Abstract
Elevated protein synthesis is an important feature of many cancer cells and often arises as a consequence of increased signaling flux channeled to eukaryotic initiation factor (eIF) 4F, the key regulator of the mRNA-ribosome recruitment phase of translation initiation. In many cellular and pre-clinical models of cancer, eIF4F deregulation results in changes in translational efficiency of specific mRNA classes. Importantly, many of these mRNAs code for proteins that potently regulate critical cellular processes such as cell growth and proliferation, enhanced cell survival, and cell migration that ultimately impinge on several hallmarks of cancer, including increased angiogenesis, deregulated growth control, enhanced cellular survival, epithelial-to-mesenchymal transition, invasion and metastasis. By being positioned as the molecular nexus downstream of key oncogenic signaling pathways (e.g. Ras, PI3K/AKT/TOR, and Myc), eIF4F serves as a direct link between important steps in cancer development and translation initiation. Identification of mRNAs particularly responsive to elevated eIF4F activity that typifies tumorigenesis underscores the critical role of eIF4F in cancer and raises the exciting possibility of developing new-in-class small molecules targeting translation initiation as anti-neoplastic agents.
Keywords: eIF4F, eIF4E, eIF4A, mTOR, Translational Control, Targeted Therapy
The Eukaryotic Initiation Factor 4F Complex
There have been tomes written on the subject of translational control under normal and pathophysiological conditions (1). In this review, we focus on the role that the “cap binding complex”, eukaryotic initiation factor (eIF) 4F, plays in mRNA discrimination and in driving tumorigenesis. We also discuss strategies aimed at therapeutically inhibiting eIF4F activity.
A salient hallmark of eukaryotic cytoplasmic, non-organellar, mRNAs is the 5′ terminal cap, a structure added to nascent mRNA templates shortly after initiation of transcription. Although the cap has been implicated in several nuclear events (splicing, polyadenylation, and nuclear/cytoplasmic transport) and plays a protective role against 5′ exonucleolytic degradation, its best documented function is in facilitating the recruitment of 43S preinitiation complexes to mRNA templates (2). Initial pioneering studies elucidating a role for the cap in translation uncovered an important conceptual point – in vitro its presence is facilitative in nature but in vivo it is absolutely essential (3).
A second important finding that emerged from these early experiments was the existence of an inverse relationship between secondary structure within the 5′ untranslated region (UTR) of mRNAs and translational efficiency. This link was deduced from experiments reporting on the translational efficiency of mRNAs with differing secondary structure, on the ATP requirement of initiation factors involved in cap recognition, and the varying degree of inhibition by cap analogues on initiation of mRNAs with differing secondary structure (4–9). An understanding of the basis of this relationship was afforded when the cytoplasmic mammalian cap binding protein eIF4E was identified and purified (10) and shown capable of stimulating translation of capped mRNA in HeLa cell extracts (11). eIF4E was subsequently found to be a component of the hetero-trimeric eIF4F complex which also contains a large ~220 kDa scaffolding protein (eIF4G) and the ATP-dependent RNA helicase, eIF4A (12).
A Molecular Commitment – Recruiting the Ribosome to the mRNA
Cap-Dependent Ribosome Recruitment
eIF4E is the least abundant of the initiation factors, present at 0.2–0.3 molecules/ribosome in reticulocytes and HeLa cells, rendering it rate-limiting for translation (13, 14). However, whether eIF4E levels are limiting at the organismal level across all cell types and cancer cells in vivo remains an outstanding question. In contrast to eIF4E, eIF4A is the most abundant initiation factor – present at ~3–6 molecules/ribosome and is solely cytoplasmic (13, 14). In mammals, there exist two highly related eIF4A homologs; eIF4AI (DDX2A) and eIF4AII (DDX2B) (the human proteins are 90% identical) (15, 16), with eIF4AI generally being the more abundantly expressed (14, 17). The majority (~90%) of eIF4A exists as a free form (eIF4Af) whilst a small proportion is present as an eIF4F subunit (eIF4Ac) (18–20). There are also two homologs of eIF4G, eIF4GI and eIF4GII, that share 46% identity, with eIF4GI being more abundant (21). eIF4G interacts with eIF4E and eIF4A through defined domains and provides the scaffold upon which other factors important for the initiation process assemble (22). In mammals there are two separate eIF4A interacting domains on eIF4G and it is generally thought that the two domains interact with different regions of the same eIF4A molecule (23). Given that eIF4AI and eIF4AII are interchangeable in the eIF4F complex (16) it would appear that mammalian cells can generate four different eIF4F complexes, the functional consequences of which remain unknown.
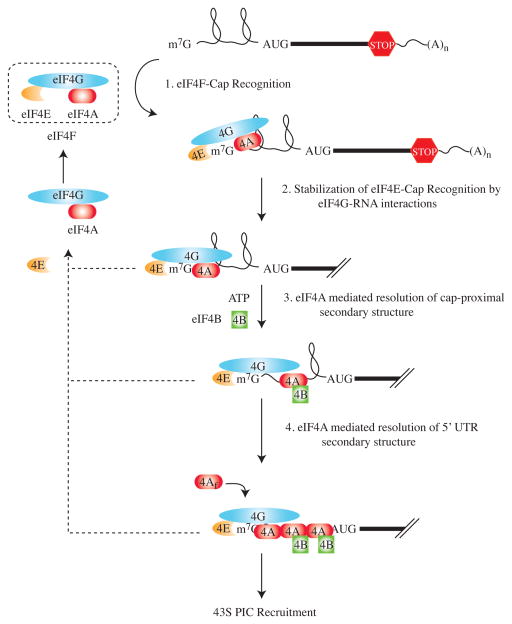
Although the involvement of eIF4B and eIF4H in translation initiation is well established, their precise roles need to be better characterized. eIF4B and eIF4H are RNA binding proteins that stimulate eIF4A helicase activity, enabling eIF4A to unwind more stable duplexes (24–27). Their interaction with eIF4A is mutually exclusive, as the two proteins share a common binding site (28). eIF4B and eIF4H modulate the affinity of eIF4A for ATP or ADP (29, 30) and RNA (31) with the interaction of eIF4B near the 5′ cap structure being ATP (and presumably eIF4A)-dependent (6), and inhibited by secondary structure (7). Through their RNA-recognition motifs, eIF4B and eIF4H may also stabilize single-stranded regions in the 5′UTR to prevent re-annealing following unwinding by eIF4A (Fig. 1). eIF4B is obligatory for 48S initiation complex formation on mRNAs possessing even modest levels of 5′ UTR complexity (32) and its depletion results in reduced proliferation rates, cell survival and enhanced sensitivity to camptothecin-induced cell death (33). These results implicate eIF4B function in controlling the translation of mRNAs critical for cell proliferation and survival.
Figure 1.
Schematic model of cap-dependent binding of eIF4F-mRNA binding and subsequent destabilization of secondary structure. For clarity, the PABP-eIF4G interaction has not been recapitulated in this figure. Shown are four steps where mRNA structural barriers may impact on initiation efficiency. See text for details.
mRNA Discrimination
eIF4F is the long-sought discriminatory factor responsible for differences in translation rates among many mRNAs. Two criteria are required for this to hold true – firstly, eIF4F had to be limiting (see above) and different mRNAs had to exhibit distinct affinities (or requirements) for recruitment of eIF4F – a feature that was linked to the degree of mRNA 5′ UTR secondary structure (34, 35). Consistent with eIF4F discriminating between different mRNAs based on secondary structure, initiation on simple, unstructured model mRNAs (containing [CAA]n as 5′ UTR) does not require eIF4F, the cap, or ATP (36). As well, a dominant-negative mutant of eIF4A capable of inhibiting eIF4F activity exhibits less potent inhibition towards translation of mRNAs with a lower degree of secondary structure compared to transcripts harboring more structure (37).
How the location of secondary structure within the 5′ UTR determines whether a particular transcript will be a “weak” or “strong” competitor is not well defined, but the net consequences may reflect the accumulated effects on various steps of the ribosome recruitment process. Binding of eIF4E to the cap structure is the first step in loading of the 43S pre-initiation complex onto the mRNA– an interaction mediated by base stacking of the positively charged N-7 methylguanosine with two tryptophans (W56 and W102) and auxiliary interactions (38–40). Three RNA-binding sites on eIF4G, necessary for efficient translation initiation, stabilize this interaction (41–43) (Fig. 1). Structure proximal to the cap (secondary structure or protein-mRNA interactions) can influence the interaction between eIF4E and mRNA in a negative manner (44–48). On the other hand, structure located distal from the cap exerts little effect on eIF4E-cap interaction but can interfere with eIF4F mediated unwinding - inhibiting the interaction of eIF4A and/or eIF4B with mRNA (7, 49). Since eIF4Af has bi-directional helicase activity, its delivery to the 5′ end of the mRNA provides forced directionality to this enzyme. Although eIF4Af has weak helicase in vitro (24, 50), its presence in the eIF4F complex leads to a ~20-fold increase in helicase activity (24, 51, 52). This has been attributed to eIF4E since its binding to eIF4G masks an eIF4A autoinhibitory domain on eIF4G (53). Hence, eIF4E is capable of promoting the helicase activity of eIF4A and increasing translation rates by a mechanism distinct from its cap-binding function. The overabundance of eIF4A relative to eIF4E (13, 54), the elevated helicase activity of eIF4Ac relative to eIF4Af (24, 51, 52), and the ability of eIF4Af to exchange with eIF4Ac (16) has led to the proposal that eIF4A recycles through the eIF4F complex during initiation (55) (Fig. 1). The idea that eIF4A/eIF4B/eIF4H may polymerize on the mRNA is consistent with the observed cap-dependent crosslinking of these factors downstream of the cap (56). As noted by Kapp and Lorsch (22), such a polymerization model is also consistent with the kinetics of unwinding at low concentrations of eIF4A where a lag in activity is often observed.
One additional interaction that impacts translation initiation, and where 5′ UTR may also exert a negative effect, is between eIF4G and the poly(A) binding protein, PABP (57). This interaction is thought to circularize the mRNA (57) and has been associated with stimulation of 48S pre-initiation complex formation (58, 59), 60S subunit joining (59), increased eIF4F cap-binding and ATPase activity (60–63), and stabilization of eIF4F-mRNA interactions in vivo (64). It is not known whether eIF4E needs to remain bound to the mRNA or is released and available for formation of new eIF4F complexes (Fig. 1). Consistent with the latter possibility is the finding that the addition of cap analogues to cell extracts after commencement of translation fails to block cap-dependent translation (65). Whereas the PABP-eIF4G interaction is stimulatory but dispensable in yeast cells (64, 66), it appears critical for translational control of maternal mRNAs during Xenopus development (67). Whether mRNAs with elevated structural barriers are less efficiently circularized or require de novo cap-recognition by eIF4E at every translational attempt is unknown, but could be an additional step that renders “weaker” mRNAs at a disadvantage over their more robust counterparts. Collectively, these studies support the notion that not all mRNAs are equally “translatable” and that the successful translation of certain mRNAs (i.e. “weak” mRNAs) is dictated by multiple and cumulative properties mostly associated with that mRNA’s 5′UTR. Importantly, translation of these “weak” mRNAs, which typically encode for the potent growth and survival factors that drive the hallmarks of cancer, is suppressed except under conditions of enhanced eIF4F activity, that is during malignant progression.
Elevated eIF4F activity selectively upregulates translation of a subset of mRNAs
eIF4E over-expression in experimental cell models elicits only small increases in overall protein synthesis rates while enabling a substantial, disproportionate and selective increase in translation of a subset of mRNAs. Early attempts at identifying these eIF4E-responsive mRNAs focused on examining candidates on a one-by-one basis. These studies revealed that the production of housekeeping proteins (e.g. actin, glyceraldehyde 3-phosphate dehydrogenase (GAPDH)] was not altered by changes in eIF4E levels, but that the translation of mRNAs harboring long, highly-structured 5′UTRs (e.g. ODC, c-Myc) was profoundly impacted by fluctuations in eIF4E levels (68–70). Subsequent genome-wide analyses of changes in translation revealed that 5′ UTR structure was one determinant of eIF4E or eIF4F sensitivity, but also uncovered a more complex situation in that eIF4E responsive mRNAs did not always appear to possess complex 5′ UTRs. Inhibition of mTOR activity (and therefore eIF4F assembly) by rapamycin (for 4 hrs) in Jurkat T cells, followed by polysome profiling, revealed that the translation of only ~6% of expressed genes was inhibited (71). Similarly, polysome analyses of NIH 3T3 cells transiently overexpressing eIF4E revealed that mRNAs encoding ribosomal proteins were prominently eIF4E-responsive (72). Interestingly, some of these mRNAs contain a distinct element in their 5′ UTR known as a 5′ TOP (5′ terminal oligopyrimidine tract) and recent evidence suggests that the RNA binding protein, LARP1, modulates the translation of this class of mRNAs (73). The relationship between LARP1 and eIF4E responsiveness remains to be fully explored.
More recently, these finding have been corroborated by ribosome profiling analyses, a methodology that relies on deep sequencing of ribosome-protected mRNAs to quantify the translation of mRNAs on a codon-by-codon resolution (74, 75). These studies further reveal a new regulatory element termed the PRTE (pyrimidine-rich translation element), which interfaces with eIF4E activity and is essential to control the translation of a subset of mRNAs encoding key proteins involved in critical steps of cancer initiation and progression (74). However, the mechanism by which PRTE elements act to cooperate with eIF4E to regulate the translation of specific mRNAs is still under investigation. Additional global approaches probing changes in mRNA distribution in polysomes as a consequence of altered eIF4F levels or activity revealed profound effects on the translation of mRNAs implicated in immune response, cell cycle progression, and metabolism (76, 77). Similar to increased eIF4E expression, eIF4E phosphorylation does not significantly increase global protein synthesis but stimulates the translation of pro-survival (e.g.- MCL) and pro-invasion mRNAs (e.g. MMP3 and Snail) (78–80). Since the cellular translatome at any given time is affected by upstream regulatory processes, relative mRNA abundance, and mRNA complexity the subset of mRNAs responsive to eIF4F can be expected to vary across tumor types and in response to different physiological stimuli.
Critical oncogenic signaling pathways converge to regulate eIF4F assembly and activity
Feed-forward loops drive malignancy - MYC and eIF4F
The MYC transcription factor is one of the most frequently activated oncogenes in human cancers (155) and has been repeatedly associated with poor prognosis and decreased patient survival (156–158). A major consequence of elevated MYC activity is a dramatic elevation in protein synthesis due to increased transcription of ribosomal RNA (rRNA) genes and genes required for rRNA processing and assembly. This increase in translational output is a critical determinant for MYC oncogenic activity in vivo (81). Importantly, elevated MYC also increases transcription of the core eIF4F components - eIF4E, eIF4AI, and eIF4GI, (but not eIF4AII or eIF4GII) (159–161). Indeed, MYC levels are elevated in pre-lymphomatous, pre-B, and pro-B cells isolated from Eμ-Myc mice, and engender an increase in eIF4F levels (162). MYC, in turn, is one of the best characterized translationally-controlled mRNAs. Hence, MYC and eIF4F work in a feed-forward loop, each enhancing the expression of one another. Indeed, elevated eIF4E consequent to MYC activity has been deemed necessary to suppress apoptosis resulting from aberrant MYC activity (169–171). This intimate relationship affords an opportunity by which part of MYC’s function can be interdicted by inhibiting eIF4F activity (162, 163).
mTOR - Regulating eIF4F assembly and activity
The assembly of the eIF4F complex is under the control of the mammalian target of rapamycin (mTOR), a serine/threonine kinase whose activity is perturbed in many human cancers (82). mTOR exists in two functionally distinct complexes. mTORC2 regulates cytoskeletal reorganization and cell survival, whereas mTORC1 controls translation initiation, ribosome synthesis, expression of metabolism-related genes and autophagy (82). mTORC1 controls eIF4F assembly by liberating both eIF4E and eIF4A from their respective inhibitory binding proteins, eIF4E binding proteins (4E-BPs) and programmed cell death 4 (PDCD4). 4E-BPs (there are three highly related proteins in mammals with the most well characterized being 4E-BP1) compete with eIF4G for a binding site on eIF4E (83), an interaction that is regulated by mTORC1-dependent phosphorylation of 4E-BP. Hypophosphorylated 4E-BP binds to eIF4E with high affinity (nanomolar range) and inhibits translation initiation, whereas hyperphosphorylated 4E-BP does not bind eIF4E, allowing eIF4F complexes to form (83).
Similarly, eIF4A availability is regulated downstream of mTORC1 by the tumor suppressor gene product PDCD4 (84–87). PDCD4 binds to eIF4A, inhibiting its helicase activity and preventing its binding to eIF4G. PDCD4 is regulated by S6 kinase which, when activated by mTORC1, phosphorylates PDCD4, marking PDCD4 for ubiquitin-mediated degradation which then leads to liberation of eIF4A and eIF4F assembly (84, 88).
As noted above, the role of eIF4B is critical to the overall helicase activity of eIF4A within the eIF4F complex. eIF4B activity is also controlled downstream of mTOR and Ras-ERK pathway signaling by p90 ribosomal S6 kinase (RSK) and p70 ribosomal S6 kinase (S6K) (89–92). Phosphorylation of eIF4B on Ser422 increases its interaction with eIF4A and eIF3 and is required for ribosome recruitment to mRNAs containing secondary structure (89–91). Moreover, recombinant eIF4B, which is presumably not phosphorylated, cannot substitute for native eIF4B in this assay. Hence, the phosphorylation status of eIF4B affects mRNA discrimination, possibly by influencing eIF4A activity.
Regulation by phosphorylation - Ras and MAPK signaling converge on eIF4F
Increased signaling flux through the Ras-Raf-Erk pathway is a frequent occurrence in many human cancer types often consequent to mutational activation of Ras or Raf oncogenes or activation of upstream receptor tyrosine kinases (e.g. EGFR). Constitutive activation of this pathway enhances the assembly and activity of the eIF4F complex in multiple ways. Activation of Erk- and p90RSK can phosphorylate TSC2 activating mTORC1, which would promote eIF4F assembly (93). Ras pathway signaling also directly impinges upon eIF4F activity. Early studies by Rinker-Schaeffer et al. ((94) revealed that oncogenic Ras dramatically increased the rate of eIF4E phosphorylation. Subsequent work has now shown that ras pathway signaling through ERK stimulates the MNK kinases to interact with eIF4G and phosphorylate eIF4E at serine 209 (95–99).
What are the consequences of eIF4E phosphorylation? At this point, we do not have a complete picture. Mouse knockout studies have revealed that MNK1 and MNK2 are dispensable for normal development (100) and eIF4ES209A/S209A knock-in mice display no obvious phenotype (79). Several studies have documented increased translation as a consequence of eIF4E phosphorylation (79, 83, 96), yet phosphorylation of eIF4E lowers its affinity for the cap structure (101–104). This conundrum can be explained if one posits that the reduced cap affinity is associated with increased recycling of eIF4E once functional 43S pre-initiation complexes have been successfully recruited to the 5′ end (105). Regardless, the phosphorylation of eIF4E at serine 209 has been shown in multiple reports to be absolutely critical to the oncogenic function of eIF4E (78, 79, 106).
Elevated eIF4F function drives cellular transformation, tumorigenesis and malignant progression
eIF4E in cellular transformation and tumorigenesis
The landmark study by Lazaris-Karatzas et al. (107) was the first to demonstrate that ectopic over-expression of eIF4E was oncogenic in NIH-3T3 cells, driving cellular transformation (focus formation, soft agar colonization) and tumorigenesis. Further highlighting the importance of eIF4E function in cellular transformation and tumorigenesis, engineered reduction of eIF4E consequent to expression of antisense eIF4E RNA profoundly suppressed the transformed phenotype of highly aggressive, Ras oncogene-transformed fibrosarcomas, reducing soft agar colonization more than 90%, increasing tumor latency periods and reducing tumor growth rates (108). Furthermore, development of an eIF4E transgenic mouse established eIF4E as a bona fide oncogene in vivo. Indeed, in vivo constitutive overexpression of eIF4E alone leads to increased cancer susceptibility, demonstrated by the wide spectrum of tumors that develop in these mice, including lymphomas, angiosarcomas, lung carcinomas, and hepatomas (109). As well, in vivo overexpression of eIF4E within the B cell compartment cooperates with c-Myc in lymphomagenesis as eIF4E counteracts MYC-induced apoptosis, a critical barrier to tumor formation (110). These and other studies showed that elevated levels of eIF4E can recapitulate key oncogenic functions of Akt (110) and antagonize the pro-apoptotic activity of c-Myc (110–112).
The phosphorylation of eIF4E at Ser209 also plays an important role in eIF4E’s oncogenic function (106). In MYC-driven lymphomas, eIF4E expression accelerates lymphomagenesis whereas overexpression of an eIF4E S209A mutant is incapable of accelerating disease in this model (78). Similarly, in a PTEN knockout model of prostate cancer, knock-in of the same non-phosphorylatable eIF4E S209A mutant delays progression of prostate cancer (79). Consistent with these data, inhibition of MNK activity delays tumor development and outgrowth of metastases (113, 114). Mice homozygously deleted for both MNK1 and MNK2 are viable (100) making the targeting of this activity attractive for the development of novel anti-neoplastic agents. Increased Ras signaling (115) or altered eIF4E phosphorylation (79, 80) enhances the translation of a subset of mRNAs, with some encoding functions in cell growth, proliferation, and metastasis (80).
The prevailing data from these studies indicate that the oncogenic effects of eIF4E are a result of activated eIF4F rather than a unique function of eIF4E overexpression (116, 117). Early work showed eIF4E-mediated transformation to be particularly dependent on the enhanced translation of ornithine decarboxylase (ODC) and cyclin D1 (68, 118–120). Subsequent work has continued to highlight the role that eIF4E plays in driving cellular transformation - selectively enhancing the translation of a limited pool of mRNAs whose protein products play an integral role in malignancy - c-MYC, cyclin D1, ODC, VEGF, among others (121). Importantly, a few common themes have consistently emerged from this body of work. Chief amongst these is that very modest changes in expression of eIF4E are required to affect malignancy. Indeed, only 2–3 fold increases in eIF4E are sufficient to drive cellular transformation whereas only a 50–60% reduction in eIF4E expression is necessary to block tumor formation and growth (108, 109, 119, 122–124).
eIF4E as a central regulator of metastatic progression
Beyond the initial changes that enable cellular transformation and expansion of a primary tumor, metastatic progression requires that tumors acquire a wide range of phenotypic characteristics, including the ability to establish autocrine growth and survival signaling, escape from the primary tumor site, invade surrounding normal tissue, disseminate through the blood or lymphatic circulation, establish and survive within the foreign microenvironment of the distal tissue site and outgrow as a metastatic colony. Though diverse stimuli that regulate the transcription of the critical gene products that drive these phenotypes, the synthesis of these proteins is coordinately elevated by eIF4E and eIF4F (125, 126).
The first evidence supporting a role for eIF4E in the metastatic process was revealed by antisense RNA studies in highly aggressive ras-transformed fibroblasts. Depletion of eIF4E by ~50% in these cells reduced the number of metastases formed in the lungs of mice following tail vein injection by up to 90% (123). Moreover, when implanted under the renal capsule, these same cells failed to invade the kidney parenchyma. Importantly, when metastases derived from these cells were examined, levels of eIF4E had been restored to that of the parental Ras-transformed cells indicating a selection in vivo for enhanced eIF4E function (123). Similarly, in breast cancer models, pharmacologic suppression of mTOR activity, which limits eIF4E availability and reduces eIF4F complex levels, diminished invasiveness and migration, as well as the formation of pulmonary metastases, thus delaying breast cancer progression (127).
Early overexpression studies have also implicated elevated eIF4E levels in driving not only tumorigenesis but also metastasis. Rat embryo fibroblasts engineered to overexpress eIF4E formed lung metastases both spontaneously after subcutaneous tumor growth, and experimentally following tail vein injection (123). Moreover, cells derived from these tumors and lung nodules showed elevated eIF4E expression levels, again indicating a selection for enhanced eIF4E function with malignant progression in vivo (123). Collectively, these data strongly implicate eIF4E, and by extension eIF4F, as a critical driver of metastatic progression.
To survive and grow in the primary tumor site, and especially within the foreign microenvironment of the metastatic site, tumor cells acquire growth factor autonomy, often through the establishment of autocrine growth factor networks. In the rat embryo fibroblast model, eIF4E overexpression enabled the establishment of an autocrine stimulatory loop involving enhanced signaling through the ERK pathway (125, 128). Importantly, the progressive selection in vivo for more aggressive tumor behavior (reduced tumor latency, enhanced metastatic potential, reduced mouse survival) selected for increased eIF4E expression as well as enhanced signaling through the ERK pathway (125, 128). In the NIH3T3 model, eIF4E overexpression was shown to drive tumorigenesis via the establishment of a Ras-dependent autocine loop (107).
Collectively, these studies indicate that eIF4E plays a key regulatory role in metastasis. Another aspect of metastatic progression is the ability of tumor cells to establish a new vascular network. In experimental models of breast and head and neck cancers, eIF4E overexpression was shown to promote the overexpression of the potent angiogenic factors VEGF (129) and FGF2 (130), in both cases by selectively enhancing translation of these mRNAs. Immunohistochemical surveys of both breast and head and neck cancers have further supported the link between eIF4E overexpression, VEGF overexpression and enhanced microvessel density (131, 132). These data suggest that eIF4E may indirectly govern tumor-related angiogenesis by enabling the enhanced translation of critical angiogenic factors from the tumor cell compartment. Indeed, treatment of nude mice bearing human breast or prostate cancer xenografts with an antisense oligonucleotide targeting eIF4E effectively reduced eIF4E expression in these tumors and profoundly reduced tumor vascularity. Importantly, in in vitro cord formation assays, depletion of eIF4E by antisense oligonucleotide transfection suppressed the ability of endothelial cells to form vessel-like structures suggesting for the first time that eIF4E may also directly govern the response of endothelial cells to angiogenic stimuli (124). Similar results were observed when the eIF4A helicase subunit of eIF4F was targeted with the small molecule inhibitor, silvestrol (133).
In addition to the establishment of a vascular network, metastatic progression requires persistent cellular survival, not only at the primary tumor site but also within the foreign microenvironment of a metastatic site. Numerous studies have linked enhanced eIF4E expression to the suppression of apoptosis. The earliest demonstration of this was that eIF4E upregulation was necessary in MYC-induced malignancies to overcome MYC-induced apoptosis (111). The mechanism could be explained by enhanced synthesis of BCL-XL and blockade of mitochondrial cytochrome C release (134). Subsequent work showed that eIF4E regulated the translation of a network of mRNAs encoding anti-apoptotic proteins (135). Most prominent amongst these was osteopontin (135), a protein that has been repeatedly implicated in metastasis (136). Additional work also highlighted the translational regulation of additional anti-apoptotic proteins by eIF4E including BI-1, dad1 and survivin (72) as well as BCL-2 (124, 137, 138). In the Eμ-myc B cell lymphoma model, enforced expression of eIF4E clearly promoted tumor cell survival and chemoresistance (122), in part by upregulating translation of the anti-apoptotic protein Mcl-1 (78).
The ability of tumor cells to break from the primary tumor mass, invade surrounding normal tissues and ultimately disseminate to distal tissue sites requires the remodeling and degradation of the extracellular matrix. This process is driven by expression and secretion of protein-degrading enzymes, most notably the Matrix Metalloproteases (MMP). In Ras-transformed rat embryo fibroblasts, reduction of eIF4E levels by only ~50% resulted in a remarkable reduction in the expression of MMP-9, concomitant with a marked diminution in invasiveness. Interestingly, the cells selected for increased aggressiveness in vivo regained eIF4E levels as well as MMP-9 expression levels (123). Similarly, in murine prostate carcinoma cells, MMP-9 is translationally controlled and associated with increased malignancy (139). More recently, in a murine model of prostate carcinoma progression, knock-in of the non-phosphorylatable eIF4E (S209A) mutant allele suppressed disease progression and specifically reduced the translation of a subset of mRNAs critical for progression including MMP-9 and MMP-3 (79). As well, heparanase production, an enzyme implicated in the metastatic process and angiogenesis due to degradation of heparin sulfate proteoglycans with subsequent destruction of the basement membrane, is eIF4E responsive and is decreased when eIF4E levels are reduced by antisense oligonucleotides (140). Further highlighting a role for eIF4E in tumor cell invasiveness, Robichaud et al (2014) have shown that eIF4E Ser209 phosphorylation plays a prominent role in regulating the TGFβ-induced epithelial-to-mesenchymal (EMT) transition by controlling the translation of a pool of mRNAs critical for EMT, including Snail and MMP-3 (80). Consistent with these genetic studies, pharmacologic inhibition of eIF4E phosphorylation profoundly suppressed the outgrowth of lung metastases in the B16 melanoma model (114).
Studies employing ribosome profiling have revealed that oncogenic eIF4E activity, downstream of mTOR signaling, has a striking effect on the translational landscape of the cancer genome, particularly in the context of metastasis (74). This study has functionally characterized a novel subset of translationally regulated mRNAs associated with cancer cell invasion and metastasis in vivo. These mRNAs include vimentin, MTA1 (metastasis associated 1), CD44, and YB-1 (Y-box binding protein 1; also called YBX1) and have critical roles in controlling cell migration, metastasis and EMT (141). Mechanistically, eIF4E regulates the translation of these mRNAs at least in part through the PRTE, a regulatory element that is present in their 5′ UTRs. Significantly, INK128, a clinical ATP-site inhibitor of mTOR, blocks the increased translation of these eIF4E sensitive mRNAs with therapeutic benefit at all stages of prostate cancer progression, including metastasis (74). Elevated expression of eIF4E is common in a wide array of human cancers (colorectal, breast, prostate, lymphoma) (69). Importantly, in many studies, elevated eIF4E expression has been linked to advanced disease and/or reduced survival (69, 142, 143). Recent work has now also shown that 4E-BP1 is hyperphosphorylated in human cancers (notably ovarian and prostate carcinomas) and also associated with reduced patient survival (69, 137, 142–144).
eIF4E and chemoresponsiveness
Altered eIF4E levels modify tumor cell drug sensitivity. Increased eIF4E levels have been associated with resistance to front line therapy (e.g. – doxorubicin) (122) and rapamycin (145) in the Eμ-Myc lymphoma model (122). Elevated eIF4E levels are also associated with resistance to PI3K/mTOR kinase inhibitors (146, 147). In a report documenting resistance to anti-BRAF and anti-MEK therapies, eIF4F levels correlated with drug response with increased levels associated with diminished efficacy (148). Increased eIF4E phosphorylation has been associated with expression of BRAFV600E in melanocytes (149). Targeting the eIF4E:eIF4G interaction or eIF4A activity synergizes with anti-BRAF therapy (148). A recent shRNA screen targeting the translatome in multiple myeloma identified all three subunits of eIF4F (eIF4E, eIF4AI, eIF4GI) as modifiers of dexamethasone response in this tumor type. Inhibition of eIF4F by small molecules synergized with dexamethasone as well as resensitized previously unresponsive cells (150). It will be important to identify the translational landscape responsible for these effects so as to obtain mechanistic insight and to distinguish between effects due to synergy versus resensitization of a previously resistant phenotype.
Breaking Bad Addictions - Targeting eIF4F
Over the last 10 years there has been significant interest in targeting the activity of the eIF4F complex. The knowledge that eIF4F assembly is under mTOR control was the first stepping stone towards this. The finding that MYC is one of the most frequently amplified genes in human cancers (151), coupled with an appreciation of its regulatory relationship with eIF4F, helped further fuel this interest. Targeting translation as an anti-neoplastic approach is not new. Depletion of asparagine pools with asparaginase inhibits translation elongation and is used to treat acute lymphoblastic leukemia and pediatric acute myeloid leukemia (152). As well, homoharringtonine, an inhibitor of translation elongation has been approved by the FDA for treatment of chronic myeloid leukemia (153). A large body of biological data ranging from cell-based and pre-clinical models assays suppressing eIF4E, eIF4A, and eIF4F support the idea that targeting eIF4F activity would be anti-neoplastic (reviewed in (154, 155)).
Experiments targeting eIF4E using antisense RNA (108, 124, 156–158), or peptides to interfere with eIF4E/eIF4G interaction (159–161), demonstrated suppression of the oncogenic properties of transformed cell lines ex vivo and/or tumor growth in vivo. Most telling, the development of a mouse model in which eIF4E expression can be inducibly suppressed by shRNAs engineered to be under doxycycline responsiveness afforded an alternative, genetic approach to pharmacologically targeting eIF4F activity in vivo (162). Using this model to suppress eIF4E in pre-lymphomatous B cells in Eμ-Myc mice revealed significantly delayed tumor onset and demonstrated a tolerance for suppressed eIF4E levels at the organismal level, thus underscoring eIF4F’s status as an important marker for tumor-specific vulnerability in vivo (162). A further key functional link between MYC and mTOR has been recently described, where MYC directs mTOR-dependent phosphorylation of 4EBP1, without affecting other mTOR substrates such as S6K (163). Taken together, these findings reveal a critical vulnerability for MYC overexpressing cancer cells that may rely on eIF4E availability for cancer cell survival (162, 163). Indeed, blocking 4E-BP phosphorylation either genetically or pharmacologically with the ATP-site inhibitor INK128, results in a dramatic reduction of lymphomagenesis in Eμ-Myc mice (225).
The eIF4F complex offers multiple possibilities for functional interdiction, some of which have been probed with small molecule inhibitors in high throughput screens. These include blocking eIF4E-cap interaction, interfering with eIF4E-eIF4G interaction, inhibiting eIF4A helicase activity, and suppressing eIF4E levels (164–166).
Targeting eIF4E function
Although cap analogues have been used to probe eIF4E-cap interaction since the late 1970s, their use has been limited to in vitro experiments since they are not readily cell permeable (167, 168)1. Therefore, efforts have recently focused on developing pro-drugs, with modifications that would allow the nucleosides to enter the cell followed by conversion to active inhibitors. Accordingly, one compound, 4Ei-1 inhibited cap-dependent translation in vitro and in vivo when injected into zebrafish embryos (150, 172). 4Ei-1 chemosensitized breast and lung cancer cells to non-toxic levels of the cytotoxic drug gemcitabine (173). 4Ei-1 reduced proliferation and repressed colony formation in mesothelioma cells and sensitized these to pemetrexed, a folate antimetabolite (174). Several inhibitors of eIF4E:eIF4G interaction have also been discovered (4EGI-1, 4E1RCat, 4E2RCat) and shown to inhibit cancer cell growth ex vivo, breast cancer xenograft growth in vivo, and reverse chemoresistance in MYC-driven murine lymphomas (166, 175–177).
Antisense oligonucleotides (ASO) against eIF4E (LY2275796 [aka ISIS EIF4E Rx]) have been tested in cell lines ex vivo and in xenograft models with promising activity (124). Here, translation of known eIF4E-specific pro-growth and pro-survival gene products (c-Myc, cyclin D1, VEGF, Bcl-2, and survivin), were reduced by LY2275796, while global protein synthesis was only modestly affected. A phase I trial demonstrated that LY2275796 could be safely administered to patients and was effective at decreasing eIF4E mRNA and protein levels in tumor cells (178). In this study, 30 patients with stage 4 disease received LY2275796 for three consecutive days and then were maintained on this compound by weekly administration for 3 additional consecutive weeks. The most common drug-related cytotoxicities reported were fatigue (47%), nausea (33%), fever (27%), and vomiting (20%) (178). The compound was effective at reducing eIF4E mRNA in vivo by 80% in post-treatment biopsies (compared to pre-treatment biopsies) (178). Phase II clinical trials are now underway combining ISIS EIF4E Rx with carboplatin and paclitaxel for non-small cell lung cancer (NCT01234038) or with docetaxel and prednisone for castration-resistant prostate cancer (NCT01234025).
Inhibiting eIF4A activity
A high throughput screen based on the differential inhibition of translation of eIF4F-dependent versus HCV IRES-dependent reporter mRNAs identified three natural products (pateamine A [Pat A], hippuristanol, and silvestrol) that selectively target eIF4A (164, 179–181). The binding site for hippuristanol has been mapped to the carboxyl terminal domain of eIF4A, but the binding sites of Pat A and silvestrol are not known. Hippuristanol prevents eIF4A from binding RNA (182), whereas Pat A and silvestrol act as chemical inducers of dimerization and force a non-sequence specific engagement between eIF4A and RNA, resulting in depletion of eIF4A from the eIF4F complex (133, 181, 183). Pat A inhibits translation irreversibly (likely as the consequence of a Michael addition site on the molecule) whereas inhibition by hippuristanol or silvestrol is readily reversible. All compounds exhibit anti-neoplastic activity in various xenograft mouse tumor models as single agents (133, 184–187). Hippuristanol and silvestrol reverse drug resistance in MYC-driven tumor models (181, 188).
Of the three eIF4A inhibitors, silvestrol and the related rocaglamide family members show the most favorable pharmacological properties for in vivo studies. Systemic availability for silvestrol when delivered intraperitoneally is 100% - with 60% of the parental compound remaining after 6 hr (189). Silvestrol does not cause weight loss, liver damage, or immunosuppression in mice (133). B-cells derived from chronic lymphocytic leukemia patients are more sensitive to silvestrol than B-cells from healthy individuals (187) suggesting preferential targeting of faster growing leukemic cells. The anti-proliferative properties of silvestrol appear to be mediated primarily through inhibition of eIF4A since silvestrol-resistant eIF4A mutants can rescue the effect (190). As expected, the translation of mRNAs with extensive secondary structure is more sensitive to inhibition by silvestrol (133, 191–193). A current barrier to the clinical development of silvestrol is that resistance can develop due to overexpression of the ABCB1/P-glycoprotein (191, 194).
Preventing eIF4E phosphorylation
Blocking eIF4E phosphorylation has been shown to prevent the oncogenic function of eIF4E in multiple experimental models (122, 123, 175) suggesting that pharmacologic inhibition of the Mnk kinases may be promising. Indeed, the Mnk inhibitors, CGP57380 and cercosporamide, have been shown to block eIF4E phosphorylation in cultured cells, limiting cellular proliferation in large part by inducing apoptosis (195–197). Similarly, (5-(2-(-(Phenylamino)pyrimidin-4-yl)thiazol-2(3H)-one derivatives have been shown to inhibit MNK2, reduce eIF4E phosphorylation and diminish Mcl-1 expression in cancer cells (198). In animal models, oral administration of cercosporamide suppressed eIF4E phosphorylation in normal mouse tissues and xenograft tumor tissues. Importantly, cercosporamide administration profoundly suppressed the outgrowth of B16 melanoma metastases (195). A more recent report also showed that cercosporamide treatment at doses which specifically inhibited eIF4E phosphorylation decreased the growth rate of AML xenografted tumors and suppressed colonization of freshly explanted AML patient samples (199). Importantly, the biologic effects of cercosporamide on AML cells reflected MNK inhibition (i.e. reduced eIF4E phosphorylation) and did not reflect inhibition of Jak3, a putative additional target of cercosporamide (199). The precise consequence of MNK inhibition on tumor cell behavior will require additional studies.
Not all Roads lead to Rome – The Differential Consequences of Targeting eIF4E, eIF4A, and eIF4G
Inhibition of eIF4E by TOR inhibitors (rapamycin, PP242, INK128) (200) or LY2275796 (124) does not dramatically reduce global protein synthesis. For example, LY2275796 causes a 80% reduction in eIF4E levels with only a modest impact on global protein synthesis (~20% change) (124). In contrast, inhibition of eIF4A has a much more profound effect on global protein synthesis (181). These results could be explained if the bulk of ongoing translation requires high concentrations of eIF4A, but not eIF4E, to be sustained. This is consistent with a model in which eIF4E recycles following the initial cap binding: eIF4G:eIF4A dimers would then be sufficient to maintain multiple subsequent rounds of initiation (Fig. 1). In this scenario, acute inhibition of eIF4A is expected to have a more immediate effect on translation than a block in eIF4E activity.
Overexpression of eIF4GI in NIH 3T3 cells is oncogenic (201). This may be a consequence of its ability to stimulate IRES-mediated translation of transcripts encoding key oncogenic functions (70, 202, 203). This is of clinical relevance since inflammatory breast cancers (IBC) display high levels of eIF4GI (with little change in eIF4E or 4E-BP1 levels). This has been linked to IRES-mediated translation of VEGFA and p120 catenin, which are required for progression to metastasis (204). Thus, in the setting of IBC, it would make more sense to target eIF4GI (and possibly eIF4A) rather than eIF4E.
Consistent with reports that document differential translational responses to eIF4E versus eIF4A inhibitors is the finding of a unique translation/transcription regulatory element, called TISU (translation initiator of short 5′ UTR; 5′SAASATGGCGGC3′, in which S is C or G) (205). TISUs are present in ~4.5% of protein-encoding genes, most of which have unusually short 5′ UTRs (~12 nt) (205). Translation of TISU-containing mRNAs is cap-dependent, but much less dependent on eIF4A (206). Perhaps, the extreme cap-proximal location of the AUG initiation codon on TISU mRNAs significantly diminishes the scanning requirement for eIF4A-dependent unwinding.
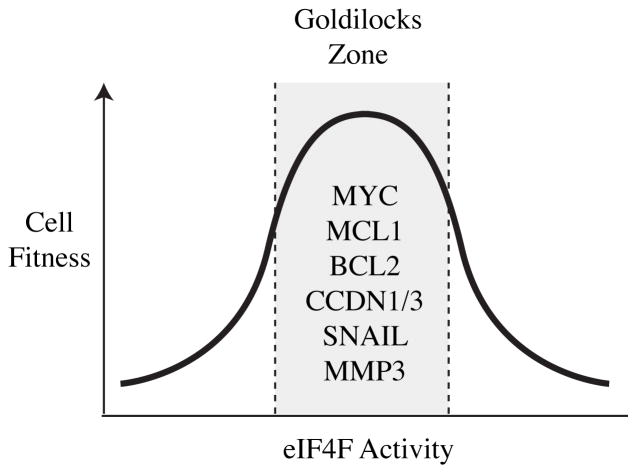
Conclusion – Pushing eIF4F out of the Goldilocks Zone
The Goldilocks principle posits that parameters to maintain a specific state need to fall within a certain margin, or zone, to favor a particular outcome. For example, the distance of the earth from the sun is within a “Goldilocks zone” and hence favorable to life as we know it. The same analogy can be applied to the relationship between eIF4F activity and tumor cell homeostasis. The translational output of a tumor cell needs to fall within a “Goldilocks zone” to optimally support its proteastasis (Fig. 2). Too much eIF4F activity is deleterious to the cell, too little eIF4F activity precludes cellular transformation. Indeed, the preponderance of experimental work has revealed that cellular transformation requires only 2–3 fold increased eIF4E expression whereas depletion of eIF4E activity by only ~50% can reverse the transformed and malignant phenotype (108, 109, 119, 122–124). Perhaps most important for the therapeutic potential of targeting the eIF4F complex is the consistent observation from many experimental studies that malignant cells seem to be tuned to this “Goldilocks zone” of eIF4F activity. That is, malignant cells have selected for a certain enhanced translational output (i.e. enhanced eIF4F activity) necessary for the manifestation of the varied phenotypes responsible for tumor formation and metastatic progression- growth factor autonomy, angiogenesis, enhanced cellular survival, invasiveness and metastatic outgrowth. The particular dependence of malignant cells on this “zone” of enhanced eIF4F activity makes these cells especially susceptible to eIF4F inhibition. Indeed, in multiple preclinical studies in vivo, inhibition of eIF4F activity (via inhibition of eIF4E, Mnk or eIF4A) profoundly impacted malignant cells, inducing tumor cell death, cessation of tumor growth, and repression of metastatic outgrowth without substantially impacting normal cellular and organismal function (114, 124, 133, 178). For example, intravenous administration of the eIF4E ASO dropped eIF4E levels in the liver of treated, xenograft-bearing mice > 80% without affecting liver enzymes or body weight. Yet, in these same animals, a reduction in eIF4E expression in the xenografted tumor of only 50–60% was sufficient to robustly induce apoptosis and flat-line xenograft tumor growth (124).
Figure 2.
Model illustrating eIF4F activity residing within a Goldilocks zone to sustain optimal tumor cell survival. Given that eIF4F activity is critical for sustained output of key players in tumor cell initiation, maintenance, and metastasis, altering this activity can dramatically impact tumor cell fitness. See text for details.
As detailed above, the sustained translation of a specific subset of mRNAs is key to enabling tumor maintenance and metastatic progression- that is to allow for the enhanced, selective expression of the potent growth and survival factors that regulate tumor cell survival, angiogenesis, growth factor autonomy, invasiveness and metastasis. The question arises as to which eIF4E-responsive mRNA(s) is/are particularly critical and whether this is expected to vary among tumor types. If the latter were true, this would entail the development of robust biomarkers to inform on eIF4F dysregulation and inhibition unique to each tumor type – a truly daunting task.
However, the situation may not be this complicated. Firstly, high resolution analysis of copy number variations (CNV) from >3000 cancer samples representing largely 26 different cancer types has documented that among the top 20 most common amplifications are four genes encoding key oncogenic drivers and tumor maintenance factors, whose mRNAs are eIF4E-responsive: MYC, MCL1, BCL2L1(BCL-xL), and CCND1 (151). Although the expression of these proteins may be elevated due to amplification events, if the translation of their mRNAs remains eIF4E-dependent, inhibition of eIF4F (coupled with the naturally rapid turnover of the MYC, MCL1, and CCND1 proteins (207, 208)), is expected to cause a rapid depletion of these proteins and have a dramatic consequence on tumor cell homeostasis- shifting cells out of the Goldilocks zone. MYC, MCL1, and CCND1 have been difficult to “drug” directly and thus inhibiting their production at the translation level is one strategy to overcome this problem.
Targeting the eIF4F complex may potentially bode well for dealing with the intratumoral heterogeneity that drives malignant progression and treatment resistance (209). Intratumoral heterogeneity arises from the diverse selection pressures imposed upon the tumor cell population and may reflect genetic, epigenetic and/or cellular changes. The manifestation of this diversity must involve changes in translational output- i.e. in eIF4F activity. Indeed, the eIF4F complex sits at the junction of numerous, potent oncogenic pathways: Ras-Raf-ERK, Myc, and PI3K-TOR pathways, which in turn may also be activated by other divergent oncogenic stimuli (e.g. - receptor tyrosine kinase activation (EGFR)). As such, these divergent pathways- which reflect the underlying cellular, genetic and epigenetic heterogeneity of the tumor- are critically reliant upon the activity of the eIF4F node for the alterations to the proteome that give rise to phenotypic heterogeneity driving malignant progression and therapeutic resistance. Hence, targeting this complex - this critical node of convergence for so many divergent oncogenic stimuli- may provide a powerful means to address the intratumoral heterogeneity that plagues current cancer therapy.
We’ve come a long way since the discovery of the mRNA cap structure and fundamental studies that defined its biochemical and biological function. One could never have predicted that these fundamental studies would have led to such a profound interest in targeting the eIF4F complex, particularly for the treatment of cancer. The thrust to “translate” these findings to the clinic provides a substantial challenge and will continue to demand rigorous, concerted scientific partnership between academia and industry to reveal the best opportunities to leverage and advance our current knowledge of the eIF4F complex in order that we might develop new therapies to inhibit the translation of mRNAs encoding oncogenic functions.
Acknowledgments
We apologize to those authors whose work we did not cite due to space constraints. Work in the authors laboratories on the role of deregulated translational control in tumorigenesis is supported by grants from the Canadian Institutes of Health Research (MOP-115126 and MOP-106530 to JP), the Canadian Cancer Society Research Institute (CCSRI to JP and NS), a Lilly LIFT award (to JP and NS), the National Institutes of Health (R01 CA140456, R01 CA154916, R01 CA184624 to DR). DR is a Leukemia & Lymphoma scholar.
Footnotes
Ribavirin has been reported to behave as a cap analogue ex vivo (169) However, this has been challenged (170, 171). Importantly, in over 100 antitumor screens involving over 10 different xenograft models, ribavirin failed to show any efficacy as a single agent (http://dtp.nci.nih.gov/dtpstandard/servlet/dwindex?searchtype=namestarts&chemnameboolean=and&outputformat=html&searchlist=ribavirin%0D%0A&Submit=Submit). Any biological outcomes observed with ribavirin are therefore unlikely to be the consequence of inhibiting translation.
References
- 1.Hershey JW, Sonenberg N, Mathews MB. Principles of translational control: an overview. Cold Spring Harbor perspectives in biology. 2012;4(12) doi: 10.1101/cshperspect.a011528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, et al. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes & Dev. 1999;13:1422–37. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horikami SM, De Ferra F, Moyer SA. Characterization of the infections of permissive and nonpermissive cells by host range mutants of vesicular stomatitis virus defective in RNA methylation. Virology. 1984;138(1):1–15. doi: 10.1016/0042-6822(84)90142-9. [DOI] [PubMed] [Google Scholar]
- 4.Pelletier J, Sonenberg N. Insertion mutagenesis to increase secondary structure within the 5′ noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell. 1985;40(3):515–26. doi: 10.1016/0092-8674(85)90200-4. [DOI] [PubMed] [Google Scholar]
- 5.Kozak M. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc Natl Acad Sci U S A. 1986;83(9):2850–4. doi: 10.1073/pnas.83.9.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sonenberg N. ATP/Mg++-dependent cross-linking of cap binding proteins to the 5′ end of eukaryotic mRNA. Nucleic Acids Res. 1981;9(7):1643–56. doi: 10.1093/nar/9.7.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pelletier J, Sonenberg N. Photochemical cross-linking of cap binding proteins to eucaryotic mRNAs: effect of mRNA 5′ secondary structure. Mol Cell Biol. 1985;5(11):3222–30. doi: 10.1128/mcb.5.11.3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kozak M. Influence of mRNA secondary structure on binding and migration of 40S ribosomal subunits. Cell. 1980;19(1):79–90. doi: 10.1016/0092-8674(80)90390-6. [DOI] [PubMed] [Google Scholar]
- 9.Morgan MA, Shatkin AJ. Initiation of reovirus transcription by inosine 5′-triphosphate and properties of 7-methylinosine-capped, inosine-substituted messenger ribonucleic acids. Biochemistry. 1980;19(26):5960–6. doi: 10.1021/bi00567a003. [DOI] [PubMed] [Google Scholar]
- 10.Sonenberg N, Rupprecht KM, Hecht SM, Shatkin AJ. Eukaryotic mRNA cap binding protein: purification by affinity chromatography on sepharose-coupled m7GDP. Proc Natl Acad Sci USA. 1979;76(9):4345–9. doi: 10.1073/pnas.76.9.4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sonenberg N, Trachsel H, Hecht S, Shatkin AJ. Differential stimulation of capped mRNA translation in vitro by cap binding protein. Nature. 1980;285(5763):331–3. doi: 10.1038/285331a0. [DOI] [PubMed] [Google Scholar]
- 12.Tahara SM, Morgan MA, Shatkin AJ. Two forms of purified m7G-cap binding protein with different effects on capped mRNA translation in extracts of uninfected and poliovirus-infected HeLa cells. J Biol Chem. 1981;256(15):7691–4. [PubMed] [Google Scholar]
- 13.Duncan R, Hershey JW. Identification and quantitation of levels of protein synthesis initiation factors in crude HeLa cell lysates by two-dimensional polyacrylamide gel electrophoresis. J Biol Chem. 1983;258(11):7228–35. [PubMed] [Google Scholar]
- 14.Galicia-Vazquez G, Cencic R, Robert F, Agenor AQ, Pelletier J. A cellular response linking eIF4AI activity to eIF4AII transcription. RNA. 2012;18(7):1373–84. doi: 10.1261/rna.033209.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conroy SC, Dever TE, Owens CL, Merrick WC. Characterization of the 46,000-dalton subunit of eIF-4F. Arch Biochem Biophys. 1990;282(2):363–71. doi: 10.1016/0003-9861(90)90130-q. [DOI] [PubMed] [Google Scholar]
- 16.Yoder-Hill J, Pause A, Sonenberg N, Merrick WC. The p46 subunit of eukaryotic initiation factor (eIF)-4F exchanges with eIF-4A. J Biol Chem. 1993;268(8):5566–73. [PubMed] [Google Scholar]
- 17.Nielsen PJ, McMaster GK, Trachsel H. Cloning of eukaryotic protein synthesis initiation factor genes: isolation and characterization of cDNA clones encoding factor eIF-4A. Nucleic Acids Res. 1985;13(19):6867–80. doi: 10.1093/nar/13.19.6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grifo JA, Tahara SM, Morgan MA, Shatkin AJ, Merrick WC. New initiation factor activity required for globin mRNA translation. J Biol Chem. 1983;258(9):5804–10. [PubMed] [Google Scholar]
- 19.Edery I, Humbelin M, Darveau A, Lee KA, Milburn S, Hershey JW, et al. Involvement of eukaryotic initiation factor 4A in the cap recognition process. J Biol Chem. 1983;258(18):11398–403. [PubMed] [Google Scholar]
- 20.Thomas A, Goumans H, Amesz H, Benne R, Voorma HO. A comparison of the initiation factors of eukaryotic protein synthesis from ribosomes and from the postribosomal supernatant. Eur J Biochem. 1979;98(2):329–37. doi: 10.1111/j.1432-1033.1979.tb13192.x. [DOI] [PubMed] [Google Scholar]
- 21.Gradi A, Imataka H, Svitkin YV, Rom E, Raught B, Morino S, et al. A novel functional human eukaryotic translation initiation factor 4G. Mol Cell Biol. 1998;18(1):334–42. doi: 10.1128/mcb.18.1.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kapp LD, Lorsch JR. The molecular mechanics of eukaryotic translation. Annu Rev Biochem. 2004;73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- 23.Imataka H, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol Cell Biol. 1997;17(12):6940–7. doi: 10.1128/mcb.17.12.6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rogers GW, Jr, Richter NJ, Merrick WC. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J Biol Chem. 1999;274(18):12236–44. doi: 10.1074/jbc.274.18.12236. [DOI] [PubMed] [Google Scholar]
- 25.Chen Y, Potratz JP, Tijerina P, Del Campo M, Lambowitz AM, Russell R. DEAD-box proteins can completely separate an RNA duplex using a single ATP. Proc Natl Acad Sci U S A. 2008;105(51):20203–8. doi: 10.1073/pnas.0811075106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ray BK, Lawson TG, Kramer JC, Cladaras MH, Grifo JA, Abramson RD, et al. ATP-dependent unwinding of messenger RNA structure by eukaryotic initiation factors. J Biol Chem. 1985;260(12):7651–8. [PubMed] [Google Scholar]
- 27.Rogers GW, Jr, Richter NJ, Lima WF, Merrick WC. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J Biol Chem. 2001;276(33):30914–22. doi: 10.1074/jbc.M100157200. [DOI] [PubMed] [Google Scholar]
- 28.Marintchev A, Edmonds KA, Marintcheva B, Hendrickson E, Oberer M, Suzuki C, et al. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell. 2009;136(3):447–60. doi: 10.1016/j.cell.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rozovsky N, Butterworth AC, Moore MJ. Interactions between eIF4AI and its accessory factors eIF4B and eIF4H. RNA. 2008;14(10):2136–48. doi: 10.1261/rna.1049608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bi X, Ren J, Goss DJ. Wheat germ translation initiation factor eIF4B affects eIF4A and eIFiso4F helicase activity by increasing the ATP binding affinity of eIF4A. Biochemistry. 2000;39(19):5758–65. doi: 10.1021/bi992322p. [DOI] [PubMed] [Google Scholar]
- 31.Abramson RD, Dever TE, Merrick WC. Biochemical evidence supporting a mechanism for cap-independent and internal initiation of eukaryotic mRNA. J Biol Chem. 1988;263(13):6016–9. [PubMed] [Google Scholar]
- 32.Dmitriev SE, Terenin IM, Dunaevsky YE, Merrick WC, Shatsky IN. Assembly of 48S translation initiation complexes from purified components with mRNAs that have some base pairing within their 5′ untranslated regions. Mol Cell Biol. 2003;23(24):8925–33. doi: 10.1128/MCB.23.24.8925-8933.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shahbazian D, Parsyan A, Petroulakis E, Topisirovic I, Martineau Y, Gibbs BF, et al. Control of cell survival and proliferation by mammalian eukaryotic initiation factor 4B. Mol Cell Biol. 2010;30(6):1478–85. doi: 10.1128/MCB.01218-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarkar G, Edery I, Gallo R, Sonenberg N. Preferential stimulation of rabbit alpha globin mRNA translation by a cap-binding protein complex. Biochim Biophys Acta. 1984;783(2):122–9. doi: 10.1016/0167-4781(84)90003-4. [DOI] [PubMed] [Google Scholar]
- 35.Edery I, Lee KA, Sonenberg N. Functional characterization of eukaryotic mRNA cap binding protein complex: effects on translation of capped and naturally uncapped RNAs. Biochemistry. 1984;23(11):2456–62. doi: 10.1021/bi00306a021. [DOI] [PubMed] [Google Scholar]
- 36.Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16(22):2906–22. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Svitkin YV, Pause A, Haghighat A, Pyronnet S, Witherell G, Belsham GJ, et al. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA. 2001;7(3):382–94. doi: 10.1017/s135583820100108x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell. 1997;89(6):951–61. doi: 10.1016/s0092-8674(00)80280-9. [DOI] [PubMed] [Google Scholar]
- 39.Tomoo K, Shen X, Okabe K, Nozoe Y, Fukuhara S, Morino S, et al. Crystal structures of 7-methylguanosine 5′-triphosphate (m(7)GTP)- and P(1)-7-methylguanosine-P(3)-adenosine-5′,5′-triphosphate (m(7)GpppA)-bound human full-length eukaryotic initiation factor 4E: biological importance of the C-terminal flexible region. Biochem J. 2002;362(Pt 3):539–44. doi: 10.1042/0264-6021:3620539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsuo H, Li H, McGuire AM, Fletcher CM, Gingras AC, Sonenberg N, et al. Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein. Nat Struct Biol. 1997;4(9):717–24. doi: 10.1038/nsb0997-717. [DOI] [PubMed] [Google Scholar]
- 41.Berset C, Zurbriggen A, Djafarzadeh S, Altmann M, Trachsel H. RNA-binding activity of translation initiation factor eIF4G1 from Saccharomyces cerevisiae. Rna. 2003;9(7):871–80. doi: 10.1261/rna.5380903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yanagiya A, Svitkin YV, Shibata S, Mikami S, Imataka H, Sonenberg N. Requirement of RNA binding of mammalian eukaryotic translation initiation factor 4GI (eIF4GI) for efficient interaction of eIF4E with the mRNA cap. Mol Cell Biol. 2009;29(6):1661–9. doi: 10.1128/MCB.01187-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haghighat A, Sonenberg N. eIF4G dramatically enhances the binding of eIF4E to the mRNA 5′-cap structure. J Biol Chem. 1997;272(35):21677–80. doi: 10.1074/jbc.272.35.21677. [DOI] [PubMed] [Google Scholar]
- 44.Godefroy-Colburn T, Ravelonandro M, Pinck L. Cap accessibility correlates with the initiation efficiency of alfalfa mosaic virus RNAs. Eur J Biochem. 1985;147(3):549–52. doi: 10.1111/j.0014-2956.1985.00549.x. [DOI] [PubMed] [Google Scholar]
- 45.Lawson TG, Cladaras MH, Ray BK, Lee KA, Abramson RD, Merrick WC, et al. Discriminatory interaction of purified eukaryotic initiation factors 4F plus 4A with the 5′ ends of reovirus messenger RNAs. J Biol Chem. 1988;263:7266–76. [PubMed] [Google Scholar]
- 46.Parkin NT, Cohen EA, Darveau A, Rosen C, Haseltine W, Sonenberg N. Mutational analysis of the 5′ non-coding region of human immunodeficiency virus type 1: effects of secondary structure on translation. EMBO J. 1988;7:2831–7. doi: 10.1002/j.1460-2075.1988.tb03139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Svitkin YV, Evdokimova VM, Brasey A, Pestova TV, Fantus D, Yanagiya A, et al. General RNA-binding proteins have a function in poly(A)-binding protein-dependent translation. EMBO J. 2009;28(1):58–68. doi: 10.1038/emboj.2008.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Svitkin YV, Ovchinnikov LP, Dreyfuss G, Sonenberg N. General RNA binding proteins render translation cap dependent. Embo J. 1996;15(24):7147–55. [PMC free article] [PubMed] [Google Scholar]
- 49.Lawson TG, Ray BK, Dodds JT, Grifo JA, Abramson RD, Merrick WC, et al. Influence of 5′ proximal secondary structure on the translational efficiency of eukaryotic mRNAs and on their interaction with initiation factors. J Biol Chem. 1986;261(30):13979–89. [PubMed] [Google Scholar]
- 50.Rogers GW, Jr, Lima WF, Merrick WC. Further characterization of the helicase activity of eIF4A. Substrate specificity. J Biol Chem. 2001;276(16):12598–608. doi: 10.1074/jbc.M007560200. [DOI] [PubMed] [Google Scholar]
- 51.Rozen F, Edery I, Meerovitch K, Dever TE, Merrick WC, Sonenberg N. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol Cell Biol. 1990;10(3):1134–44. doi: 10.1128/mcb.10.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pause A, Sonenberg N. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 1992;11(7):2643–54. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feoktistova K, Tuvshintogs E, Do A, Fraser CS. Human eIF4E promotes mRNA restructuring by stimulating eIF4A helicase activity. Proc Natl Acad Sci U S A. 2013;110(33):13339–44. doi: 10.1073/pnas.1303781110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Duncan R, Milburn SC, Hershey JW. Regulated phosphorylation and low abundance of HeLa cell initiation factor eIF-4F suggest a role in translational control. Heat shock effects on eIF-4F. J Biol Chem. 1987;262(1):380–8. [PubMed] [Google Scholar]
- 55.Sonenberg N. Cap-binding proteins of eukaryotic messenger RNA: functions in initiation and control of translation. Prog Nucleic Acid Res Mol Biol. 1988;35:173–207. doi: 10.1016/s0079-6603(08)60614-5. [DOI] [PubMed] [Google Scholar]
- 56.Lindqvist L, Imataka H, Pelletier J. Cap-dependent eukaryotic initiation factor-mRNA interactions probed by cross-linking. RNA. 2008;14(5):960–9. doi: 10.1261/rna.971208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tarun SZ, Jr, Sachs AB. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. Embo J. 1996;15(24):7168–77. [PMC free article] [PubMed] [Google Scholar]
- 58.Tarun SZ, Jr, Sachs AB. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev. 1995;9(23):2997–3007. doi: 10.1101/gad.9.23.2997. [DOI] [PubMed] [Google Scholar]
- 59.Kahvejian A, Svitkin YV, Sukarieh R, M’Boutchou M-N, Sonenberg N. Mammalian poly(A)-binding protien is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes & Dev. 2005;19:104–13. doi: 10.1101/gad.1262905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luo Y, Goss DJ. Homeostasis in mRNA initiation: wheat germ poly(A)-binding protein lowers the activation energy barrier to initiation complex formation. J Biol Chem. 2001;276(46):43083–6. doi: 10.1074/jbc.M104970200. [DOI] [PubMed] [Google Scholar]
- 61.Borman AM, Michel YM, Kean KM. Biochemical characterisation of cap-poly(A) synergy in rabbit reticulocyte lysates: the eIF4G-PABP interaction increases the functional affinity of eIF4E for the capped mRNA 5′-end. Nucleic Acids Res. 2000;28(21):4068–75. doi: 10.1093/nar/28.21.4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Borman AM, Michel YM, Malnou CE, Kean KM. Free poly(A) stimulates capped mRNA translation in vitro through the eIF4G-poly(A)-binding protein interaction. J Biol Chem. 2002;277(39):36818–24. doi: 10.1074/jbc.M205065200. [DOI] [PubMed] [Google Scholar]
- 63.Bi X, Goss DJ. Wheat germ poly(A)-binding protein increases the ATPase and the RNA helicase activity of translation initiation factors eIF4A, eIF4B, and eIF-iso4F. J Biol Chem. 2000;275(23):17740–6. doi: 10.1074/jbc.M909464199. [DOI] [PubMed] [Google Scholar]
- 64.Park EH, Walker SE, Lee JM, Rothenburg S, Lorsch JR, Hinnebusch AG. Multiple elements in the eIF4G1 N-terminus promote assembly of eIF4G1*PABP mRNPs in vivo. EMBO J. 2011;30(2):302–16. doi: 10.1038/emboj.2010.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Asselbergs FA, Peters W, Venrooij WJ, Bloemendal H. Diminished sensitivity of re-initiation of translation to inhibition by cap analogues in reticulocyte lysates. Eur J Biochem. 1978;88(2):483–8. doi: 10.1111/j.1432-1033.1978.tb12473.x. [DOI] [PubMed] [Google Scholar]
- 66.Park EH, Zhang F, Warringer J, Sunnerhagen P, Hinnebusch AG. Depletion of eIF4G from yeast cells narrows the range of translational efficiencies genome-wide. BMC Genomics. 2011;12:68. doi: 10.1186/1471-2164-12-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wakiyama M, Imataka H, Sonenberg N. Interaction of eIF4G with poly(A)-binding protein stimulates translation and is critical for Xenopus oocyte maturation. Curr Biol. 2000;10(18):1147–50. doi: 10.1016/s0960-9822(00)00701-6. [DOI] [PubMed] [Google Scholar]
- 68.Shantz LM, Pegg AE. Overproduction of ornithine decarboxylase caused by relief of translational repression is associated with neoplastic transformation. Cancer Res. 1994;54(9):2313–6. [PubMed] [Google Scholar]
- 69.De Benedetti A, Graff JR. eIF-4E expression and its role in malignancies and metastases. Oncogene. 2004;23(18):3189–99. doi: 10.1038/sj.onc.1207545. [DOI] [PubMed] [Google Scholar]
- 70.Silvera D, Formenti SC, Schneider RJ. Translational control in cancer. Nat Rev Cancer. 2010;10(4):254–66. doi: 10.1038/nrc2824. [DOI] [PubMed] [Google Scholar]
- 71.Grolleau A, Bowman J, Pradet-Balade B, Puravs E, Hanash S, Garcia-Sanz JA, et al. Global and specific translational control by rapamycin in T cells uncovered by microarrays and proteomics. J Biol Chem. 2002;277(25):22175–84. doi: 10.1074/jbc.M202014200. [DOI] [PubMed] [Google Scholar]
- 72.Mamane Y, Petroulakis E, Martineau Y, Sato TA, Larsson O, Rajasekhar VK, et al. Epigenetic activation of a subset of mRNAs by eIF4E explains its effects on cell proliferation. PLoS ONE. 2007;2(2):e242. doi: 10.1371/journal.pone.0000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tcherkezian J, Cargnello M, Romeo Y, Huttlin EL, Lavoie G, Gygi SP, et al. Proteomic analysis of cap-dependent translation identifies LARP1 as a key regulator of 5′TOP mRNA translation. Genes Dev. 2014;28(4):357–71. doi: 10.1101/gad.231407.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485(7396):55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485(7396):109–13. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Morita M, Gravel SP, Chenard V, Sikstrom K, Zheng L, Alain T, et al. mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab. 2013;18(5):698–711. doi: 10.1016/j.cmet.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 77.Piccirillo CA, Bjur E, Topisirovic I, Sonenberg N, Larsson O. Translational control of immune responses: from transcripts to translatomes. Nature immunology. 2014;15(6):503–11. doi: 10.1038/ni.2891. [DOI] [PubMed] [Google Scholar]
- 78.Wendel HG, Silva RL, Malina A, Mills JR, Zhu H, Ueda T, et al. Dissecting eIF4E action in tumorigenesis. Genes Dev. 2007;21(24):3232–7. doi: 10.1101/gad.1604407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Furic L, Rong L, Larsson O, Koumakpayi IH, Yoshida K, Brueschke A, et al. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc Natl Acad Sci U S A. 2010;107(32):14134–9. doi: 10.1073/pnas.1005320107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Robichaud N, Del Rincon SV, Huor B, Alain T, Petruccelli LA, Hearnden J, et al. Phosphorylation of eIF4E promotes EMT and metastasis via translational control of SNAIL and MMP-3. Oncogene. 2014 doi: 10.1038/onc.2014.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Barna M, Pusic A, Zollo O, Costa M, Kondrashov N, Rego E, et al. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature. 2008;456(7224):971–5. doi: 10.1038/nature07449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12(1):9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 83.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–63. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 84.Yang HS, Jansen AP, Komar AA, Zheng X, Merrick WC, Costes S, et al. The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol Cell Biol. 2003;23(1):26–37. doi: 10.1128/MCB.23.1.26-37.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rotili D, Tarantino D, Artico M, Nawrozkij MB, Gonzalez-Ortega E, Clotet B, et al. Diarylpyrimidine-dihydrobenzyloxopyrimidine hybrids: new, wide-spectrum anti-HIV-1 agents active at (sub)-nanomolar level. J Med Chem. 2011;54(8):3091–6. doi: 10.1021/jm101626c. [DOI] [PubMed] [Google Scholar]
- 86.Suzuki C, Garces RG, Edmonds KA, Hiller S, Hyberts SG, Marintchev A, et al. PDCD4 inhibits translation initiation by binding to eIF4A using both its MA3 domains. Proc Natl Acad Sci U S A. 2008;105(9):3274–9. doi: 10.1073/pnas.0712235105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Maga G, Falchi F, Radi M, Botta L, Casaluce G, Bernardini M, et al. Toward the discovery of novel anti-HIV drugs. Second-generation inhibitors of the cellular ATPase DDX3 with improved anti-HIV activity: synthesis, structure-activity relationship analysis, cytotoxicity studies, and target validation. ChemMedChem. 2011;6(8):1371–89. doi: 10.1002/cmdc.201100166. [DOI] [PubMed] [Google Scholar]
- 88.Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M. S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science. 2006;314(5798):467–71. doi: 10.1126/science.1130276. [DOI] [PubMed] [Google Scholar]
- 89.Shahbazian D, Roux PP, Mieulet V, Cohen MS, Raught B, Taunton J, et al. The mTOR/PI3K and MAPK pathways converge on eIF4B to control its phosphorylation and activity. Embo J. 2006;25(12):2781–91. doi: 10.1038/sj.emboj.7601166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Holz MK, Ballif BA, Gygi SP, Blenis J. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell. 2005;123(4):569–80. doi: 10.1016/j.cell.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 91.Kroczynska B, Kaur S, Katsoulidis E, Majchrzak-Kita B, Sassano A, Kozma SC, et al. Interferon-dependent engagement of eukaryotic initiation factor 4B via S6 kinase (S6K)- and ribosomal protein S6K-mediated signals. Mol Cell Biol. 2009;29(10):2865–75. doi: 10.1128/MCB.01537-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.van Gorp AG, van der Vos KE, Brenkman AB, Bremer A, van den Broek N, Zwartkruis F, et al. AGC kinases regulate phosphorylation and activation of eukaryotic translation initiation factor 4B. Oncogene. 2009;28(1):95–106. doi: 10.1038/onc.2008.367. [DOI] [PubMed] [Google Scholar]
- 93.Ma L, Chen Z, Erdjument-Bromage H, Tempst P, Pandolfi PP. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell. 2005;121(2):179–93. doi: 10.1016/j.cell.2005.02.031. [DOI] [PubMed] [Google Scholar]
- 94.Rinker-Schaeffer CW, Austin V, Zimmer S, Rhoads RE. Ras transformation of cloned rat embryo fibroblasts results in increased rates of protein synthesis and phosphorylation of eukaryotic initiation factor 4E. J Biol Chem. 1992;267(15):10659–64. [PubMed] [Google Scholar]
- 95.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10(5):307–18. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 96.Flynn A, Proud CG. Serine 209, not serine 53, is the major site of phosphorylation in initiation factor eIF-4E in serum-treated Chinese hamster ovary cells. J Biol Chem. 1995;270(37):21684–8. doi: 10.1074/jbc.270.37.21684. [DOI] [PubMed] [Google Scholar]
- 97.Joshi B, Cai AL, Keiper BD, Minich WB, Mendez R, Beach CM, et al. Phosphorylation of eukaryotic protein synthesis initiation factor 4E at Ser-209. J Biol Chem. 1995;270(24):14597–603. doi: 10.1074/jbc.270.24.14597. [DOI] [PubMed] [Google Scholar]
- 98.Waskiewicz AJ, Johnson JC, Penn B, Mahalingam M, Kimball SR, Cooper JA. Phosphorylation of the cap-binding protein eukaryotic translation initiation factor 4E by protein kinase Mnk1 in vivo. Mol Cell Biol. 1999;19(3):1871–80. doi: 10.1128/mcb.19.3.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pyronnet S, Imataka H, Gingras AC, Fukunaga R, Hunter T, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. EMBO J. 1999;18(1):270–9. doi: 10.1093/emboj/18.1.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ueda T, Watanabe-Fukunaga R, Fukuyama H, Nagata S, Fukunaga R. Mnk2 and Mnk1 Are Essential for Constitutive and Inducible Phosphorylation of Eukaryotic Initiation Factor 4E but Not for Cell Growth or Development. Mol Cell Biol. 2004;24(15):6539–49. doi: 10.1128/MCB.24.15.6539-6549.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Scheper GC, van Kollenburg B, Hu J, Luo Y, Goss DJ, Proud CG. Phosphorylation of eukaryotic initiation factor 4E markedly reduces its affinity for capped mRNA. J Biol Chem. 2002;277(5):3303–9. doi: 10.1074/jbc.M103607200. [DOI] [PubMed] [Google Scholar]
- 102.Slepenkov SV, Darzynkiewicz E, Rhoads RE. Stopped-flow kinetic analysis of eIF4E and phosphorylated eIF4E binding to cap analogs and capped oligoribonucleotides: evidence for a one-step binding mechanism. J Biol Chem. 2006;281(21):14927–38. doi: 10.1074/jbc.M601653200. [DOI] [PubMed] [Google Scholar]
- 103.Zuberek J, Jemielity J, Jablonowska A, Stepinski J, Dadlez M, Stolarski R, et al. Influence of Electric Charge Variation at Residues 209 and 159 on the Interaction of eIF4E with the mRNA 5′ Terminus. Biochemistry. 2004;43(18):5370–9. doi: 10.1021/bi030266t. [DOI] [PubMed] [Google Scholar]
- 104.Zuberek J, Wyslouch-Cieszynska A, Niedzwiecka A, Dadlez M, Stepinski J, Augustyniak W, et al. Phosphorylation of eIF4E attenuates its interaction with mRNA 5′ cap analogs by electrostatic repulsion: intein-mediated protein ligation strategy to obtain phosphorylated protein. RNA. 2003;9(1):52–61. doi: 10.1261/rna.2133403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Scheper GC, Proud CG. Does phosphorylation of the cap-binding protein eIF4E play a role in translation initiation? Eur J Biochem. 2002;269(22):5350–9. doi: 10.1046/j.1432-1033.2002.03291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Topisirovic I, Ruiz-Gutierrez M, Borden KL. Phosphorylation of the eukaryotic translation initiation factor eIF4E contributes to its transformation and mRNA transport activities. Cancer Res. 2004;64(23):8639–42. doi: 10.1158/0008-5472.CAN-04-2677. [DOI] [PubMed] [Google Scholar]
- 107.Lazaris-Karatzas A, Montine KS, Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature. 1990;345(6275):544–7. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- 108.Rinker-Schaeffer CW, Graff JR, De Benedetti A, Zimmer SG, Rhoads RE. Decreasing the level of translation initiation factor 4E with antisense RNA causes reversal of ras-mediated transformation and tumorigenesis of cloned rat embryo fibroblasts. Int J Cancer. 1993;55(5):841–7. doi: 10.1002/ijc.2910550525. [DOI] [PubMed] [Google Scholar]
- 109.Ruggero D, Montanaro L, Ma L, Xu W, Londei P, Cordon-Cardo C, et al. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat Med. 2004;10(5):484–6. doi: 10.1038/nm1042. [DOI] [PubMed] [Google Scholar]
- 110.Wendel H-G, de Stanchina E, Fridman JS, Malina A, Ray S, Kogan S, et al. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 2004;428:332–7. doi: 10.1038/nature02369. [DOI] [PubMed] [Google Scholar]
- 111.Polunovsky VA, Rosenwald IB, Tan AT, White J, Chiang L, Sonenberg N, et al. Translational control of programmed cell death: eukaryotic translation initiation factor 4E blocks apoptosis in growth-factor-restricted fibroblasts with physiologically expressed or deregulated Myc. Mol Cell Biol. 1996;16(11):6573–81. doi: 10.1128/mcb.16.11.6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tan A, Bitterman P, Sonenberg N, Peterson M, Polunovsky V. Inhibition of Myc-dependent apoptosis by eukaryotic translation initiation factor 4E requires cyclin D1. Oncogene. 2000;19(11):1437–47. doi: 10.1038/sj.onc.1203446. [DOI] [PubMed] [Google Scholar]
- 113.Ueda T, Sasaki M, Elia AJ, Chio II, Hamada K, Fukunaga R, et al. Combined deficiency for MAP kinase-interacting kinase 1 and 2 (Mnk1 and Mnk2) delays tumor development. Proc Natl Acad Sci U S A. 2010;107(32):13984–90. doi: 10.1073/pnas.1008136107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Konicek BW, Stephens JR, McNulty AM, Robichaud N, Peery RB, Dumstorf CA, et al. Therapeutic inhibition of MAP kinase interacting kinase blocks eukaryotic initiation factor 4E phosphorylation and suppresses outgrowth of experimental lung metastases. Cancer Res. 2011;71(5):1849–57. doi: 10.1158/0008-5472.CAN-10-3298. [DOI] [PubMed] [Google Scholar]
- 115.Rajasekhar VK, Viale A, Socci ND, Wiedmann M, Hu X, Holland EC. Oncogenic Ras and Akt signaling contribute to glioblastoma formation by differential recruitment of existing mRNAs to polysomes. Mol Cell. 2003;12(4):889–901. doi: 10.1016/s1097-2765(03)00395-2. [DOI] [PubMed] [Google Scholar]
- 116.Avdulov S, Li S, Michalek V, Burrichter D, Peterson M, Perlman DM, et al. Activation of translation complex eIF4F is essential for the genesis and maintenance of the malignant phenotype in human mammary epithelial cells. Cancer Cell. 2004;5(6):553–63. doi: 10.1016/j.ccr.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 117.Nasr Z, Robert F, Porco JA, Jr, Muller WJ, Pelletier J. eIF4F suppression in breast cancer affects maintenance and progression. Oncogene. 2012 doi: 10.1038/onc.2012.105. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rosenwald IB, Lazaris-Karatzas A, Sonenberg N, Schmidt EV. Elevated levels of cyclin D1 protein in response to increased expression of eukaryotic initiation factor 4E. Mol Cell Biol. 1993;13(12):7358–63. doi: 10.1128/mcb.13.12.7358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rousseau D, Kaspar R, Rosenwald I, Gehrke L, Sonenberg N. Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc Natl Acad Sci U S A. 1996;93(3):1065–70. doi: 10.1073/pnas.93.3.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shantz LM, Coleman CS, Pegg AE. Expression of an ornithine decarboxylase dominant-negative mutant reverses eukaryotic initiation factor 4E-induced cell transformation. Cancer Res. 1996;56(22):5136–40. [PubMed] [Google Scholar]
- 121.Graff JR, Konicek BW, Carter JH, Marcusson EG. Targeting the eukaryotic translation initiation factor 4E for cancer therapy. Cancer Res. 2008;68(3):631–4. doi: 10.1158/0008-5472.CAN-07-5635. [DOI] [PubMed] [Google Scholar]
- 122.Wendel HG, De Stanchina E, Fridman JS, Malina A, Ray S, Kogan S, et al. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 2004;428(6980):332–7. doi: 10.1038/nature02369. [DOI] [PubMed] [Google Scholar]
- 123.Graff JR, Boghaert ER, De Benedetti A, Tudor DL, Zimmer CC, Chan SK, et al. Reduction of translation initiation factor 4E decreases the malignancy of ras-transformed cloned rat embryo fibroblasts. Int J Cancer. 1995;60(2):255–63. doi: 10.1002/ijc.2910600221. [DOI] [PubMed] [Google Scholar]
- 124.Graff JR, Konicek BW, Vincent TM, Lynch RL, Monteith D, Weir SN, et al. Therapeutic suppression of translation initiation factor eIF4E expression reduces tumor growth without toxicity. J Clin Invest. 2007;117(9):2638–48. doi: 10.1172/JCI32044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Graff JR, Zimmer SG. Translational control and metastatic progression: Enhanced activity of the mRNA cap-binding protein eIF-4E selectively enhances translation of metastasis-related mRNAs. Clin Exp Met. 2003;20:265–73. doi: 10.1023/a:1022943419011. [DOI] [PubMed] [Google Scholar]
- 126.Nasr Z, Pelletier J. Tumor progression and metastasis: role of translational deregulation. Anticancer Res. 2012;32(8):3077–84. [PubMed] [Google Scholar]
- 127.Nasr Z, Robert F, Porco JA, Jr, Muller WJ, Pelletier J. eIF4F suppression in breast cancer affects maintenance and progression. Oncogene. 2013;32(7):861–71. doi: 10.1038/onc.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zimmer SG, DeBenedetti A, Graff JR. Translational control of malignancy: the mRNA cap-binding protein, eIF4E, as a central regulator of tumor formation, growth, invasion and metastasis. Anticancer Res. 2000;20:1343–1351. [PubMed] [Google Scholar]
- 129.Kevil CG, De Benedetti A, Payne DK, Coe LL, Laroux FS, Alexander JS. Translational regulation of vascular permeability factor by eukaryotic initiation factor 4E: implications for tumor angiogenesis. Int J Cancer. 1996;65(6):785–90. doi: 10.1002/(SICI)1097-0215(19960315)65:6<785::AID-IJC14>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 130.Kevil C, Carter P, Hu B, DeBenedetti A. Translational enhancement of FGF-2 by eIF-4 factors, and alternate utilization of CUG and AUG codons for translation initiation. Oncogene. 1995;11(11):2339–48. [PubMed] [Google Scholar]
- 131.Zhou S, Wang GP, Liu C, Zhou M. Eukaryotic initiation factor 4E (eIF4E) and angiogenesis: prognostic markers for breast cancer. BMC Cancer. 2006;6:231. doi: 10.1186/1471-2407-6-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nathan CA, Franklin S, Abreo FW, Nassar R, de Benedetti A, Williams J, et al. Expression of eIF4E during head and neck tumorigenesis: possible role in angiogenesis. Laryngoscope. 1999;109(8):1253–8. doi: 10.1097/00005537-199908000-00013. [DOI] [PubMed] [Google Scholar]
- 133.Cencic R, Carrier M, Galicia-Vazquez G, Bordeleau ME, Sukarieh R, Bourdeau A, et al. Antitumor activity and mechanism of action of the cyclopenta[b]benzofuran, silvestrol. PLoS ONE. 2009;4(4):e5223. doi: 10.1371/journal.pone.0005223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Li S, Takasu T, Perlman DM, Peterson MS, Burrichter D, Avdulov S, et al. Translation factor eIF4E rescues cells from Myc-dependent apoptosis by inhibiting cytochrome c release. J Biol Chem. 2003;278(5):3015–22. doi: 10.1074/jbc.M208821200. [DOI] [PubMed] [Google Scholar]
- 135.Larsson O, Perlman DM, Fan D, Reilly CS, Peterson M, Dahlgren C, et al. Apoptosis resistance downstream of eIF4E: posttranscriptional activation of an anti-apoptotic transcript carrying a consensus hairpin structure. Nucleic Acids Res. 2006;34(16):4375–86. doi: 10.1093/nar/gkl558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wai PY, Kuo PC. Osteopontin: regulation in tumor metastasis. Cancer metastasis reviews. 2008;27(1):103–18. doi: 10.1007/s10555-007-9104-9. [DOI] [PubMed] [Google Scholar]
- 137.Graff JR, Konicek BW, Lynch RL, Dumstorf CA, Dowless MS, McNulty AM, et al. eIF4E activation is commonly elevated in advanced human prostate cancers and significantly related to reduced patient survival. Cancer Res. 2009;69(9):3866–73. doi: 10.1158/0008-5472.CAN-08-3472. [DOI] [PubMed] [Google Scholar]
- 138.Pardo OE, Arcaro A, Salerno G, Raguz S, Downward J, Seckl MJ. Fibroblast growth factor-2 induces translational regulation of Bcl-XL and Bcl-2 via a MEK-dependent pathway: correlation with resistance to etoposide-induced apoptosis. J Biol Chem. 2002;277(14):12040–6. doi: 10.1074/jbc.M109006200. [DOI] [PubMed] [Google Scholar]
- 139.Jiang Y, Muschel RJ. Regulation of matrix metalloproteinase-9 (MMP-9) by translational efficiency in murine prostate carcinoma cells. Cancer Res. 2002;62(6):1910–4. [PubMed] [Google Scholar]
- 140.Yang YJ, Zhang YL, Li X, Dan HL, Lai ZS, Wang JD, et al. Contribution of eIF-4E inhibition to the expression and activity of heparanase in human colon adenocarcinoma cell line: LS-174T. World journal of gastroenterology : WJG. 2003;9(8):1707–12. doi: 10.3748/wjg.v9.i8.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Evdokimova V, Tognon C, Ng T, Ruzanov P, Melnyk N, Fink D, et al. Translational activation of snail1 and other developmentally regulated transcription factors by YB-1 promotes an epithelial-mesenchymal transition. Cancer Cell. 2009;15(5):402–15. doi: 10.1016/j.ccr.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 142.Haydon MS, Googe JD, Sorrells DS, Ghali GE, Li BD. Progression of eIF4e gene amplification and overexpression in benign and malignant tumors of the head and neck. Cancer. 2000;88(12):2803–10. doi: 10.1002/1097-0142(20000615)88:12<2803::aid-cncr20>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 143.Rosenwald IB, Chen JJ, Wang S, Savas L, London IM, Pullman J. Upregulation of protein synthesis initiation factor eIF-4E is an early event during colon carcinogenesis. Oncogene. 1999;18(15):2507–17. doi: 10.1038/sj.onc.1202563. [DOI] [PubMed] [Google Scholar]
- 144.Armengol G, Rojo F, Castellvi J, Iglesias C, Cuatrecasas M, Pons B, et al. 4E-binding protein 1: a key molecular “funnel factor” in human cancer with clinical implications. Cancer Res. 2007;67(16):7551–5. doi: 10.1158/0008-5472.CAN-07-0881. [DOI] [PubMed] [Google Scholar]
- 145.Wendel HG, Malina A, Zhao Z, Zender L, Kogan SC, Cordon-Cardo C, et al. Determinants of sensitivity and resistance to rapamycin-chemotherapy drug combinations in vivo. Cancer Res. 2006;66(15):7639–46. doi: 10.1158/0008-5472.CAN-06-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ilic N, Utermark T, Widlund HR, Roberts TM. PI3K-targeted therapy can be evaded by gene amplification along the MYC-eukaryotic translation initiation factor 4E (eIF4E) axis. Proc Natl Acad Sci U S A. 2011;108(37):E699–708. doi: 10.1073/pnas.1108237108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cope CL, Gilley R, Balmanno K, Sale MJ, Howarth KD, Hampson M, et al. Adaptation to mTOR kinase inhibitors by amplification of eIF4E to maintain cap-dependent translation. J Cell Sci. 2014;127(Pt 4):788–800. doi: 10.1242/jcs.137588. [DOI] [PubMed] [Google Scholar]
- 148.Boussemart L, Malka-Mahieu H, Girault I, Allard D, Hemmingsson O, Tomasic G, et al. eIF4F is a nexus of resistance to anti-BRAF and anti-MEK cancer therapies. Nature. 2014 doi: 10.1038/nature13572. [DOI] [PubMed] [Google Scholar]
- 149.Croft A, Tay KH, Boyd SC, Guo ST, Jiang CC, Lai F, et al. Oncogenic activation of MEK/ERK primes melanoma cells for adaptation to endoplasmic reticulum stress. The Journal of investigative dermatology. 2014;134(2):488–97. doi: 10.1038/jid.2013.325. [DOI] [PubMed] [Google Scholar]
- 150.Robert F, Roman W, Bramoulle A, Fellmann C, Roulston A, Shustik C, et al. Translation initiation factor eIF4F modifies the dexamethasone response in multiple myeloma. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1402650111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463(7283):899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hill JM, Roberts J, Loeb E, Khan A, MacLellan A, Hill RW. L-asparaginase therapy for leukemia and other malignant neoplasms. Remission in human leukemia. JAMA : the journal of the American Medical Association. 1967;202(9):882–8. [PubMed] [Google Scholar]
- 153.O’Brien S, Kantarjian H, Keating M, Beran M, Koller C, Robertson LE, et al. Homoharringtonine therapy induces responses in patients with chronic myelogenous leukemia in late chronic phase. Blood. 1995;86(9):3322–6. [PubMed] [Google Scholar]
- 154.Malina A, Mills JR, Pelletier J. Emerging therapeutics targeting mRNA translation. Cold Spring Harbor perspectives in biology. 2012;4(4):a012377. doi: 10.1101/cshperspect.a012377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Chu J, Pelletier J. Targeting the eIF4A RNA helicase as an anti-neoplastic approach. Biochim Biophys Acta. 2014 doi: 10.1016/j.bbagrm.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 156.DeFatta RJ, Nathan CO, De Benedetti A. Antisense RNA to eIF4E suppresses oncogenic properties of a head and neck squamous cell carcinoma cell line. Laryngoscope. 2000;110(6):928–33. doi: 10.1097/00005537-200006000-00007. [DOI] [PubMed] [Google Scholar]
- 157.De Benedetti A, Joshi-Barve S, Rinker-Schaeffer C, Rhoads RE. Expression of antisense RNA against initiation factor eIF-4E mRNA in HeLa cells results in lengthened cell division times, diminished translation rates, and reduced levels of both eIF-4E and the p220 component of eIF-4F. Mol Cell Biol. 1991;11(11):5435–45. doi: 10.1128/mcb.11.11.5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.DeFatta RJ, Nathan CA, De Benedetti A. Antisense RNA to eIF4E suppresses oncogenic properties of a head and neck squamous cell carcinoma cell line. Laryngoscope. 2000;110(6):928–33. doi: 10.1097/00005537-200006000-00007. [DOI] [PubMed] [Google Scholar]
- 159.Li S, Sonenberg N, Gingras AC, Peterson M, Avdulov S, Polunovsky VA, et al. Translational control of cell fate: availability of phosphorylation sites on translational repressor 4E-BP1 governs its proapoptotic potency. Mol Cell Biol. 2002;22(8):2853–61. doi: 10.1128/MCB.22.8.2853-2861.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Herbert TP, Fahraeus R, Prescott A, Lane DP, Proud CG. Rapid induction of apoptosis mediated by peptides that bind initiation factor eIF4E. Curr Biol. 2000;10(13):793–6. doi: 10.1016/s0960-9822(00)00567-4. [DOI] [PubMed] [Google Scholar]
- 161.Brown CJ, Lim JJ, Leonard T, Lim HC, Chia CS, Verma CS, et al. Stabilizing the eIF4G1 alpha-helix increases its binding affinity with eIF4E: implications for peptidomimetic design strategies. J Mol Biol. 2011;405(3):736–53. doi: 10.1016/j.jmb.2010.10.045. [DOI] [PubMed] [Google Scholar]
- 162.Lin CJ, Nasr Z, Premsrirut PK, Porco JA, Jr, Hippo Y, Lowe SW, et al. Targeting Synthetic Lethal Interactions between Myc and the eIF4F Complex Impedes Tumorigenesis. Cell Reports. 2012;1(4):325–33. doi: 10.1016/j.celrep.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Pourdehnad M, Truitt ML, Siddiqi IN, Ducker GS, Shokat KM, Ruggero D. Myc and mTOR converge on a common node in protein synthesis control that confers synthetic lethality in Myc-driven cancers. Proc Natl Acad Sci U S A. 2013;110(29):11988–93. doi: 10.1073/pnas.1310230110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Novac O, Guenier AS, Pelletier J. Inhibitors of protein synthesis identified by a high throughput multiplexed translation screen. Nucleic Acids Res. 2004;32(3):902–15. doi: 10.1093/nar/gkh235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Cencic R, Hall DR, Robert F, Du Y, Min J, Li L, et al. Reversing chemoresistance by small molecule inhibition of the translation initiation complex eIF4F. Proc Natl Acad Sci U S A. 2011;108(3):1046–51. doi: 10.1073/pnas.1011477108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Moerke NJ, Aktas H, Chen H, Cantel S, Reibarkh MY, Fahmy A, et al. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell. 2007;128(2):257–67. doi: 10.1016/j.cell.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 167.Jemielity J, Kowalska J, Rydzik AM, Darzynkiewicz E. Synthetic mRNA cap analogs with a modified triphosphate bridge – synthesis, applications and prospects. New Journal of Chemistry. 2010;34:829–44. [Google Scholar]
- 168.Wagner CR, Iyer VV, McIntee EJ. Pronucleotides: toward the in vivo delivery of antiviral and anticancer nucleotides. Med Res Rev. 2000;20(6):417–51. doi: 10.1002/1098-1128(200011)20:6<417::aid-med1>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 169.Kentsis A, Topisirovic I, Culjkovic B, Shao L, Borden KL. Ribavirin suppresses eIF4E-mediated oncogenic transformation by physical mimicry of the 7-methyl guanosine mRNA cap. Proc Natl Acad Sci U S A. 2004;101(52):18105–10. doi: 10.1073/pnas.0406927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yan Y, Svitkin Y, Lee JM, Bisaillon M, Pelletier J. Ribavirin is not a functional mimic of the 7-methyl guanosine mRNA cap. Rna. 2005;11(8):1238–44. doi: 10.1261/rna.2930805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Westman B, Beeren L, Grudzien E, Stepinski J, Worch R, Zuberek J, et al. The antiviral drug ribavirin does not mimic the 7-methylguanosine moiety of the mRNA cap structure in vitro. Rna. 2005;11(10):1505–13. doi: 10.1261/rna.2132505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ghosh B, Benyumov AO, Ghosh P, Jia Y, Avdulov S, Dahlberg PS, et al. Nontoxic chemical interdiction of the epithelial-to-mesenchymal transition by targeting cap-dependent translation. ACS Chem Biol. 2009;4(5):367–77. doi: 10.1021/cb9000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Li S, Jia Y, Jacobson B, McCauley J, Kratzke R, Bitterman PB, et al. Treatment of breast and lung cancer cells with a N-7 benzyl guanosine monophosphate tryptamine phosphoramidate pronucleotide (4Ei-1) results in chemosensitization to gemcitabine and induced eIF4E proteasomal degradation. Molecular pharmaceutics. 2013;10(2):523–31. doi: 10.1021/mp300699d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Chen EZ, Jacobson BA, Patel MR, Okon AM, Li S, Xiong K, et al. Small-molecule inhibition of oncogenic eukaryotic protein translation in mesothelioma cells. Investigational new drugs. 2014 doi: 10.1007/s10637-014-0076-7. [DOI] [PubMed] [Google Scholar]
- 175.Chen L, Aktas BH, Wang Y, He X, Sahoo R, Zhang N, et al. Tumor suppression by small molecule inhibitors of translation initiation. Oncotarget. 2012;3(8):869–81. doi: 10.18632/oncotarget.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Cencic R, Desforges M, Hall DR, Kozakov D, Du Y, Min J, et al. Blocking eIF4E-eIF4G Interaction as a Strategy To Impair Coronavirus Replication. J Virol. 2011;85(13):6381–9. doi: 10.1128/JVI.00078-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Mahalingam P, Takrouri K, Chen T, Sahoo R, Papadopoulos E, Chen L, et al. Synthesis of Rigidified eIF4E/eIF4G Inhibitor-1 (4EGI-1) Mimetic and Their in Vitro Characterization as Inhibitors of Protein-Protein Interaction. J Med Chem. 2014 doi: 10.1021/jm401733v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hong DS, Kurzrock R, Oh Y, Wheler J, Naing A, Brail L, et al. A phase 1 dose escalation, pharmacokinetic, and pharmacodynamic evaluation of eIF-4E antisense oligonucleotide LY2275796 in patients with advanced cancer. Clin Cancer Res. 2011;17(20):6582–91. doi: 10.1158/1078-0432.CCR-11-0430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Bordeleau ME, Matthews J, Wojnar JM, Lindqvist L, Novac O, Jankowsky E, et al. Stimulation of mammalian translation initiation factor eIF4A activity by a small molecule inhibitor of eukaryotic translation. Proc Natl Acad Sci U S A. 2005;102(30):10460–5. doi: 10.1073/pnas.0504249102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bordeleau M-E, Mori A, Oberer M, Lindqvist L, Chard LS, Higa T, et al. Functional Characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat Chem Biol. 2006;2:213–20. doi: 10.1038/nchembio776. [DOI] [PubMed] [Google Scholar]
- 181.Bordeleau ME, Robert F, Gerard B, Lindqvist L, Chen SM, Wendel HG, et al. Therapeutic suppression of translation initiation modulates chemosensitivity in a mouse lymphoma model. J Clin Invest. 2008;118(7):2651–60. doi: 10.1172/JCI34753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Sun Y, Atas E, Lindqvist LM, Sonenberg N, Pelletier J, Meller A. Single-Molecule Kinetics of the Eukaryotic Initiation Factor 4AI upon RNA Unwinding. Structure. 2014 doi: 10.1016/j.str.2014.04.014. [DOI] [PubMed] [Google Scholar]
- 183.Bordeleau ME, Cencic R, Lindqvist L, Oberer M, Northcote P, Wagner G, et al. RNA-mediated sequestration of the RNA helicase eIF4A by Pateamine A inhibits translation initiation. Chemistry & biology. 2006;13(12):1287–95. doi: 10.1016/j.chembiol.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 184.Tsumuraya T, Ishikawa C, Machijima Y, Nakachi S, Senba M, Tanaka J, et al. Effects of hippuristanol, an inhibitor of eIF4A, on adult T-cell leukemia. Biochem Pharmacol. 2011;81(6):713–22. doi: 10.1016/j.bcp.2010.12.025. [DOI] [PubMed] [Google Scholar]
- 185.Kuznetsov G, Xu Q, Rudolph-Owen L, Tendyke K, Liu J, Towle M, et al. Potent in vitro and in vivo anticancer activities of des-methyl, des-amino pateamine A, a synthetic analogue of marine natural product pateamine A. Mol Cancer Ther. 2009;8(5):1250–60. doi: 10.1158/1535-7163.MCT-08-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Kogure T, Kinghorn AD, Yan I, Bolon B, Lucas DM, Grever MR, et al. Therapeutic potential of the translation inhibitor silvestrol in hepatocellular cancer. PLoS One. 2013;8(9):e76136. doi: 10.1371/journal.pone.0076136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lucas DM, Edwards RB, Lozanski G, West DA, Shin JD, Vargo MA, et al. The novel plant-derived agent silvestrol has B-cell selective activity in chronic lymphocytic leukemia and acute lymphoblastic leukemia in vitro and in vivo. Blood. 2009;113(19):4656–66. doi: 10.1182/blood-2008-09-175430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Cencic R, Robert F, Galicia-Vazquez G, Malina A, Ravindar K, Somaiah R, et al. Modifying chemotherapy response by targeted inhibition of eukaryotic initiation factor 4A. Blood cancer journal. 2013;3:e128. doi: 10.1038/bcj.2013.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Saradhi UV, Gupta SV, Chiu M, Wang J, Ling Y, Liu Z, et al. Characterization of silvestrol pharmacokinetics in mice using liquid chromatography-tandem mass spectrometry. AAPS J. 2011;13(3):347–56. doi: 10.1208/s12248-011-9273-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Sadlish H, Galicia-Vazquez G, Paris CG, Aust T, Bhullar B, Chang L, et al. Evidence for a Functionally Relevant Rocaglamide Binding Site on the eIF4A-RNA Complex. ACS Chem Biol. 2013 doi: 10.1021/cb400158t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Liu T, Nair SJ, Lescarbeau A, Belani J, Peluso S, Conley J, et al. Synthetic silvestrol analogues as potent and selective protein synthesis inhibitors. J Med Chem. 2012;55(20):8859–78. doi: 10.1021/jm3011542. [DOI] [PubMed] [Google Scholar]
- 192.Wolfe AL, Singh K, Zhong Y, Drewe P, Rajasekhar VK, Sanghvi VR, et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature. 2014 doi: 10.1038/nature13485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Rubio CA, Weisburd B, Holderfield M, Arias C, Fang E, DeRisi JL, et al. Transcriptome-wide characterization of the eIF4A signature highlights plasticity in translation regulation. Genome Biol. 2014;15(10):476. doi: 10.1186/s13059-014-0476-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Gupta SV, Sass EJ, Davis ME, Edwards RB, Lozanski G, Heerema NA, et al. Resistance to the Translation Initiation Inhibitor Silvestrol is Mediated by ABCB1/P-Glycoprotein Overexpression in Acute Lymphoblastic Leukemia Cells. AAPS J. 2011;13(3):357–64. doi: 10.1208/s12248-011-9276-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Konicek BW, Stephens JR, McNulty AM, Robichaud N, Peery RB, Dumstorf CA, et al. Therapeutic inhibition of MAP kinase interacting kinase blocks eukaryotic initiation factor 4E phosphorylation and suppresses outgrowth of experimental lung metastases. Cancer research. 2011;71(5):1849–57. doi: 10.1158/0008-5472.CAN-10-3298. [DOI] [PubMed] [Google Scholar]
- 196.Tschopp C, Knauf U, Brauchle M, Zurini M, Ramage P, Glueck D, et al. Phosphorylation of eIF-4E on Ser 209 in response to mitogenic and inflammatory stimuli is faithfully detected by specific antibodies. Molecular cell biology research communications : MCBRC. 2000;3(4):205–11. doi: 10.1006/mcbr.2000.0217. [DOI] [PubMed] [Google Scholar]
- 197.Knauf U, Tschopp C, Gram H. Negative regulation of protein translation by mitogen-activated protein kinase-interacting kinases 1 and 2. Mol Cell Biol. 2001;21(16):5500–11. doi: 10.1128/MCB.21.16.5500-5511.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Diab S, Teo T, Kumarasiri M, Li P, Yu M, Lam F, et al. Discovery of 5-(2-(Phenylamino)pyrimidin-4-yl)thiazol-2(3H)-one Derivatives as Potent Mnk2 Inhibitors: Synthesis, SAR Analysis and Biological Evaluation. ChemMedChem. 2014;9(5):962–72. doi: 10.1002/cmdc.201300552. [DOI] [PubMed] [Google Scholar]
- 199.Altman JK, Szilard A, Konicek BW, Iversen PW, Kroczynska B, Glaser H, et al. Inhibition of Mnk kinase activity by cercosporamide and suppressive effects on acute myeloid leukemia precursors. Blood. 2013;121(18):3675–81. doi: 10.1182/blood-2013-01-477216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Alain T, Morita M, Fonseca BD, Yanagiya A, Siddiqui N, Bhat M, et al. eIF4E/4E-BP ratio predicts the efficacy of mTOR targeted therapies. Cancer Res. 2012;72(24):6468–76. doi: 10.1158/0008-5472.CAN-12-2395. [DOI] [PubMed] [Google Scholar]
- 201.Fukuchi-Shimogori T, Ishii I, Kashiwagi K, Mashiba H, Ekimoto H, Igarashi K. Malignant transformation by overproduction of translation initiation factor eIF4G. Cancer Res. 1997;57(22):5041–4. [PubMed] [Google Scholar]
- 202.Ramirez-Valle F, Braunstein S, Zavadil J, Formenti SC, Schneider RJ. eIF4GI links nutrient sensing by mTOR to cell proliferation and inhibition of autophagy. J Cell Biol. 2008;181(2):293–307. doi: 10.1083/jcb.200710215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Hayashi S, Nishimura K, Fukuchi-Shimogori T, Kashiwagi K, Igarashi K. Increase in cap- and IRES-dependent protein synthesis by overproduction of translation initiation factor eIF4G. Biochem Biophys Res Commun. 2000;277(1):117–23. doi: 10.1006/bbrc.2000.3637. [DOI] [PubMed] [Google Scholar]
- 204.Silvera D, Arju R, Darvishian F, Levine PH, Zolfaghari L, Goldberg J, et al. Essential role for eIF4GI overexpression in the pathogenesis of inflammatory breast cancer. Nat Cell Biol. 2009;11(7):903–8. doi: 10.1038/ncb1900. [DOI] [PubMed] [Google Scholar]
- 205.Elfakess R, Dikstein R. A translation initiation element specific to mRNAs with very short 5′UTR that also regulates transcription. PLoS One. 2008;3(8):e3094. doi: 10.1371/journal.pone.0003094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Elfakess R, Sinvani H, Haimov O, Svitkin Y, Sonenberg N, Dikstein R. Unique translation initiation of mRNAs-containing TISU element. Nucleic Acids Res. 2011;39(17):7598–609. doi: 10.1093/nar/gkr484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Hann SR, Eisenman RN. Proteins encoded by the human c-myc oncogene: differential expression in neoplastic cells. Mol Cell Biol. 1984;4(11):2486–97. doi: 10.1128/mcb.4.11.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Muise-Helmericks RC, Grimes HL, Bellacosa A, Malstrom SE, Tsichlis PN, Rosen N. Cyclin D expression is controlled post-transcriptionally via a phosphatidylinositol 3-kinase/Akt-dependent pathway. J Biol Chem. 1998;273(45):29864–72. doi: 10.1074/jbc.273.45.29864. [DOI] [PubMed] [Google Scholar]
- 209.Ramon YCS, De Mattos-Arruda L, Sonenberg N, Cortes J, Peg V. The intra-tumor heterogeneity of cell signaling factors in breast cancer: p4E-BP1 and peIF4E are diffusely expressed and are real potential targets. Clinical & translational oncology : official publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico. 2014 doi: 10.1007/s12094-014-1203-9. [DOI] [PubMed] [Google Scholar]