; 2010. [Google Scholar]
- Hallström T, Siegel C, Mörgelin M, Kraiczy P, Skerka C, Zipfel PF. CspA from Borrelia burgdorferi inhibits the terminal complement pathway. mBio. 2013a;4 doi: 10.1128/mBio.00481-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallström T, Siegel C, Mörgelin M, Kraiczy P, Skerka C, Zipfel PF. CspA from Borrelia burgdorferi inhibits the terminal complement pathway. mBio. 2013b;4 doi: 10.1128/mBio.00481-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerschmidt C, Koenigs A, Siegel C, Hallström T, Skerka C, Wallich R, Zipfel PF, Kraiczy P. Versatile Roles of CspA Orthologs in Complement Inactivation of Serum-Resistant Lyme Disease Spirochetes. Infect. Immun. 2014;82:380–392. doi: 10.1128/IAI.01094-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanincova K, Mukherjee P, Ogden NH, Margos G, Wormser GP, Reed KD, Meece JK, Vandermause MF, Schwartz I. Multilocus Sequence Typing of Borrelia burgdorferi Suggests Existence of Lineages with Differential Pathogenic Properties in Humans. PLoS ONE. 2013;8:e73066. doi: 10.1371/journal.pone.0073066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl DL, Clark AG. Principles of Population Genetics. Sinauer Associates, Incorporated. 2007 [Google Scholar]
- Harvey PH, Pagel MD. The comparative method in evolutionary biology. Oxford University Press; 1998. [Google Scholar]
- Haven J, Vargas LC, Mongodin EF, Xue V, Hernandez Y, Pagan P, Fraser-Liggett CM, Schutzer SE, Luft BJ, Casjens SR, Qiu W-G. Pervasive Recombination and Sympatric Genome Diversification Driven by Frequency-Dependent Selection in Borrelia burgdorferi, the Lyme Disease Bacterium. Genetics. 2011;189:951–966. doi: 10.1534/genetics.111.130773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG, Robertson A. The effect of linkage on limits to artificial selection. Genet. Res. 1966;8:269–294. [PubMed] [Google Scholar]
- Hoen AG, Margos G, Bent SJ, Diuk-Wasser MA, Barbour A, Kurtenbach K, Fish D. Phylogeography of Borrelia burgdorferi in the eastern United States reflects multiple independent Lyme disease emergence events. Proc. Natl. Acad. Sci. 2009;106:15013–15018. doi: 10.1073/pnas.0903810106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD. The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. TIG. 2002;18:486. doi: 10.1016/s0168-9525(02)02722-1. [DOI] [PubMed] [Google Scholar]
- Ivanova LB, Tomova A, González-Acuña D, Murúa R, Moreno CX, Hernández C, Cabello J, Cabello C, Daniels TJ, Godfrey HP, Cabello FC. Borrelia chilensis, a new member of the Borrelia burgdorferi sensu lato complex that extends the range of this genospecies in the Southern Hemisphere. Environ. Microbiol. 2013 doi: 10.1111/1462-2920.12310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang B, Yao H, Tong Y, Yang X, Huang Y, Jiang J, Cao W. Genome Sequence of Borrelia garinii Strain NMJW1, Isolated from China. J. Bacteriol. 2012;194:6660–6661. doi: 10.1128/JB.01844-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang B-G, Zheng Y-C, Tong Y-G, Jia N, Huo Q-B, Fan H, Ni X-B, Ma L, Yang XF, Jiang J-F, Cao W-C. Genome sequence of Borrelia afzelii Strain HLJ01, isolated from a patient in China. J. Bacteriol. 2012;194:7014–7015. doi: 10.1128/JB.01863-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katara P, Grover A, Sharma V. Phylogenetic footprinting: a boost for microbial regulatory genomics. Protoplasma. 2012;249:901–907. doi: 10.1007/s00709-011-0351-9. [DOI] [PubMed] [Google Scholar]
- Keesing F, Belden LK, Daszak P, Dobson A, Harvell CD, Holt RD, Hudson P, Jolles A, Jones KE, Mitchell CE, Myers SS, Bogich T, Ostfeld RS. Impacts of biodiversity on the emergence and transmission of infectious diseases. Nature. 2010;468:647–652. doi: 10.1038/nature09575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. The Neutral Theory of Molecular Evolution. Cambridge University Press; 1984. [Google Scholar]
- Koenigs A, Hammerschmidt C, Jutras BL, Pogoryelov D, Barthel D, Skerka C, Kugelstadt D, Wallich R, Stevenson B, Zipfel PF, Kraiczy P. BBA70 of Borrelia burgdorferi is a novel plasminogen-binding protein. J. Biol. Chem. 2013;288:25229–25243. doi: 10.1074/jbc.M112.413872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Filipski AJ, Battistuzzi FU, Pond SLK, Tamura K. Statistics and Truth in Phylogenomics. Mol. Biol. Evol. 2012;29:457–472. doi: 10.1093/molbev/msr202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtenbach K, Hanincová K, Tsao JI, Margos G, Fish D, Ogden NH. Fundamental processes in the evolutionary ecology of Lyme borreliosis. Nat. Rev. Microbiol. 2006;4:660–669. doi: 10.1038/nrmicro1475. [DOI] [PubMed] [Google Scholar]
- Li YF, Costello JC, Holloway AK, Hahn MW. “Reverse ecology” and the power of population genomics. Evol. Int. J. Org. Evol. 2008;62:2984–2994. doi: 10.1111/j.1558-5646.2008.00486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang FT, Caimano MJ, Radolf JD, Fikrig E. Borrelia burgdorferi outer surface protein (osp) B expression independent of ospA. Microb. Pathog. 2004;37:35–40. doi: 10.1016/j.micpath.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Liu L, Yu L, Kubatko L, Pearl DK, Edwards SV. Coalescent methods for estimating phylogenetic trees. Mol. Phylogenet. Evol. 2009;53:320–328. doi: 10.1016/j.ympev.2009.05.033. [DOI] [PubMed] [Google Scholar]
- Margos G, Gatewood AG, Aanensen DM, Hanincová K, Terekhova D, Vollmer SA, Cornet M, Piesman J, Donaghy M, Bormane A, Hurn MA, Feil EJ, Fish D, Casjens S, Wormser GP, Schwartz I, Kurtenbach K. MLST of housekeeping genes captures geographic population structure and suggests a European origin of Borrelia burgdorferi. Proc. Natl. Acad. Sci. 2008;105:8730–8735. doi: 10.1073/pnas.0800323105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margos G, Hojgaard A, Lane RS, Cornet M, Fingerle V, Rudenko N, Ogden N, Aanensen DM, Fish D, Piesman J. Multilocus sequence analysis of Borrelia bissettii strains from North America reveals a new Borrelia species, Borrelia kurtenbachii. Ticks Tick-Borne Dis. 2010;1:151–158. doi: 10.1016/j.ttbdis.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margos G, Tsao JI, Castillo-Ramírez S, Girard YA, Hamer SA, Hoen AG, Lane RS, Raper SL, Ogden NH. Two Boundaries Separate Borrelia burgdorferi Populations in North America. Appl. Environ. Microbiol. 2012;78:6059–6067. doi: 10.1128/AEM.00231-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margos G, Vollmer SA, Ogden NH, Fish D. Population genetics, taxonomy, phylogeny and evolution of Borrelia burgdorferi sensu lato. Infect. Genet. Evol. 2011;11:1545–1563. doi: 10.1016/j.meegid.2011.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- McVean G, Awadalla P, Fearnhead P. A Coalescent-Based Method for Detecting and Estimating Recombination From Gene Sequences. Genetics. 2002;160:1231–1241. doi: 10.1093/genetics/160.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R. The microbial pan-genome. Curr. Opin. Genet. Dev. 2005;15:589–594. doi: 10.1016/j.gde.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Milkman R, Bridges MM. Molecular evolution of the Escherichia coli chromosome. III. Clonal frames. Genetics. 1990;126:505–517. doi: 10.1093/genetics/126.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongodin EF, Casjens SR, Bruno JF, Xu Y, Drabek EF, Riley DR, Cantarel BL, Pagan PE, Hernandez YA, Vargas LC, Dunn JJ, Schutzer SE, Fraser CM, Qiu W-G, Luft BJ. Inter- and intra-specific pan-genomes of Borrelia burgdorferi sensu lato: genome stability and adaptive radiation. BMC Genomics. 2013;14:693. doi: 10.1186/1471-2164-14-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlon H, Kemps BD, Plotkin JB, Brisson D. Explosive radiation of a bacterial species group. Evol. Int. J. Org. Evol. 2012;66:2577–2586. doi: 10.1111/j.1558-5646.2012.01598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HJ. The relation of recombination to muational advance. Mutat. Res. 1964;106:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- Norris SJ, Lin T. Out of the Woods: the Remarkable Genomes of the Genus Borrelia. J. Bacteriol. 2011;193:6812–6814. doi: 10.1128/JB.06317-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden NH, Mechai S, Margos G. Changing geographic ranges of ticks and tick-borne pathogens: drivers, mechanisms and consequences for pathogen diversity. Front. Cell. Infect. Microbiol. 2013;3:46. doi: 10.3389/fcimb.2013.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostfeld R. Lyme Disease: The Ecology of a Complex System. Oxford: University Press; 2010. [Google Scholar]
- Pagani I, Liolios K, Jansson J, Chen I-MA, Smirnova T, Nosrat B, Markowitz VM, Kyrpides NC. The Genomes OnLine Database (GOLD) v.4: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2012;40:D571–579. doi: 10.1093/nar/gkr1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Pritchard JK. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012;8:e1002967. doi: 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Przeworski M. Linkage Disequilibrium in Humans: Models and Data. Am. J. Hum. Genet. 2001;69:1–14. doi: 10.1086/321275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüfer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, Heinze A, Renaud G, Sudmant PH, de Filippo C, Li H, Mallick S, Dannemann M, Fu Q, Kircher M, Kuhlwilm M, Lachmann M, Meyer M, Ongyerth M, Siebauer M, Theunert C, Tandon A, Moorjani P, Pickrell J, Mullikin JC, Vohr SH, Green RE, Hellmann I, Johnson PLF, Blanche H, Cann H, Kitzman JO, Shendure J, Eichler EE, Lein ES, Bakken TE, Golovanova LV, Doronichev VB, Shunkov MV, Derevianko AP, Viola B, Slatkin M, Reich D, Kelso J, Pääbo S. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu W-G. Wide Distribution of a High-Virulence Borrelia burgdorferi Clone in Europe and North America. Emerg. Infect. Dis. 2008;14:1097–1104. doi: 10.3201/eid1407.070880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu W-G, Schutzer SE, Bruno JF, Attie O, Xu Y, Dunn JJ, Fraser CM, Casjens SR, Luft BJ. Genetic exchange and plasmid transfers in Borrelia burgdorferi sensu stricto revealed by three-way genome comparisons and multilocus sequence typing. Proc. Natl. Acad. Sci. U. S. A. 2004;101:14150–14155. doi: 10.1073/pnas.0402745101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. 2013 [Google Scholar]
- Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- Roux C, Tsagkogeorga G, Bierne N, Galtier N. Crossing the species barrier: genomic hotspots of introgression between two highly divergent Ciona intestinalis species. Mol. Biol. Evol. 2013;30:1574–1587. doi: 10.1093/molbev/mst066. [DOI] [PubMed] [Google Scholar]
- Rudenko N, Golovchenko M, Grubhoffer L, Oliver JH. Borrelia carolinensis sp.nov., a New (14th) Member of the Borrelia burgdorferi Sensu Lato Complex from the Southeastern Region of the United States. J. Clin. Microbiol. 2009;47:134–141. doi: 10.1128/JCM.01183-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko N, Golovchenko M, Hönig V, Mallátová N, Krbková L, Mikulásek P, Fedorova N, Belfiore NM, Grubhoffer L, Lane RS, Oliver JH., Jr. Detection of Borrelia burgdorferi sensu stricto ospC alleles associated with human lyme borreliosis worldwide in non-human-biting tick Ixodes affinis and rodent hosts in Southeastern United States. Appl. Environ. Microbiol. 2013;79:1444–1453. doi: 10.1128/AEM.02749-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels DS. Gene Regulation in Borrelia burgdorferi. Annu. Rev. Microbiol. 2011;65:479–499. doi: 10.1146/annurev.micro.112408.134040. [DOI] [PubMed] [Google Scholar]
- Schmit VL, Patton TG, Gilmore RD., Jr. Analysis of Borrelia burgdorferi Surface Proteins as Determinants in Establishing Host Cell Interactions. Front. Microbiol. 2011;2:141. doi: 10.3389/fmicb.2011.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutzer SE, Fraser-Liggett CM, Casjens SR, Qiu W-G, Dunn JJ, Mongodin EF, Luft BJ. Whole-Genome Sequences of Thirteen Isolates of Borrelia burgdorferi. J. Bacteriol. 2011;193:1018–1020. doi: 10.1128/JB.01158-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutzer SE, Fraser-Liggett CM, Qiu W-G, Kraiczy P, Mongodin EF, Dunn JJ, Luft BJ, Casjens SR. Whole-Genome Sequences of Borrelia bissettii, Borrelia valaisiana, and Borrelia spielmanii. J. Bacteriol. 2012;194:545–546. doi: 10.1128/JB.06263-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seinost G, Dykhuizen DE, Dattwyler RJ, Golde WT, Dunn JJ, Wang IN, Wormser GP, Schriefer ME, Luft BJ. Four clones of Borrelia burgdorferi sensu stricto cause invasive infection in humans. Infect. Immun. 1999;67:3518–3524. doi: 10.1128/iai.67.7.3518-3524.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanek G, Wormser GP, Gray J, Strle F. Lyme borreliosis. The Lancet. 2012;379:461–473. doi: 10.1016/S0140-6736(11)60103-7. [DOI] [PubMed] [Google Scholar]
- Stoletzki N, Eyre-Walker A. Estimation of the neutrality index. Mol. Biol. Evol. 2011;28:63–70. doi: 10.1093/molbev/msq249. [DOI] [PubMed] [Google Scholar]
- Su F, Ou H-Y, Tao F, Tang H, Xu P. PSP: rapid identification of orthologous coding genes under positive selection across multiple closely related prokaryotic genomes. BMC Genomics. 2013;14:924. doi: 10.1186/1471-2164-14-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N. Evolutionary genetics of human paleo-population, in: Mechanisms of Molecular Evolution: Introduction to Molecular Paleopopulation Biology. Japan Scientific Societies Press; 1993. pp. 1–21. [Google Scholar]
- Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, Deboy RT, Davidsen TM, Mora M, Scarselli M, Margarit y Ros I, Peterson JD, Hauser CR, Sundaram JP, Nelson WC, Madupu R, Brinkac LM, Dodson RJ, Rosovitz MJ, Sullivan SA, Daugherty SC, Haft DH, Selengut J, Gwinn ML, Zhou L, Zafar N, Khouri H, Radune D, Dimitrov G, Watkins K, O’Connor KJB, Smith S, Utterback TR, White O, Rubens CE, Grandi G, Madoff LC, Kasper DL, Telford JL, Wessels MR, Rappuoli R, Fraser CM. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. U. S. A. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H, Riley D, Cattuto C, Medini D. Comparative genomics: the bacterial pan-genome. Curr. Opin. Microbiol. 2008;11:472–477. doi: 10.1016/j.mib.2008.09.006. [DOI] [PubMed] [Google Scholar]
- Tilly K, Bestor A, Rosa PA. Lipoprotein succession in Borrelia burgdorferi: similar but distinct roles for OspC and VlsE at different stages of mammalian infection. Mol. Microbiol. 2013;89:216–227. doi: 10.1111/mmi.12271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, Bingen E, Bonacorsi S, Bouchier C, Bouvet O, Calteau A, Chiapello H, Clermont O, Cruveiller S, Danchin A, Diard M, Dossat C, Karoui ME, Frapy E, Garry L, Ghigo JM, Gilles AM, Johnson J, Le Bouguénec C, Lescat M, Mangenot S, Martinez-Jéhanne V, Matic I, Nassif X, Oztas S, Petit MA, Pichon C, Rouy Z, Ruf CS, Schneider D, Tourret J, Vacherie B, Vallenet D, Médigue C, Rocha EPC, Denamur E. Organised Genome Dynamics in the Escherichia coli Species Results in Highly Diverse Adaptive Paths. PLoS Genet. 2009;5:e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernot B, Akey JM. Resurrecting Surviving Neandertal Lineages from Modern Human Genomes. Science. 2014 doi: 10.1126/science.1245938. 1245938. [DOI] [PubMed] [Google Scholar]
- Vollmer SA, Feil EJ, Chu C-Y, Raper SL, Cao W-C, Kurtenbach K, Margos G. Spatial spread and demographic expansion of Lyme borreliosis spirochaetes in Eurasia. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2013;14:147–155. doi: 10.1016/j.meegid.2012.11.014. [DOI] [PubMed] [Google Scholar]
- Vos M, te Beek TAH, van Driel MA, Huynen MA, Eyre-Walker A, van Passel MWJ. ODoSE: a webserver for genome-wide calculation of adaptive divergence in prokaryotes. PLoS ONE. 2013;8:e62447. doi: 10.1371/journal.pone.0062447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong HB, Canham CD, Fonseca DM, Brisson D, Morin PJ, Smouse PE, Ostfeld RS. Occurrence and transmission efficiencies of Borrelia burgdorferi ospC types in avian and mammalian wildlife. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2013 doi: 10.1016/j.meegid.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang I-N, Dykhuizen DE, Qiu W, Dunn JJ, Bosler EM, Luft BJ. Genetic Diversity of ospC in a Local Population of Borrelia burgdorferi sensu stricto. Genetics. 1999;151:15–30. doi: 10.1093/genetics/151.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Hey J. Estimating divergence parameters with small samples from a large number of loci. Genetics. 2010;184:363–379. doi: 10.1534/genetics.109.110528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattam AR, Abraham D, Dalay O, Disz TL, Driscoll T, Gabbard JL, Gillespie JJ, Gough R, Hix D, Kenyon R, Machi D, Mao C, Nordberg EK, Olson R, Overbeek R, Pusch GD, Shukla M, Schulman J, Stevens RL, Sullivan DE, Vonstein V, Warren A, Will R, Wilson MJC, Yoo HS, Zhang C, Zhang Y, Sobral BW. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt1099. gkt1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker RJ, Banfield JF. Population genomics in natural microbial communities. Trends Ecol. Evol. 2006;21:508–516. doi: 10.1016/j.tree.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Wickham H. Ggplot2 elegant graphics for data analysis. New York: Springer, Dordrecht; 2009. [Google Scholar]
- Wiehe T, Mountain J, Parham P, Slatkin M. Distinguishing recombination and intragenic gene conversion by linkage disequilibrium patterns. Genet. Res. 2000;75:61–73. doi: 10.1017/s0016672399004036. [DOI] [PubMed] [Google Scholar]
- Wiuf C, Hein J. The Coalescent With Gene Conversion. Genetics. 2000;155:451–462. doi: 10.1093/genetics/155.1.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood CL, Lafferty KD. Biodiversity and disease: a synthesis of ecological perspectives on Lyme disease transmission. Trends Ecol. Evol. 2013;28:239–247. doi: 10.1016/j.tree.2012.10.011. [DOI] [PubMed] [Google Scholar]
- Wu D, Jospin G, Eisen JA. Systematic Identification of Gene Families for Use as “Markers” for Phylogenetic and Phylogeny-Driven Ecological Studies of Bacteria and Archaea and Their Major Subgroups. PLoS ONE. 2013;8:e77033. doi: 10.1371/journal.pone.0077033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Eisen JA. A simple, fast, and accurate method of phylogenomic inference. Genome Biol. 2008;9:R151. doi: 10.1186/gb-2008-9-10-r151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wywial E, Haven J, Casjens SR, Hernandez YA, Singh S, Mongodin EF, Fraser-Liggett CM, Luft BJ, Schutzer SE, Qiu W-G. Fast, adaptive evolution at a bacterial host-resistance locus: the PFam54 gene array in Borrelia burgdorferi. Gene. 2009;445:26–37. doi: 10.1016/j.gene.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, He M, He JJ, Yang XF. Role of the surface lipoprotein BBA07 in the enzootic cycle of Borrelia burgdorferi. Infect. Immun. 2010a;78:2910–2918. doi: 10.1128/IAI.00372-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, He M, Pang X, Xu ZC, Piesman J, Yang XF. Characterization of the highly regulated antigen BBA05 in the enzootic cycle of Borrelia burgdorferi. Infect. Immun. 2010b;78:100–107. doi: 10.1128/IAI.01008-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Qin J, Promnares K, Kariu T, Anderson JF, Pal U. Novel microbial virulence factor triggers murine lyme arthritis. J. Infect. Dis. 2013;207:907–918. doi: 10.1093/infdis/jis930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]