Abstract
Brain atrophy has been identified as an important contributing factor to the development of disability in multiple sclerosis (MS). In this respect, more and more interest is focussing on the role of deep grey matter (DGM) areas. Novel data analysis pipelines are available for the automatic segmentation of DGM using three-dimensional (3D) MRI data. However, in clinical trials, often no such high-resolution data are acquired and hence no conclusions regarding the impact of new treatments on DGM atrophy were possible so far. In this work, we used FMRIB's Integrated Registration and Segmentation Tool (FIRST) to evaluate the possibility of segmenting DGM structures using standard two-dimensional (2D) T1-weighted MRI. In a cohort of 70 MS patients, both 2D and 3D T1-weighted data were acquired. The thalamus, putamen, pallidum, nucleus accumbens, and caudate nucleus were bilaterally segmented using FIRST. Volumes were calculated for each structure and for the sum of basal ganglia (BG) as well as for the total DGM. The accuracy and reliability of the 2D data segmentation were compared with the respective results of 3D segmentations using volume difference, volume overlap and intra-class correlation coefficients (ICCs). The mean differences for the individual substructures were between 1.3% (putamen) and −25.2% (nucleus accumbens). The respective values for the BG were −2.7% and for DGM 1.3%. Mean volume overlap was between 89.1% (thalamus) and 61.5% (nucleus accumbens); BG: 84.1%; DGM: 86.3%. Regarding ICC, all structures showed good agreement with the exception of the nucleus accumbens. The results of the segmentation were additionally validated through expert manual delineation of the caudate nucleus and putamen in a subset of the 3D data. In conclusion, we demonstrate that subcortical segmentation of 2D data are feasible using FIRST. The larger subcortical GM structures can be segmented with high consistency. This forms the basis for the application of FIRST in large 2D MRI data sets of clinical trials in order to determine the impact of therapeutic interventions on DGM atrophy in MS.
Keywords: Segmentation, Basal ganglia, T1-weighted data, Two-dimensional data, Multiple sclerosis, FMRIB's Integrated Registration and Segmentation Tool
Highlights
-
•
Segmentation of deep grey matter (DGM) using 2-dimensional MRI data is challenging.
-
•
We performed a systematic comparison between 2 and 3D data segmentation using FIRST.
-
•
The DGM volumes of 70 multiple sclerosis patients were statistically compared.
-
•
We found a good agreement of the segmentation results for the large DGM structures.
-
•
Our results indicate that FIRST is suitable for the segmentation of 2D data sets acquired in multicentre studies.
1. Introduction
Several grey matter nuclei are located in the depth of the brain and hence being termed deep grey matter (DGM). These nuclei include the thalamus, globus pallidus (pallidum), putamen, nucleus accumbens and caudate nucleus. All these areas serve as relay stations to and between cortical brain areas. The pallidum, putamen and caudate nucleus are usually grouped as basal ganglia (BG), because of a common involvement in extrapyramidal motor functions. These areas receive inputs from multiple cortical areas and the thalamus and project mostly to the premotor, supplementary motor and prefrontal areas. They are implicated in voluntary and automatic motor responses, predicting future events, suppressing unwanted behaviour, shifting attention and are involved in spatial working memory (Herrero et al., 2002). The thalamus is a complex of 50–60 nuclei that serves as a relay and filter centre for both sensory and motor pathways. First order nuclei receive input mostly from ascending sensory pathways, whereas higher order nuclei receive primary afferents from the cerebral cortex (Herrero et al., 2002). Thalamic damage can be associated with motor and somatosensory disturbances, cognitive decline, fatigue and central pain.
DGM involvement is increasingly recognized as an important pathophysiological feature of multiple sclerosis (MS). Histopathological studies have shown demyelination, inflammation, iron accumulation and neurodegeneration of DGM in MS patients (Bagnato et al., 2011; Vercellino et al., 2009). Moreover, DGM abnormalities have been reported using magnetic resonance (MR) imaging. Using arterial spin labelling (Rashid et al., 2004), it was possible to demonstrate DGM hypoperfusion in MS patients, a finding that was corroborated by dynamic contrast-enhanced perfusion MR (Inglese et al., 2007; Varga et al., 2009). Increased iron accumulation has been shown by means of susceptibility weighted imaging as well as by quantitative T2 studies (Bakshi et al., 2002; Drayer et al., 1987; Haacke et al., 2009) and may precede DGM atrophy (Hagemeier et al., 2012).
Several studies demonstrated a close relationship between DGM atrophy and the clinical manifestation of MS. For example, Houtchens et al. (2007) found significant correlation between cognitive performance and thalamic volume in MS patients and a reduction of thalamic volume compared to healthy controls. A correlation between atrophy of the putamen and the clinical outcome has been reported by Henry et al. (2008) in a cohort of patients with clinically isolated syndrome suggestive of multiple sclerosis (CIS). Atrophy of DGM structures may predict conversion of CIS to definite MS (Calabrese et al., 2010), correlates with memory impairment (Benedict et al., 2009), information processing slowing (Batista et al., 2012) and may be associated with development of fatigue (Calabrese et al., 2010). There is some evidence that DGM atrophy correlates with (Tao et al., 2009) or is even driven by WM lesions (Muhlau et al., 2013).
Several methods have been previously used to assess DGM atrophy: 1. indirect measures such as the bicaudate ratio (Bermel et al., 2002; Caon et al., 2003); 2. morphometric methods including voxel based morphometry (Audoin et al., 2010; Henry et al., 2008; Lansley et al., 2013; Prinster et al., 2006; Riccitelli et al., 2012) and tensor based morphometry (Tao et al., 2009) and 3. volumetric methods with manual (Bermel et al., 2003), semiautomatic (Houtchens et al., 2007) or fully automatic DGM segmentation (Calabrese et al., 2010; Schoonheim et al., 2012; Zivadinov et al., 2013).
A dedicated and frequently used method to automatically segment subcortical grey matter structures based on three-dimensional (3D) T1 weighted (T1w) MRI data is FMRIB's Integrated Registration and Segmentation Tool (FIRST) (Patenaude et al., 2011), which is part of the FMRIB Software Library (FSL) (Jenkinson et al., 2012; Smith et al., 2004; Woolrich et al., 2009). FIRST uses a shape model obtained from manually segmented images to guide the segmentation of the subcortical structures including the thalamus, basal ganglia, hippocampus and amygdala. In brief, the individual 3D data set is registered by a two-stage affine transformation to a standard space (MNI space). Then, FIRST fits a mesh to the registered data set. This mesh represents the surfaces of the subcortical structures. It is modelled using a multivariate Gaussian model based on shape and intensity information (for a further description see Patenaude et al. (2011)). FIRST has been extensively evaluated for segmenting 3D data in terms of scan–rescan reliability (Morey et al., 2010) as well as interscanner reliability (Nugent et al., 2013). Furthermore, results have been compared with manual segmentations (Morey et al., 2009; Nugent et al., 2013) and other automated segmentation tools (Morey et al., 2010; Nugent et al., 2013). To date, FIRST has been applied successfully not only in studies covering various neurological and psychiatric disorders such as epilepsy (Lin et al., 2012), schizophrenia (Felsky et al., 2012), mild cognitive impairment (Alves et al., 2012; Zhuang et al., 2013), Alzheimer's disease (Alves et al., 2012; Cheng et al., 2013; Park et al., 2012) and depression (Turner et al., 2012), but also in research focussing on normal aging (Hughes et al., 2012), cognitive flexibility in older adults (Verstynen et al., 2012) and topographical memory in young adults (Hartley and Harlow, 2012).
In the aforementioned studies, FIRST was applied to 3D data sets. The three-dimensional MRI acquisition provides data with high spatial resolution as well as isotropic voxels and allows whole head coverage. To date, however, no 3D data are acquired in many multicentre clinical MS trials (e.g. (Barkhof et al., 2007; Khatri et al., 2011; Radue et al., 2012)). Instead, only two-dimensional (2D) T1w data are acquired. This relates to specific challenges in the multicentre setting, where shorter scanning times are advantageous and not all sites may have access to state-of-the-art MRI scanner hardware. Moreover, study sponsors often aim at comparing data of new trials to previous studies favouring the continued use of older sequences. Finally, 3D sequences are prone to movement artefacts. On the other hand, segmentation of DGM using 2D MRI data using existing automatic segmenting tools seems to be challenging. One reason is that two dimensional data acquisition provides an anisotropic spatial resolution with relatively large voxel dimensions in the slice direction, typically in the range of 3–5 mm. Additionally, 2D T1w data may suffer from suboptimal tissue contrast between the DGM and the adjacent white matter (WM) structures.
Altogether, segmentation of DGM using 2D MRI data seems to be challenging using automatic segmentation tools. In the original paper of Patenaude et al. (2011), a training data set was used for FIRST, which also encompassed 2D data. However to our knowledge, no systematic comparison between 2 and 3D segmentation has been carried out so far. In this work, we investigated the feasibility and reliability of applying the FIRST segmentation tool on 2D T1w data sets. We did so by comparing DGM volumes of 70 MS patients obtained by 2D data segmentation with the respective results of 3D data segmentation in the same subjects. The results of the segmentation were additionally validated through expert manual delineation of the caudate nucleus and putamen in a subset of the 3D data.
In our opinion, the validation of FIRST on 2D data sets is important as a large amount of such data has been collected in the last two decades in most clinical multiple sclerosis trials (Gold et al., 2013; Kappos et al., 2008; Kappos et al., 2010; Kappos et al., 2014; Miller et al., 2003; Molyneux et al., 2000; Rovaris et al., 2007; Rudick et al., 2006). A successful validation would form the basis for the application of FIRST in these trials in order to determine the impact of therapeutic interventions on DGM atrophy in MS.
2. Methods
2.1. Patients and image acquisition
Seventy consecutive patients with MS (47 women, 23 men) who underwent clinical routine MRI head examinations at our institution were included in this study. The study was approved by the local ethics committee (Ethikkommission beider Basel, EKBB) and the participants gave written informed consent before study entry. Sixty-two patients had relapsing–remitting MS, three patients had secondary progressive MS, one patient had primary progressive MS, and four patients had a clinically isolated syndrome suggestive of MS. The mean age of the patients was 45.3 years (range 24 years–74 years, standard deviation: 11.2 years). The MR scans were performed on either one of the following three 1.5 Tesla scanners (all Siemens Medical, Erlangen, Germany): 47 patients on Magnetom Avanto, 16 patients on Magnetom Symphony, and 7 patients on Magnetom Espree. The standard scanning protocol on each scanner included two pre-contrast T1w scans: an isotropic 3D MPRAGE sequence (TR/TI/TE/α = 2.7 s/950 ms/5 ms/8°) and a 2D T1 weighted spin echo (SE) sequence with (TR/TE = 552 ms/17 ms) for the Magnetom Avanto and Magnetom Espree; for the Magnetom Symphony, the sequence parameters were slightly different (TR/TE = 450 ms/10 ms) to gain optimal image quality. In the MPRAGE sequence, 192 sagittal slices with a spatial resolution of 1 × 1 × 1 mm3 were acquired, in the T1 weighted SE, 40 slices parallel to the inferior borders of the corpus callosum were scanned with two interleaved concatenations (spatial resolution 0.9 × 0.9 × 3 mm3). All head scans were acquired with a standard 12-element head matrix coil.
2.2. Data processing
All data were processed using FIRST as part of FSL version 5.0. The initial step of the FIRST data processing pipeline is a two-stage affine registration to a standard space template (MNI space) with 1 mm resolution using 12 degrees of freedom. The whole brain is registered to the template and thereafter the registration is refined for the subcortical structures using a mask. To enable this two-stage registration, the 2D data were interpolated with a sinc function to an isotropic resolution of 1 × 1 × 1 mm3 using FLIRT (FMRIB's Linear Image Registration Tool, also part of FSL (Jenkinson et al., 2002; Jenkinson and Smith, 2001)). Besides this data interpolation, a slight modification of the FIRST data processing pipeline was necessary as the spatial head coverage of the 2D slices was less than that of the original MNI template with only partial inclusion of scalp and medulla oblongata. Therefore, we removed the twenty most superior axial slices as well as the twenty most inferior slices in both MNI template and subcortical mask (see Fig. 1) and used these two modified templates in the further processing of the interpolated 2D data sets. Except for this modification, both 2D and 3D data were processed identically. In our study, the following subcortical structures were segmented bilaterally: thalamus, caudate, pallidum, putamen and nucleus accumbens. Although it is possible to segment the amygdala and hippocampus with FIRST, we decided not to include these two structures in the analysis as it is challenging to control their anatomical boundaries visually. Also the brainstem and 4th ventricle were excluded from evaluation as these structures were only partially covered in the 2D data.
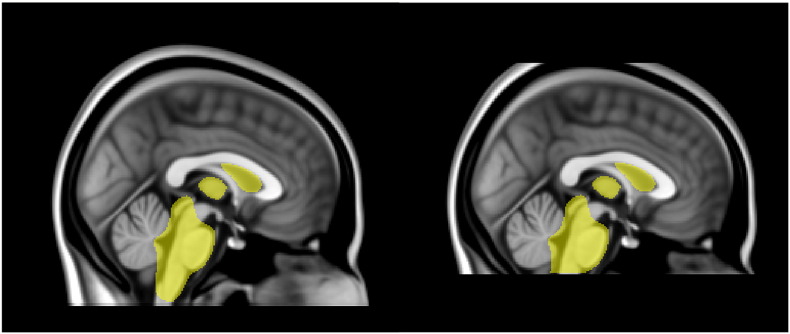
Fig. 1.
Modified MNI template: Left — original MNI 1 mm T1w template used in FIRST, in yellow the subcortical mask; right — modified “cut” templates.
Even after spatial interpolation, the 2D data demonstrated partial volume effects, which were most obvious at the borders of the lateral ventricles to thalamus bilaterally (see Fig. 6A). To reduce these partial volume effects, a ventricle mask was calculated for both the 2D and the 3D data sets using SIENAX (Smith et al., 2001; Smith et al., 2002) and applied to the subcortical segmentation. SIENAX is also part of FSL and has been extensively used for cross-sectional estimation of brain atrophy. It offers the possibility to segment a ventricular CSF mask (option “–r”), which was used in this work.
Fig. 6.
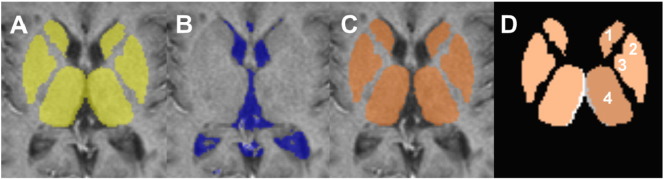
CSF mask effect: The effect of applying a CSF mask onto 2D data; A: original DGM mask; B: CSF mask; C: DGM mask corrected for CSF; D: difference between original mask (grey) and corrected mask (orange). Only the thalamus (4) was significantly impacted by the CSF mask. The other DGM structures in this figure are (1) caudate nucleus; (2): putamen; and (3): pallidum.
The processed data sets were visually inspected for quality control at three stages of the FIRST routine: First, the results of the two-step registration of the head were checked. If either the brain outline or the subcortical structures were not well aligned to the MNI template, the respective data sets were considered as having failed the analysis. Second, the subcortical segmentation was screened for apparent inaccuracies. Third, the ventricle masks were checked for correctness.
For each bilateral subcortical structure, we obtained two volumes (left and right) for each data set (in units of mm3). In the statistical evaluation of these data, we found no significant effects of laterality (results not shown). Therefore, left- and right-sided volumes were pooled for each structure in the statistical evaluation. This means that for N successfully segmented data sets, statistical analyses were carried out over 2 ∗ N volumes for each bilateral structure.
The results of the FIRST segmentation were additionally validated through expert manual delineation. An experienced rater segmented caudate and putamen manually according to Nugent et al. (2013). The manual segmentations were restricted to a subset of twenty randomly selected 3D data sets (9 from Avanto, 5 from Symphony, and 6 from Espree), because the manual segmentation of brain structures is very time consuming. The pallidum, thalamus and nucleus accumbens were not outlined as the majority of their boundaries possess very low tissue contrast to the adjacent WM and GM structures and a manual segmentation using T1 data sets is therefore very difficult in these structures. As the 2D SE data demonstrate even less tissue contrast than the 3D MPRAGE data in all DGM structures, manual segmentation was not feasible in these data sets.
2.3. Statistical analysis
In the statistical analyses, only patients whose 2D and 3D data sets survived all three visual quality control steps were included. The analyses were carried out using IBM SPSS Statistics 21 (IBM Corporation Software Group, Somers, USA).
The agreement between the automatic segmentation results of the 2D and 3D data and the validation of FIRST with manual segmentation were tested by means of: 1. Percentage difference in volume (ΔV), 2. Percentage overlap in volume (Dice coefficient s) and 3. Intraclass correlation coefficients.
Percentage differences in volume between the two segmentations were calculated for each subcortical structure in all patients according to Eq. (1):
| (1) |
With
V(2D): volume of the segmented substructure in the 2D data
V(3D): volume of the segmented substructure in the 3D data.
With this definition, ΔV is the difference between the two volumes normalized to the mean volume of each methods.
Additionally, also the percentage volume differences between all basal ganglia (BG; sum of caudate, pallidum, putamen and nucleus accumbens) and between the whole deep grey matter (DGM; sum of BG and thalamus) were calculated, as these overall measures are often surveyed in clinical trials.
Another very common measure of similarity between two structures is the Dice's similarity coefficient (Dice, 1945). In our case Dice's coefficient equals the percentage overlap of the volumes and is given by Eq. (2). A value of 0 means no overlap, whereas a value of 1 represents a complete agreement.
| (2) |
With ‖V(2D)∩V(3D)‖: modulus of overlap between V(2D) and V(3D).
In this study, the subcortical segmentation was performed on data sets which were acquired in different orientations. Therefore, computation of Dice's coefficients is not possible per se. To enable the use of this measure, we aligned the 2D data sets onto the respective 3D data sets using FLIRT. The resulting affine transformation parameters were then applied to the segmented subcortical structures. Dice's coefficients between realigned 2D structures and 3D structures were calculated using the routine @DiceMetric of AFNI (Cox, 1996).
The accuracy and reliability of the 2D data segmentation in comparison to the 3D data segmentation were further tested by calculating intra-class correlation coefficients (ICCs). More specifically, we calculated ICC for the particular subcortical structures, for BG, and for the whole DGM. A two-way mixed model for single measures was used with data acquisition (2D versus 3D) as fixed factor and the subjects as random factor. ICCs for consistency and for absolute agreement were calculated. A high ICC for consistency means that if a volume is largest in the 3D segmentation, it is highly probable that it is also largest in the 2D segmentation. A high ICC in absolute value means in addition that the volumes in both segmentations are nearly the same in terms of absolute numbers (no systematic errors). High ICC in either case means that both segmentation results have a similar predictive value in regression analyses. In line with previous publications by Nugent et al. (2013) and Innes and Straker (1999), we considered ICC > 0.7 as acceptable agreement and ICC > 0.9 as strong agreement.
3. Results
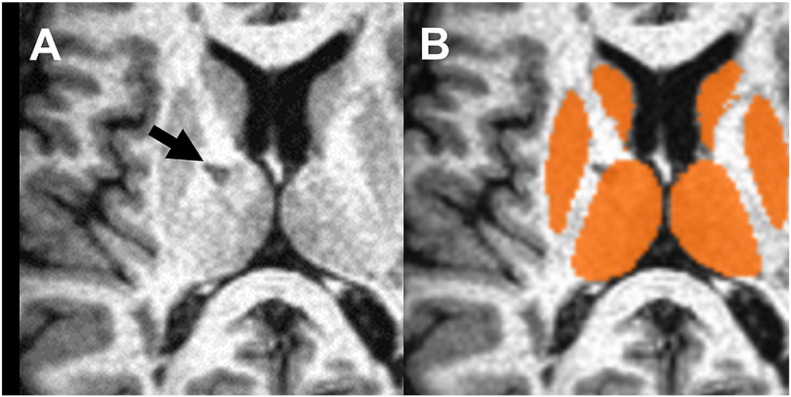
After visual inspection of the registration to MNI space, the data of eight patients (11.4%) had to be discarded (seven from the Avanto and one from the Symphony). In four of these eight cases (5.7%; three on the Avanto, one on the Symphony), both the registration of the 3D and the 2D data failed. These patients demonstrated severe global brain atrophy with much enlarged ventricles (Fig. 2 shows a typical example). In the other four patients (5.7%; all Avanto), only the registration of the interpolated 2D data failed. Visual inspection of these data sets revealed errors in the angulation of the co-registered data sets to the MNI templates and/or gross scaling errors. Whereas in two of these four patients no obvious reason was found, the other two patients showed motion artefacts in the original 2D data sets. As the 2D data were acquired in two interleaved concatenations, motion becomes apparent in these cases as a displacement between the even and the odd slices. The displacement artefacts were slightly blurred due to the spatial interpolation, but were still manifest in the interpolated data (see Fig. 3). None of the remaining patients (N = 62; 88.6%) showed either major inaccuracies of the subcortical segmentation or failures in the CSF mask. Furthermore, hypo-intense MS lesions located at the borders of the particular subcortical structures had no visible impact on segmentation (see Fig. 4). Therefore, all of the remaining data sets were included in the statistical analysis.
Fig. 2.
Impact of atrophy: In this case, both registration of the 2D and 3D data to MNI space failed because of severely enlarged ventricles. A: Original axial view of the 3D data; B: same data registered to MNI space (slice is not identical but similar); C: 50% overlay to the MNI template D: MNI template (for comparison).
Fig. 3.
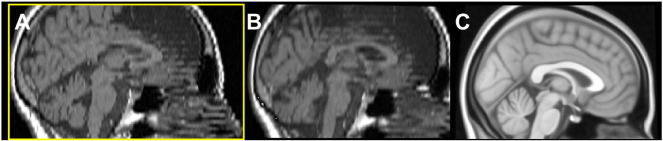
Impact of motion artefacts: Motion artefacts can hamper the registration to MNI space. A: The 2D data set show step-like artefacts due to motion between the two concatenated acquisitions. The registration to MNI space fails in this case as can be seen of the deviation of the pitch angle of the registered data set (B) compared to the MNI template (C).
Fig. 4.
Impact of MS lesions: The subcortical segmentation of FIRST is robust even in presence of MS lesions. The black arrow in (A) points to a hypo-intense lesion at the border of white matter and thalamus. (B) The segmentation (orange) is not impacted by the lesion.
Percentage volume differences between the segmentation of 2D and of 3D data are shown in Table 1. The mean percentage differences for the individual substructures were between 1.3% (putamen) and −25.2% (nucleus accumbens). Maximal individual differences were up to 107% (nucleus accumbens). Mean volume difference was −2.7% for BG (with individual differences up to −16.6%) and 1.3% for the DGM (individual differences up to 15.9%). The individual differences of BG and DGM are shown in Fig. 5.
Table 1.
Percentage volume differences for the different subcortical grey matter regions (number of subjects N = 62). Basal ganglia: sum over caudate, putamen, pallidum, and nucleus accumbens; deep grey matter (DGM): basal ganglia plus thalamus. Positive difference means that the segmented structures are larger in the 2D data sets.
| Thalamus | Putamen | Pallidum | Caudatus | Nucleus accumbens | Basal ganglia | DGM | |
|---|---|---|---|---|---|---|---|
| Mean difference | 7.035 | 1.341 | 2.608 | −8.303 | −25.164 | −2.705 | 1.348 |
| Standard deviation | 5.878 | 7.683 | 10.061 | 8.198 | 28.031 | 4.718 | 4.270 |
| Most negative difference | −16.320 | −13.383 | −29.778 | −32.651 | −106.977 | −16.642 | −15.921 |
| Most positive difference | 23.951 | 25.190 | 32.467 | 14.074 | 46.859 | 13.739 | 12.718 |
Fig. 5.
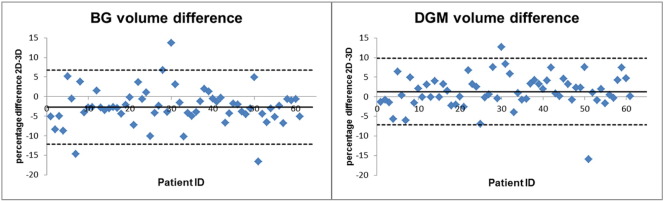
Volume differences between 2 and 3D segmentation: Percentage volume difference between 2 and 3D data for the basal ganglia (BG) and for deep grey matter (DGM; BG + thalamus). The 2D data were additionally corrected with a CSF mask (see Fig. 6). Solid line: mean difference for all patients; broken lines: mean difference ± 2 ∗ standard deviation.
The impact of the CSF masks was evaluated separately. From all substructures, only the thalamus and the caudate nucleus were affected by this correction as only these two substructures border to the lateral ventricles (see also Fig. 6). Without the CSF masks, the mean difference for the thalamus was 10.4 ± 6.7% (range −11.8%–27.6%); it dropped to 7.0 ± 5.9% (range −16.3% to +24.0%) using the CSF masks. For the caudate nucleus, the effects of the CSF masking were negligible (with and without CSF masks: 8.3 ± 8.2%, range −14.1% to +32.6%). For the DGM volume, mean difference dropped from 2.8 ± 4.8% (without CSF masks) to 1.3 ± 4.3% (with CSF masks). The individual differences ranged from −14.0% to 16.0% (without CSF masks) and from −15.9% to 12.7% respectively (with CSF masks). In line with the prior results, the CSF masks had only negligible impact on the BG volume.
Dice's similarity coefficients (percentage volume overlap) are shown in Table 2. The highest mean volume overlap was found for the thalamus (89.1 ± 2.9%), whereas the lowest was found for nucleus accumbens (61.5 ± 9.1%). The respective values for the BG were 84.1 ± 3.3% and for DGM 86.3 ± 3.0%. Again, the CSF masks changed only the volume overlap of the thalamus (without: 88.5 ± 3.3%; with: 89.1 ± 2.9%) and the volume overlap of DGM (without: 86.1 ± 3.0%; with: 86.3 ± 3.0%). The individual Dice's coefficients for BG and DGM using the CSF masks are depicted in Fig. 7.
Table 2.
Volume overlap for the different subcortical grey matter regions (N = 62). Basal ganglia: sum over caudate, putamen, pallidum, and nucleus accumbens; deep grey matter (DGM): basal ganglia plus thalamus.
| Thalamus | Putamen | Pallidum | Caudatus | Nucleus accumbens | BG | DGM | |
|---|---|---|---|---|---|---|---|
| Mean overlap | 0.891 | 0.857 | 0.796 | 0.752 | 0.615 | 0.841 | 0.863 |
| Standard deviation | 0.029 | 0.040 | 0.077 | 0.057 | 0.091 | 0.033 | 0.030 |
| Minimal overlap | 0.724 | 0.679 | 0.574 | 0.543 | 0.336 | 0.763 | 0.765 |
| Maximal overlap | 0.935 | 0.921 | 0.912 | 0.841 | 0.769 | 0.896 | 0.911 |
Fig. 7.
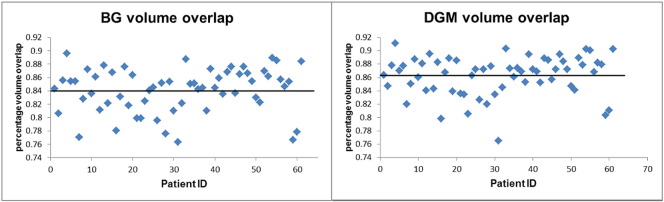
Volume overlap between 2 and 3D segmentation: Percentage volume overlap (Dice's coefficient) between 2 and 3D data for BG and for DGM. Solid line: mean overlap for all patients.
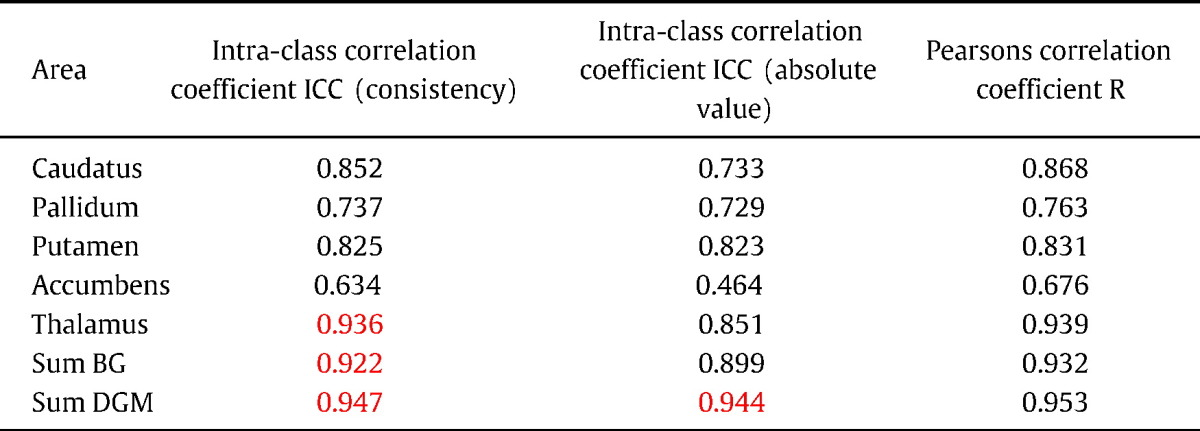
ICCs for the particular subcortical structures and for BG and for DGM are summarized in Table 3. All structures showed acceptable agreement both for consistency and for absolute value except the nucleus accumbens. Regarding consistency, strong agreement was found in the thalamus (ICC = 0.936), BG (ICC = 0.922) and in DGM (ICC = 0.947); regarding absolute value, total DGM volume (ICC = 0.944) showed strong agreement. Again, the CSF mask had significant impact only on the ICC of the thalamus (consistency: ICC increased from 0.907 to 0.936; total agreement: ICC increased from 0.731 to 0.851) and on the ICC of the total volume of DGM (consistency: ICC increased from 0.931 to 0.947; absolute value: ICC increased from 0.913 to 0.944).
Table 3.
Intra-class coefficients between 2 and 3D data volumes (N = 62). ICC values > 0.7 (bold) are considered as acceptable agreement, ICC values > 0.9 (red) as strong agreement. Pearson's correlation coefficients are shown for comparison.
The validation results of FIRST against the manual segmentation are shown in Table 4 for the caudate and putamen. The mean volume difference between 3D FIRST and manual segmentation was −11.03% for the caudate (FIRST segmentation smaller) with individual differences between −22.5% and +4.6%. The respective results for the putamen were 15.6% (FIRST segmentation larger) with individual differences between −4.5% and +34.2%. Mean overlap was 83.3% for the caudate (with an individual range between 73.2% and 87.0%) and 85.8% for the putamen (78.5%–90.1%). The respective ICCs were: caudate 0.92 (consistency) and 0.74 (absolute value); putamen 0.87 (con) and 0.52 (abs).
Table 4.
Comparison between the results of FIRST segmentation (both 2D and 3D) and manual segmentation for a subset of 20 data sets. Manual segmentation was applied only to 3D data.
| Mean difference | Mean overlap | ICC con | ICC abs | ||
|---|---|---|---|---|---|
| Caudatus | 3D | −11.031 ± 6.000 | 0.833 ± 0.0273 | 0.915 | 0.743 |
| 2D | −20.110 ± 7.926 | 0.686 ± 0.0678 | 0.827 | 0.468 | |
| Putamen | 3D | 15.569 ± 7.330 | 0.858 ± 0.0270 | 0.872 | 0.517 |
| 2D | 14.217 ± 9.745 | 0.794 ± 0.0420 | 0.718 | 0.440 |
For the 2D data, ICCs to manual segmentation were generally smaller: caudate 0.83 (cons) and 0.52 (abs); putamen 0.72 (con) and 0.44 (abs). Mean difference was −20.1% for caudate (−33.8% to + 2.4%) and +14.2% for putamen (−4.5%–34.2%), mean volume overlap was 68.6% for caudate (52.1%–77.5%) and 79.4% for putamen (67.7%–85.0%).
4. Discussion
In this study, we systematically evaluated the possibility of segmenting subcortical grey matter structures using two-dimensional T1-weighted MRI data sets with FIRST, a software tool that has been optimized for the segmentation of subcortical brain structures. Up to now, only 2D T1-weighted data are acquired in most clinical trials on MS and hence, a successful extension of the FIRST algorithm to this type of data would enable to study subcortical changes in large patient cohorts and to better understand effects of therapeutic interventions on DGM in MS.
4.1. Registration
The initial step when using FIRST is a registration to a common stereotaxic space, which is defined by the isotropic 1 mm3 MNI 152 template (Fonov et al., 2011). Two problems arise by registering 2D data sets onto this template. Namely, the 2D data sets have an anisotropic spatial resolution with a relative large voxel size in slice direction (in our case 3 mm) and, the spatial coverage is in general more limited than that of the original 3D template, e.g. the medulla is usually not covered. Our preliminary tests with non-interpolated 2D data showed that registration failed when using the original voxel size of 0.9 × 0.9 × 3 mm3. Therefore, the spatial interpolation of the original 2D data to the voxel size of the MNI template (or at least to similar voxel size) seems to be a prerequisite.
In an initial evaluation, we processed 25 data sets with the original MNI template. The registration failed in approximately one third (eight) of the interpolated 2D data sets because of large deviations in angulation and/or gross scaling errors. The modification of the original MNI templates (whole brain template and template of the subcortical structures) by “cutting off” the most superior and most inferior axial slices reduced the number of failed registrations to MNI space to two cases. In both of these cases, not only the registration of 2D data sets, but also the registration of the 3D data sets failed. Both patients had severe global brain atrophy with enlarged lateral ventricles. The fact that severe atrophy hampers registration independent of the dimensionality of data acquisition was reproduced for the rest of the patient cohort as we found two other cases in which both the registration for the 2D and the 3D data failed due to severe atrophy. It is possible that replacing the original MNI 152 template with a new template with advanced atrophy may further reduce registration failures. However, the shape variance modelling of the subcortical structures in FIRST is specific to the original template. Slight modifications of the original template (at least when they are distant from the central structures of interest) should not affect the modelling of the residual shape variance whereas a replacement with an “atrophic” template would probably do so. Potential segmentation failures due to advanced atrophy may be a limitation of the FIRST routine, i.e. in longitudinal studies with a long follow-up duration. However, as we could demonstrate here, this problem occurs in both 2D and 3D data.
Based on our observations, motion artefacts can also impair the registration process. In 2D data acquisition, slices are often scanned in two or more interleaved concatenations. In the case of two concatenations, motion becomes apparent as a displacement between the even and the odd slices, which can be observed as step-wise artefacts in slice direction (Fig. 3). Spatial interpolation can reduce such artefacts, but they still remain apparent. These artefacts may not only impair registration, but also affect tissue type segmentation and/or segmentation of specific brain structures. One possibility to overcome this problem could be the registration of the concatenations on each other before any further processing. However, a careful fixation of the patient's head to avoid motion artefacts is certainly preferable to any post-processing.
In our study, the registration errors observed in two 2D data sets (3%) could neither be explained by atrophy nor by motion artefacts. The low number of such failures without clear reason makes it impossible to calculate any reasonable statistics concerning the influence of different MR scanner types onto data registration.
4.2. FIRST segmentation
In the data sets where MNI registration was working properly, we did not find major segmentation errors either in the 2D or in the 3D data sets. Moreover, the subcortical segmentation using deformable shape models in FIRST seems to be robust even in the presence of hypointense MS lesions at the DGM/WM borders or within the structures itself (Fig. 4). Recently, it has been shown that the misclassification of voxels in intensity-based segmentation methods can impact the measurement of DGM volume (Gelineau-Morel et al., 2012; Nakamura and Fisher, 2009). On the one hand, the DGM volume may be underestimated in that WM lesions change the intensity threshold for tissue segmentation. On the other hand, WM lesions adjacent to DGM border may falsely increase the DGM volume due to neighbouring effects. Based on our qualitative observations, we consider the second effect as minor in our data. It cannot be ruled out that the DGM volume is underestimated due to the impact of WM lesions, but both 2D and 3D data should be affected in a similar way.
4.3. Agreement between FIRST 2D and 3D segmentation
We calculated three quality measures comparing the subcortical segmentation of (interpolated) 2D data to the standard segmentation of 3D data: volume difference, volume overlap and intra-class correlation coefficients. Considering all three measures, total DGM volume as well as the total volume of the BG and the thalamic volume can be reliably estimated on 2D data sets with FIRST. In general, the thalamic volume is somewhat overestimated using the 2D data due to partial volume effects caused by the relatively large slice thickness. This overestimation can be reduced by calculating an additional CSF mask. In our study, the mean volume difference was reduced from 10.4% to 7.0%, although in few cases the difference became larger after CSF correction (mainly in those cases where the 2D segmentation yielded lower thalamic volumes than the 3D segmentation). In contrast to the thalamus, the segmentation of other structures did not benefit from the correction using the CSF mask.
The segmentation results for the putamen, pallidum and caudate nucleus were performed with sufficient quality on 2D data using FIRST, with a slight volume overestimation of putamen and pallidum compared to the respective volumes in 3D data and an underestimation of the caudate volume. Such systematic variations will not affect statistical evaluations, if all study data are two-dimensional.
The nucleus accumbens was segmented only with unsatisfactory quality, not only in individual cases but also generally. This certainly relates not only to the small size of the nucleus accumbens (the mean volume of bilateral nucleus accumbens was 780 mm3 in our patient cohort, the mean volume of bilateral thalamus was 14,450 mm3 for comparison), but also to the fact that the nucleus accumbens abuts to other deep brain nuclei and therefore the delineation of its exact borders is difficult. Our results are in line with those of Morey et al. (2010) evaluating scan–rescan reliability of FIRST. In this work, the authors also found poor reliability for small subcortical structures such as the nucleus accumbens, but higher reliability in caudate, pallidum, putamen and thalamus. Similar results were found for interscanner reliability and in the comparison to manual segmentation in the work of Nugent et al. (2013).
4.4. Agreement between FIRST and manual segmentation
We used manual segmentation for validation of the FIRST results, as it has been demonstrated that manual segmentations can account for the variability of the underlying brain structures not only at a subcortical (Nugent et al., 2013), but also at a cortical level (Wengenroth et al., 2014) (Seither-Preisler et al., 2014). Compared to the manual segmentation, the volume of the caudate was systematically underestimated with FIRST whereas the volume of the putamen was overestimated. This is also reflected by the relatively low ICCs for absolute value. However, the ICCs for consistency reveal strong to moderate agreement between FIRST and the manual segmentation for both structures. Compared to Nugent et al. (2013), the ICCs for consistency between the manual segmentation and 3D FIRST segmentation were in the same range, whereas the ICCs for absolute value were slightly higher in our work.
For both caudate and putamen, mean overlap and ICCs were lower in 2D versus manual segmentation than in 3D versus manual segmentation. This was expected, as the manual segmentation was based on the 3D data. However, the ICCs for consistency demonstrate still moderate agreement between 2D and manual segmentation.
In contrast, the mean volume difference between the 3D and 2D FIRST segmentation was smaller than between the manual and either FIRST segmentation in both structures, indicating the intra-model consistency of FIRST independent from the input data.
4.5. Relevance of our results
The validation of FIRST on 2D data sets appears important in the light of large amounts of existing 2D data, which has been collected in the last two decades in most clinical multiple sclerosis trials. In these trials 2D data are preferred as semi-automatic data analysis (e.g. lesion marking and segmentation) of 3D data is more time-consuming.
The present results are significant in at least two aspects:
1. Up to now, only 2D T1-weighted data are acquired in most clinical trials on MS (i.e. glatiramer acetate (Rovaris et al., 2007), Interferone-beta in secondary progressive MS (Molyneux et al., 2000), Daclizumab (Gold et al., 2013), Natalizumab (Miller et al., 2003; Rudick et al., 2006), Dimethyl fumarate (Kappos et al., 2008), Atacicept (Kappos et al., 2014), or Fingolimod (Kappos et al., 2010)) and hence, a successful extension of the FIRST algorithm to this type of data would enable to study subcortical changes in large patient cohorts and to better understand effects of therapeutic interventions on DGM in MS.
Although the majority of studies on Alzheimer disease, Parkinson disease or psychiatric disorders uses protocols with three-dimensional T1 images, there are two-dimensional data on other neurological disorders where FIRST could be applied, i.e. in studies with Neuromyelitis optica (Kiyat-Atamer et al., 2013) or in MS natural history studies (Pagani et al., 2005; Zivadinov et al., 2001).
2. Applying FIRST on (MS) patients at later stages of the disease with pronounced degrees of brain atrophy (severely enlarged lateral ventricles) results in registration failure in both 2D and 3D data sets. This problem may be critical in applying FIRST to conditions with pronounced brain atrophy, i.e. Alzheimer disease, Parkinson's disease, or vascular dementia as for the registration step, brain templates averaged from brain images of young healthy subjects are used. This issue raises a question whether a new “atrophic” template and the re-adjustment of the FIRST algorithm (residual shape variance modelling) to such a template would be needed in studies on primarily neurodegenerative conditions. This issue may at least partially be resolved (and hence percentage of successfully registered images be increased) by applying the method suggested by Goodro et al. (2012) who registered the MNI152 standard space template using FSL's FNIRT (FMRIB's Nonlinear Image Registration Tool), then processed the registered 3D-MRIs with FIRST and finally transformed the registered images back to the original MRI-space (details in Goodro et al. (2012)).
5. Conclusion
In conclusion, we were able to demonstrate that subcortical GM segmentation of interpolated 2D data with FIRST is comparable with the segmentation of 3D data for larger deep grey matter areas. The results of the total volume of the DGM and of the BG agree well with those determined using the 3D data. Two slight modifications of the FIRST data processing pipeline, namely a minor modification of the MNI templates as well as an additional spatial interpolation of the 2D data, improved the number of successfully segmented data sets. The comparison between FIRST and manual segmentation revealed similar agreement between the two methods as between FIRST segmentation of 2D and 3D data. Taken together, our data indicate that FIRST is suitable for the segmentation of 2D data sets acquired in multicentre studies in MS if appropriate quality control assessments are performed.
Acknowledgements
The authors want to thank Inga Fellner for the organization of the MRI scans.
References
- Alves G.S., O'Dwyer L., Jurcoane A., Oertel-Knöchel V., Knöchel C., Prvulovic D., Sudo F., Alves C.E., Valente L., Moreira D., Fußer F., Fuβer F., Karakaya T., Pantel J., Engelhardt E. Different patterns of white matter degeneration using multiple diffusion indices and volumetric data in mild cognitive impairment and Alzheimer patients. PLoS ONE. 2012;7(12):e52859. doi: 10.1371/journal.pone.0052859. 23300797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audoin B., Zaaraoui W., Reuter F., Rico A., Malikova I., Confort-Gouny S., Cozzone P.J., Pelletier J., Ranjeva J.P. Atrophy mainly affects the limbic system and the deep grey matter at the first stage of multiple sclerosis. J. Neurol. Neurosurg. Psychiatr. 2010;81(6):690–695. doi: 10.1136/jnnp.2009.188748. 20392976 [DOI] [PubMed] [Google Scholar]
- Bagnato F., Hametner S., Yao B., van Gelderen P., Merkle H., Cantor F.K., Lassmann H., Duyn J.H. Tracking iron in multiple sclerosis: a combined imaging and histopathological study at 7 Tesla. Brain. 2011;134(12):3602–3615. doi: 10.1093/brain/awr278. 22171355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakshi R., Benedict R.H., Bermel R.A., Caruthers S.D., Puli S.R., Tjoa C.W., Fabiano A.J., Jacobs L. T2 hypointensity in the deep gray matter of patients with multiple sclerosis: a quantitative magnetic resonance imaging study. Arch. Neurol. 2002;59(1):62–68. doi: 10.1001/archneur.59.1.62. 11790232 [DOI] [PubMed] [Google Scholar]
- Barkhof F., Polman C.H., Radue E.W., Kappos L., Freedman M.S., Edan G., Hartung H.P., Miller D.H., Montalbán X., Poppe P., de Vos M., Lasri F., Bauer L., Dahms S., Wagner K., Pohl C., Sandbrink R. Magnetic resonance imaging effects of interferon beta-1b in the BENEFIT study: integrated 2-year results. Arch. Neurol. 2007;64(9):1292–1298. doi: 10.1001/archneur.64.9.1292. 17846268 [DOI] [PubMed] [Google Scholar]
- Batista S., Zivadinov R., Hoogs M., Bergsland N., Heininen-Brown M., Dwyer M.G., Weinstock-Guttman B., Benedict R.H. Basal ganglia, thalamus and neocortical atrophy predicting slowed cognitive processing in multiple sclerosis. J. Neurol. 2012;259(1):139–146. doi: 10.1007/s00415-011-6147-1. 21720932 [DOI] [PubMed] [Google Scholar]
- Benedict R.H., Ramasamy D., Munschauer F., Weinstock-Guttman B., Zivadinov R. Memory impairment in multiple sclerosis: correlation with deep grey matter and mesial temporal atrophy. J. Neurol. Neurosurg. Psychiatr. 2009;80(2):201–206. doi: 10.1136/jnnp.2008.148403. 18829629 [DOI] [PubMed] [Google Scholar]
- Bermel R.A., Bakshi R., Tjoa C., Puli S.R., Jacobs L. Bicaudate ratio as a magnetic resonance imaging marker of brain atrophy in multiple sclerosis. Arch. Neurol. 2002;59(2):275–280. doi: 10.1001/archneur.59.2.275. 11843699 [DOI] [PubMed] [Google Scholar]
- Bermel R.A., Innus M.D., Tjoa C.W., Bakshi R. Selective caudate atrophy in multiple sclerosis: a 3D MRI parcellation study. Neuroreport. 2003;14(3):335–339. doi: 10.1097/00001756-200303030-00008. 12634479 [DOI] [PubMed] [Google Scholar]
- Calabrese M., Rinaldi F., Grossi P., Mattisi I., Bernardi V., Favaretto A., Perini P., Gallo P. Basal ganglia and frontal/parietal cortical atrophy is associated with fatigue in relapsing–remitting multiple sclerosis. Mult. Scler. 2010;16(10):1220–1228. doi: 10.1177/1352458510376405. 20670981 [DOI] [PubMed] [Google Scholar]
- Caon C., Zvartau-Hind M., Ching W., Lisak R.P., Tselis A.C., Khan O.A. Intercaudate nucleus ratio as a linear measure of brain atrophy in multiple sclerosis. Neurol. 2003;60(2):323–325. doi: 10.1212/01.wnl.0000042094.91478.4a. 12552053 [DOI] [PubMed] [Google Scholar]
- Cheng W.C., Cheng P.E., Liou M. Group factor analysis for Alzheimer's disease. Comput. Math. Methods Med. 2013;2013:428385. doi: 10.1155/2013/428385. 23533539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R.W. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput. Biomed. Res. 1996;29(3):162–173. doi: 10.1006/cbmr.1996.0014. 8812068 [DOI] [PubMed] [Google Scholar]
- Dice L.R. Measures of the amount of ecologic association between species. Ecology. 1945;26(3):297–302. [Google Scholar]
- Drayer B.P., Burger P., Hurwitz B., Dawson D., Cain J., Leong J., Herfkens R., Johnson G.A. Magnetic resonance imaging in multiple sclerosis: decreased signal in thalamus and putamen. Ann. Neurol. 1987;22(4):546–550. doi: 10.1002/ana.410220418. 3435073 [DOI] [PubMed] [Google Scholar]
- Felsky D., Voineskos A.N., Lerch J.P., Nazeri A., Shaikh S.A., Rajji T.K., Mulsant B.H., Kennedy J.L. Myelin-associated glycoprotein gene and brain morphometry in schizophrenia. Front. Psychiatry. 2012;3:40. doi: 10.3389/fpsyt.2012.00040. 22563322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonov V., Evans A.C., Botteron K., Almli C.R., McKinstry R.C., Collins D.L. Unbiased average age-appropriate atlases for pediatric studies. Neuroimage. 2011;54(1):313–327. doi: 10.1016/j.neuroimage.2010.07.033. 20656036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelineau-Morel R., Tomassini V., Jenkinson M., Johansen-Berg H., Matthews P.M., Palace J. The effect of hypointense white matter lesions on automated gray matter segmentation in multiple sclerosis. Hum. Brain Mapp. 2012;33(12):2802–2814. doi: 10.1002/hbm.21402. 21976406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold R., Giovannoni G., Selmaj K., Havrdova E., Montalban X., Radue E.W., Stefoski D., Robinson R., Riester K., Rana J., Elkins J., O'Neill G. Daclizumab high-yield process in relapsing–remitting multiple sclerosis (SELECT): a randomised, double-blind, placebo-controlled trial. Lancet. 2013;381(9884):2167–2175. doi: 10.1016/S0140-6736(12)62190-4. 23562009 [DOI] [PubMed] [Google Scholar]
- Goodro M., Sameti M., Patenaude B., Fein G. Age effect on subcortical structures in healthy adults. Psychiatry Res. 2012;203(1):38–45. doi: 10.1016/j.pscychresns.2011.09.014. 22863654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haacke E.M., Makki M., Ge Y., Maheshwari M., Sehgal V., Hu J., Selvan M., Wu Z., Latif Z., Xuan Y., Khan O., Garbern J., Grossman R.I. Characterizing iron deposition in multiple sclerosis lesions using susceptibility weighted imaging. J. Magn. Reson. Imaging. 2009;29(3):537–544. doi: 10.1002/jmri.21676. 19243035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeier J., Weinstock-Guttman B., Bergsland N., Heininen-Brown M., Carl E., Kennedy C., Magnano C., Hojnacki D., Dwyer M.G., Zivadinov R. Iron deposition on SWI-filtered phase in the subcortical deep gray matter of patients with clinically isolated syndrome may precede structure-specific atrophy. AJNR Am. J. Neuroradiol. 2012;33(8):1596–1601. doi: 10.3174/ajnr.A3030. 22460343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley T., Harlow R. An association between human hippocampal volume and topographical memory in healthy young adults. Front. Hum. Neurosci. 2012;6:338. doi: 10.3389/fnhum.2012.00338. 23293595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry R.G., Shieh M., Okuda D.T., Evangelista A., Gorno-Tempini M.L., Pelletier D. Regional grey matter atrophy in clinically isolated syndromes at presentation. J. Neurol. Neurosurg. Psychiatr. 2008;79(11):1236–1244. doi: 10.1136/jnnp.2007.134825. 18469033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero M.T., Barcia C., Navarro J.M. Functional anatomy of thalamus and basal ganglia. Childs Nerv. Syst. 2002;18(8):386–404. doi: 10.1007/s00381-002-0604-1. 12192499 [DOI] [PubMed] [Google Scholar]
- Houtchens M.K., Benedict R.H., Killiany R., Sharma J., Jaisani Z., Singh B., Weinstock-Guttman B., Guttmann C.R., Bakshi R. Thalamic atrophy and cognition in multiple sclerosis. Neurology. 2007;69(12):1213–1223. doi: 10.1212/01.wnl.0000276992.17011.b5. 17875909 [DOI] [PubMed] [Google Scholar]
- Hughes E.J., Bond J., Svrckova P., Makropoulos A., Ball G., Sharp D.J., Edwards A.D., Hajnal J.V. Regional changes in thalamic shape and volume with increasing age. Neuroimage. 2012;63(3):1134–1142. doi: 10.1016/j.neuroimage.2012.07.043. 22846656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglese M., Park S.J., Johnson G., Babb J.S., Miles L., Jaggi H., Herbert J., Grossman R.I. Deep gray matter perfusion in multiple sclerosis: dynamic susceptibility contrast perfusion magnetic resonance imaging at 3 T. Arch. Neurol. 2007;64(2):196–202. doi: 10.1001/archneur.64.2.196. 17296835 [DOI] [PubMed] [Google Scholar]
- Innes E., Straker L. Validity of work-related assessments. Work. 1999;13(2):125–152. 12441557 [PubMed] [Google Scholar]
- Jenkinson M., Bannister P., Brady M., Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17(2):825–841. doi: 10.1016/s1053-8119(02)91132-8. 12377157 [DOI] [PubMed] [Google Scholar]
- Jenkinson M., Beckmann C.F., Behrens T.E., Woolrich M.W., Smith S.M. Fsl. Neuroimage. 2012;62(2):782–790. doi: 10.1016/j.neuroimage.2011.09.015. 21979382 [DOI] [PubMed] [Google Scholar]
- Jenkinson M., Smith S. A global optimisation method for robust affine registration of brain images. Med. Image Anal. 2001;5(2):143–156. doi: 10.1016/s1361-8415(01)00036-6. 11516708 [DOI] [PubMed] [Google Scholar]
- Kappos L., Gold R., Miller D.H., Macmanus D.G., Havrdova E., Limmroth V., Polman C.H., Schmierer K., Yousry T.A., Yang M., Eraksoy M., Meluzinova E., Rektor I., Dawson K.T., Sandrock A.W., O'Neill G.N. Efficacy and safety of oral fumarate in patients with relapsing–remitting multiple sclerosis: a multicentre, randomised, double-blind, placebo-controlled phase IIb study. Lancet. 2008;372(9648):1463–1472. doi: 10.1016/S0140-6736(08)61619-0. 18970976 [DOI] [PubMed] [Google Scholar]
- Kappos L., Hartung H.P., Freedman M.S., Boyko A., Radü E.W., Mikol D.D., Lamarine M., Hyvert Y., Freudensprung U., Plitz T., van Beek J. Atacicept in multiple sclerosis (ATAMS): a randomised, placebo-controlled, double-blind, phase 2 trial. Lancet Neurol. 2014;13(4):353–363. doi: 10.1016/S1474-4422(14)70028-6. 24613349 [DOI] [PubMed] [Google Scholar]
- Kappos L., Radue E.W., O'Connor P., Polman C., Hohlfeld R., Calabresi P., Selmaj K., Agoropoulou C., Leyk M., Zhang-Auberson L., Burtin P. A placebo-controlled trial of oral fingolimod in relapsing multiple sclerosis. N. Engl. J. Med. 2010;362(5):387–401. doi: 10.1056/NEJMoa0909494. 20089952 [DOI] [PubMed] [Google Scholar]
- Khatri B., Barkhof F., Comi G., Hartung H.P., Kappos L., Montalban X., Pelletier J., Stites T., Wu S., Holdbrook F., Zhang-Auberson L., Francis G., Cohen J.A. Comparison of fingolimod with interferon beta-1a in relapsing–remitting multiple sclerosis: a randomised extension of the TRANSFORMS study. Lancet Neurol. 2011;10(6):520–529. doi: 10.1016/S1474-4422(11)70099-0. 21571593 [DOI] [PubMed] [Google Scholar]
- Kıyat-Atamer A., Ekizoğlu E., Tüzün E., Kürtüncü M., Shugaiv E., Akman-Demir G., Eraksoy M. Long-term MRI findings in neuromyelitis optica: seropositive versus seronegative patients. Eur. J. Neurol. 2013;20(5):781–787. doi: 10.1111/ene.12058. 23279782 [DOI] [PubMed] [Google Scholar]
- Lansley J., Mataix-Cols D., Grau M., Radua J., Sastre-Garriga J. Localized grey matter atrophy in multiple sclerosis: a meta-analysis of voxel-based morphometry studies and associations with functional disability. Neurosci. Biobehav. Rev. 2013;37(5):819–830. doi: 10.1016/j.neubiorev.2013.03.006. 23518268 [DOI] [PubMed] [Google Scholar]
- Lin J.J., Riley J.D., Hsu D.A., Stafstrom C.E., Dabbs K., Becker T., Seidenberg M., Hermann B.P. Striatal hypertrophy and its cognitive effects in new-onset benign epilepsy with centrotemporal spikes. Epilepsia. 2012;53(4):677–685. doi: 10.1111/j.1528-1167.2012.03422.x. 22360313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D.H., Khan O.A., Sheremata W.A., Blumhardt L.D., Rice G.P., Libonati M.A., Willmer-Hulme A.J., Dalton C.M., Miszkiel K.A., O'Connor P.W. A controlled trial of natalizumab for relapsing multiple sclerosis. N. Engl. J. Med. 2003;348(1):15–23. doi: 10.1056/NEJMoa020696. 12510038 [DOI] [PubMed] [Google Scholar]
- Molyneux P.D., Kappos L., Polman C., Pozzilli C., Barkhof F., Filippi M., Yousry T., Hahn D., Wagner K., Ghazi M., Beckmann K., Dahlke F., Losseff N., Barker G.J., Thompson A.J., Miller D.H. The effect of interferon beta-1b treatment on MRI measures of cerebral atrophy in secondary progressive multiple sclerosis. European Study Group on interferon beta-1b in secondary progressive multiple sclerosis. Brain J. Neurol. 2000;123(11):2256–2263. doi: 10.1093/brain/123.11.2256. [DOI] [PubMed] [Google Scholar]
- Morey R.A., Petty C.M., Xu Y., Hayes J.P., Wagner H.R., Lewis D.V., LaBar K.S., Styner M., McCarthy G. A comparison of automated segmentation and manual tracing for quantifying hippocampal and amygdala volumes. Neuroimage. 2009;45(3):855–866. doi: 10.1016/j.neuroimage.2008.12.033. 19162198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey R.A., Selgrade E.S., Wagner H.R., Huettel S.A., Wang L., McCarthy G. Scan-rescan reliability of subcortical brain volumes derived from automated segmentation. Hum Brain Mapp. 2010;31(11):1751–1762. doi: 10.1002/hbm.20973. 20162602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlau M., Buck D., Förschler A., Boucard C.C., Arsic M., Schmidt P., Gaser C., Berthele A., Hoshi M., Jochim A., Kronsbein H., Zimmer C., Hemmer B., Ilg R. White-matter lesions drive deep gray-matter atrophy in early multiple sclerosis: support from structural MRI. Mult. Scler. 2013;19(11):1485–1492. doi: 10.1177/1352458513478673. 23462349 [DOI] [PubMed] [Google Scholar]
- Nakamura K., Fisher E. Segmentation of brain magnetic resonance images for measurement of gray matter atrophy in multiple sclerosis patients. Neuroimage. 2009;44(3):769–776. doi: 10.1016/j.neuroimage.2008.09.059. 19007895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent A.C., Luckenbaugh D.A., Wood S.E., Bogers W., Zarate C.A., Jr., Drevets W.C. Automated subcortical segmentation using FIRST: test–retest reliability, interscanner reliability, and comparison to manual segmentation. Hum. Brain Mapp. 2013;34(9):2313–2329. doi: 10.1002/hbm.22068. 22815187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani E., Rocca M.A., Gallo A., Rovaris M., Martinelli V., Comi G., Filippi M. Regional brain atrophy evolves differently in patients with multiple sclerosis according to clinical phenotype. AJNR Am. J. Neuroradiol. 2005;26(2):341–346. 15709132 [PMC free article] [PubMed] [Google Scholar]
- Park B., Lee W., Han K. Modeling the interactions of Alzheimer-related genes from the whole brain microarray data and diffusion tensor images of human brain. B.M.C. Bioinformatics. 2012;13(Suppl. 7):S10. doi: 10.1186/1471-2105-13-S7-S10. 22594996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patenaude B., Smith S.M., Kennedy D.N., Jenkinson M. A Bayesian model of shape and appearance for subcortical brain segmentation. Neuroimage. 2011;56(3):907–922. doi: 10.1016/j.neuroimage.2011.02.046. 21352927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinster A., Quarantelli M., Orefice G., Lanzillo R., Brunetti A., Mollica C., Salvatore E., Morra V.B., Coppola G., Vacca G., Alfano B., Salvatore M. Grey matter loss in relapsing–remitting multiple sclerosis: a voxel-based morphometry study. Neuroimage. 2006;29(3):859–867. doi: 10.1016/j.neuroimage.2005.08.034. 16203159 [DOI] [PubMed] [Google Scholar]
- Radue E.W., O'Connor P., Polman C.H., Hohlfeld R., Calabresi P., Selmaj K., Mueller-Lenke N., Agoropoulou C., Holdbrook F., de Vera A., Zhang-Auberson L., Francis G., Burtin P., Kappos L. Impact of fingolimod therapy on magnetic resonance imaging outcomes in patients with multiple sclerosis. Arch. Neurol. 2012;69(10):1259–1269. doi: 10.1001/archneurol.2012.1051. 22751847 [DOI] [PubMed] [Google Scholar]
- Rashid W., Parkes L.M., Ingle G.T., Chard D.T., Toosy A.T., Altmann D.R., Symms M.R., Tofts P.S., Thompson A.J., Miller D.H. Abnormalities of cerebral perfusion in multiple sclerosis. J. Neurol. Neurosurg. Psychiatr. 2004;75(9):1288–1293. doi: 10.1136/jnnp.2003.026021. 15314117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccitelli G., Rocca M.A., Pagani E., Martinelli V., Radaelli M., Falini A., Comi G., Filippi M. Mapping regional grey and white matter atrophy in relapsing–remitting multiple sclerosis. Mult. Scler. 2012;18(7):1027–1037. doi: 10.1177/1352458512439239. 22422807 [DOI] [PubMed] [Google Scholar]
- Rovaris M., Comi G., Rocca M.A., Valsasina P., Ladkani D., Pieri E., Weiss S., Shifroni G., Wolinsky J.S., Filippi M. Long-term follow-up of patients treated with glatiramer acetate: a multicentre, multinational extension of the European/Canadian double-blind, placebo-controlled, MRI-monitored trial. Mult. Scler. 2007;13(4):502–508. doi: 10.1177/1352458506070704. 17483532 [DOI] [PubMed] [Google Scholar]
- Rudick R.A., Stuart W.H., Calabresi P.A., Confavreux C., Galetta S.L., Radue E.W., Lublin F.D., Weinstock-Guttman B., Wynn D.R., Lynn F., Panzara M.A., Sandrock A.W. Natalizumab plus interferon beta-1a for relapsing multiple sclerosis. N. Engl. J. Med. 2006;354(9):911–923. doi: 10.1056/NEJMoa044396. 16510745 [DOI] [PubMed] [Google Scholar]
- Schoonheim M.M., Popescu V., Rueda Lopes F.C., Wiebenga O.T., Vrenken H., Douw L., Polman C.H., Geurts J.J., Barkhof F. Subcortical atrophy and cognition: sex effects in multiple sclerosis. Neurology. 2012;79(17):1754–1761. doi: 10.1212/WNL.0b013e3182703f46. 23019265 [DOI] [PubMed] [Google Scholar]
- Seither-Preisler A., Parncutt R., Schneider P. Size and synchronization of auditory cortex promotes musical, literacy, and attentional skills in children. J. Neurosci. 2014;34(33):10937–10949. doi: 10.1523/JNEUROSCI.5315-13.2014. 25122894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.M., De Stefano N., Jenkinson M., Matthews P.M. Normalized accurate measurement of longitudinal brain change. J. Comput. Assist. Tomogr. 2001;25(3):466–475. doi: 10.1097/00004728-200105000-00022. 11351200 [DOI] [PubMed] [Google Scholar]
- Smith S.M., Jenkinson M., Woolrich M.W., Beckmann C.F., Behrens T.E., Johansen-Berg H., Bannister P.R., De Luca M., Drobnjak I., Flitney D.E., Niazy R.K., Saunders J., Vickers J., Zhang Y., De Stefano N., Brady J.M., Matthews P.M. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23(Suppl. 1):S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. 15501092 [DOI] [PubMed] [Google Scholar]
- Smith S.M., Zhang Y., Jenkinson M., Chen J., Matthews P.M., Federico A., De Stefano N. Accurate, robust, and automated longitudinal and cross-sectional brain change analysis. Neuroimage. 2002;17(1):479–489. doi: 10.1006/nimg.2002.1040. 12482100 [DOI] [PubMed] [Google Scholar]
- Tao G., Datta S., He R., Nelson F., Wolinsky J.S., Narayana P.A. Deep gray matter atrophy in multiple sclerosis: a tensor based morphometry. J. Neurol. Sci. 2009;282(1-2):39–46. doi: 10.1016/j.jns.2008.12.035. 19168189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner A.D., Furey M.L., Drevets W.C., Zarate C., Jr., Nugent A.C. Association between subcortical volumes and verbal memory in unmedicated depressed patients and healthy controls. Neuropsychologia. 2012;50(9):2348–2355. doi: 10.1016/j.neuropsychologia.2012.06.003. 22714007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga A.W., Johnson G., Babb J.S., Herbert J., Grossman R.I., Inglese M. White matter hemodynamic abnormalities precede sub-cortical gray matter changes in multiple sclerosis. J. Neurol. Sci. 2009;282(1-2):28–33. doi: 10.1016/j.jns.2008.12.036. 19181347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercellino M., Masera S., Lorenzatti M., Condello C., Merola A., Mattioda A., Tribolo A., Capello E., Mancardi G.L., Mutani R., Giordana M.T., Cavalla P. Demyelination, inflammation, and neurodegeneration in multiple sclerosis deep gray matter. J. Neuropathol. Exp. Neurol. 2009;68(5):489–502. doi: 10.1097/NEN.0b013e3181a19a5a. 19525897 [DOI] [PubMed] [Google Scholar]
- Verstynen T.D., Lynch B., Miller D.L., Voss M.W., Prakash R.S., Chaddock L., Basak C., Szabo A., Olson E.A., Wojcicki T.R., Fanning J., Gothe N.P., McAuley E., Kramer A.F., Erickson K.I. Caudate nucleus volume mediates the Link between cardiorespiratory fitness and cognitive flexibility in older adults. J Aging Res. 2012;2012:939285. doi: 10.1155/2012/939285. 22900181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengenroth M., Blatow M., Heinecke A., Reinhardt J., Stippich C., Hofmann E., Schneider P. Increased volume and function of right auditory cortex as a marker for absolute pitch. Cereb. Cortex. 2014;24(5):1127–1137. doi: 10.1093/cercor/bhs391. 23302811 [DOI] [PubMed] [Google Scholar]
- Woolrich M.W., Jbabdi S., Patenaude B., Chappell M., Makni S., Behrens T., Beckmann C., Jenkinson M., Smith S.M. Bayesian analysis of neuroimaging data in FSL. Neuroimage. 2009;45(1 Suppl):S173–S186. doi: 10.1016/j.neuroimage.2008.10.055. 19059349 [DOI] [PubMed] [Google Scholar]
- Zhuang L., Sachdev P.S., Trollor J.N., Reppermund S., Kochan N.A., Brodaty H., Wen W. Microstructural white matter changes, not hippocampal atrophy, detect early amnestic mild cognitive impairment. PLoS ONE. 2013;8(3):e58887. doi: 10.1371/journal.pone.0058887. 23516569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivadinov R., Havrdová E., Bergsland N., Tyblova M., Hagemeier J., Seidl Z., Dwyer M.G., Vaneckova M., Krasensky J., Carl E., Kalincik T., Horáková D. Thalamic atrophy is associated with development of clinically definite multiple sclerosis. Radiology. 2013;268(3):831–841. doi: 10.1148/radiol.13122424. 23613615 [DOI] [PubMed] [Google Scholar]
- Zivadinov R., Sepcic J., Nasuelli D., De Masi R., Bragadin L.M., Tommasi M.A., Zambito-Marsala S., Moretti R., Bratina A., Ukmar M., Pozzi-Mucelli R.S., Grop A., Cazzato G., Zorzon M. A longitudinal study of brain atrophy and cognitive disturbances in the early phase of relapsing–remitting multiple sclerosis. J. Neurol. Neurosurg. Psychiatr. 2001;70(6):773–780. doi: 10.1136/jnnp.70.6.773. 11385012 [DOI] [PMC free article] [PubMed] [Google Scholar]