Abstract
The development of tools in computational pathology to assist physicians and biomedical scientists in the diagnosis of disease requires access to high-quality annotated images for algorithm learning and evaluation. Generating high-quality expert-derived annotations is time-consuming and expensive. We explore the use of crowdsourcing for rapidly obtaining annotations for two core tasks in computational pathology: nucleus detection and nucleus segmentation. We designed and implemented crowdsourcing experiments using the CrowdFlower platform, which provides access to a large set of labor channel partners that accesses and manages millions of contributors worldwide. We obtained annotations from four types of annotators and compared concordance across these groups. We obtained: crowdsourced annotations for nucleus detection and segmentation on a total of 810 images; annotations using automated methods on 810 images; annotations from research fellows for detection and segmentation on 477 and 455 images, respectively; and expert pathologist-derived annotations for detection and segmentation on 80 and 63 images, respectively. For the crowdsourced annotations, we evaluated performance across a range of contributor skill levels (1, 2, or 3). The crowdsourced annotations (4,860 images in total) were completed in only a fraction of the time and cost required for obtaining annotations using traditional methods. For the nucleus detection task, the research fellow-derived annotations showed the strongest concordance with the expert pathologist-derived annotations (F−M =93.68%), followed by the crowd-sourced contributor levels 1,2, and 3 and the automated method, which showed relatively similar performance (F−M = 87.84%, 88.49%, 87.26%, and 86.99%, respectively). For the nucleus segmentation task, the crowdsourced contributor level 3-derived annotations, research fellow-derived annotations, and automated method showed the strongest concordance with the expert pathologist-derived annotations (F−M = 66.41%, 65.93%, and 65.36%, respectively), followed by the contributor levels 2 and 1 (60.89% and 60.87%, respectively). When the research fellows were used as a gold-standard for the segmentation task, all three contributor levels of the crowdsourced annotations significantly outperformed the automated method (F−M = 62.21%, 62.47%, and 65.15% vs. 51.92%). Aggregating multiple annotations from the crowd to obtain a consensus annotation resulted in the strongest performance for the crowd-sourced segmentation. For both detection and segmentation, crowd-sourced performance is strongest with small images (400 × 400 pixels) and degrades significantly with the use of larger images (600 × 600 and 800 × 800 pixels). We conclude that crowdsourcing to non-experts can be used for large-scale labeling microtasks in computational pathology and offers a new approach for the rapid generation of labeled images for algorithm development and evaluation.
Keywords: Crowdsourcing, Annotation, Nuclei Detection, Nuclei Segmentation, Digital Pathology, Computational Pathology, Histopathology
1. Introduction
Cancer is diagnosed based on a pathologist’s interpretation of the nuclear and architectural features of a microscopic image of a histopathological section of tissue removed from a patient. Over the past several decades, computational methods have been developed to enable pathologists to develop and apply quantitative methods for the analysis and interpretation of histopathological images of cancer.1 These methods can be used to automate standard methods of histopathological analysis (e.g. nuclear grading),2 as well as to discover novel morphological characteristics predictive of clinical outcome (e.g. relational features and stromal attributes), which are difficult or impossible to measure using standard manual approaches.3
Accurate nuclear detection and segmentation is an important image processing step prior to feature extraction for most computational pathology analyses. In the past decade a large number of methods have been proposed for automated nuclear detection and segmentation.4 However, despite the generation of a large number of competing approaches for these tasks, the comparative performance of nuclei detection and segmentation methods has not been evaluated rigorously.
A major barrier to rigorous comparative evaluation of existing methods is the time and expense required to obtain expert-derived labeled images. Using traditional approaches, obtaining labeled images requires enlisting the support of a trained research fellow and/or pathologist to annotate microscopic images. Most computational labs do not have access to support from highly trained physicians and research staff to annotate images for algorithm development and evaluation, and even in pathology research laboratories, obtaining high-quality hand-labeled images is a significant challenge, as the task is time-consuming and can be tedious.
These challenges are exacerbated when attempting large-scale image annotation projects of hundreds-to-thousands of images. Further, recent advances in whole slide imaging are enabling the generation of large archives of whole slide images (WSIs) of disease. In contrast to images obtained from a standard microscope camera (which will capture a single region-of-interest (ROI) per image), WSIs are large and capture tissue throughout the entire slide, which typically contains thousands of ROIs and tens-of-thousands of nuclei per WSI.5 Thus, it is not feasible to obtain comprehensive annotation from pathologists or research fellows in a single research laboratory for large sets of WSIs.
In this project, we explore the use of crowdsourcing as an alternative method for obtaining large-scale image annotations for nucleus detection and segmentation. In recent years, crowd-sourcing has been increasingly used for bioinformatics, with image annotation representing an important application area.6 Crowdsourced image annotation has been successfully used to serve a diverse set of scientific goals, including: classification of galaxy morphology,7 the mapping of neuron connectivity in the mouse retina,8 the detection of sleep spindles from EEG data,9 and the detection of malaria from blood smears.10,11 To our knowledge, no prior published studies have used non-expert crowdsourced image annotation for nucleus detection and segmentation from histopathological images of cancer. The Cell Slider project by the Cancer Research UK (http://www.cellslider.net/) launched in October 2012 is attempting to use crowdsourcing to annotate cell types in histopathological images of breast cancer; however, to our knowledge results of this study have not yet been released.
Here, we provide a framework for understanding and applying crowdsourcing to the annotation of histopathological images obtained from a large-scale WSI dataset. We develop and evaluate this framework in the setting of nucleus detection and segmentation from a set of WSIs obtained from renal cell carcinoma cases that previously underwent comprehensive molecular profiling as part of The Cancer Genome Atlas (TCGA) project.12
We performed a set of experiments to compare the annotations achieved by: pathologists, trained research fellows, and non-expert crowdsourced annotators. For the crowdsourced image annotators, we performed additional experiments to gain insight into factors that influence contributor performance, including: assessing the relationship of the contributor’s pre-defined skill level with the contributor’s performance on the nucleus detection and segmentation tasks; assessing the influence of image size on contributor performance; and comparing performance based on a single annotation-per-image versus aggregating multiple contributor annotations-per-image.
The remainder of the paper is organized as follows. Section 2 describes the dataset used for the study, and the proposed framework for evaluating performance of nucleus detection and segmentation. Experimental results are presented in Section 3, and concluding remarks and proposed future work are presented in Section 4.
2. Method
In this section, we describe the dataset, CrowdFlower platform, and design of our experiments.
2.1. Dataset
The images used in our study come from WSIs of kidney renal clear cell carcinoma (KIRC) from the TCGA data portal. TCGA represents a large-scale initiative funded by the National Cancer Institute and National Human Genome Research Institute. TCGA has performed comprehensive molecular profiling on a total of approximately ten-thousand cancers, spanning the 25 most common cancer types. In addition to the collection of molecular and clinical data, TCGA has collected WSIs from most study participants. Thus, TCGA represents a major resource for projects in computational pathology aiming at linking morphological, molecular, and clinical characteristics of disease.13,14
We selected 10 KIRC whole slide images (WSI) from the TCGA data portal (https://tcga-data.nci.nih.gov/tcga/), representing a range of histologic grades of KIRC. From these WSIs, we identified nucleus-rich ROIs and extracted 400 × 400 pixel size images (98.24 µm × 98.24 µm) for each ROI at 40× magnification. The total number of images per region of interest is 81. Finally, we obtained a total of 810 images from the 10 KIRC WSIs.
2.2. CrowdFlower Platform
We employ the CrowdFlower platform to design jobs, access and manage contributors, and obtain results for the nucleus detection and segmentation image annotation jobs. CrowdFlower is a crowdsourcing service that works with over 50 labor channel partners to enable access to a network of more than 5 million contributors worldwide. The CrowdFlower platform provides several features aimed at increasing the likelihood of obtaining high-quality work from contributors. Jobs are served to contributors in tasks. Each task is a collection of one or more images sampled from the data set. Prior to completing a job, the platform requires contributors to complete job-specific training. In addition, contributors must complete test questions both before (quiz mode) and throughout (judgment mode) the course of the job. Test questions serve the dual purpose of training contributors and monitoring their performance. Contributors must obtain a minimum level of accuracy on the test questions to be permitted to complete the job. CrowdFlower categorizes contributors into three skill levels (1,2,3) based on performance on other jobs, and when designing a job the job designer may target a specific contributor skill level. In addition, the job designer specifies the payment per task and the number of annotations desired per image. After job completion, CrowdFlower provides the job designers with a confidence map for each annotated image. The confidence map is an image in the same dimension as the input image, but the pixel intensity now represents an aggregation of annotations to that image, which is weighted by both the annotation agreement among contributors and each contributor’s trust level. Additional information on the CrowdFlower platform is available at www.crowdower.com.
2.3. Job Design
Our study includes two types of image annotation jobs: nucleus detection and segmentation. The contributors used a dot operator (by clicking at the center of a nucleus) for nucleus detection and a polygon operator (by drawing a line around the nucleus) for nuclei segmentation. Each job contains instructions, which provide examples of expert-derived annotations and guidance to assist the contributor in learning the process of nuclear annotation. These instructions are followed by a set of test questions. Test questions are presented to the contributor in one of two modes: quiz mode and judgment mode. Quiz mode occurs at the beginning of a job (immediately following the instructions), while judgment mode test questions are interspersed throughout the course of completing a job. In our experiments, contributors were required to achieve at least 40% accuracy on five test questions in quiz mode in order to qualify for annotation of unlabeled images from the job during judgment mode. In judgment mode, each task consists of four unlabeled images and one test question image, which is presented to the contributor in the same manner as the unlabeled images, such that the contributor is unaware if he/she is annotating an unlabeled image or a test question. The total pool of quiz and judgment mode test questions used in our study was based on 20 images, which had been annotated by medical experts. If the contributor’s accuracy decreased to below 40% during judgment mode, the contributor was barred from completion of additional annotations for the job.
There are several additional job design options provided by the CrowdFlower platform which may influence annotation performance. The CrowdFlower platform divides the contributors into three skill levels based on their performance on prior jobs, and the job designer can target jobs to specific contributor skill levels. In our experiments, we compared performance when targeting jobs to each skill level. The job designer must specify the number of annotations to collect per image. For most of our experiments, we used a single annotation per image. In addition, we conducted an experiment for the image segmentation job, in which the number of contributors per image ranged from 1 to 3 to 5, and we compared performance across these three levels of redundancy.
In addition to the annotations obtained from the non-expert crowd, we obtained annotations from three additional types of labelers: published state-of-the-art automated nucleus detection and segmentation algorithms;15 research fellows trained for these specific jobs; and MD-trained surgical pathologists, who have completed residency in Anatomic Pathology.
3. Experiments
3.1. Performance Metrics
Detection Metrics
A detected nucleus was accepted as correctly detected if the coordinates of its centroid were within a range of 15 pixels (3.75 µm) from the centroid of a ground truth nucleus. The metrics used to evaluate nucleus detection include: number of true positives (TP), number of false positives (FP), number of false negatives (FN), sensitivity or true positive rate , precision or positive predictive value and F-Measure . The TPR and PPV are presented in Tables 1 – 4 with their 95% Confidence Intervals, computed using the prop.test function in the stats package in R.
Table 1.
Detection results on 80 images (Pathologists’ annotation as GT) GT nuclei = 4436
| Annotations | TP | FN | FP | TPR % | PPV % | F-M % |
|---|---|---|---|---|---|---|
| Research Fellow | 4109 | 327 | 227 | 92.63 ± 0.8 | 94.76 ± 0.7 | 93.68 |
| Automated Method | 3735 | 701 | 416 | 84.20 ± 1.1 | 89.98 ± 1.0 | 86.99 |
| Contributor Level 1 | 3814 | 622 | 434 | 85.98 ± 1.1 | 89.78 ± 1.0 | 87.84 |
| Contributor Level 2 | 4016 | 420 | 625 | 90.53 ± 0.9 | 86.53 ± 1.0 | 88.49 |
| Contributor Level 3 | 3787 | 649 | 457 | 85.37 ± 1.1 | 89.23 ± 0.9 | 87.26 |
Table 4.
Detection results on different image sizes (Automated Method as GT) GT nuclei = 44281
| Annotations | TP | FN | FP | TPR % | PPV % | F-M |
|---|---|---|---|---|---|---|
| Image Size 400 × 400 (810 images) | 36191 | 8090 | 5705 | 81.73 ± 0.4 | 86.38 ± 0.3 | 83.99 |
| Image Size 600 × 600 (380 images) | 24870 | 19411 | 16993 | 56.16 ± 0.5 | 59.41 ± 0.5 | 57.74 |
| Image Size 800 × 800 (170 images) | 12144 | 32137 | 21842 | 27.42 ± 0.4 | 35.73 ± 0.5 | 31.03 |
Segmentation Metrics
The metrics used to evaluate segmentation annotation include: sensitivity ( - proportion of nucleus pixels that are correctly labeled as positive), specificity or true negative rate ( - proportion of non-nucleus pixels that are correctly labeled as negative), precision , F-Measure, and ; where I is the image, A(S) is the area of the segmented nuclei, A(G) is the area of the ground truth nuclei.
3.2. Detection Results
In the first experiment, we considered pathologist’s annotations as ground truth (GT). Pathologists provided annotations on a total of 80 study images. For these 80 images, we assessed the performance of research fellows, the automated method, and non-expert contributors from three skill levels as shown in Table 1. Focusing on the F-M measure (which incorporates both TPR and PPV), we observe the strongest performance for the research fellow, followed by similar performance for the three other annotation groups (FM between 86.99% and 88.49%) as shown in Table 1.
In the second experiment, we considered annotations from research fellows as GT. The research fellows provided annotations for a total of 477 images containing 25,323 annotated nuclei, considered as GT nuclei in this experiment. Thus, the dataset for this evaluation is significantly larger than that for the initial analysis, which used the pathologist annotation as the GT. In this experiment, all four groups showed similar performance, with F-M scores between 83.94% and 85.32%, as shown in Table 2.
Table 2.
Detection results on 477 images (Research fellows’ annotation as GT) GT nuclei = 25323
| Annotations | TP | FN | FP | TPR % | PPV % | F-M % |
|---|---|---|---|---|---|---|
| Automated Method | 21177 | 4146 | 3955 | 83.63 ± 0.5 | 84.26 ± 0.5 | 83.94 |
| Contributor Level 1 | 21495 | 3828 | 3982 | 84.88 ± 0.4 | 84.37 ± 0.5 | 84.63 |
| Contributor Level 2 | 22488 | 2835 | 4904 | 88.80 ± 0.4 | 82.10 ± 0.5 | 85.32 |
| Contributor Level 3 | 21788 | 3535 | 4049 | 86.04 ± 0.4 | 84.33 ± 0.5 | 85.18 |
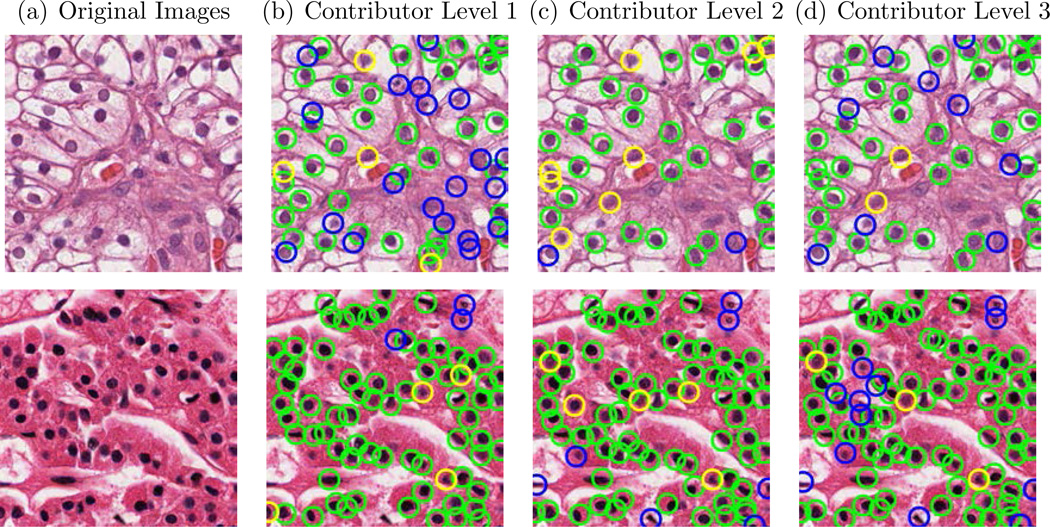
In the third experiment, we used the annotations produced by the automated method as the GT. The automated method was run on all 810 study images and detected a total of 44,281 nuclei which were considered as GT nuclei in this experiment. We compared these GT nuclei with the three crowdsourced contributor levels across all 810 images and results are shown in Table 3. Overall, the three contributor levels achieved similar TPR levels, with a significantly higher PPV for Contributor Level 2, resulting in the highest F-M for Contributor Level 2(83.99%), with slightly lower F-M’s achieved by Contributor Levels 1 and 2, as shown in Table 3. Visual examples of nucleus detection by different level of contributors are shown in Figure 1.
Table 3.
Detection results on 810 images (Automated Method as GT) GT nuclei = 44281
| Annotations | TP | FN | FP | TPR % | PPV % | F-M % |
|---|---|---|---|---|---|---|
| Contributor Level 1 | 35823 | 8458 | 7792 | 80.90 ± 0.4 | 82.13 ± 0.4 | 81.51 |
| Contributor Level 2 | 36191 | 8090 | 5705 | 81.73 ± 0.4 | 86.38 ± 0.3 | 83.99 |
| Contributor Level 3 | 36125 | 8156 | 6874 | 81.58 ± 0.4 | 84.01 ± 0.4 | 82.78 |
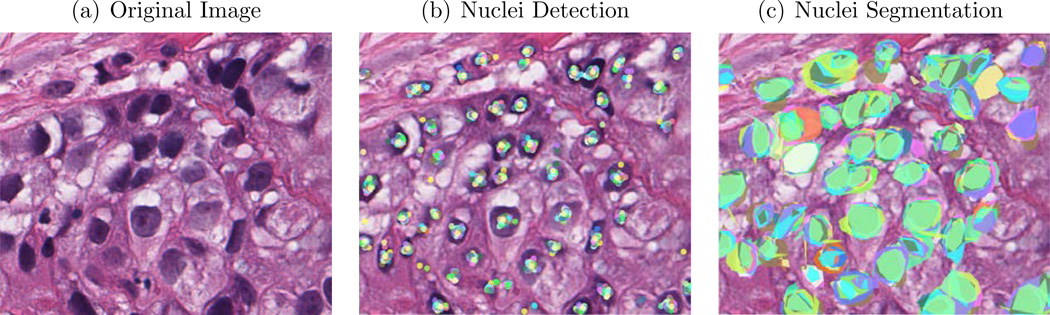
Fig. 1.
Examples of nucleus detection results produced by different contributor levels (Green circle indicates TP nuclei, yellow circle indicates FN nuclei and blue circle indicates FP). The automated detected nuclei were used as ground truth.
On the CrowdFlower platform interface, individual nuclei are rendered at relatively larger size on smaller images as compared to larger images. Further, smaller images contain fewer nuclei per image. To assess the influence of image size on contributor performance, we performed an experiment in which we extracted images of three different sizes (400 × 400, 600 × 600 and 800 × 800) from the same ROIs. We collected annotations with Contributor Level 2 and compared the annotations with those obtained with automated methods as shown in Table 4. The image size 400 × 400 performed significantly better than the larger image sizes. These results suggest that defining a small image size is important for obtaining optimal performance when using crowdsourced microtasks for image annotation for complex and tedious work, such as nucleus detection.
3.3. Segmentation Results
Like nucleus detection, we also performed four experiments for nuclear segmentation. In the first experiment, we considered pathologist’s nuclear segmentation as GT segmentation. Pathologists provided annotation on 63 images. We compared those 63 segmented images with the segmentations produced by research fellows, an automated method and three different level of contributors’ annotation as shown in Table 5. The strongest performance was achieved by Contributor Level 3, research fellow, and the automated method, which all achieved F-M scores between 65.93% and 66.41%. The Contributor Levels 1 and 2 showed slightly worse performance with F-M scores of 60.9% as shown in Table 5.
Table 5.
Segmentation results on 63 images (Pathologists’ annotation as GT)
| Annotations | TPR % | PPV % | F-M % | TNR % | Overlap % |
|---|---|---|---|---|---|
| Research Fellow | 60.40 | 79.80 | 65.93 | 97.68 | 49.66 |
| Automated Method | 76.22 | 62.26 | 65.36 | 96.34 | 49.87 |
| Contributor Level 1 | 56.95 | 71.47 | 60.87 | 97.08 | 44.30 |
| Contributor Level 2 | 59.02 | 71.19 | 60.89 | 97.04 | 44.46 |
| Contributor Level 3 | 67.73 | 69.07 | 66.41 | 96.86 | 50.14 |
In this experiment, we considered the research fellow-derived nuclear segmentation as GT segmentation, which we obtained on 455 images. On these 455 images, all three levels of crowdsourced non-expert annotations significantly outperformed the automated method, as shown in Table 6. Overall, Contributor Level 3 achieved the highest TPR (76.47%), F-Measure (65.15%) and overlap (48.68%).
Table 6.
Segmentation results on 455 images (Research Fellows’ annotation as GT)
| Annotations | TPR % | PPV % | F-M % | TNR % | Overlap % |
|---|---|---|---|---|---|
| Automated Method | 60.28 | 48.01 | 51.92 | 69.04 | 40.33 |
| Contributor Level 1 | 70.13 | 60.73 | 62.21 | 93.58 | 45.85 |
| Contributor Level 2 | 68.93 | 63.98 | 62.47 | 94.19 | 45.95 |
| Contributor Level 3 | 76.47 | 59.23 | 65.15 | 93.69 | 48.68 |
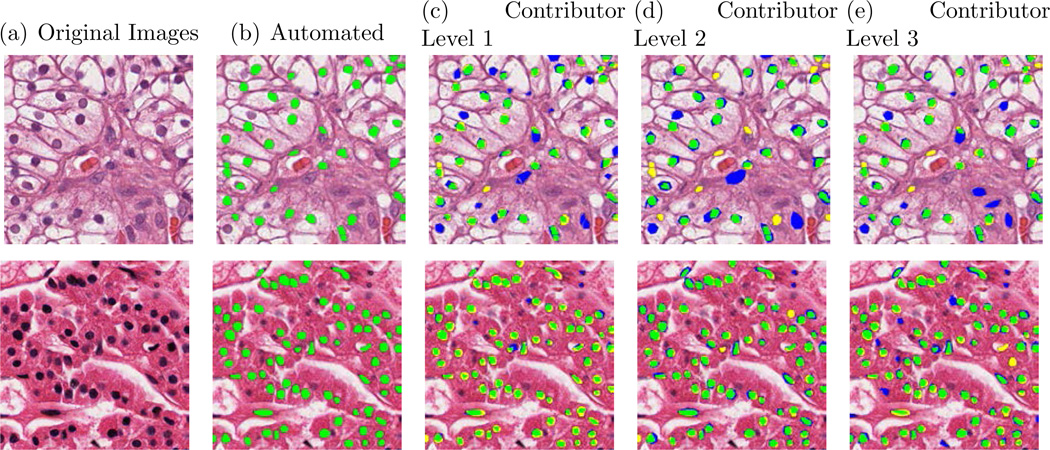
In our next experiment, we compared the annotations of different contributor levels and used the automated method annotations as the GT across all 810 study images, as shown Table 7. Contributor Level 3 achieved the highest TPR(75.78%), PPV(57.83%), F-Measure(62.10%) and overlap (46.75%), significantly outperforming Contributor Levels 1 and 2. Visual examples of different level of contributor annotations are shown in Figure 2.
Table 7.
Segmentation results on 810 images (Automated Method’s annotation as GT)
| Annotations | TPR % | PPV % | F-M % | TNR % | Overlap % |
|---|---|---|---|---|---|
| Contributor Level 1 | 74.17 | 52.49 | 57.34 | 93.10 | 41.80 |
| Contributor Level 2 | 74.14 | 49.31 | 54.17 | 91.54 | 38.97 |
| Contributor Level 3 | 75.78 | 57.83 | 62.10 | 95.21 | 46.75 |
Fig. 2.
Examples of nuclear segmentation using an automated method and increasing contributor skill level, ranging from 1 to 3. (Green region indicates TP region, yellow region indicates FN region and blue region indicates FP region). The automated nuclei segmentation used as ground truth.
Next, we assessed the relationship of contributor performance with image size for the job of nuclear segmentation. As we did for the nucleus detection experiment, we extracted three different image sizes(400 × 400, 600 × 600 and 800 × 800) from the same ROIs. We collected annotations from Contributor Level 2 and compared them with automated methods as shown in Table 8. As we observed for the nucleus detection job, annotation performance for the nuclear segmentation job was highest for the image size 400 × 400 and degraded significantly when image size was increased, as shown in Table 8.
Table 8.
Segmentation results on 63 images (Automated Method as GT)
| Annotations | TPR % | PPV % | F-M % | TNR % | Overlap % |
|---|---|---|---|---|---|
| Image Size 400 × 400 | 74.14 | 49.31 | 54.17 | 91.54 | 38.97 |
| Image Size 600 × 600 | 69.27 | 30.68 | 36.75 | 84.96 | 24.06 |
| Image Size 800 × 800 | 44.65 | 42.10 | 25.32 | 80.65 | 15.87 |
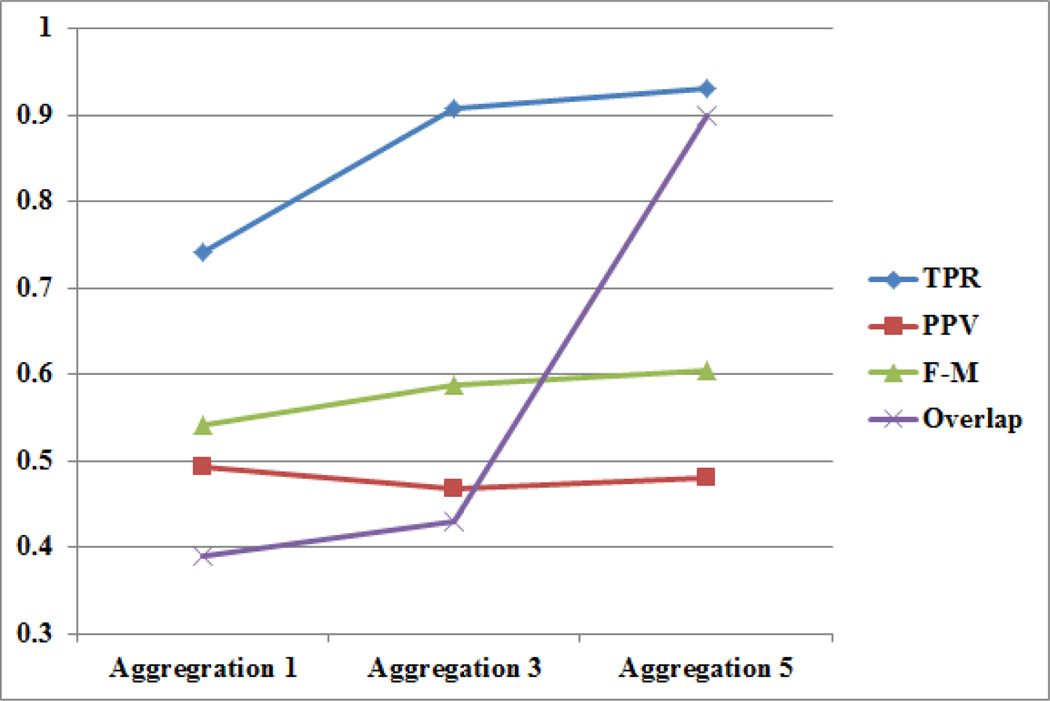
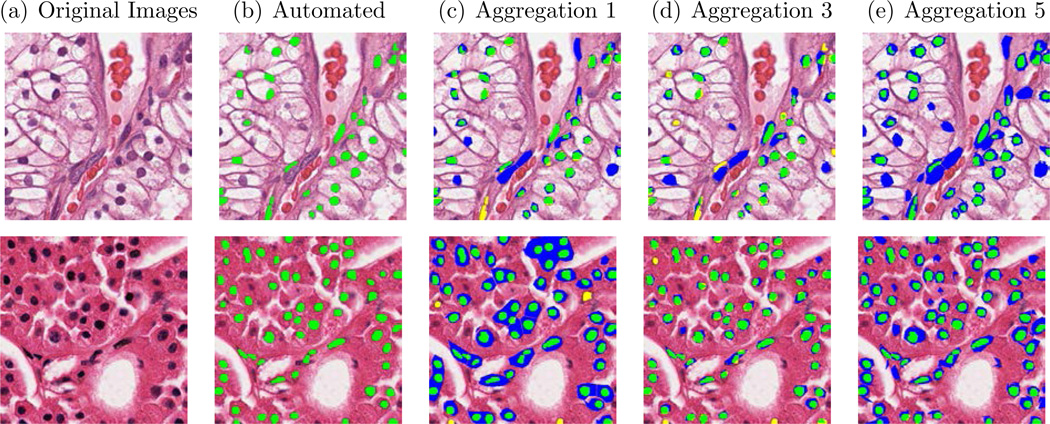
In addition to single contributor annotation, we collected multiple contributor annotations-per-image for the segmentation job. As shown in Figure 3, nuclei segmentation performance improved with increasing levels of annotation aggregation. A visual example of nuclei segmentation performance with multiple annotators is shown in Figure 4. Figure 5 shows the aggregated results of the contributors on a test question for both the nuclei detection and segmentation jobs.
Fig. 3.
Graph showing TPR, PPV, F-M and overlap curves for nuclear segmentation results using increasing numbers of aggregated contributor (level 2) annotations (from 1 to 3 to 5). The automated segmentation used as ground truth.
Fig. 4.
Examples of nuclear segmentation using an automated method and increasing levels of aggregation from Contributor Level 2, ranging from 1 to 3 to 5. (Green region indicates TP region, yellow region indicates FN region and blue region indicates FP region). The automated nuclei segmentation was used as ground truth.
Fig. 5.
Aggregation of results of the contributors on a test question. Different colors of dots and region represent different contributor annotations.
3.4. Cost and Time Analysis, and the Heterogeneity of the Crowd
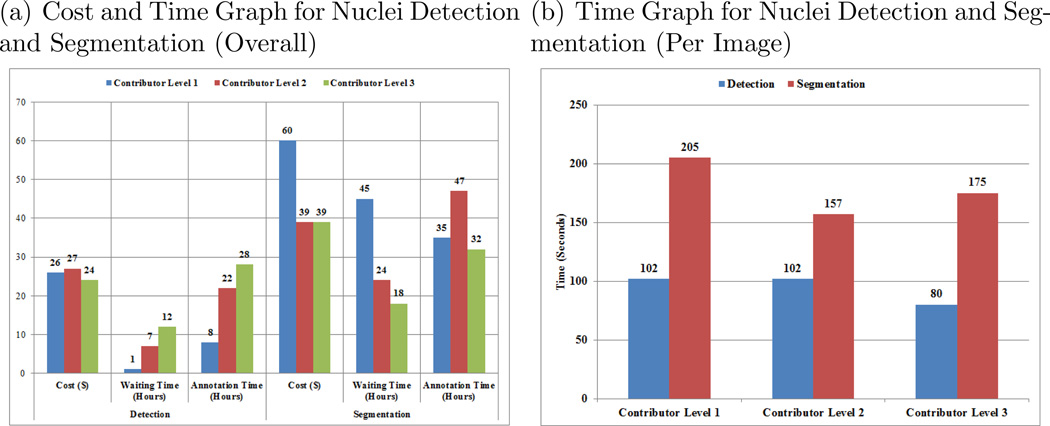
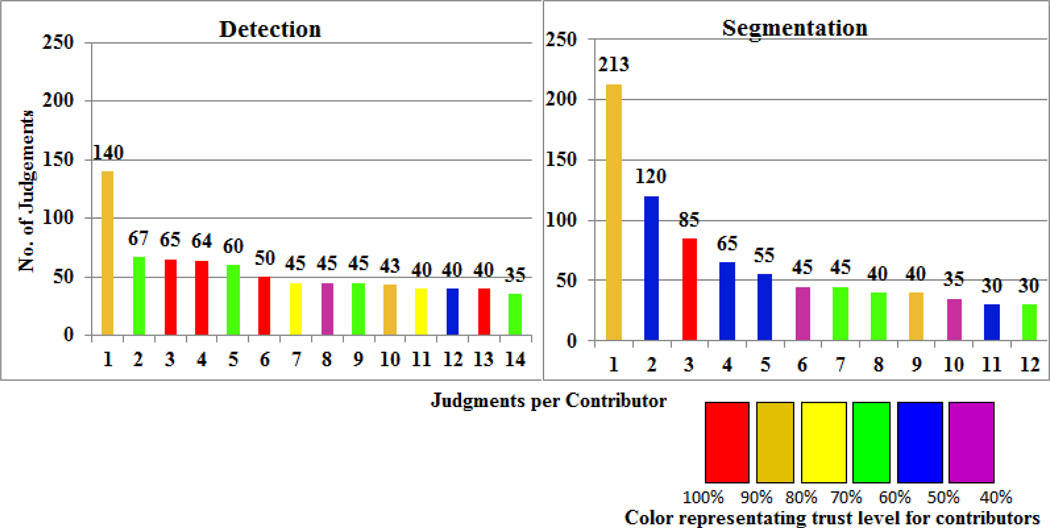
The Cost and Time analysis aggregated across all images for nucleus detection and segmentation and stratified by Contributor Level are shown on the left-panel in Figure 6, and the time analysis for one image across different contributor levels is shown on the right-panel in Figure 6. These data show that the segmentation job accounts for significantly more time per task and time overall, suggesting that nuclei segmentation is the more complex of the jobs. For the nucleus detection job, the waiting time required for attracting contributors to the job was significantly longer for the higher skill level contributors and the annotation time spent completing the job was also longer for the more skilled workers. For the segmentation job (which is the more complex job), the overall waiting time and annotation time were shorter for the Contributor Level 3 workers as compared with the Level 1 and 2 workers, likely owing to the fact that the Contributor Level 3 workers may have been more attracted to the higher complexity job. The distribution of contributor judgment trust level, which reflects contributor performance on test questions, is displayed in Figure 7. These plots show that although the highest proportion of judgments come from contributors with moderate-to-high trust levels (80% – 90% trust level), there is a wide distribution of contributor trust levels with a significant number of judgments derived from contributors with only moderate-to-poor trust levels. These results suggest the value of targeting specific jobs to specific crowd skill levels, and that by better targeting jobs to the appropriate crowds, we may obtain improvements in performance.
Fig. 6.
Time and Cost Analysis for Nucleus Detection and Segmentation for Different Contributor Levels.
Fig. 7.
Distribution of contributor judgments and trust level.
4. Conclusions
Our experiments show that crowdsourced non-expert-derived scores perform at a similar level to research fellow-derived scores and automated methods for nucleus detection and segmentation, with the research fellow annotations showing the strongest performance for detection, and the crowdsourced level 3 scores showing the strongest performance for segmentation. We conclude that crowdsourced image annotation is a highly-scalable and effective method for obtaining nuclear annotations for large-scale projects in computational pathology. Our results show that performance may be improved further by aggregating multiple crowd-sourced annotations per image, and by targeting jobs to specific crowds based on the complexity of the job and the skill level of the contributors. Ultimately, we expect that large-scale crowdsourced image annotations will lead to the creation of massive, high-quality annotated histopathological image datasets, which will support the improvement of supervised machine learning algorithms for computational pathology and will enable the design of systematic and rigorous comparative analyses of competing approaches, ultimately leading to the identification of top-performing methods, which will power the next generation of computational pathology research and practice.
Acknowledgements
Research reported in this publication was supported by the National Library of Medicine of the National Institutes of Health under Award Number K22LM011931. We thank Ari Klein, Nathan Zukoff, Sam Rael and the CrowdFlower team for their support, and we thank all the image annotation contributors for making this work possible.
Contributor Information
H. Irshad, Email: hirshad@bidmc.harvard.edu.
L. Montaser-Kouhsari, Email: lmontase@bidmc.harvard.edu.
G. Waltz, Email: gwaltz@bidmc.harvard.edu.
O. Bucur, Email: obucur@bidmc.harvard.edu.
J.A. Nowak, Email: janowak@partners.org.
F. Dong, Email: fdong1@partners.org.
N.W. Knoblauch, Email: nknoblau@bidmc.harvard.edu.
A. H. Beck, Email: abeck2@bidmc.harvard.edu.
References
- 1.Gurcan MN, Boucheron LE, Can A, Madabhushi A, Rajpoot NM, Yener B. Biomedical Engineering, IEEE Reviews in. 2009;2:147. doi: 10.1109/RBME.2009.2034865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dawson A, Austin R, Jr, Weinberg D. American journal of clinical pathology. 1991;95:S29. [PubMed] [Google Scholar]
- 3.Beck AH, Sangoi AR, Leung S, Marinelli RJ, Nielsen TO, van de Vijver MJ, West RB, van de Rijn M, Koller D. Science translational medicine. 2011;3:108ra113. doi: 10.1126/scitranslmed.3002564. [DOI] [PubMed] [Google Scholar]
- 4.Irshad H, Veillard A, Roux L, Racoceanu D. Biomedical Engineering, IEEE Reviews in. 2014;7:97. doi: 10.1109/RBME.2013.2295804. [DOI] [PubMed] [Google Scholar]
- 5.Ghaznavi F, Evans A, Madabhushi A, Feldman M. Annual Review of Pathology: Mechanisms of Disease. 2013;8:331. doi: 10.1146/annurev-pathol-011811-120902. [DOI] [PubMed] [Google Scholar]
- 6.Good BM, Su AI. Bioinformatics. 2013:btt333. doi: 10.1093/bioinformatics/btt333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lintott CJ, Schawinski K, Slosar A, Land K, Bamford S, Thomas D, Raddick MJ, Nichol RC, Szalay A, Andreescu D, et al. Monthly Notices of the Royal Astronomical Society. 2008;389:1179. [Google Scholar]
- 8.Kim JS, Greene MJ, Zlateski A, Lee K, Richardson M, Turaga SC, Purcaro M, Balkam M, Robinson A, Behabadi BF, et al. Nature. 2014;509:331. doi: 10.1038/nature13240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warby SC, Wendt SL, Welinder P, Munk EG, Carrillo O, Sorensen HB, Jennum P, Peppard PE, Perona P, Mignot E. Nature methods. 2014;11:385. doi: 10.1038/nmeth.2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mavandadi S, Dimitrov S, Feng S, Yu F, Sikora U, Yaglidere O, Padmanabhan S, Nielsen K, Ozcan A. PLoS One. 2012;7:e37245. doi: 10.1371/journal.pone.0037245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luengo-Oroz MA, Arranz A, Frean J. Journal of medical Internet research. 2012;14 doi: 10.2196/jmir.2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Network CGAR, et al. Nature. 2013;499:43. [Google Scholar]
- 13.Kong J, Cooper LA, Wang F, Gutman DA, Gao J, Chisolm C, Sharma A, Pan T, Van Meir EG, Kurc TM, et al. Biomedical Engineering, IEEE Transactions on. 2011;58:3469. doi: 10.1109/TBME.2011.2169256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang H, Fontenay GV, Han J, Cong G, Baehner FL, Gray JW, Spellman PT, Parvin B. BMC bioinformatics. 2011;12:484. doi: 10.1186/1471-2105-12-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cataldo SD, Ficarra E, Acquaviva A, Macii E. Computer Methods and Programs in Biomedicine. 2010;100:1. doi: 10.1016/j.cmpb.2010.02.002. [DOI] [PubMed] [Google Scholar]