Abstract
Big data bring new opportunities for methods that efficiently summarize and automatically extract knowledge from such compendia. While both supervised learning algorithms and unsupervised clustering algorithms have been successfully applied to biological data, they are either dependent on known biology or limited to discerning the most significant signals in the data. Here we present denoising autoencoders (DAs), which employ a data-defined learning objective independent of known biology, as a method to identify and extract complex patterns from genomic data. We evaluate the performance of DAs by applying them to a large collection of breast cancer gene expression data. Results show that DAs successfully construct features that contain both clinical and molecular information. There are features that represent tumor or normal samples, estrogen receptor (ER) status, and molecular subtypes. Features constructed by the autoencoder generalize to an independent dataset collected using a distinct experimental platform. By integrating data from ENCODE for feature interpretation, we discover a feature representing ER status through association with key transcription factors in breast cancer. We also identify a feature highly predictive of patient survival and it is enriched by FOXM1 signaling pathway. The features constructed by DAs are often bimodally distributed with one peak near zero and another near one, which facilitates discretization. In summary, we demonstrate that DAs effectively extract key biological principles from gene expression data and summarize them into constructed features with convenient properties.
Keywords: Denoising Autoencoders, Breast Cancer, Feature Construction, Functional Genomics
1. Introduction
Modern genomic technologies have dramatically reduced the cost of comprehensively interrogating biological systems, which has made genome-scale measurements a routine part of biology. This has produced vast amounts of data from which we can extract complex biological principles, potentially broadening our horizon from examining a single pathway to extracting and summarizing the global principles that underlie complex biological systems.
While “big data” introduce new opportunities, they also raise new computational challenges. Even though hardware advances will play a role, new algorithms that can extract, represent, and reason about the principles embedded in such data are also needed. Supervised machine learning algorithms have been developed to predict gene functions and interacting partners1 or to identify new disease-related genes.2 These methods identify new genes or interactions by building upon known examples and consequently are well suited to discovering key biological features, but are not well suited to discovering new processes. Unsupervised algorithms such as clustering can identify key relationships embedded within data, e.g. the existence of tumor subtypes,3 but tend to identify only the strongest signals in the data.
To best utilize big data in reasoning systems, the feature extraction method should allow for the discovery of new pathways and principles, construct features with amenable distributions, and build features that generalize across datasets. Based on these key factors, we identified Denoising Autoencoders (DAs) as a promising approach. DAs are a variant of Artificial Neural Networks (ANNs), but unlike ANNs, which are frequently used for classification, the goal of DAs is to learn compact and efficient representations from input data.4 DAs improve upon the classic autoencoder by incorporating noise during training, a procedure which generates robust features.5 The training objective for DAs is to build features that reconstruct initial input data from corrupted data, i.e. input data with random noise added. DAs depart from supervised ANNs whose performance heavily relies on the quality of gold standards, and because the data define the objective function for the algorithms, DAs can directly use unlabeled data. DAs also serve as building blocks for deep networks6 which have gained popularity in fields such as image and audio processing for their high performance. Compared with commonly used feature extraction approaches such as PCA or ICA that linearly map input to features , DAs extract features in the non-linear space.
In this paper, we introduce an unsupervised feature construction approach based on DAs that summarizes available genomic data and extracts useful features. We apply this approach to a large compendium of breast cancer gene expression data. We demonstrate that the DA trained on one breast cancer dataset captures the same biological features in an independent dataset measured by a different technology. We use the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) cohort7 as a training set and the cohort from The Cancer Genome Atlas (TCGA)8 as an independent evaluation set. We demonstrate an approach that uses known characteristics to perform post-hoc interpretation of these features. The DA constructs features that distinguish tumor from normal samples, classify patients’ estrogen receptor (ER) status, summarize intrinsic subtypes, and identify the activity of key transcription factors (TFs). We also observe a feature that is highly predictive of patient survival. This constructed feature is more predictive of survival than commonly used markers such as tumor grade or ER status. These results suggest that constructed features from denoising autoencoders shed light on unexplored knowledge in breast cancer and provide a fruitful ground for approaches that aim to reason from such data.
2. Methods
2.1. Construction of Denoising Autoencoders from Genomic Data
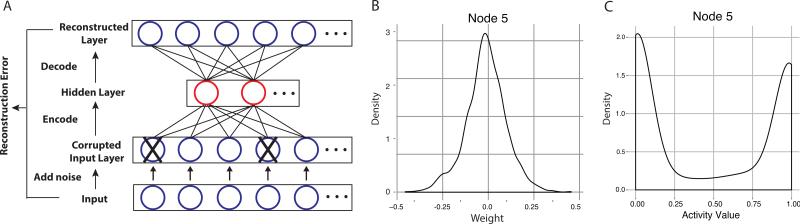
DAs provide a powerful means to analyze audio or image data where measurements are temporally or spatially linked.4,9 Here we describe a technique that allowed construction of DAs from genomic data, which do not have temporal or spatial linkage. These DAs summarized the gene expression characteristics of both breast cancers and normal tissues to automatically extract biologically and clinically relevant features. The constructed DA networks contained three layers: an input layer, a hidden layer and a reconstructed layer (Fig. 1A). The hidden layer represented the constructed features, with each node corresponding to one feature. The reconstructed layer and the input layer had the same dimensions, and the objective function for the algorithm was to minimize the difference between the two layers. Noise was added to the input data during training to construct robust, high-quality features.6
Fig. 1.
A) The network structure of denoising autoencoders. B) The distribution of one node's weight vector. C) The distribution of activity values for a node are bimodally distributed. Here we use Node5 as an example.
Here we describe the detailed training process for DAs from genome-wide transcriptome measurements. We define the term “sample” to represent the complete gene expression vector for each collected tissue biopsy. To facilitate training, samples were randomly grouped into batches, and the number of samples contained in a batch was termed the batch size. For each sample in a batch, a set of genes matching a defined proportion of genes (termed the corruption level) were randomly chosen. The expression values for these genes in this sample were set to zero. Like other feed-forward ANNs, the hidden layer y was constructed by multiplying one sample x with a weight matrix W. A bias vector, b, was added before transformation by the sigmoid function (Formula 1). The value contained in the hidden vector y for each node was termed the activity value of that node. The reconstructed layer was generated from the hidden layer in a similar manner (Formula 2). We used tied weights, which meant that the transpose of W was used for W′. Cross-entropy (Formula 3) was used to measure the difference between the input layer (x) and the reconstructed layer (z). Thus, the problem became fitting appropriate weights and bias terms to minimize the cross-entropy. This optimization was achieved by stochastic gradient descent using the Theano10 library , with weight and bias being updated after each batch and the size of each update is controlled by learning rate. Training proceeded through epochs, and samples were rebatched at the beginning of each epoch. Training was stopped after a specified number of epochs (termed epoch size) was reached.
| (1) |
| (2) |
| (3) |
To determine the appropriate parameter setting for the METABRIC dataset described in Section 2.3, we carried out a parameter sweep with 10-fold cross validation. We performed a full factorial design over all combinations of the following parameters: epoch size of 100, 200, 500; batch size of 1, 10, 20, 50; corruption level of 0.0, 0.1, 0.2; learning rate of 0.005, 0.01, 0.05. The dimension of hidden layer was manually set as 100 since it is a amenable size for interpretation and could be trained efficiently. With the selected parameters, all samples in the METABRIC dataset were used to train a DA network. Then we fixed the optimized weight matrix and bias vectors and calculated the activity values for each node in the hidden layer for each sample. We used the weights and bias terms derived from METABRIC to directly calculate the activity values for the same nodes for each sample in an independent set characterized by TCGA (Section 2.3). Specifically, the weight matrix, W, is derived during training the METABRIC dataset, and we calculate activity values for each node over each sample in both the METABRIC and TCGA datasets based on the same weight matrix W .
2.2. Interpretation of Constructed Features
A major weakness of traditional ANNs has been the difficulty of interpreting the constructed models. DAs have largely been used in image processing, where these algorithms construct features that recognize key components of images, for example diagonal lines. Unlike pixels in image data, genes are not linked to their neighbors, and unlike audio data, they are not linked temporally. Instead they are linked by their transcription factors, their pathway membership, and other biological properties. To address this interpretation challenge, we developed strategies that allow constructed features to be linked to clinical and molecular features of the underlying samples.
2.2.1. Linking Constructed Features to Sample Characteristics
We first linked constructed features to specific sample characteristics. This included the identification of features that categorize tumor and normal samples, molecular subtypes, and ER status. We divided the METABRIC samples into two parts: two thirds of the sample set (1424 samples) was used for a discovery set, and one third of the sample set (712 samples) was reserved as a test set. For these tasks, we binarized each node activity by identifying each node's highest and lowest activity values among the samples in the discovery set and defined 10 equally spaced activation thresholds between these values. We evaluated the balanced accuracy for each node at each threshold to predict the desired sample characteristic. For each task, we chose the nodes with the highest balanced classification accuracies and recorded the corresponding thresholds. These high-accuracy nodes were tested on the test set using the activation threshold identified in the discovery set. To avoid sampling bias, the above procedure was repeated ten times with random partitioning of the discovery and testing sets. The final reported discovery/test accuracy for a node represents these accuracies averaged over the ten runs. We applied this procedure to identify nodes that could stratify tumor/normal samples, ER+/− samples, and categorize samples into molecular subtypes (i.e. Luminal A, Luminal B, Basal-like, HER2-enriched, and Normal-like). We verify that this procedure avoids overestimating the performance of constructed features by evaluating these features in the independent TCGA dataset without retraining. This separate dataset was never used at any prior stages, and in this dataset gene expression values were measured by an entirely different experimental platform. To evaluate each node from METABRIC in TCGA, we used the average of the thresholds identified over the ten METABRIC partitions as the activation threshold for TCGA samples. We termed this the “independent evaluation” performance of the node.
2.2.2. Linking Constructed Features to Transcription Factors
To interpret selected features in the context of transcriptional programs that they summarized, we developed an approach to link transcription factors to constructed features. The weights connecting the input and hidden layers determined how each gene in the input layer influenced the activity values of each node in the hidden layer. The distribution of weights for all genes to a single node approximately resembled a normal distribution centered at zero (Fig. 1B). Most genes gave zero or low weight to a hidden node; while a small number of genes gave high positive or high negative weights. Hence, we defined “high-weight genes” as those exhibiting weight values that lie outside two standard deviations from the mean of the weight distribution. We identified nodes whose high-weight genes were overrepresented by genes bound by one transcription factor and calculated the odds ratio. We employed the transcription factor ChIP-seq data from ENCODE via the UCSC genome browser at http://genome.ucsc.edu/ENCODE/downloads.html.11,12 ChIP-seq data were derived from experiments in T47d and MCF7, a pair of breast cancer cell lines that were expected to best match these breast cancer datasets. All transcription factor target genes were determined using a probabilistic model called Target Identification for Profiles (TIP).13
2.2.3. Linking Constructed Features to Patient Survival
We linked features constructed by the DAs to patient survival. We evaluated patient survival using the METABRIC dataset because it contained up to 15 years of follow-up on each patient. Because the distribution of activity values for each node is bimodal with one peak close to 0 and another close to 1 (Fig. 1C), we defined 0.5 as the activation cutoff to divide samples into two groups. We assessed the differences of these groups for each node by Kaplan-Meier curves and the non-parametric logrank test using the survival package in R.14 The node whose activities best separated two high and low survival groups was the one with the most prognostic power. To evaluate the importance of this constructed feature, we further compared this feature with frequently-used clinical markers of survival including tumor grade, molecular subtype and ER status.
2.2.4. Linking Specific Features to Biological Pathways
To interpret selected constructed features in the context of biological pathways, we developed an approach that leverages gene set enrichment analysis (GSEA)15 to associate known pathways to constructed features. GSEA was developed to identify enrichment within the distribution of differentially expressed genes from microarray experiments. We applied GSEA to a node's weight vector, which identified pathways associated with genes that consistently gave high positive or high negative weight to a node. These genes also exhibited the most influence over the activity value of that node. We defined significantly associated pathways using a false discovery rate (FDR) q-value threshold of 0.1. We used cancer pathways available from the Pathway Interaction Database (PID) curated by the National Cancer Institute and Nature Publishing Group16 as gene sets for GSEA. In total, this set contained 196 pathways downloaded from the Molecular Signatures Database (MSigDB).15
2.3. The METABRIC and TCGA Breast Cancer Compendium
We constructed a compendium consisting of the two largest molecular characterizations of breast cancer to date. The first dataset, METABRIC, was collected from tumor banks in the UK and Canada and contained 1992 clinically annotated breast cancer specimens and 144 tumor-adjacent normal tissues.7 We used this dataset to train DAs and identify predictive features. The gene expression data from this study were downloaded from the European Genomephenome Archive after approval by the specified Data Access Committee. The transcriptomes of tumor and normal tissue samples were profiled on the Illumina HT-12 v3 platform. We converted Illumina identifiers to gene symbols, and the expression values for identifiers that mapped to the same gene symbol were averaged. Missing values were imputed using KNNImputer from the Sleipnir library17 using 10 neighbors as recommended by Troyanskaya et al.18 We used median absolute deviation (MAD) to filter genes with invariant expression across samples and retained the top 3000 genes with the highest MAD values.
The second dataset was obtained from The Cancer Genome Atlas (TCGA). It consisted of 522 primary tumors, 3 metastatic tumors, and 22 tumor-adjacent normal samples.8 These data were obtained by measurement on three distinct platforms, none of which match the METABRIC platform. The TCGA consortium constructed a unified expression collection that summarized genes’ measurements from three platforms into one mean-centered expression value per gene. In this unified expression dataset, genes were identified by their symbol. This dataset served as our independent evaluation dataset, and no training, discovery, or threshold selection was performed on this dataset.
In order to evaluate DAs constructed from the METABRIC dataset directly on the independent TCGA dataset, we removed genes that were not measured by TCGA. This results in a METABRIC set containing 2520 genes measured for 2136 samples and a TCGA set containing the same 2520 genes measured for 547 samples. DAs use values between zero and one in the input vector, so the range of expression values for each gene were linearly transformed to this range.
3. Results and Discussion
To summarize our breast cancer gene expression compendium and construct biologically meaningful features, DAs were trained using the METABRIC dataset and evaluated on the TCGA dataset. We interpreted the constructed features by sample characteristics classification, transcription factor enrichment, survival analysis, and pathway analysis. We identified features representing a variety of important clinical or molecular characters of breast cancer, including sample type (tumor or normal tissue), ER status, and intrinsic subtype. They were shown to be robust across datasets. In addition, breast cancer related transcription factors were found to be linked to these features. Finally, DAs constructed a novel feature that was highly predictive of patient survival, and from this we uncovered a variety of biological processes enriched in that feature.
3.1. Construction of a Denoising Autoencoder from Genomic Data
To apply DAs to genomic data for the first time, we performed a full factorial parameter sweep over the METABRIC dataset. We fixed the number of nodes in the hidden layer to 100 to fix the number of weights the algorithm is allowed to fit under each parameter setting. The remaining parameters were evaluated by the ability of autoencoders generated using each set to reconstruct held out test data. The parameters that we selected were: a batch size of 10, an epoch size of 500, a corruption level of 0.1, and a learning rate of 0.01. We note that by keeping other parameters constant while setting the corruption level to 0.0 (no noise added) results in the best performance. We chose a corruption level of 0.1 to avoid the risk of overfitting and to improve the quality and robustness of constructed features. Although METABRIC dataset is one of the largest available expression datasets of breast cancer, they have fewer examples than most datasets used for training neural networks. We observe that DAs still effectively summarize dataset of this size. After selecting these parameters, DAs were built by training on the METABRIC dataset. We named the constructed features “Node##” based on the order in which they appear in the hidden layer. The DAs trained on the METABRIC dataset were directly applied to the TCGA dataset to generate activity values for each already constructed feature in each sample. The results of our parameter sweep, as well as the activity scores for each feature in each sample, are available for download from the online supplement.
3.2. Features constructed by DAs represent clinical characteristics
In order to examine whether the features constructed by DAs exhibit clinical significance, we assessed the ability of hidden nodes to classify two important clinical features: sample type and ER status. We first identified hidden nodes that best classified whether samples were obtained from tumor or normal tissue using two thirds of the METABRIC samples (discovery set) and then tested the performance of these nodes in the remaining samples (test set). Tumor and normal tissues are very distinct from each other, and consequently there should be at least one feature that separates these with high accuracy. Table 1 shows the balanced classification accuracies calculated during discovery, testing, and an independent evaluation dataset. The top 5 hidden nodes are ordered by their performance in the discovery set. Nodes 64 and 99 achieved very high accuracy in distinguishing tumor from normal samples in the METABRIC dataset. More interestingly, Node64 and Node99 also classified TCGA samples with near-perfect accuracy. This indicates that training the weights matrix using all of the METABRIC samples does not appear to lead to test set contamination during the feature interpretation phase.
Table 1.
Performance of hidden nodes in classifying tumor from normal samples.
| METABRIC |
TCGA |
||
|---|---|---|---|
| Node | Discovery | Test | Evaluation |
| 64 | 0.970 | 0.968 | 0.996 |
| 99 | 0.957 | 0.959 | 0.998 |
| 38 | 0.879 | 0.887 | 0.911 |
| 43 | 0.873 | 0.873 | 0.750 |
| 69 | 0.871 | 0.872 | 0.906 |
Next we sought to apply our framework to the more challenging task of constructing features with activities that reflect the ER status of breast cancer samples. The estrogen receptor plays an important role in the progression of breast cancer, as the majority of breast cancers start out as estrogen dependent and overexpress the estrogen receptor.19 A tumor's expression status (ER+ vs. ER−) serves as an immunohistological biomarker that helps determine whether patients will benefit from endocrine therapy.20 Table 2 shows that Node30 and Node58 achieve the highest accuracy for ER status classification. As expected, the accuracy associated with stratifying samples into ER categories is not as high as when stratifying tumor and normal samples. This is because ER signaling is a complex biological process involving many co-regulatory proteins and can be activated by both estrogen ligands and a variety of other pathways.20 Thus, the expression pattern for ER positive samples and ER negative samples is more complicated, making classification more challenging. Again, the results for ER status stratification in METABRIC and TCGA datasets indicate that features constructed by DAs maintain robust performance across datasets. This is consistent with our observations from the tumor/normal separation and further indicates that the unsupervised training using all of METABRIC does not lead to contamination during the discovery/test phase.
Table 2.
Performance of hidden nodes in classifying ER + from ER - samples.
| METABRIC |
TCGA |
||
|---|---|---|---|
| Node | Discovery | Test | Evaluation |
| 89 | 0.848 | 0.833 | 0.749 |
| 30 | 0.824 | 0.822 | 0.856 |
| 58 | 0.808 | 0.801 | 0.828 |
| 6 | 0.798 | 0.799 | 0.771 |
| 69 | 0.784 | 0.779 | 0.820 |
Transcription factors FOXA1 and GATA3 are two key determinants of ER function and endocrine response. FOXA1 facilitates ER-chromatin interactions that are necessary for ER-mediated gene regulation,21 and GATA3 acts upstream of FOXA1 in modulating the accessibility of enhancers bound by ER.22 Therefore, to understand whether these nodes were capturing underlying biological principles, we evaluated whether these TFs were strongly associated with nodes categorizing ER status. We calculated odds ratios for each node/TF pair as described in section 2.2.2. We found that Node58 achieved the highest odds ratios for both FOXA1 and GATA3 among all nodes. Specifically, it was enriched of genes regulated by FOXA1 with an odds ratio of 4.02 and of genes regulated by GATA3 with an odds ratio of 3.16. These results suggest that Node58 was able to distinguish ER+ from ER- breast cancers with high accuracy because it contained genes that reflect the activity of key ER-associated TFs.
3.3. Features constructed by DAs recapitulate molecular subtypes
Breast cancer is a complex and heterogeneous disease caused by various molecular alterations in distinct signaling pathways. Breast cancer patients respond differently to treatment and show diverse clinical outcomes. Categorizing breast cancers into molecular subtypes that exhibit similar characteristics opens the door to the development of subtype-specific prognostic markers and treatments. Parker et al. developed a 50-gene subtype predictor (PAM50) based on gene expression data.23 PAM50 subtypes are available for both the METABRIC and TCGA cancers. Here, we evaluated whether features constructed by DAs can also predict these subtypes. We defined each subtype prediction as a binary classification problem and show the results in Table 3. The table contains the node that achieves the highest accuracy in the discovery set. In general, we observed that only one node was predictive of each sub-type. There were two exceptions: for the Normal-like subtype Node38 performed similarly to Node42, and for the Luminal A subtype Node23 performed similarly to Node5. We observed that the Basal-like subtype reached the highest accuracy, which is consistent with clustering results showing Basal as the most distinct cluster.8 Luminal A (LumA) and Luminal B (LumB) subtypes were mixed together in the clustering analysis as well. Combining LumA and LumB into one subtype identified features that obtained higher accuracies. Both Node30 (Basal) and Node6 (LumA/B), identified here as subtype nodes, were also predictive of ER status. This is because ER positive patients usually fall into the luminal subtypes, and ER negative patients usually fall into the Basal subtype. In METABRIC, 77.6% of ER positive samples are LumA or LumB, while only 3.9% of ER positive samples are from the Basal subtype. Therefore the ER status signal is sufficient to identify these groups, but not differentiate between, for example, the two luminal subtypes. The specific LumA and LumB subtypes are best captured by different nodes. Methods that aim to reason based upon features constructed from the DA may be able to exploit these distinct but complementary features to obtain superior accuracy and better reflect the underlying complex biology of breast cancer.
Table 3.
Performance of hidden nodes in classifying each intrinsic subtype.
| Subtype | Basal | Her2 | LumA | LumB | Normal | LumA/B |
|---|---|---|---|---|---|---|
| Node | 30 | 29 | 5 | 66 | 42 | 6 |
| METABRIC Discovery | 0.929 | 0.761 | 0.780 | 0.755 | 0.750 | 0.849 |
| METABRIC Test | 0.918 | 0.741 | 0.777 | 0.750 | 0.748 | 0.849 |
| TCGA Evaluation | 0.992 | 0.712 | 0.800 | 0.717 | 0.733 | 0.825 |
3.4. Features constructed by DAs are associated with prognosis
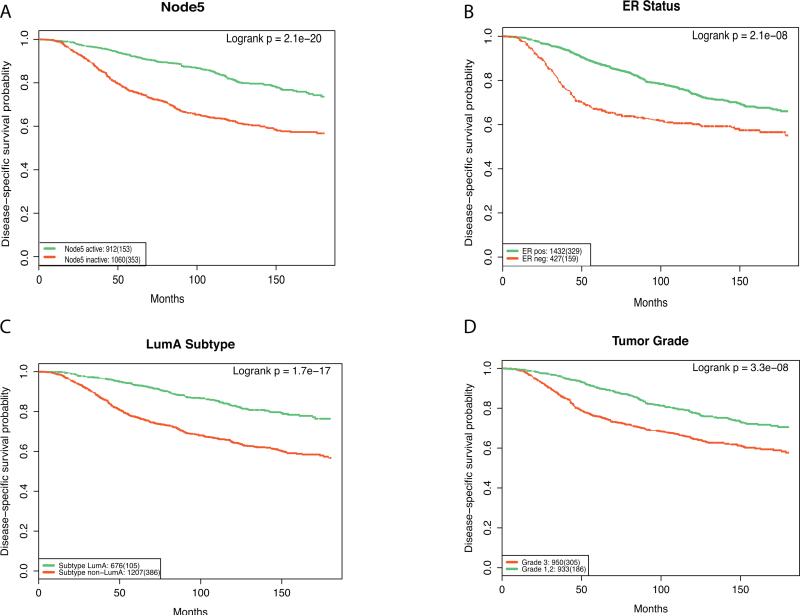
Many factors have been correlated to prognosis, such as histologic grade,24 ER status,25 lymph node metastases,26 and intrinsic molecular subtypes.27 Here we assessed the correlation between features constructed by DAs and patient prognosis. As shown in Fig. 1C, the distribution of node activity was bimodal. We separated patients into two groups based on their node activity using a cutoff of 0.5 and then correlated activity values of each node to patient survival time. The node that best separated good and poor prognosis was Node5 (logrank p-value of 2.1e−20; Fig. 2A). To compare the performance of Node5 with other clinical or molecular features, we carried out survival analysis for ER status (Fig. 2B), each tumor grade, and each intrinsic molecular subtype. The LumA subtype had a significantly better prognosis than other subtypes and provided the strongest subtype-survival association in the METABRIC dataset (Fig. 2C). A tumor of grade 3 (clinically termed high-grade) was a sign of poor prognosis when compared to grades 1 and 2 (Fig. 2D). Comparing the ability of these survival-associated breast cancer features to Node5, Node5 was more strongly associated with survival. Interestingly, Node5 was also the node that best classifies the LumA subtype (Table 3). We investigated the associations of each node with tumor grade and found that Node5 was also most strongly associated with grade (online supplement). These results suggest that Node5 learned a combined expression pattern that captures features of both the LumA subtype and a tumor's grade, which contributed to its strong association with survival.
Fig. 2.
Kaplan-Meier plots of disease-specific survival for Node5 (A), ER status (B), Luminal A subtype (C), and Tumor Grade (D) demonstrate that the constructed feature outperforms the other predictors.
To further investigate how Node5 learned this combined pattern, we performed pathway analysis using a modified GSEA. GSEA identified pathways that defined the activity values of Node5 (Table 4). The most significant pathway was the FOXM1 transcription factor network. FOXM1 is one of the most overexpressed genes in breast cancer28 and its down-regulation has been shown to inhibit proliferation, migration and invasion of breast cancer cells.29 The Aurora Kinase A, Aurora Kinase B, and PLK1 pathways affect cell cycle progression.30,31 Misregulation of the cell cycle is related to both tumor formation and growth and is consistent with FOXM1's role in modulating cell cycle progression. The number of cells undergoing division is used in determination of a tumor's grade, indicating a potential relation for Node5 to grade based on the number of mitotic events. The other two pathways have also been demonstrated to be key players in breast cancer,32,33 and c-Myb is a known marker of the luminal subtypes34 indicating a potential connection between Node5 and the Luminal A subtype.
Table 4.
PID pathways enriched in Node5.
| Pathway | FDR q-value |
|---|---|
| FOXM1 transcription factor network | < 1e–4 |
| Aurora B signaling | 4.93e–4 |
| Aurora A signaling | 0.001 |
| PLK1 signaling | 0.003 |
| Integrin-linked kinase signaling | 0.068 |
| C-MYB transcription factor network | 0.074 |
4. Conclusion
While machine learning has made key contributions to biology, a gap still exists between data integration methods, which are largely supervised, and discovery-oriented approaches, which are unsupervised and don't condition on known biology. We have evaluated DAs as a means to fill this gap allowing us to develop unsupervised methods for data integration. We found that DAs effectively summarize key features in breast cancer data. We identified features that stratify tumor/normal samples, ER+/− samples, and molecular subtypes, in addition to identifying transcription factor activities. Moreover, DAs constructed a feature significantly associated with the FOXM1 pathway that was highly predictive of patient survival by combining information from the Luminal A subtype and tumor grade. A DA constructed from one dataset identifies the same features in an independent dataset, even though the strongest principle component of a combined dataset captures the underlying study highlighting the major methodological differences between these studies.35
Future work will focus on developing new approaches to interpret features, especially features that cannot be mapped to existing knowledge but may represent new signals. We will also evaluate deep network architectures with multiple layers of stacked DAs. We anticipate that employing features generated by DAs in supervised learning will improve prediction accuracies, as we have shown that these features comprise clinical information and molecular patterns. Because the patterns observed are consistent across datasets, we also anticipate that DAs will provide a fruitful mechanism for data integration. Future work should explore the scope and limitations of this approach for large-scale data integration. Overall, we anticipate that DAs can provide a new mechanism to effectively summarize and integrate large compendia of genomic data in an unsupervised manner.
Supplementary Material
Acknowledgements
This work was supported in part by P20 GM103534 from the NIH and an American Cancer Society Research Grant, #IRG-82-003-27. JT is a Neukom Graduate Fellow supported by the William H. Neukom 1964 Institute for Computational Science.
Footnotes
Supplement: http://discovery.dartmouth.edu/~cgreene/da-psb2015/.
References
- 1.Wong AK, Park CY, Greene CS, Bongo LA, Guan Y, Troyanskaya OG. Nucleic Acids Research. 2012;40:W484. doi: 10.1093/nar/gks458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, GuhaThakurta D, Sieberts SK, Monks S, Reitman M, Zhang C, et al. Nature Genetics. 2005;37:710. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sørlie T, Tibshirani R, Parker J, Hastie T, Marron J, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, et al. Proceedings of the National Academy of Sciences. 2003;100:8418. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vincent P, Larochelle H, Bengio Y, Manzagol P-A. Proceedings of the 25th International Conference on Machine Learning. ACM; New York, NY, USA: 2008. Extracting and composing robust features with denoising autoencoders. [Google Scholar]
- 5.Bengio Y. Foundations and trends in Machine Learning. 2009;2:1. [Google Scholar]
- 6.Vincent P, Larochelle H, Lajoie I, Bengio Y, Manzagol P-A. The Journal of Machine Learning Research. 2010;11:3371. [Google Scholar]
- 7.Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. Nature. 2012;486:346. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The Cancer Genome Atlas Network Nature. 2012;490:61. [Google Scholar]
- 9.Dahl GE, Yu D, Deng L, Acero A. Audio, Speech, and Language Processing. IEEE Transactions on. 2012;20:30. [Google Scholar]
- 10.Bergstra J, Breuleux O, Bastien F, Lamblin P, Pascanu R, Desjardins G, Turian J, Warde-Farley D, Bengio Y. Theano: a CPU and GPU math expression compiler; Proceedings of the Python for Scientific Computing Conference (SciPy); June 2010; Oral Presentation. [Google Scholar]
- 11.Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan K-K, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, et al. Nature. 2012;489:91. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, et al. Genome Research. 2012;22:1813. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng C, Min R, Gerstein M. Bioinformatics. 2011;27:3221. doi: 10.1093/bioinformatics/btr552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Therneau TM. A Package for Survival Analysis in S. R package version 2.37-7. 2014.
- 15.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Proceedings of the National Academy of Sciences. 2005;102:15545. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, Buetow KH. Nucleic Acids Research. 2009;37:D674. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huttenhower C, Schroeder M, Chikina MD, Troyanskaya OG. Bioinformatics. 2008;24:1559. doi: 10.1093/bioinformatics/btn237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Troyanskaya O, Cantor M, Sherlock G, Brown P, Hastie T, Tibshirani R, Botstein D, Altman RB. Bioinformatics. 2001 Jun;17:520. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- 19.Saha Roy S, Vadlamudi RK. International Journal of Breast Cancer. 2011;2012 doi: 10.1155/2012/654698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shao W, Brown M, et al. Breast Cancer Research. 2004;6:39. doi: 10.1186/bcr742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. Nature Genetics. 2011;43:27. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Theodorou V, Stark R, Menon S, Carroll JS. Genome Research. 2013;23:12. doi: 10.1101/gr.139469.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, et al. Journal of Clinical Oncology. 2009;27:1160. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elston C, Ellis I. Histopathology. 1991;19:403. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 25.Lower EE, Glass EL, Bradley DA, Blau R, Heffelfinger S. Breast Cancer Research and Treatment. 2005;90:65. doi: 10.1007/s10549-004-2756-z. [DOI] [PubMed] [Google Scholar]
- 26.Fisher ER, Costantino J, Fisher B, Palekar AS, Redmond C. Cancer. 1995;75:1310. doi: 10.1002/1097-0142(19950315)75:6<1310::aid-cncr2820750613>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 27.Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Proceedings of the National Academy of Sciences. 2001;98:10869. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bektas N, Ten Haaf A, Veeck J, Wild PJ, Lüscher-Firzlaff J, Hartmann A, Knüchel and R, Dahl E. BMC Cancer. 2008;8:42. doi: 10.1186/1471-2407-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahmad A, Wang Z, Kong D, Ali S, Li Y, Banerjee S, Ali R, Sarkar FH. Breast Cancer Research and Treatment. 2010;122:337. doi: 10.1007/s10549-009-0572-1. [DOI] [PubMed] [Google Scholar]
- 30.Vader G, Lens S. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer. 2008;1786:60. doi: 10.1016/j.bbcan.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 31.Takai N, Hamanaka R, Yoshimatsu J, Miyakawa I. Oncogene. 2005;24:287. doi: 10.1038/sj.onc.1208272. [DOI] [PubMed] [Google Scholar]
- 32.Troussard AA, McDonald PC, Wederell ED, Mawji NM, Filipenko NR, Gelmon KA, Kucab JE, Dunn SE, Emerman JT, Bally MB, et al. Cancer Research. 2006;66:393. doi: 10.1158/0008-5472.CAN-05-2304. [DOI] [PubMed] [Google Scholar]
- 33.Gonda T, Leo P, Ramsay R. Expert Opinion on Biological Therapy. 2008;8:713. doi: 10.1517/14712598.8.6.713. [DOI] [PubMed] [Google Scholar]
- 34.Thorner AR, Parker JS, Hoadley KA, Perou CM. PLOS ONE. 2010;5:e13073. doi: 10.1371/journal.pone.0013073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greene CS, Tan J, Ung M, Moore JH, Cheng C. Journal of Cellular Physiology. 2014 doi: 10.1002/jcp.24662. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.