Abstract
Substantial progress has been made in identifying susceptibility variants for age-related macular degeneration (AMD). The majority of research to identify genetic variants associated with AMD has focused on nuclear genetic variation. While there is some evidence that mitochondrial genetic variation contributes to AMD susceptibility, to date, these studies have been limited to populations of European descent resulting in a lack of data in diverse populations. A major goal of the Epidemiologic Architecture for Genes Linked to Environment (EAGLE) study is to describe the underlying genetic architecture of common, complex diseases across diverse populations. This present study sought to determine if mitochondrial genetic variation influences risk of AMD across diverse populations. We performed a genetic association study to investigate the contribution of mitochondrial DNA variation to AMD risk. We accessed samples from the National Health and Nutrition Examination Surveys, a U.S population-based, cross-sectional survey collected without regard to health status. AMD cases and controls were selected from the Third NHANES and NHANES 2007-2008 datasets which include non-Hispanic whites, non-Hispanic blacks, and Mexican Americans. AMD cases were defined as those > 60 years of age with early/late AMD, as determined by fundus photography. Targeted genotyping was performed for 63 mitochondrial SNPs and participants were then classified into mitochondrial haplogroups. We used logistic regression assuming a dominant genetic model adjusting for age, sex, body mass index, and smoking status (ever vs. never). Regressions and meta-analyses were performed for individual SNPs and mitochondrial haplogroups J, T, and U. We identified five SNPs associated with AMD in Mexican Americans at p < 0.05, including three located in the control region (mt16111, mt16362, and mt16319), one in MT-RNR2 (mt1736), and one in MT-ND4 (mt12007). No mitochondrial variant or haplogroup was significantly associated in non-Hispanic blacks or non-Hispanic whites in the final meta-analysis. This study provides further evidence that mitochondrial variation plays a role in susceptibility to AMD and contributes to the knowledge of the genetic architecture of AMD in Mexican Americans.
1. Introduction
Vision loss and blindness are powerful detriments to the economic and social well-being of individuals. Economically it costs the United States upwards of $51 billion a year in medical expenses and lost worker productivity.1 More importantly, loss of vision has been ranked as one condition with the greatest impact on an individual’s day-to-day life.2 Age-related macular degeneration (AMD), a disease of the retina, afflicts more seniors in developed countries than any other form of blindness. The number of incident AMD cases is expected to grow from 11 million today to approximately 22 million by the year 2050.3
AMD is a late-onset, multifactorial disease that eventually results in loss of central vision. The retina is a photosensitive tissue that lines the back of the human eye. It collects visual data in the form of photons and through a series of chemical and electrical events, sends a neurological impulse to regions in the brain which can then interpret them as images.4 Central to these images is a region of the retina called the macula, responsible for center vision, which contains a high concentration of photoreceptor cells responsible for color vision (i.e. cones).
Clinically AMD is defined as the degeneration of the macula in which central vision becomes impaired. It occurs in two subtypes termed early and late AMD. Early AMD is typically defined as eyes that present with mild to moderate drusen deposits and pigment abnormalities. Late stage AMD is subdivided into dry (atrophic) and wet (exudative) AMD. Dry AMD is the result of geographic atrophy of the retinal pigment epithelial (RPE) layer which lies directly under the retina and provides metabolic and cellular support to the retina and photoreceptors. Exudative AMD leads to vision loss due to abnormal blood vessel growth. These abnormal blood vessels can break and cause blood to leak below the surface of the retina leading to irreversible damage to the macula and subsequent blindness.
Although AMD has a complex etiology, many environmental and genetic variables have been identified that alter the risk of this disease. Modifiable factors include smoking,5-9 body-mass-index (BMI), and blood lipid10-12 levels such as high density lipoprotein cholesterol (HDL-C). Non-modifiable factors include race/ethnicity, age,13 gender, and genetics. Individuals of European-descent are at highest risk with prevalence rates in individuals over the age of 40 years reaching 7.3% compared to African Americans (2.4%), Asian Americans (6.8%),14 and Mexican Americans (5.1%).15 In European Americans over the age of 80, nearly 1 out of 10 is likely to be diagnosed with some form of AMD.16 Major genetic risk loci include CFH,17-19 ARMS2,20,21 and C2/C322 which account for most of the known heritable risk of AMD. In all, 20 risk loci have been associated with risk of AMD, however, these only account for upwards of 60% of the heritable risk.23 Nearly all of these loci are located within the nuclear genome as there have been few studies investigating the potential role that mitochondrial genetic variation plays in the development of AMD.
In vitro studies have found that mitochondrial DNA (mtDNA) variants can affect the replication rate of the mitochondrial genome and thus mtDNA copy number.24 Mitochondria are both particularly sensitive to and a major contributor of cellular reactive oxygen species (ROS), which are a byproduct of oxidative phosphorylation. These free radicals play a large role in chronic inflammation, the complement system pathways, and cardiovascular disease.25 Exposure to excessive oxidative stress can lead to mitochondrial dysfunction in the RPE layer,26,27 a decrease in cellular bioenergetics imperative to photoreceptor initiation/maintenance,24,28,29 and susceptibility to apoptosis. Additionally, mitochondrial genetic variation has been associated with AMD risk in European Americans. MtDNA variants on mitochondrial haplogroup H, the most common European haplogroup, have been associated with decreased risk of AMD,30-32 while mitochondrial haplogroups J33 and T34 are associated with increased risk. Collectively, these data suggest that the health of ocular mitochondria may play a role in AMD pathology.
Determining the role that mitochondrial genetic variation plays in AMD risk across populations may provide new insights into the underlying disease pathology. More research is necessary to understand the genetic architecture of disease states in diverse populations. This study explores the contribution of mitochondrial genetic variation to AMD risk not only for European Americans, but also in African Americans and Mexican Americans, populations for which studies are few or missing.
2. Materials and Methods
2.1. Study population and genotyping
The National Health and Nutrition Examination Surveys (NHANES) are conducted by the National Center for Health Statistics (NCHS) at the Centers for Disease Control and Prevention (CDC). NHANES are U.S. population-based, cross-sectional surveys designed to ascertain non-institutionalized Americans without regard to health status. All NHANES include detailed demographic, health, lifestyle, laboratory, clinical, and physical examination data from study participants. Genetic NHANES consists of DNA samples collected from the Third NHANES (III) and subsets of Continuous NHANES (1999-2002 and 2007-2008), which began collection in 1999 and occurs annually. NHANES III was conducted in two phases between 1988-1994 and 1991-1994, with DNA samples having been collected in the second phase.35,36 The method of collection for Genetic NHANES has been previously described.37,38 We, as the Epidemiologic Architecture for Genes Linked to Environment (EAGLE) study, targeted a total of 63 mitochondrial SNPs for genotyping using Sequenom iPLEX® Gold MassArray as previously described.39
We accessed study participant data exclusively from NHANES III and NHANES 2007-2008, as NHANES 1999-2002 did not collect ophthalmologic data. NHANES III, of which 3,131 participants had available fundus photographs and laboratory measurements of serum cotinine (ng/mL), oversampled non-Hispanic blacks and Mexican-Americans. NHANES 2007-2008 oversampled Mexican Americans and other-Hispanic blacks and had a total of 3,172 participants who completed the fundus exam. Current smokers were defined as those responding “yes” to the question “do you smoke cigarettes now?” or those with serum cotinine levels > 15ng/ml.
All procedures were approved by the CDC Ethics Review Board and written informed consent was obtained from all participants. Because no identifying information is available to the investigators, Vanderbilt University’s Institutional Review Board determined that this study met the criteria of “non-human subjects.”
2.2 AMD phenotype definition
Participants over the age of 40 were selected to have a non-stereoscopic, 45° color fundus photograph taken of one randomly selected eye in NHANES III and a 45° non-mydriatic digital photo taken of both eyes in NHANES 2007-2008. Fundus photographs were graded according to a modified version of the Wisconsin Age-related Maculopathy Scale.40 Early AMD cases were at least 60 years of age and included participants with: 1) soft drusen, 2) depigmentation of the retinal pigment epithelium in the presence of soft and/or hard drusen, or 3) hyperpigmentation in the presence of soft and/or hard drusen. Late AMD included participants 60 years of age and older with 1) geographic atrophy, 2) sub-retinal hemorrhage, 3) sub-retinal fibrous scarring, or 4) sensory serous sub-retinal detachments. Controls were at least 60 years old with gradable retinal photographs showing an absence of hallmark AMD features. Controls were not excluded if they presented with another retinal disease.
2.3 Statistical analysis
We tested for an association between each individual mtSNP and mitochondrial haplogroups J, T, and U with AMD. Given that AMD largely occurs on a disease spectrum, data for early and late AMD was pooled for analyses so as to increase power to detect an association. Each mitochondrial variant was tested for an association with AMD using logistic regression assuming a dominant genetic model stratified by self-described race/ethnicity (e.g. non-Hispanic white, non-Hispanic black, and Mexican American). Of the SNPs that passed quality control (QC) standards (call rate >95%), a total of 55 SNPs were included in analyses for NHANES III and 60 SNPs were analyzed in NHANES 2007-2008. A total of 50 SNPs were available for meta-analysis. Haplogroups were assigned to each NHANES participant as previously described.39 Haplogroup analyses were conducted in the same manner as the individual mtSNPs but with participants identified as having either haplogroup J, T, and U each being compared to participants in all other haplogroups. All models were adjusted for age, sex, BMI, and smoking status (current versus ever/never). Analyses were conducted using SAS v9.2 via the Analytic Data Research by Email (ANDRE) portal of the CDC Research Data Center in Hyattsville, MD. All p-values presented are uncorrected for multiple testing.
3. Results
3.1 Population
The study population consisted of a total of 416 AMD cases (312 non-Hispanic whites, 37 non-Hispanic blacks, and 67 Mexican Americans) and 2,200 controls (1,349 non-Hispanic whites, 430 non-Hispanic blacks, and 421 Mexican Americans) 60 years or older at the time of examination (Table 1). In the combined NHANES III/2007-2008 dataset, cases were generally female, except in Mexican Americans (49% female), and overweight defined as BMI >25 kg/m2. Non-Hispanic black cases were nearly twice as likely to be smokers (62% smokers) compared to non-Hispanic white (29% smokers) and Mexican American (36%) cases. On average, controls were younger compared to cases across all race/ethnicities and were nearly as likely to be smokers compared to cases with the exception of non-Hispanic black cases (62% smokers) versus controls (50% smokers).
Table 1.
Combined NHANES III and 2007-2008 study population demographics by case/control status and race/ethnicity
| non-Hispanic whites | non-Hispanic blacks | Mexican Americans | ||||
|---|---|---|---|---|---|---|
|
|
||||||
| Case | Control | Case | Control | Case | Control | |
|
|
||||||
| N | 312 | 1349 | 37 | 430 | 67 | 421 |
| Age (years) | 76.0 | 71.0 | 69.2 | 67.8 | 69.0 | 67.1 |
| % Female | 60 | 51 | 57 | 49 | 49 | 48 |
| % Smoker | 29 | 30 | 62 | 50 | 36 | 31 |
| BMI (kg/m2) | 27.2 | 27.8 | 31.1 | 29.0 | 29.2 | 28.9 |
Means are presented unless otherwise noted.
3.2 Meta-analysis of mitochondrial variants
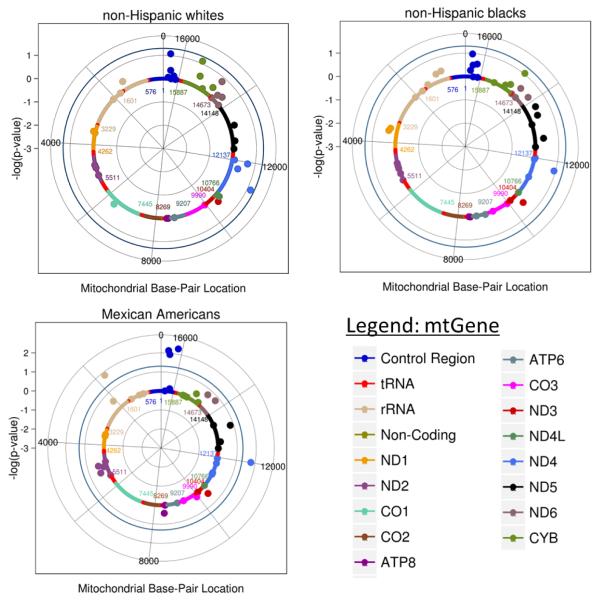
A total of 50 mitochondrial SNPs passed QC and were tested across the three race/ethnicities in NHANES III and NHANES 2007-2008. Not all SNPs were available across each population as some SNPs were monomorphic in one population or did not pass QC. Of these 50 SNPs, 41 were available for analysis in non-Hispanic whites, 44 in non-Hispanic blacks, and 42 Mexican Americans (Figure 1).
Figure 1. Meta-analysis single SNP association results by race/ethnicity.
Log(p) values were plotted using R. The outer blue circle represents a significance threshold of p = 0.05. SNPs are color coded by mitochondrial gene/regions as denoted in the legend. Cases/controls for the three populations are as follows: non-Hispanic whites (case = 312, control = 1,349), non-Hispanic blacks (case = 37, control = 430), and Mexican Americans (case = 67, control = 421).
In Mexican Americans, five mtSNPs were associated with AMD at p < 0.05 (Table 2). Of these, three are located in the mitochondrial control region, the non-coding region responsible for the initiation of transcription of the MT-genome: mt16111 (p = 0.005; OR = 2.90; 95% CI 1.38 – 6.11), mt16362 (p = 0.007; OR = 2.80; 95% CI 1.32 – 5.95), and mt16319 (p = 0.01; OR = 2.59; 95% CI 1.24 – 5.42). A synonymous variant, mt12007, located within the NADH dehydrogenase subunit 4 (MT-ND4) gene, was also found to increase risk of AMD in Mexican Americans (p = 0.018; OR = 2.47; CI 1.18 – 5.18). Lastly, mt1736 located in the mitochondrial 16S ribosomal RNA (MT-RNR2) gene, was found to be protective (p = 0.01; OR = 0.40; CI 0.19 – 0.83) in this population.
Table 2.
Mitochondrial genetic variants associated with AMD risk in Mexican Americans
| SNPID | Gene | OR | lower CI | upper CI | p-value | CA | CAF (%) |
Race/Ethnicity |
|---|---|---|---|---|---|---|---|---|
| mt16111 | control region | 2.90 | 1.38 | 6.11 | 0.005 | A | 0.31 | Mexican American |
| — | — | — | 0.98 | A | 0.01 | non-Hispanic white | ||
| — | — | — | 0.98 | A | 0.02 | non-Hispanic black | ||
| mt16362 | control region | 2.80 | 1.32 | 5.95 | 0.01 | C | 0.42 | Mexican American |
| 1.70 | 0.93 | 3.10 | 0.08 | C | 0.08 | non-Hispanic white | ||
| 2.29 | 0.84 | 6.27 | 0.10 | C | 0.13 | non-Hispanic black | ||
| mt16319 | control region | 2.59 | 1.24 | 5.42 | 0.01 | A | 0.34 | Mexican American |
| 0.42 | 0.05 | 3.52 | 0.43 | A | 0.01 | non-Hispanic white | ||
| 3.23 | 0.33 | 31.60 | 0.31 | A | 0.02 | non-Hispanic black | ||
| mt1736 | MT-RNR2 | 0.40 | 0.19 | 0.83 | 0.01 | A | 0.65 | Mexican American |
| — | — | — | 0.98 | A | 0.99 | non-Hispanic white | ||
| — | — | — | 0.99 | A | — | non-Hispanic black | ||
| mt12007 | MT-ND4 | 2.47 | 1.18 | 5.18 | 0.01 | A | 0.34 | Mexican American |
| 1.18 | 0.31 | 4.50 | 0.81 | A | 0.02 | non-Hispanic white | ||
| 0.91 | 0.11 | 7.50 | 0.93 | A | 0.06 | non-Hispanic black |
Most significant meta-analysis results for the model adjusted by age, sex, body mass index, and smoking status.
Results are listed for tests with the smallest p-value
Abbreviations: Odds Ratio (OR), confidence interval (CI), coded allele (CA), coded allele frequency (CAF) “—“ denotes genetic association tests with uninterpretable results due to very few case counts or monomorphic allele
In non-Hispanic blacks and non-Hispanic whites, no test of association was significant at p < 0.05 in the adjusted model following inclusion of individual study results into the meta-analysis.
3.3 Mitochondrial Haplogroups
Previous studies have suggested that mitochondrial haplogroups J, T, and U are associated with AMD.30-34 In NHANES III, none of the three haplogroups were associated with AMD at p < 0.05 although haplogroup J was associated in non-Hispanic whites at p = 0.057 (OR = 2.03; 95% CI 0.98 – 4.20). In NHANES 2007-2008, only haplogroup T was significantly associated at p < 0.05 in non-Hispanic whites (OR = 2.50; 95% CI 1.17 – 5.33). No haplogroup was found to be associated with AMD at p < 0.05 in any of the racial/ethnic groups in the NHANES III/2007-2008 meta-analysis.
4. Discussion
In this study, haplogroup analyses did not replicate previous associations of the European haplogroups J, T, and U with risk of AMD in non-Hispanic whites.31,33,34 Although a little surprising, other studies have not always replicated these associations,30,41 which may be due in part to heterogeneity across these studies or else suggesting a weak role of mitochondrial variation in the risk of AMD. However, we did observe that individual variants on the T haplogroup in the NHANES 2007-2008 non-Hispanic whites were associated with AMD risk in this population. Unsurprisingly, neither individual variants nor haplogroups that were previously associated with AMD in Europeandescent populations generalized to non-Hispanic blacks. African-descent populations suffer from a lesser burden of AMD, and previous studies have suggested that African-descent populations may have a different genetic architecture contributing to AMD etiology42-45 compared with other populations. We observed that mitochondrial variants mt16111, mt16362, mt16319, mt1736, and mt12007 were associated with AMD risk within the meta-analyzed Mexican American population after adjusting for well-known non-modifiable factors and environmental modifiers. The direction of the genetic effect was the same across the individual NHANES analyses for these SNPs in the Mexican American populations as follows: mt16111 OR = 2.90 and 2.33; mt16362 OR = 2.49 and 3.35; mt16319 OR = 2.17 and 3.72; mt1736 OR = 0.55 and 0.22; mt12007 OR = 1.83 and 4.26 in NHANES III and NHANES 2007-2008 respectively.
Limited studies have been performed to assess the genetic factors of AMD in Mexican or Latino populations. A handful of studies have examined whether CFH, ARMS2, and C2/C3, strong risk loci in European populations, contribute to risk of AMD in Mexican-descent populations.43,46,47 These studies, although limited in case size, did find a correlation between these European-derived variants and risk of advanced AMD in Mexicans and Latinos, suggesting that risk of AMD is being driven in part by European risk variants in these admixed populations. All five of the mtSNPs associated with AMD in Mexican Americans in this study are located on the A-A2 haplogroup background. Haplogroup A developed in Asia over 30,000 years ago and occurs most frequently in the Indigenous peoples of the Americas, with its subgroup A2 found to be the most common haplogroup in many of the indigenous ethnic groups of Central and North America.48 In the combined NHANES III, 1999-2002, and 2007-2008 populations, haplogroup A is the most prevalent among Mexican Americans39 with a frequency of 34.2% while composing less than 1% in non-Hispanic whites and non-Hispanic blacks. This observation is interesting given that Mexican Americans, who experience similar rates of AMD as that observed in European-descent groups (5.1% vs 7.3%),15 may contain a set of genetic risk factors on this haplogroup that are driving AMD risk in addition to or in combination with the already known European-derived variants.
Three of the significant mtSNPs (mt1611, mt16362, and mt16319) are located within the control region of the mitochondrial genome containing the origin of replication and the origin of transcription. These SNPs have not previously been associated with AMD but have been identified as contributors to various forms of cancer. A high load of somatic mtSNPs in the mitochondrial control region (i.e. mt16111) was found in patients suffering from prostate cancer.49 In a study examining the effect of mtSNPs located more specifically within the D-loop of the control region on risk for renal cell carcinoma, mt16319 was specifically found to be reduced in cases of clear cell renal cell carcinoma.50 Lastly, mt16362 was found to be a risk factor associated with familial breast cancer.51
Strengths of this study include the systematic fashion in which all participants over the age of 40 years were included in ophthalmologic exams to ascertain eye health and AMD status. This ensures a strong degree of homogeneity in case and control status across the various NHANES cohorts and minimizes between study heterogeneity. Over sampling of minority groups also likely increased the number of cases available for study in these underrepresented groups. Limitations include differences in data collection between NHANES III and NHANES 2007-2008, as NHANES III only performed fundus photography on one randomly selected eye whereas NHANES 2007-2008 performed fundus photography on both eyes. Other limitations include low statistical power and lack of correction for multiple hypothesis testing. When considering a significance threshold for mitochondrial variation analyses it should be noted that independence of each test is questionable as all variation is inherited as a whole. Lastly, we relied on self-reported race/ethnicity as opposed to genetic ancestry of the mitochondrial genome which may lead to false positive associations that are in actuality identifying differences in haplogroup ancestry. Future studies are needed to validate the results of the present study.
Despite limited sample sizes available for AMD analyses, NHANES is one of only a few surveys to include ophthalmologic exams of minority populations. As large scale epidemiology surveys become more cost prohibitive, a stronger emphasis on the utilization of electronic medical records (EMR) to identify cases and controls for inclusion in future studies will become more pronounced. Given that many of these EMR systems are still predominately composed of European-descent patients, a concerted effort must be made to increase the number of minorities with access to routine, continuous health care.
5. Conclusion
In conclusion, we identified potential novel associations between mitochondrial variants and risk of AMD in the NHANES Mexican Americans. All of the associated mitochondrial variants are found on the A-A2 haplogroup which is common among this racial/ethnic group and varies from the haplogroups previously reported to influence risk in European-descent populations. Future studies in larger Mexican-descent datasets may clarify some of these findings. These new findings may offer insight into the cellular mechanics underlying disease etiology.
Table 3.
Haplogroup frequencies in the combined NHANES III, NHANES 1999-2002, and NHANES 2007-2008 populations as previously published39
| Haplogroup | non-Hispanic whites | non-Hispanic blacks | Mexican Americans |
|---|---|---|---|
| J | 9.2 % | 0.4% | 1.4% |
| T | 9.6% | 0.4% | 0.9% |
| U | 13.6% | 1.4% | 1.6% |
6. Acknowledgments
This work was supported by NIH U01 HG004798 and its ARRA supplements. The Vanderbilt University Center for Human Genetics Research, Computational Genomics Core provided computational and/or analytical support for this work. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
7. References
- 1.National Alliance for Eye and Vision Research The Silver Book: Vision Loss: Chronic Disease and Medical Innovationn in an Aging nation. 2006 http://www.eyeresearch.org/pdf/VisionLossSilverbook.pdf. [Google Scholar]
- 2.Lions Clubs International Foundation N. E. I. 2005 Survey of public knowledge, attitudes, and practices related to eye health and disease. 2007 http://www.nei.nih.gov/nehep/kap. [Google Scholar]
- 3.Friedman David S., O’Colmain B, Mestril Ilona. 2012 Fifth Edition of Vision Problems in the U.S. 2012 http://www.visionproblemsus.org/introduction/acknowledgments.html. [Google Scholar]
- 4.Luo D-G, Xue T, Yau K-W. How vision begins: an odyssey. Proc. Natl. Acad. Sci. U. S. A. 2008;105:9855–9862. doi: 10.1073/pnas.0708405105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Risk factors associated with age-related macular degeneration. A case-control study in the age-related eye disease study: Age-Related Eye Disease Study Report Number 3. Ophthalmology. 2000;107:2224–2232. doi: 10.1016/s0161-6420(00)00409-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chakravarthy U, et al. Clinical risk factors for age-related macular degeneration: a systematic review and meta-analysis. BMC Ophthalmol. 2010;10:31. doi: 10.1186/1471-2415-10-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmidt S, et al. Cigarette smoking strongly modifies the association of LOC387715 and age-related macular degeneration. Am. J. Hum. Genet. 2006;78:852–864. doi: 10.1086/503822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schaumberg DA, Hankinson SE, Guo Q, Rimm E, Hunter DJ. A prospective study of 2 major age-related macular degeneration susceptibility alleles and interactions with modifiable risk factors. Arch. Ophthalmol. 2007;125:55–62. doi: 10.1001/archopht.125.1.55. [DOI] [PubMed] [Google Scholar]
- 9.Neuner B, et al. LOC387715, smoking and their prognostic impact on visual functional status in age-related macular degeneration-The Muenster Aging and Retina Study (MARS) cohort. Ophthalmic Epidemiol. 2008;15:148–154. doi: 10.1080/09286580802105830. [DOI] [PubMed] [Google Scholar]
- 10.Reynolds R, Rosner B, Seddon JM. Serum lipid biomarkers and hepatic lipase gene associations with age-related macular degeneration. Ophthalmology. 2010;117:1989–1995. doi: 10.1016/j.ophtha.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Leeuwen R, et al. Cholesterol and age-related macular degeneration: is there a link? Am. J. Ophthalmol. 2004;137:750–752. doi: 10.1016/j.ajo.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 12.Klein R, Klein BEK, Tomany SC, Cruickshanks KJ. The association of cardiovascular disease with the long-term incidence of age-related maculopathy: the Beaver Dam Eye Study. Ophthalmology. 2003;110:1273–1280. doi: 10.1016/S0161-6420(03)00599-2. [DOI] [PubMed] [Google Scholar]
- 13.Klein R, et al. The prevalence of age-related macular degeneration and associated risk factors. Arch. Ophthalmol. 2010;128:750–758. doi: 10.1001/archophthalmol.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawasaki R, et al. The Prevalence of Age-Related Macular Degeneration in Asians: A Systematic Review and Meta-Analysis. Ophthalmology. 2010;117:921–927. doi: 10.1016/j.ophtha.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 15.Klein R, et al. Prevalence of age-related macular degeneration in the US population. Arch. Ophthalmol. 2011;129:75–80. doi: 10.1001/archophthalmol.2010.318. [DOI] [PubMed] [Google Scholar]
- 16.Friedman DS, et al. Prevalence of age-related macular degeneration in the United States. Arch. Ophthalmol. 2004;122:564–572. doi: 10.1001/archopht.122.4.564. [DOI] [PubMed] [Google Scholar]
- 17.Haines JL, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 18.Klein RJ, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edwards AO, et al. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308:421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- 20.Jakobsdottir J, et al. Susceptibility genes for age-related maculopathy on chromosome 10q26. Am. J. Hum. Genet. 2005;77:389–407. doi: 10.1086/444437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rivera A, et al. Hypothetical LOC387715 is a second major susceptibility gene for age-related macular degeneration, contributing independently of complement factor H to disease risk. Hum. Mol. Genet. 2005;14:3227–3236. doi: 10.1093/hmg/ddi353. [DOI] [PubMed] [Google Scholar]
- 22.Gold B, et al. Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nat. Genet. 2006;38:458–462. doi: 10.1038/ng1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fritsche LG, et al. Age-Related Macular Degeneration: Genetics and Biology Coming Together. Annu. Rev. Genomics Hum. Genet. 2014;15:151–171. doi: 10.1146/annurev-genom-090413-025610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kenney MC, et al. Molecular and bioenergetic differences between cells with African versus European inherited mitochondrial DNA haplogroups: implications for population susceptibility to diseases. Biochim. Biophys. Acta. 2014;1842:208–219. doi: 10.1016/j.bbadis.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cristina Kenney M, et al. Inherited mitochondrial DNA variants can affect complement, inflammation and apoptosis pathways: insights into mitochondrial-nuclear interactions. Hum. Mol. Genet. 2014;23:3537–3551. doi: 10.1093/hmg/ddu065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang F-Q, Godley BF. Oxidative stress-induced mitochondrial DNA damage in human retinal pigment epithelial cells: a possible mechanism for RPE aging and age-related macular degeneration. Exp. Eye Res. 2003;76:397–403. doi: 10.1016/s0014-4835(03)00023-x. [DOI] [PubMed] [Google Scholar]
- 27.Cano M, et al. Oxidative stress induces mitochondrial dysfunction and a protective unfolded protein response in RPE cells. Free Radic. Biol. Med. 2014;69:1–14. doi: 10.1016/j.freeradbiomed.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheu S-J, et al. Resveratrol stimulates mitochondrial bioenergetics to protect retinal pigment epithelial cells from oxidative damage. Invest. Ophthalmol. Vis. Sci. 2013;54:6426–6438. doi: 10.1167/iovs.13-12024. [DOI] [PubMed] [Google Scholar]
- 29.Kenney MC, et al. Mitochondrial DNA variants mediate energy production and expression levels for CFH, C3 and EFEMP1 genes: implications for age-related macular degeneration. PloS One. 2013;8:e54339. doi: 10.1371/journal.pone.0054339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones MM, et al. Mitochondrial DNA haplogroups and age-related maculopathy. Arch. Ophthalmol. 2007;125:1235–1240. doi: 10.1001/archopht.125.9.1235. [DOI] [PubMed] [Google Scholar]
- 31.Udar N, et al. Mitochondrial DNA Haplogroups Associated with Age-Related Macular Degeneration. Invest. Ophthalmol. Vis. Sci. 2009;50:2966–2974. doi: 10.1167/iovs.08-2646. [DOI] [PubMed] [Google Scholar]
- 32.SanGiovanni JP, et al. Mitochondrial DNA variants of respiratory complex I that uniquely characterize haplogroup T2 are associated with increased risk of age-related macular degeneration. PloS One. 2009;4:e5508. doi: 10.1371/journal.pone.0005508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mueller EE, et al. Mitochondrial Haplogroups and Control Region Polymorphisms in Age-Related Macular Degeneration: A Case-Control Study. PLoS ONE. 2012;7:e30874. doi: 10.1371/journal.pone.0030874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Canter JA, et al. Mitochondrial DNA Polymorphism A4917G Is Independently Associated with Age-Related Macular Degeneration. PLoS ONE. 2008;3:e2091. doi: 10.1371/journal.pone.0002091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.US Department of Health and Human Services (DHHS). Centers for Disease Control and Prevention Third National Health and Nutrition Examination Survey, 1988-94 . Plan and Operations Procedures Manuals[CD-ROM] National Center for Health Statistics, Centers for Disease Control and Prevention; Hyattsville, MD: 1996. [Google Scholar]
- 36.Centers for Disease Control and Prevention Plan and Operation of the Third National Health and Nutrition Examination Survey 2004. 1988-94 Bethesda, MD.
- 37.Steinberg KK, et al. DNA banking in epidemiologic studies. Epidemiol. Rev. 1997;19:156–162. doi: 10.1093/oxfordjournals.epirev.a017938. [DOI] [PubMed] [Google Scholar]
- 38.Chang M-H, et al. Prevalence in the United States of selected candidate gene variants: Third National Health and Nutrition Examination Survey, 1991-1994. Am. J. Epidemiol. 2009;169:54–66. doi: 10.1093/aje/kwn286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitchell SL, et al. Characterization of mitochondrial haplogroups in a large population-based sample from the United States. Hum. Genet. 2014;133:861–868. doi: 10.1007/s00439-014-1421-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klein R, et al. The Wisconsin age-related maculopathy grading system. Ophthalmology. 1991;98:1128–1134. doi: 10.1016/s0161-6420(91)32186-9. [DOI] [PubMed] [Google Scholar]
- 41.Tilleul J, et al. Genetic association study of mitochondrial polymorphisms in neovascular age-related macular degeneration. Mol. Vis. 2013;19:1132–1140. [PMC free article] [PubMed] [Google Scholar]
- 42.Sadigh S, et al. Drusen and Photoreceptor Abnormalities in African-Americans with Intermediate Non-neovascular Age-related Macular Degeneration. Curr. Eye Res. 2014:1–9. doi: 10.3109/02713683.2014.925934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spencer KL, Glenn K, Brown-Gentry K, Haines JL, Crawford DC. Population differences in genetic risk for age-related macular degeneration and implications for genetic testing. Arch. Ophthalmol. 2012;130:116–117. doi: 10.1001/archopthalmol.2011.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Friedman DS, Katz J, Bressler NM, Rahmani B, Tielsch JM. Racial differences in the prevalence of age-related macular degeneration: the Baltimore Eye Survey. Ophthalmology. 1999;106:1049–1055. doi: 10.1016/S0161-6420(99)90267-1. [DOI] [PubMed] [Google Scholar]
- 45.Klein R, et al. Subclinical atherosclerotic cardiovascular disease and early age-related macular degeneration in a multiracial cohort: the Multiethnic Study of Atherosclerosis. Arch. Ophthalmol. 2007;125:534–543. doi: 10.1001/archopht.125.4.534. [DOI] [PubMed] [Google Scholar]
- 46.Buentello-Volante B, et al. Susceptibility to advanced age-related macular degeneration and alleles of complement factor H, complement factor B, complement component 2, complement component 3, and age-related maculopathy susceptibility 2 genes in a Mexican population. Mol. Vis. 2012;18:2518–2525. [PMC free article] [PubMed] [Google Scholar]
- 47.Contreras AV, et al. CFH haplotypes and ARMS2, C2, C3, and CFB alleles show association with susceptibility to age-related macular degeneration in Mexicans. Mol. Vis. 2014;20:105–116. [PMC free article] [PubMed] [Google Scholar]
- 48.Fagundes NJR, et al. Mitochondrial population genomics supports a single pre-Clovis origin with a coastal route for the peopling of the Americas. Am. J. Hum. Genet. 2008;82:583–592. doi: 10.1016/j.ajhg.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen JZ, Gokden N, Greene GF, Mukunyadzi P, Kadlubar FF. Extensive somatic mitochondrial mutations in primary prostate cancer using laser capture microdissection. Cancer Res. 2002;62:6470–6474. [PubMed] [Google Scholar]
- 50.Zhang J, et al. Identification of sequence polymorphisms in the displacement loop region of mitochondrial DNA as a risk factor for renal cell carcinoma. Biomed. Rep. 2013;1:563–566. doi: 10.3892/br.2013.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng M, et al. Identification of sequence polymorphisms in the mitochondrial displacement loop as risk factors for sporadic and familial breast cancer. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2014;35:4773–4777. doi: 10.1007/s13277-014-1626-5. [DOI] [PubMed] [Google Scholar]