Abstract
Maternal obesity and excess gestational weight gain with compromised metabolic fitness predispose offspring to lifelong obesity and its comorbidities. We demonstrated that compared to offspring born before maternal gastrointestinal bypass surgery (BMS) those born after (AMS) were less obese, with less cardiometabolic risk reflected in the expression and methylation of diabetes, immune and inflammatory pathway genes. Here we examine relationships between gestational obesity and offspring gene variations on expression levels.
Methods
Whole-genome genotyping and gene expression analyses in blood of 22 BMS and 23 AMS offspring from 19 mothers were conducted using Illumina HumanOmni-5-Quad and HumanHT-12 v4 Expression BeadChips, respectively. Using PLINK we analyzed interactions between offspring gene variations and maternal surgical status on offspring gene expression levels. Altered biological functions and pathways were identified and visualized using DAVID and Ingenuity Pathway Analysis.
Results
Significant interactions (p ≤ 1.22x10-12) were found for 525 among the 16,060 expressed transcripts: 1.9% of tested SNPs were involved. Gene function and pathway analysis demonstrated enrichment of transcription and of cellular metabolism functions and overrepresentation of cellular stress and signaling, immune response, inflammation, growth, proliferation and development pathways.
Conclusion
We suggest that impaired maternal gestational metabolic fitness interacts with offspring gene variations modulating gene expression levels, providing potential mechanisms explaining improved cardiometabolic risk profiles of AMS offspring related to ameliorated maternal lipid and carbohydrate metabolism.
Introduction
Epidemiological studies demonstrate that parental obesity increases obesity risk in offspring and suggest an important role of the intrauterine environment owing to stronger associations between maternal than paternal body mass index (BMI) with offspring obesity [1, 2]. Maternal obesity, excess gestational weight gain, high inter-pregnancy BMI and gestational diabetes increase risks of offspring obesity, type 2 diabetes mellitus (T2DM), cardiovascular disease (CVD) and fatty liver [3–5]. Environmental and genetic factors mediate the link between parental obesity and increased risk of obesity in offspring [2, 6]; family and twin studies demonstrate heritability of obesity and CVD risk factors [7, 8].
Large-scale genome-wide association studies (GWAS) have consistently revealed the presence of specific genes in metabolic diseases such as type 1 and T2DM [9, 10] and obesity [7, 11]. GWAS on expression traits identified variations regulating gene expression (expression quantitative trait loci; eQTL) and demonstrated that gene expression levels show complex inheritance patterns [12, 13]. Such studies elucidate basic processes of gene regulation and may identify the pathogenesis of prevalent diseases adding information to associations identified by GWAS.
Gene expression levels are greatly affected by genetic and environmental factors [14] where gene variations have the potential to attenuate or amplify environmental effects. An adverse intrauterine environment has long been known to contribute to metabolic and cardiovascular diseases [15] where differences in expression levels between offspring born under different maternal conditions were reported for specific genes and at genome-wide level [16–19]. Several loci associated with specific traits interact with intrauterine environment [20–22]. A striking example of such gene-environment interaction is the association of SIRT1 SNPs with lower prevalence of type 2 diabetes observed in individuals prenatally exposed to famine in utero but not in those not exposed to famine.
Bariatric bypass operations improve glucose and lipid metabolism and treat and/or prevent hypertension, dyslipidemia, T2DM and fatty liver disease [23–25]. Similar to weight loss [26, 27], bariatric surgery results in changes in gene expression levels [28, 29]. Our studies uniquely demonstrated that offspring born after maternal gastrointestinal bypass surgery (AMS) exhibit lower prevalence of severe obesity, greater insulin sensitivity and improved lipid profiles compared to offspring born before maternal surgery (BMS) [30, 31]. Recently, we demonstrated that these improvements are associated with differences in gene expression and methylation of genes involved in diabetes and immune and inflammatory pathways [17, 32].
In order to further explore the role of the intrauterine environment in the determination of offspring phenotype and to provide molecular mechanisms explaining changes in cardiometabolic risk markers of AMS vs. BMS offspring, we studied the combined influence of maternal surgical status and offspring gene variations on offspring gene expression levels.
Materials and Methods
Subjects
Women from Quebec City and surrounding areas (administrative regions of Capitale-Nationale, Mauricie and Chaudière-Appalaches) who had given birth before and after biliopancreatic diversion with duodenal switch [25] for severe obesity were eligible. We recruited a subset of 19 unrelated mothers aged 34–51 years having offspring aged 2–23 years, 22 born before and 23 after maternal operations. Between July and October 2010 mothers and offspring visited the Quebec Heart and Lung Institute (Quebec City, Quebec, Canada) or a regional hospital for clinical evaluation and blood sampling. There were 15 mothers with siblings born before and after surgery (21 BMS and 18 AMS), one with BMS offspring only (1 BMS) and 3 mothers with only AMS offspring (5 AMS).
Maternal pre-surgical data were obtained from medical records. At the office visit weight and percent body fat were determined for individuals aged 6 years or more (BMS, N = 21; AMS, N = 15) using bioelectric impedance analysis (Tanita; Arlington Heights, IL). Height and resting systolic (SBP) and diastolic (DBP) blood pressure were obtained using standardized procedures. BMI was calculated for mothers and adults and BMI percentiles for children were obtained from the National Health and Nutrition Examination Survey 2000 chart [33]. BMI Z-score was calculated for children using charts from the Centers for Disease Control and Prevention [34]. Fasting whole blood samples were collected from an antecubital vein into tubes containing EDTA and PAXgene Blood RNA collection tubes (Qiagen, Valencia, CA, USA). Plasma lipid, glucose and insulin concentrations were measured as previously described [35]. Lipid and glucose levels values from 3 AMS non-fasting offspring were excluded. The homeostatic model assessment of insulin resistance (HOMA-IR) index was calculated as fasting glucose x insulin/22.5. Levels of high-sensitivity C-reactive protein (CRP) were measured with a BN ProSpec nephelometer (Siemens Canada Limited, Oakville, Ontario, Canada) [36]. CRP values under the detection limit (< 0.17 mg/L) were arbitrarily set at detection limit.
Ethics Statement
This study was approved by the Quebec Heart and Lung Institute Ethics Committee. Written informed consent was obtained from mothers and adult offspring and assent from minor offspring were obtained from mothers.
Gene expression analysis
Gene expression levels of the 45 offspring analyzed here were obtained from previous studies from our group evaluating differences in gene expression and methylation of genes involved in diabetes, immune and inflammatory pathways [17, 32]. Briefly, total RNA was isolated and purified from offspring whole blood using PAXgene Blood RNA Kit (Qiagen). The quality and integrity of the purified RNA was assessed using both the NanoDrop (Thermo Scientific, Wilmington, DE, USA) and the 2100 Bioanalyzer (Agilent Technologies, Cedar Creek, TX, USA). Expression levels were measured using the HumanHT-12 v4 Expression BeadChip (Illumina Inc., San Diego, CA) with 250 ng of total RNA and processed at the McGill University and Genome Quebec Innovation Centre (Montreal, Canada). Expression data were visualized and analyzed using the FlexArray software [37] (version 1.6) and the lumi R package was used for expression data analysis and normalization. To be considered as expressed, a probe had to show a detection p-value ≤ 0.05 in at least 25% of samples of a group. Among the 47,323 probes on the microarray, 16,060 (33.9%) showed significant gene expression in blood and were used as dependent expression phenotypes (expression traits) for analysis of interactions between offspring gene variations and maternal obesity status (GEO accession number GSE44407).
DNA extraction and genome-wide genotyping
Genomic DNA was isolated from offspring blood buffy coat using the GenElute Blood Genomic DNA kit (Sigma, St Louis, MO, USA). Quantification and verification of DNA quality were conducted via both NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE, USA) and PicoGreen DNA methods. Genotyping was performed at McGill University and Genome Quebec Innovation Centre (Montreal, Canada) using Illumina HumanOmni-5-Quad BeadChip (Illumina Inc., San Diego, CA), according to the manufacturer’s instructions. Each HumanOmni-5-Quad BeadChip contained 4,301,331 markers.
SNPs and sample quality control
Calculations of allele frequencies and tests of SNP data for Hardy-Weinberg equilibrium (HWE) were performed using PLINK [38] (version 1.07). Standard quality control exclusion criteria for the SNPs were used: call rate < 95%, genotype distribution deviating from Hardy-Weinberg Equilibrium (p-values less than 10–7) and monomorphic SNPs or those with a minor allele frequency (MAF) < 0.01 [39]. A total of 1,751,034 SNPs were excluded leaving 2,550,297 SNPs for statistical analyses. All samples were tested for call rate (> 90%), ethnicity (Caucasian; HapMap) and gender mismatch based on genotyping data. No subjects were excluded: all 45 samples were used in further analysis.
Statistical analysis
Anthropometric and clinical data were expressed as mean ± SD. Maternal treatment effect on anthropometric-, blood pressure-, lipid profile- and glucose-related variables was assessed using a within-subject, paired t-test. Differences between BMS and AMS offspring were tested using analysis of variance (general linear model, type III sum of squares) and adjusted for the effects of sex and puberty. BMI percentile and BMI Z-score being obtained from age- and sex-specific charts, no further adjustments for age and sex were made to test for differences between BMS and AMS offspring for those adiposity measurements. Severe obesity in offspring was defined as BMI percentile > 98% and Z-score > 3. Transformations were applied to non-normally distributed variables (log10 transformed for insulin and HOMA-IR; negative inverse transformed for C-reactive protein). In the absence of Tanner scores, we arbitrarily defined puberty as 12 years for female and 14 years for male offspring based on Canadian sex-specific probabilities of having entered puberty [40]. Differences in severe obesity between BMS and AMS offspring were evaluated using BMI percentile and BMI Z-score and tested using Fisher’s exact test. P-values for CRP were adjusted for the effects of sex, puberty and BMI percentile. Statistical analyses were done using the SAS software version 9.2 (SAS Institute Inc). Statistical significance was defined as p ≤ 0.05. Interactions between offspring gene variations and maternal surgical status were tested on offspring gene expression levels in whole blood using PLINK. Differences in regression slopes obtained from additive model were then tested between BMS and AMS offspring. Bonferroni correction was applied to correct for multiple testing of offspring gene variation x environment interactions thus leading to a p-value cutoff of p ≤ 1.22x10–12 (as calculated with 0.05/ (2,550,297 SNPs x 16,060 transcripts)) to claim statistical significance. Linkage disequilibrium (LD; r2) between SNPs demonstrating significant interactions was calculated using Haploview [41] to assess the number of independent (non-linked) SNPs. The tagger algorithm implemented in Haploview was used to identify tag SNPs among the significant polymorphisms (r2 threshold = 0.8).
Gene functions and pathways analysis
Two independent function and pathway analysis tools were employed to identify potentially enriched functions and overrepresented pathways from the list of transcripts demonstrating significant interactions, namely the Database for Annotation, Visualization, and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov) bioinformatics resources [42, 43] and Ingenuity Pathway Analysis (IPA). DAVID provided annotation for the list of transcripts and computed annotation term enrichment to highlight the most relevant functions from the list of transcripts. Similarly, IPA classified each transcript from this list according to function and pathway. Using a right-tailed Fisher’s exact test, IPA measured the likelihood that transcripts from the list participate in each function/pathway solely due to chance and calculated p-values. Enriched functions and overrepresented pathways were then obtained from transcripts showing interactions.
Results
Characteristics of 19 mothers and their 45 offspring
Mean postoperative follow-up for the mothers after bilio-pancreatic diversion surgery was 12 years 7 months (range: 4 years 11 months to 22 years 4 months). Preoperative weight was 121.6 ± 18.7 kg (BMI = 45.1 ± 7.4) and 74.9 ± 12.2 kg (BMI = 27.6 ± 4.9) at follow-up, a mean loss of 46.7 kg, associated with significant, clinically important improvements in fasting plasma lipids (p ≤ 0.005 for TG, HDL-C, LDL-C, total-C and total-C/HDL-C ratio), glucose levels (5.81 ± 2.41 vs. 4.68 ± 0.32; p = 0.048) and blood pressure (SBP and DBP; p ≤ 0.001) were observed (S1 Table).
Offspring ages varied between 2 years 8 months and 23 years 9 months, with similar sex distributions in the two groups (41% male in BMS vs. 43% in AMS; Table 1). BMS offspring were born 3 years 4 months (40.2 ± 28.0 months) before and AMS 3 years 9 months (44.9 ± 26.6 months) after maternal surgery. BMS offspring were older than AMS at follow-up (14.5 ± 5.7 vs. 9.0 ± 5.0 years; p = 0.001; BMS range: 5 years 9 months to 23 years 9 months; AMS range: 2 years 8 months to 19 years 6 months). Severe obesity was less prevalent in AMS using BMI percentile (p = 0.01) or BMI Z-score (p = 0.02). Adjusting for sex and puberty, AMS offspring exhibited trends toward lower fasting insulin levels and HOMA-IR index, and lower diastolic blood pressure (p < 0.10 for all).
Table 1. Offspring characteristics.
| BMS | AMS | p-values 1 | |
|---|---|---|---|
| N (males) | 22 (9) | 23 (10) | |
| Age (years) | 14.5 ± 5.7 | 9.0 ± 5.0 | 0.001 |
| Anthropometric data | |||
| Fat percent 2 | 29.6 ± 14.4 | 22.7 ± 10.3 | 0.28 |
| BMI percentile | 68.7 ± 41.5 | 69.7 ± 30.9 | 0.93 |
| BMI Z-score 3 | 1.93 ± 2.18 | 0.90 ± 1.48 | 0.08 |
| Severe obesity | |||
| BMI percentile > 98% (N) | 11 | 3 | 0.01 |
| BMI Z-score > 3 (N) | 7 | 1 | 0.02 |
| Blood pressure | |||
| SBP (mm Hg) | 110.5 ± 14.9 | 96.7 ± 14.8 | 0.13 |
| DBP (mm Hg) | 64.3 ± 10.5 | 52.8 ± 13.2 | 0.06 |
| Lipid profile 4 | |||
| TG (mmol/l) | 1.03 ± 0.44 | 0.81 ± 0.38 | 0.29 |
| LDL-C (mmol/l) | 2.66 ± 0.56 | 2.53 ± 0.59 | 0.67 |
| HDL-C (mmol/l) | 1.30 ± 0.31 | 1.30 ± 0.26 | 0.76 |
| Total-C (mmol/l) | 4.44 ± 0.67 | 4.20 ± 0.59 | 0.39 |
| Total-C / HDL-C | 3.58 ± 0.97 | 3.37 ± 0.87 | 0.76 |
| Glucose metabolism 4 | |||
| Fasting glucose (mmol/l) | 4.94 ± 0.44 | 4.77 ± 0.37 | 0.54 |
| Insulin (μU/ml) | 19.98 ± 12.54 | 11.45 ± 7.50 | 0.06 |
| Homa-IR | 4.55 ± 3.34 | 2.49 ± 1.74 | 0.08 |
| CRP (mg/L) 5 | 5.54 ± 8.34 | 1.54 ± 3.69 | 0.12 |
Values are presented as mean ± SD.
1 P-values adjusted for sex and puberty except for BMI percentile and BMI Z-score and obtained from comparison of all BMS (N = 22) and AMS (N = 23) offspring.
2 Fat percent at 6 years or more (BMS, N = 21; AMS, N = 15).
3 BMS, N = 19; AMS, N = 22.
4 BMS, N = 22; AMS, N = 20.
5 P-values for CRP were adjusted for the effects of sex, puberty and BMI percentile. Abbreviations: BMS, before maternal surgery; AMS, after maternal surgery; BMI, body mass index; SBP and DBP, systolic diastolic and systolic blood pressure; TG, triglycerides; LDL-C; low-density lipoprotein cholesterol; HDL-C, high-density lipoprotein cholesterol; Total-C, total cholesterol; CRP, C-reactive protein; SD, standard deviation.
Interactions effects between offspring gene variations and maternal surgical status
Testing interactions between 2,550,297 offspring SNPs and maternal status on 16,060 expression phenotypes in offspring demonstrated 102,129 interactions reaching statistical significance (representing 0.00025% of all tests; p ≤ 1.22x10–12). The top significant interactions are shown in Table 2. These represent 48,156 unique SNPs (1.9% of the SNPs tested), 33.6% (16,184) of which demonstrated statistically significant interactions for multiple transcripts. Identified SNPs were mainly in intergenic regions (61%) while intronic, exonic and untranslated region SNPs represented a minority (35%, 2% and 2% respectively). SNP rs35447805 (kgp1360358) located at chr8:85131756 within the RALYL gene (NM_001100391) demonstrated the most statistically significant interaction with treatment for expression level of SEPT4 (NM_080415) and IFI35 (NM_005533), both located on chromosome 17 (Table 2). As an example, interaction effect for rs35447805 on SEPT4 expression levels manifested in a 2.5 fold increase in expression for AMS heterozygous carriers and slight decrease (0.9 fold) for BMS heterozygous carriers (S2 Table). SNPs located in the interferon (alpha, beta and omega) receptor 1 (IFNAR1)—interleukin 10 receptor, beta (IL10RB) gene region and those near transmembrane protein, adipocyte associated 1 (GPR175, also known as TPRA1), BARX homeobox 2 (BARX2) transcription factor and heparan sulfate (glucosamine) 3-O-sulfotransferase 4 (HS3ST4) demonstrated several significant interactions (S3 Table).
Table 2. Most significant SNP-by-maternal treatment interactions.
| SNP | Transcript | ||||||
|---|---|---|---|---|---|---|---|
| SNP ID 1 | rs number | Chr | Position 2 | Accession | Gene 3 | Chr | P-value 4 |
| kgp11796917 | rs146836802 | 14 | 30685942 | NM_194323 | OTOF | 2 | 1.09x10–57 |
| kgp30566776 | rs73229805 | X | 67014865 | NM_001010927 | TIAM2 | 6 | 3.53x10–45 |
| kgp8430951 | rs78240927 | 6 | 163766098 | NM_001100422 | SPATS2L | 2 | 2.37x10–37 |
| kgp11155608 | rs12059564 | 1 | 159000779 | NM_015913 | TXNDC12 | 1 | 7.81x10–32 |
| kgp1360358 | rs35447805 | 8 | 85131756 | NM_080415 | SEPT4 | 17 | 4.70x10–30 |
| kgp6389054 | rs78957751 | 16 | 34886264 | NM_001006630 | CHRM2 | 7 | 4.14x10–27 |
| rs4764191 | rs4764191 | 12 | 15560196 | NM_001032295 | SERPING1 | 11 | 4.43x10–27 |
| kgp11709552 | rs62227091 | 22 | 47811418 | NM_001011724 | HNRNPA1L2 | 13 | 3.15x10–26 |
| kgp30620929 | rs140547157 | X | 27936207 | XM_945614 | PMS2L1 | 7 | 3.32x10–26 |
| kgp11042611 | rs79592528 | 3 | 122772698 | XM_938400 | LOC142937 | 10 | 3.97x10–26 |
| kgp8787288 | rs940676 | 2 | 121235234 | NM_003733 | OASL | 12 | 8.00x10–25 |
| kgp4545030 | rs78211484 | 12 | 59934484 | CK299576 | HS.528210 | 3 | 5.46x10–24 |
| kgp18518 | rs17757256 | 2 | 19650629 | NM_001007524 | F8A3 | X | 1.19x10–23 |
| kgp2333358 | rs1461092 | 12 | 41556961 | NM_005419 | STAT2 | 12 | 4.89x10–23 |
| kgp8581526 | rs36117773 | 5 | 11939359 | NM_025126 | RNF34 | 12 | 1.15x10–22 |
| kgp494941 | rs79270474 | 12 | 12338554 | NM_002535 | OAS2 | 12 | 1.59x10–22 |
| kgp6448962 | rs57573303 | 9 | 28322259 | NM_001012978 | BEX5 | X | 5.84x10–22 |
| rs3803712 | rs3803712 | 16 | 26074500 | NM_001712 | CEACAM1 | 19 | 7.21x10–22 |
| kgp36317 | rs2247335 | 6 | 106992592 | NM_001007234 | ERCC8 | 5 | 8.83x10–22 |
| kgp4630305 | rs59557850 | 8 | 78148122 | NM_003728 | UNC5C | 4 | 1.10x10–21 |
| rs9655226 | rs9655226 | 7 | 22598742 | NM_170695 | TGIF1 | 18 | 3.07x10–21 |
| kgp10592712 | rs73063011 | 19 | 52338183 | NM_024032 | C17ORF53 | 17 | 8.71x10–21 |
| kgp237322 | rs539822 | 5 | 176310164 | XM_001721497 | LOC100132457 | 2 | 1.02x10–20 |
| rs11053624 | rs11053624 | 12 | 10283711 | NM_015589 | SAMD4A | 14 | 1.51x10–20 |
| kgp11540460 | rs144298037 | 6 | 45434746 | NM_001004349 | FLJ45422 | 6 | 1.56x10–20 |
| kgp11526978 | rs4711691 | 6 | 12404895 | NM_014453 | CHMP2A | 19 | 3.55x10–20 |
| kgp11514400 | rs35833993 | 6 | 165457847 | NM_006918 | SC5D | 11 | 5.65x10–20 |
| kgp3001132 | rs9606166 | 22 | 19811720 | NM_006704 | SUGT1 | 13 | 6.90x10–20 |
| kgp9305036 | rs943009 | 6 | 11243891 | NR_002940 | LRRC37A4 | 17 | 1.59x10–19 |
| kgp8789955 | rs62576233 | 9 | 84337468 | NM_005792 | MPHOSPH6 | 16 | 1.63x10–19 |
| kgp1173427 | rs61733660 | 6 | 151148947 | XM_925998 | SRA1 | 5 | 1.82x10–19 |
| rs2968402 5 | rs2968402 4 | 16 | 21947480 | NM_014598 | SOCS7 | 17 | 3.16x10–19 |
| kgp12304307 | rs13298711 | 9 | 83820909 | NM_002256 | KISS1 | 1 | 3.45x10–19 |
| rs11636802 | rs11636802 | 15 | 56775597 | DA276856 | HS.576243 | 1 | 3.53x10–19 |
| kgp12481432 | rs4407201 | 2 | 130522894 | NM_016134 | CPQ | 8 | 4.88x10–19 |
| kgp31122632 | rs138131809 | X | 116143012 | NR_024524 | LOC100129055 | 10 | 6.25x10–19 |
| rs7149078 | rs7149078 | 14 | 32844576 | CD369504 | HS.540642 | 16 | 6.57x10–19 |
| kgp4554682 | rs1108962 | 4 | 100663492 | AI274046 | HS.555512 | 14 | 8.86x10–19 |
| kgp8947215 | rs73030956 | 12 | 1675847 | NM_018271 | FLJ10916 | 2 | 9.40x10–19 |
| kgp7059559 | rs1831464 | 13 | 92902619 | NM_019062 | RNF186 | 1 | 9.67x10–19 |
| kgp4609959 | rs13284671 | 9 | 824742 | NM_002720 | PPP4C | 16 | 1.02x10–18 |
| kgp2913569 | rs36067040 | 1 | 224238495 | NM_001017977 | DCAF6 | 1 | 1.32x10–18 |
| kgp1360358 | rs35447805 | 8 | 85131756 | NM_005533 | IFI35 | 17 | 1.67x10–18 |
| kgp6643156 | rs79321471 | 14 | 101498881 | NM_017831 | RNF125 | 18 | 1.81x10–18 |
| kgp619464 | rs4292995 | 1 | 30738298 | NM_022148 | CRLF2 | Y | 2.36x10–18 |
| rs6074541 | rs6074541 | 20 | 12978517 | NM_006286 | TFDP2 | 3 | 2.87x10–18 |
| kgp12299095 | rs35007051 | 2 | 188415764 | NM_002164 | IDO1 | 8 | 3.76x10–18 |
| kgp12307971 | rs76107005 | 10 | 85534815 | NM_002201 | ISG20 | 15 | 4.36x10–18 |
| kgp10871570 | rs115462216 | 21 | 22882926 | U43604 | HS.550193 | 5 | 4.78x10–18 |
| rs3025651 | rs3025651 | 6 | 29539914 | NM_003646 | DGKZ | 11 | 6.03x10–18 |
List of the top 50 significant interactions. Regulated transcripts and respective p-values are shown.
SNP with the most significant association obtained from comparison of all BMS (N = 22) and AMS (N = 23) offspring was shown for each transcript.
1 SNP ID as defined by Illumina HumanOmni-5-Quad BeadChip annotation.
2 Genome build 37.
3 RefSeq or UniGene nomenclature.
4 P-values for differences between regression slopes (BMS vs. AMS) obtained from an additive model.
5 SNP mapped at two locations (chr16:21947480 and chr16:29119905). Abbreviations: SNP, single nucleotide polymorphism; Chr, chromosome.
Statistically significant offspring SNP-by-maternal surgical status interactions identified for the 48,156 unique SNPs involved 525 unique transcripts (3.3%) thus implying that a single transcript might be under multiple genetic constraints. Indeed, 375 of these 525 unique transcripts (71.4%) demonstrated multiple (≥2) significant interactions. The most highly represented transcripts from significant interactions are shown in Table 3, including transcripts encoding genes involved in regulation of gene expression per se, immune and inflammatory responses and lipid biosynthesis from which examples for STAT2 (NM_005419), IFI35 (NM_005533) and DGKZ (NM_003646) are shown in S1 Fig. LD was found between SNPs, resulting in multiple significant interactions with an identical transcript: 14,676 interaction tagging SNPs (tSNPs) were identified among the 48,156 unique SNPs showing significant interactions. Limiting analysis to these tSNPs led to the identification of 56.2% of the transcripts showing multiple statistically significant interactions.
Table 3. Most represented transcripts from the list of significant interactions.
| Transcript 1 | Accession | Significant interactions (N) | Most significant p-value 2 |
|---|---|---|---|
| SEPT4 | NM_080415 | 8492 | 4.70x10–30 |
| OTOF | NM_194323 | 8473 | 1.09x10–57 |
| SPATS2L | NM_001100422 | 8465 | 2.37x10–37 |
| TXNDC12 | NM_015913 | 8172 | 7.81x10–32 |
| LOC142937 | XM_938400 | 6690 | 3.97x10–26 |
| HNRNPA1L2 | NM_001011724 | 6151 | 3.15x10–26 |
| HS.528210 | CK299576 | 5804 | 5.46x10–24 |
| TIAM2 | NM_001010927 | 5204 | 3.53x10–45 |
| OAS2 | NM_002535 | 4396 | 1.59x10–22 |
| SERPING1 | NM_001032295 | 4168 | 4.43x10–27 |
| OASL | NM_003733 | 2964 | 8.00x10–25 |
| HS.550193 | U43604 | 2886 | 4.78x10–18 |
| FLJ45422 | NM_001004349 | 2659 | 1.56x10–20 |
| TCP1 | NM_030752 | 1823 | 3.15x10–17 |
| RNF125 | NM_017831 | 1762 | 1.81x10–18 |
| CEACAM1 | NM_001712 | 1728 | 7.21x10–22 |
| SRA1 | XM_925998 | 1545 | 1.82x10–19 |
| IFI35 | NM_005533 | 1474 | 1.67x10–18 |
| STAT2 | NM_005419 | 1371 | 4.89x10–23 |
| HS.391327 | BX110374 | 1352 | 1.39x10–16 |
| PMS2L1 | XM_945614 | 1080 | 3.32x10–26 |
| SAMD4A | NM_015589 | 949 | 1.51x10–20 |
| DGKZ | NM_003646 | 840 | 6.03x10–18 |
| ISG20 | NM_002201 | 798 | 4.36x10–18 |
| LOC649009 | XM_941706 | 701 | 2.41x10–17 |
| HS.553068 | BX103476 | 533 | 1.32x10–15 |
| F8A3 | NM_001007524 | 502 | 1.19x10–23 |
| BEX5 | NM_001012978 | 475 | 5.84x10–22 |
| LRRC37A4 | NR_002940 | 462 | 1.59x10–19 |
| LOC401525 | XM_376869 | 447 | 8.58x10–18 |
| LOC649143 | XM_944822 | 403 | 6.06x10–18 |
| SUGT1 | NM_006704 | 400 | 6.90x10–20 |
| FANCA | NM_000135 | 366 | 6.50x10–18 |
| PPP4C | NM_002720 | 321 | 1.02x10–18 |
| GOLM1 | NM_177937 | 317 | 5.85x10–17 |
| PIGC | NM_153747 | 295 | 4.51x10–17 |
| TAPBP | NM_172209 | 274 | 1.56x10–16 |
| ATF3 | NM_001040619 | 216 | 3.66x10–17 |
| RNF34 | NM_025126 | 194 | 1.15x10–22 |
| HS.539736 | AI979341 | 192 | 9.90x10–18 |
| LOC100132347 | XM_001713703 | 186 | 2.06x10–15 |
| LOC100132457 | XM_001721497 | 168 | 1.02x10–20 |
| CPQ | NM_016134 | 165 | 4.88x10–19 |
| UBXD7 | XM_936412 | 146 | 4.18x10–15 |
| FAT3 | XM_926199 | 145 | 1.15x10–15 |
| RNF186 | NM_019062 | 139 | 9.67x10–19 |
| ZBP1 | NM_030776 | 134 | 1.16x10–14 |
| OPRL1 | NM_000913 | 131 | 9.08x10–15 |
| LOC645253 | XM_944197 | 127 | 1.79x10–15 |
| HIST1H4H | NM_003543 | 125 | 7.54x10–16 |
1 RefSeq or UniGene nomenclature.
2 P-values for differences between regression slopes (BMS vs. AMS) obtained from an additive model. Abbreviation: N, number.
Gene functions and pathways
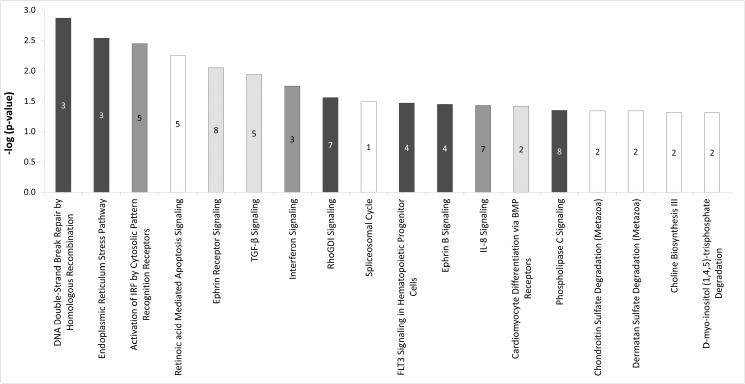
Clustering of the 525 transcripts showing significant interactions based on ontology using DAVID identified 5 over-represented functional categories (-log group enrichment score > 1.30; p-value for group enrichment score < 0.05): 1) transcription, 2) metabolic process, 3) guanine nucleotide exchange factor, 4) death/ZU5 domain and 5) zinc finger domain (S4 Table). IPA analysis revealed infectious disease, inflammatory response, gene expression, and cellular growth and proliferation among the over-represented functional categories from the list of transcripts demonstrating statistically significant interactions, thus highlighting transcription and cellular metabolism functions using both function analysis tools. Similarly, IPA revealed 18 pathways enriched for these transcripts, including 12 related to cellular stress and signaling, immune response and inflammation, and growth, proliferation and development. Importantly, DNA Double-Strand Break Repair by Homologous Recombination was the most overrepresented pathway (p = 0.001) and carbohydrate and lipid biosynthesis/degradation pathways were also identified (Fig. 1).
Figure 1. Pathways enriched from transcripts demonstrating significant interactions.
The number of submitted transcripts in each pathway is reported in histogram bars. Histogram bars for pathways related to cellular stress and signaling (black), immune response and inflammation (dark grey), and growth, proliferation and development (light grey) are highlighted. Unrelated pathways are shown in white.
Discussion
In a unique offspring cohort born discordant for maternal biliopancreatic bypass surgery affecting maternal metabolic fitness we extend observations that an adverse dysmetabolic intrauterine environment is associated with subsequent obesity and cardiometabolic risk [30, 31] related to gene expression levels [16, 18, 19]. Using a cohort in which gene expression and methylation levels were previously evaluated in regards to metabolic differences between BMS and AMS offspring [17, 32], the current study focusing on a different aspect (gene-environment interactions) demonstrated modulatory effects of maternal fitness on the association between genotype and gene expression in offspring. In this study we tested gene expression levels (expression traits) for interactions between offspring SNPs and maternal treatment. By analyzing the offspring genotypes and gene expression at the genome-wide level combined with the impact of maternal status, we provide an objective insight into the relation between maternal status and offspring cardiometabolic risk profile and have the potential to elucidate the functional basis of known associations identified previously.
Overlapping SNPs identified here with those previously associated with specific phenotypes and metabolic variables in GWAS has the potential to elucidate mechanisms for previously reported associations. Systematic comparison of SNPs and regulated transcripts with results from GWAS was then conducted. Our top interactions (Table 2) identified SNP rs57573303 in the gene LINGO2 and SNP rs3803712 located in HS3ST4. The former was associated with BEX5 expression levels, a brain expressed X-linked gene family member regulating differentiation of dopamine neurons involved in food reward signaling [44]. The latter we found was associated with CEACAM1 gene expression levels encoding a cell-cell adhesion molecule involved in differentiation, apoptosis and modulation of innate and adaptive immune response consistent with decreased liver CEACAM1 expression reported in severely obese patients [45]. In regards to the interaction identified here, increased CEACAM1 expression was observed in heterozygous AMS offspring while BMS rare allele carriers demonstrated lower CEACAM1 expression. SNPs in LINGO2 and HS3ST4 were previously found to be associated with children BMI z-score and % fat mass, respectively [11]. Associations between rs10968576, also in LINGO2, and fasting plasma cholesterol levels and BMI were found in different populations [46, 47]. We identify here rs115462216 SNP (kgp10871570) in NCAM2 for which rs11088859 has been associated with waist circumference [48]. In addition, the presence of neurological (ERCC8, KISS1, IDO1, OTOF, SEPT4, SERPING1, SOCS7, STAT2, TGIF1, TIAM2) and endocrine system development (CHRM2, SC5D, SOCS7) genes among the regulated transcripts (Table 2) suggest a mechanistic role in offspring programming under varying maternal conditions [19, 49]. Taken together, the identification of SNPs located in genes previously reported by others to be associated with obesity traits support our results and is consistent with studies showing that SNPs associated with complex traits are more likely to be eQTLs [50, 51].
The limited number of transcripts for which we found offspring gene variation-by-maternal status interactions and the large number of SNPs suggest that single transcripts may be under multiple genetic constraints, coherent with previous studies conducted on larger cohorts [52–54]. Among the numerous SNPs we identified with significant interactions, many were specifically associated with the inflammatory, insulin-resistant, dysmetabolic diathesis of diabesity such as SNPs in the IFNAR1—IL10RB gene region forming the class II cytokine receptor gene cluster (S3 Table). Those genes involved in IL10-induced signal transductions were associated with inflammatory diseases and ischemic stroke with hypertension [55–57]. We also observed significant interactions for SNPs located near the GPR175 (TPRA1) gene, expression level of which was previously demonstrated to be associated with plasma lipid levels [58]. Furthermore, we found interactions of SNPs near BARX2, a member of the homeobox transcription factor family known to influence cellular processes controlling cell adhesion and remodeling actin cytoskeleton. Others previously found such associations with T2DM and end-stage renal disease [59].
Gene function analysis conducted with two independent tools highlighted genes related to transcription and cellular metabolism. Combined with other overrepresented functional categories (guanine nucleotide exchange factor, death/ZU5 domain, zinc finger domain), these results suggested some potential effects of maternal metabolic fitness on offspring at both the cellular and transcriptional levels. Our pathway analysis also identified pathways related to cellular growth, proliferation and development, stress and signaling as well as carbohydrate and lipid metabolism similar to others’ findings. Global gene expression analysis of amniotic fluid cell-free fetal RNA identified lipid (apolipoprotein D) and transcriptional regulators (FOS and STAT3) as well as apoptotic cell death-related genes among differentially expressed genes between fetuses of obese vs. lean pregnant women [16]. In conjunction with influences of maternal obesity before conception reported on gene expression profiles of rat embryos, genes related to cell cycle, carbohydrate metabolism, DNA repair and transcriptional regulator were altered [60]. These results strengthen our observations pertaining to an overrepresentation of cellular processes (cellular growth, proliferation and development), carbohydrate and lipid metabolism pathways identified. The study on rat embryos [60] also supports overrepresentation of immune response and inflammatory genes as well as DNA double-strand break repair-related genes among transcripts with significant interactions. Similar to changes in expression previously observed in obese patients early after having undergone weight loss or bariatric surgery [26, 27, 29], we found that inflammation-related transcripts were overrepresented. Globally, overrepresentation of cellular signaling, carbohydrate metabolism and inflammatory pathways identified here from the list of transcripts showing significant offspring gene variations by maternal surgical status interactions are in line with previous results from our group comparing BMS and AMS offspring at gene methylation and expression levels [17, 32]. In addition, our results are consonant with murine studies showing effects of maternal gestational obesity and high-fat diet on offspring with differences in gene expression levels for genes related to inflammation and glucose homeostasis [61, 62] and for pathways related to cellular stress, signaling, growth, proliferation, development and regulation of lipogenic pathways [63] of significant pathogenic importance for the dysmetabolic diathesis of diabesity. Our pathway analysis demonstrated involvement of carbohydrate and lipid metabolism pathways, in agreement with results from maternal weight loss studies in sheep demonstrating an impact on insulin signaling, glucose transport and glycogen synthesis pathways in offspring’ skeletal muscle [64] as well as with previous studies demonstrating gene-by-maternal diet interactions in offspring [21, 65].
Some of the modulating effects of gestational metabolic fitness may be confounded. Young age of the offspring limits the potential contribution of different postnatal environments. However, it does not allow extrapolation over the life span stretching into mature adulthood when most pathology emerges through the cumulative effects of environmental exposures. Nevertheless, the preponderance of literature on developmental origins of adult disease, specifically for cardio-metabolic outcomes related to our findings demonstrates durability of effects over the life-span [15]. The rarity of gastrointestinal biliopancreatic bypass surgery, low pregnancy rates before and after maternal surgery, constraints of study design and the exclusive nature of the molecular analyses all limited the size of our offspring population. The size of the study sample limited the number of adjustments made to correct for confounding factors relating to offspring and maternal condition during pregnancy (breastfeeding, smoking, etc.). Adjustments for confounding factors were thus limited to sex and puberty. Although metabolic parameters in the offspring cohorts were not statistically significantly different owing to sample size, the differences were clinically significant particularly for insulin resistance, dyslipidemia and CRP. Studies from our group conducted on larger cohorts have previously demonstrated robust group differences between BMS and AMS offspring [30, 31]. Our gene analyses were performed on blood, more convenient to obtain and to justify sampling than other tissues in healthy juvenile offspring. We and others have reported partial inter-tissue correlations [54, 66–68]. The multicellular nature of blood constitutes an inherent limitation in our study; tissue heterogeneity potentially influenced measurement of gene expression levels [69, 70]. Nonetheless, we assessed gene expression as representative of systemic biological differences between BMS and AMS offspring to which multiple organs and tissues have contributed. Causality of identified variants cannot be determined: identified variants might be markers of genomic regions or loci in which causal variants lie and allelic heterogeneity cannot be ruled out, together necessitating much larger population studies than our unique but relatively small cohort study.
Strengths of our study are the unique genetically and phenotypically characterized offspring cohort discordant for maternal gestational metabolic fitness, the efficacious standardized currently performed metabolic operation as a tool to alter the intrauterine milieu and an exceptionally high follow-up rate enabled by the national health insurance system. The biliopancreatic bypass operation selectively increases steatorrhea, lowering maternal plasma free fatty acids, reducing fatty infiltration of metabolically active tissues, reducing lipid peroxidation and systemic lipotoxic inflammation altogether improving insulin action and glucose disposal approximating pre-obese levels. Although these durable effects were not replicated quantitatively or qualitatively by other current bariatric operations, they add critical insight into molecular mechanisms associated with the gene expression levels presented here.
Our results demonstrated influences of the intrauterine metabolic environment on associations between offspring genotype and gene expression levels. The lower prevalence of obesity and cardiometabolic risk observed in AMS offspring argues for implementation of maternal weight loss and improved metabolic fitness before pregnancy and provides potential mechanisms for physiological improvements through regulation of gene expression in offspring.
Supporting Information
Panel A, STAT2 (NM_005419). Panel B, IFI35 (NM_005533). Panel C, DGKZ (NM_003646).
(TIF)
(DOCX)
SNPs, regulated transcripts and genotype-specific gene expression levels are shown. Expression values (means) relative to common homozygotes from the BMS group.
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We thank all the families who participated in the study for their excellent collaboration, and the members of the department of bariatric surgery for their direct or indirect involvement in clinical care and patient recruitment (Laurent Biertho, Simon Biron, Frédéric-Simon Hould, Stéfane Lebel, Odette Lescelleur, Simon Marceau). We express our gratitude to Suzy Laroche for help in sample and clinical information collection and Paule Marceau for subject recruitment, data management and project coordination. We acknowledge the contribution of the McGill University and Genome Quebec Innovation Centre for gene expression and genotyping array analyses. F.G. is a recipient of a studentship award from the Heart and Stroke Foundation of Canada. Y.B. is the recipient of a Junior 2 Research Scholar award from the Fonds de recherche Québec—Santé (FRQS). M.C.V. is a Tier 1 Canada Research Chair in Genomics Applied to Nutrition and Health.
Data Availability
Gene expression data has been deposited in the Gene Expression Omnibus (GEO) database (accession number GSE44407).
Funding Statement
This study was supported by a grant from the Canadian Institutes of Health Research (CIHR MOP-209380; http://www.cihr-irsc.gc.ca). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Lawlor DA, Smith GD, O'Callaghan M, Alati R, Mamun AA et al. (2007) Epidemiologic evidence for the fetal overnutrition hypothesis: findings from the mater-university study of pregnancy and its outcomes. Am J Epidemiol 165: 418–424. 10.1093/aje/kwk030 [DOI] [PubMed] [Google Scholar]
- 2. Whitaker KL, Jarvis MJ, Beeken RJ, Boniface D, Wardle J (2010) Comparing maternal and paternal intergenerational transmission of obesity risk in a large population-based sample. Am J Clin Nutr 91: 1560–1567. 10.3945/ajcn.2009.28838 [DOI] [PubMed] [Google Scholar]
- 3. Dabelea D, Mayer-Davis EJ, Lamichhane AP, D'Agostino RB Jr., Liese AD et al. (2008) Association of intrauterine exposure to maternal diabetes and obesity with type 2 diabetes in youth: the SEARCH Case-Control Study. Diabetes Care 31: 1422–1426. 10.2337/dc07-2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dello RM, Ahrens W, De VT, Marild S, Molnar D, et al. (2013) Gestational weight gain and adiposity, fat distribution, metabolic profile, and blood pressure in offspring: the IDEFICS project. Int J Obes (Lond) 37: 914–919. 10.1038/ijo.2013.35 [DOI] [PubMed] [Google Scholar]
- 5. Hochner H, Friedlander Y, Calderon-Margalit R, Meiner V, Sagy Y, et al. (2012) Associations of maternal prepregnancy body mass index and gestational weight gain with adult offspring cardiometabolic risk factors: the Jerusalem Perinatal Family Follow-up Study. Circulation 125: 1381–1389. 10.1161/CIRCULATIONAHA.111.070060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Alfaradhi MZ, Ozanne SE (2011) Developmental programming in response to maternal overnutrition. Front Genet 2: 27 10.3389/fgene.2011.00027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xia Q, Grant SF (2013) The genetics of human obesity. Ann N Y Acad Sci 1281: 178–190. 10.1111/nyas.12020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zarkesh M, Daneshpour MS, Faam B, Fallah MS, Hosseinzadeh N, et al. (2012) Heritability of the metabolic syndrome and its components in the Tehran Lipid and Glucose Study (TLGS). Genet Res (Camb) 94: 331–337. 10.1017/S001667231200050X [DOI] [PubMed] [Google Scholar]
- 9. Steck AK, Rewers MJ (2011) Genetics of type 1 diabetes. Clin Chem 57: 176–185. 10.1373/clinchem.2010.148221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vassy JL, Meigs JB (2012) Is genetic testing useful to predict type 2 diabetes? Best Pract Res Clin Endocrinol Metab 26: 189–201. 10.1016/j.beem.2011.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Comuzzie AG, Cole SA, Laston SL, Voruganti VS, Haack K, et al. (2012) Novel genetic loci identified for the pathophysiology of childhood obesity in the Hispanic population. PLoS One 7: e51954 10.1371/journal.pone.0051954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brem RB, Yvert G, Clinton R, Kruglyak L (2002) Genetic dissection of transcriptional regulation in budding yeast. Science 296: 752–755. 10.1126/science.1069516 [DOI] [PubMed] [Google Scholar]
- 13. Dimas AS, Deutsch S, Stranger BE, Montgomery SB, Borel C, et al. (2009) Common regulatory variation impacts gene expression in a cell type-dependent manner. Science 325: 1246–1250. 10.1126/science.1174148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cheung VG, Spielman RS (2009) Genetics of human gene expression: mapping DNA variants that influence gene expression. Nat Rev Genet 10: 595–604. 10.1038/nrg2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hales CN, Barker DJ (1992) Type 2 (non-insulin-dependent) diabetes mellitus: the thrifty phenotype hypothesis. Diabetologia 35: 595–601. 10.1007/BF00400248 [DOI] [PubMed] [Google Scholar]
- 16. Edlow AG, Vora NL, Hui L, Wick HC, Cowan JM, et al. (2014) Maternal obesity affects fetal neurodevelopmental and metabolic gene expression: a pilot study. PLoS One 9: e88661 10.1371/journal.pone.0088661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guenard F, Deshaies Y, Cianflone K, Kral JG, Marceau P, et al. (2013) Differential methylation in glucoregulatory genes of offspring born before vs. after maternal gastrointestinal bypass surgery. Proc Natl Acad Sci U S A 110: 11439–11444. 10.1073/pnas.1216959110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. O'Shaughnessy PJ, Monteiro A, Bhattacharya S, Fowler PA (2011) Maternal smoking and fetal sex significantly affect metabolic enzyme expression in the human fetal liver. J Clin Endocrinol Metab 96: 2851–2860. 10.1210/jc.2011-1437 [DOI] [PubMed] [Google Scholar]
- 19. O'Shaughnessy PJ, Monteiro A, Bhattacharya S, Fraser MJ, Fowler PA (2013) Steroidogenic enzyme expression in the human fetal liver and potential role in the endocrinology of pregnancy. Mol Hum Reprod 19: 177–187. 10.1093/molehr/gas059 [DOI] [PubMed] [Google Scholar]
- 20. Botden IP, Zillikens MC, de Rooij SR, Langendonk JG, Danser AH et al. (2012) Variants in the SIRT1 gene may affect diabetes risk in interaction with prenatal exposure to famine. Diabetes Care 35: 424–426. 10.2337/dc11-1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. de Rooij SR, Painter RC, Phillips DI, Osmond C, Tanck MW, et al. (2006) The effects of the Pro12Ala polymorphism of the peroxisome proliferator-activated receptor-gamma2 gene on glucose/insulin metabolism interact with prenatal exposure to famine. Diabetes Care 29: 1052–1057. 10.2337/dc05-1993 [DOI] [PubMed] [Google Scholar]
- 22. Paus T, Bernard M, Chakravarty MM, Davey SG, Gillis J, et al. (2012) KCTD8 gene and brain growth in adverse intrauterine environment: a genome-wide association study. Cereb Cortex 22: 2634–2642. 10.1093/cercor/bhr350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Scopinaro N, Marinari GM, Camerini GB, Papadia FS, Adami GF (2005) Specific effects of biliopancreatic diversion on the major components of metabolic syndrome: a long-term follow-up study. Diabetes Care 28: 2406–2411. 10.2337/diacare.28.10.2406 [DOI] [PubMed] [Google Scholar]
- 24. Sjostrom L, Lindroos AK, Peltonen M, Torgerson J, Bouchard C, et al. (2004) Lifestyle, diabetes, and cardiovascular risk factors 10 years after bariatric surgery. N Engl J Med 351: 2683–2693. 10.1056/NEJMoa035622 [DOI] [PubMed] [Google Scholar]
- 25. Kral JG, Thung SN, Biron S, Hould FS, Lebel S, et al. (2004) Effects of surgical treatment of the metabolic syndrome on liver fibrosis and cirrhosis. Surgery 135: 48–58. 10.1016/j.surg.2003.10.003 [DOI] [PubMed] [Google Scholar]
- 26. Clement K, Viguerie N, Poitou C, Carette C, Pelloux V, et al. (2004) Weight loss regulates inflammation-related genes in white adipose tissue of obese subjects. FASEB J 18: 1657–1669. 10.1096/fj.04-2204com [DOI] [PubMed] [Google Scholar]
- 27. Moschen AR, Molnar C, Geiger S, Graziadei I, Ebenbichler CF, et al. (2010) Anti-inflammatory effects of excessive weight loss: potent suppression of adipose interleukin 6 and tumour necrosis factor alpha expression. Gut 59: 1259–1264. 10.1136/gut.2010.214577 [DOI] [PubMed] [Google Scholar]
- 28. Park JJ, Berggren JR, Hulver MW, Houmard JA, Hoffman EP (2006) GRB14, GPD1, and GDF8 as potential network collaborators in weight loss-induced improvements in insulin action in human skeletal muscle. Physiol Genomics 27: 114–121. 10.1152/physiolgenomics.00045.2006 [DOI] [PubMed] [Google Scholar]
- 29. Berisha SZ, Serre D, Schauer P, Kashyap SR, Smith JD (2011) Changes in whole blood gene expression in obese subjects with type 2 diabetes following bariatric surgery: a pilot study. PLoS One 6: e16729 10.1371/journal.pone.0016729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kral JG, Biron S, Simard S, Hould FS, Lebel S, et al. (2006) Large maternal weight loss from obesity surgery prevents transmission of obesity to children who were followed for 2 to 18 years. Pediatrics 118: e1644–e1649. 10.1542/peds.2006-1379 [DOI] [PubMed] [Google Scholar]
- 31. Smith J, Cianflone K, Biron S, Hould FS, Lebel S, et al. (2009) Effects of maternal surgical weight loss in mothers on intergenerational transmission of obesity. J Clin Endocrinol Metab 94: 4275–4283. 10.1210/jc.2009-0709 [DOI] [PubMed] [Google Scholar]
- 32. Guenard F, Tchernof A, Deshaies Y, Cianflone K, Kral JG, et al. (2013) Methylation and expression of immune and inflammatory genes in the offspring of bariatric bypass surgery patients. J Obes 2013: 492170 10.1155/2013/492170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. National Center for Health Statistics (2007) NHANES III clinical growth charts; children 2 to 20 years 5th to 95th percentile: boys' BMI for age and girls' BMI for age. Hyattsville, MD: National Center for Health Statistics. [Google Scholar]
- 34. National Center for Health Statistics (2007) NHANES III Zscore data files: BMI for age and sex 2 to 20 years. Hyattsville, MD: National Center for Health Statistics. [Google Scholar]
- 35. Vohl MC, Sladek R, Robitaille J, Gurd S, Marceau P, et al. (2004) A survey of genes differentially expressed in subcutaneous and visceral adipose tissue in men. Obes Res 12: 1217–1222. 10.1038/oby.2004.153 [DOI] [PubMed] [Google Scholar]
- 36. Ledue TB, Weiner DL, Sipe JD, Poulin SE, Collins MF, et al. (1998) Analytical evaluation of particle-enhanced immunonephelometric assays for C-reactive protein, serum amyloid A and mannose-binding protein in human serum. Ann Clin Biochem 35 (Pt 6): 745–753. 10.1177/000456329803500607 [DOI] [PubMed] [Google Scholar]
- 37. Blazejczyk M, Miron M, Nadon R (2007) FlexArray: A statistical data analysis software for gene expression microarrays. Genome Quebec, Montreal, Canada. [Google Scholar]
- 38. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Laurie CC, Doheny KF, Mirel DB, Pugh EW, Bierut LJ, et al. (2010) Quality control and quality assurance in genotypic data for genome-wide association studies. Genet Epidemiol 34: 591–602. 10.1002/gepi.20516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Arim RG, Shapka JD, Dahinten VS, Willms JD (2007) Patterns and correlates of pubertal development in Canadian youth: effects of family context. Can J Public Health 98: 91–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265. 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- 42. Huang dW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 43. Huang dW, Sherman BT, Lempicki RA (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13. 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Baik JH (2013) Dopamine signaling in reward-related behaviors. Front Neural Circuits 7: 152 10.3389/fncir.2013.00152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee W (2011) The CEACAM1 expression is decreased in the liver of severely obese patients with or without diabetes. Diagn Pathol 6: 40 10.1186/1746-1596-6-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kitamoto A, Kitamoto T, Mizusawa S, Teranishi H, So R, et al. (2013) NUDT3 rs206936 is associated with body mass index in obese Japanese women. Endocr J 60: 991–1000. 10.1507/endocrj.EJ13-0100 [DOI] [PubMed] [Google Scholar]
- 47. Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, et al. (2010) Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet 42: 937–948. 10.1038/ng.686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang K, Li WD, Zhang CK, Wang Z, Glessner JT, et al. (2011) A genome-wide association study on obesity and obesity-related traits. PLoS One 6: e18939 10.1371/journal.pone.0018939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vucetic Z, Kimmel J, Totoki K, Hollenbeck E, Reyes TM (2010) Maternal high-fat diet alters methylation and gene expression of dopamine and opioid-related genes. Endocrinology 151: 4756–4764. 10.1210/en.2010-0505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hao K, Bosse Y, Nickle DC, Pare PD, Postma DS, et al. (2012) Lung eQTLs to help reveal the molecular underpinnings of asthma. PLoS Genet 8: e1003029 10.1371/journal.pgen.1003029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, et al. (2010) Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet 6: e1000888 10.1371/journal.pgen.1000888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, et al. (2007) A genome-wide association study of global gene expression. Nat Genet 39: 1202–1207. 10.1038/ng2109 [DOI] [PubMed] [Google Scholar]
- 53. Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F et al. (2008) Genetics of gene expression and its effect on disease. Nature 452: 423–428. 10.1038/nature06758 [DOI] [PubMed] [Google Scholar]
- 54. Grundberg E, Small KS, Hedman AK, Nica AC, Buil A, et al. (2012) Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat Genet 44: 1084–1089. 10.1038/ng.2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kotlarz D, Beier R, Murugan D, Diestelhorst J, Jensen O, et al. (2012) Loss of interleukin-10 signaling and infantile inflammatory bowel disease: implications for diagnosis and therapy. Gastroenterology 143: 347–355. 10.1053/j.gastro.2012.04.045 [DOI] [PubMed] [Google Scholar]
- 56. Moran CJ, Walters TD, Guo CH, Kugathasan S, Klein C, et al. (2013) IL-10R polymorphisms are associated with very-early-onset ulcerative colitis. Inflamm Bowel Dis 19: 115–123. 10.1002/ibd.22974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Park HK, Kim DH, Yun DH, Ban JY (2013) Association between IL10, IL10RA, and IL10RB SNPs and ischemic stroke with hypertension in Korean population. Mol Biol Rep 40: 1785–1790. 10.1007/s11033-012-2232-5 [DOI] [PubMed] [Google Scholar]
- 58. Ma J, Dempsey AA, Stamatiou D, Marshall KW, Liew CC (2007) Identifying leukocyte gene expression patterns associated with plasma lipid levels in human subjects. Atherosclerosis 191: 63–72. 10.1016/j.atherosclerosis.2006.05.032 [DOI] [PubMed] [Google Scholar]
- 59. Palmer ND, McDonough CW, Hicks PJ, Roh BH, Wing MR, et al. (2012) A genome-wide association search for type 2 diabetes genes in African Americans. PLoS One 7: e29202 10.1371/journal.pone.0029202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shankar K, Zhong Y, Kang P, Lau F, Blackburn ML, et al. (2011) Maternal obesity promotes a proinflammatory signature in rat uterus and blastocyst. Endocrinology 152: 4158–4170. 10.1210/en.2010-1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cannon MV, Buchner DA, Hester J, Miller H, Sehayek E, et al. (2014) Maternal nutrition induces pervasive gene expression changes but no detectable DNA methylation differences in the liver of adult offspring. PLoS One 9: e90335 10.1371/journal.pone.0090335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Murabayashi N, Sugiyama T, Zhang L, Kamimoto Y, Umekawa T, et al. (2013) Maternal high-fat diets cause insulin resistance through inflammatory changes in fetal adipose tissue. Eur J Obstet Gynecol Reprod Biol 169: 39–44. 10.1016/j.ejogrb.2013.02.003 [DOI] [PubMed] [Google Scholar]
- 63. Borengasser SJ, Zhong Y, Kang P, Lindsey F, Ronis MJ, et al. (2013) Maternal obesity enhances white adipose tissue differentiation and alters genome-scale DNA methylation in male rat offspring. Endocrinology 154: 4113–4125. 10.1210/en.2012-2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nicholas LM, Morrison JL, Rattanatray L, Ozanne SE, Kleemann DO, et al. (2013) Differential effects of exposure to maternal obesity or maternal weight loss during the periconceptional period in the sheep on insulin signalling molecules in skeletal muscle of the offspring at 4 months of age. PLoS One 8: e84594 10.1371/journal.pone.0084594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. van HM, Langendonk JG, de Rooij SR, Sijbrands EJ, Roseboom TJ (2009) Genetic variant in the IGF2BP2 gene may interact with fetal malnutrition to affect glucose metabolism. Diabetes 58: 1440–1444. 10.2337/db08-1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ding J, Gudjonsson JE, Liang L, Stuart PE, Li Y, et al. (2010) Gene expression in skin and lymphoblastoid cells: Refined statistical method reveals extensive overlap in cis-eQTL signals. Am J Hum Genet 87: 779–789. 10.1016/j.ajhg.2010.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Fu J, Wolfs MG, Deelen P, Westra HJ, Fehrmann RS, et al. (2012) Unraveling the regulatory mechanisms underlying tissue-dependent genetic variation of gene expression. PLoS Genet 8: e1002431 10.1371/journal.pgen.1002431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rudkowska I, Raymond C, Ponton A, Jacques H, Lavigne C, et al. (2011) Validation of the use of peripheral blood mononuclear cells as surrogate model for skeletal muscle tissue in nutrigenomic studies. OMICS 15: 1–7. 10.1089/omi.2010.0073 [DOI] [PubMed] [Google Scholar]
- 69. Gerrits A, Li Y, Tesson BM, Bystrykh LV, Weersing E, et al. (2009) Expression quantitative trait loci are highly sensitive to cellular differentiation state. PLoS Genet 5: e1000692 10.1371/journal.pgen.1000692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Smirnov DA, Morley M, Shin E, Spielman RS, Cheung VG (2009) Genetic analysis of radiation-induced changes in human gene expression. Nature 459: 587–591. 10.1038/nature07940 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Panel A, STAT2 (NM_005419). Panel B, IFI35 (NM_005533). Panel C, DGKZ (NM_003646).
(TIF)
(DOCX)
SNPs, regulated transcripts and genotype-specific gene expression levels are shown. Expression values (means) relative to common homozygotes from the BMS group.
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
Gene expression data has been deposited in the Gene Expression Omnibus (GEO) database (accession number GSE44407).