Abstract
The conceptual complexity of a domain can make it difficult for users of information systems to comprehend and interact with the knowledge embedded in those systems. The Unified Medical Language System® (UMLS) currently integrates over 730,000 biomedical concepts from more than fifty biomedical vocabularies. The UMLS semantic network reduces the complexity of this construct by grouping concepts according to the semantic types that have been assigned to them. For certain purposes, however, an even smaller and coarser-grained set of semantic type groupings may be desirable. In this paper, we discuss our approach to creating such a set. We present six basic principles, and then apply those principles in aggregating the existing 134 semantic types into a set of 15 groupings. We present some of the difficulties we encountered and the consequences of the decisions we have made. We discuss some possible uses of the semantic groups, and we conclude with implications for future work.
Keywords: Unified Medical Language System, Knowledge Representation, Medical Informatics
Introduction
The conceptual complexity of a domain can make it difficult for users of information systems to comprehend and interact with the knowledge embedded in those systems [1]. The UMLS semantic network is a high-level structure for organizing a large number of concepts in the biomedical domain [2]. As such, it helps clarify the structure of the domain, an important property of ontologies in general. Chandrasekaran notes [3:21]:
Given a domain, its ontology forms the heart of any system of knowledge representation for that domain. Without ontologies, or the conceptualizations that underlie knowledge, there cannot be a vocabulary for representing knowledge …The ontology captures the intrinsic conceptual structure of the domain.
In addition to allowing computer applications to reason about the concepts in the domain, explicit and well-formed ontologies may be used for a variety of other purposes. Pratt [4], for example, has experimented with displaying literature search results using the UMLS semantic types, and Pisanelli et al [5], believing that ontologies can support more effective knowledge sharing in medicine, partition the UMLS according to the semantic types that have been assigned to each concept. Gu et al [6] and Chen et al [7] note that while the UMLS is a valuable knowledge resource, its size and complexity make it difficult to understand and visualize. They develop methods to partition the UMLS conceptual space to aid in comprehension. The first release of the UMLS knowledge sources included the UMLS semantic network, as well as a broad grouping of semantic types for more readily displaying MEDLINE® co-occurrence information in a HyperCard application called MetaCard [8:79, 9]. In the following, we discuss our review of this original grouping of semantic types, including a set of principles we developed to aid in analysis and validation. Based on these principles, we created a revised set of semantic groupings that may be useful for a variety of purposes.
Methods
We developed a methodology for aggregating semantic types into a small number of groups based on the following general principles:
Semantic validity – the groups must be semantically coherent
Parsimony – the number of groups should be as small as possible
Completeness – the groups must cover the full domain
Exclusivity – each concept in the domain must belong to only one group
Naturalness – the groups must characterize the domain in a way that is acceptable to a domain expert
Utility – the groups must be useful for some purpose
The original 1990 semantic groupings were examined to assess their adherence to the six general principles. Since the primary motivation for creating the groups was for visualizing data in an application program, the principle of utility was immediately met. Once each semantic type had been assigned to one of the semantic groups, the completeness principle was also automatically met for the entire UMLS because every UMLS concept is assigned at least one semantic type from the network. In addition, the naturalness principle was met, since the groupings were easily understood by domain specialists in the context of the application without any additional documentation or training. The original set consisted of a small number of groupings (14) for the 131 semantic types, so the principle of parsimony also applied. We closely reviewed the groupings for adherence to the remaining two principles, semantic validity and exclusivity, and we made a number of changes to the groupings based on the results of our analysis.
One way to measure semantic validity is to assess the degree to which the types in a group are hierarchically related to each other. This is so, since parents and children in a hierarchy share essential properties. For example, in Figure 1 below, any grouping that includes anatomical abnormalities, is, at least as a first hypothesis, expected to also include congenital abnormalities and acquired abnormalities. Further, semantic types that belong to distinct and distant branches of the network are not expected to cluster together. However, in some cases, such a grouping does result in a valid categorization. For example, the semantic type “Body Location or Region”, is, strictly speaking, a conceptual notion and is classed as a subtype of “Spatial Concept”. Since body locations share many semantic features with anatomical concepts, the “Body Location or Region” semantic type was, in fact, grouped with the other anatomical types. Analogously, semantic types belonging to the same branch of the semantic network or even having the same parent may be better clustered into distinct semantic groups. For example, although the semantic types “Gene or Genome” and “Body Part, Organ, or Organ Component” are both subtypes of “Fully Formed Anatomical Structure”, only the former belongs to the semantic group “Anatomy”, while the latter is placed in the “Genes & Molecular Sequences” group. Figure 1 illustrates this partitioning.
Figure 1.
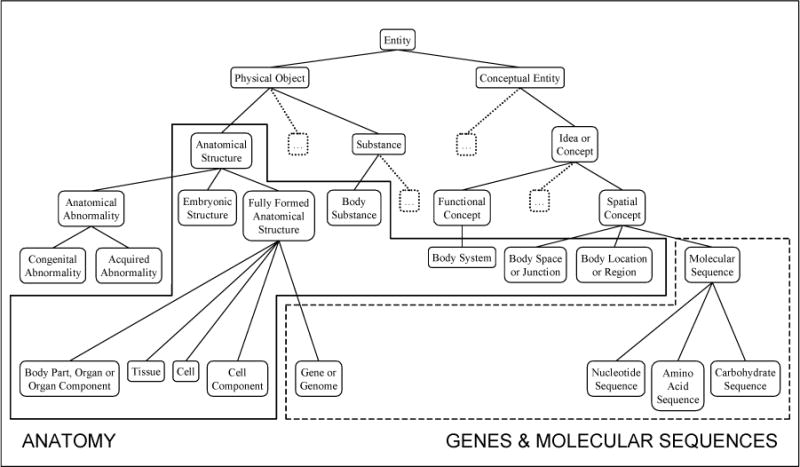
The UMLS semantic network clustered into semantic groups (partial representation)
The semantic types represent intensional, or definitional, knowledge, while the UMLS concepts that are assigned to those types represent extensional knowledge. In reviewing the members of the semantic groups, we looked not only at the definitions of the semantic types, but also at the concepts that had been assigned to those types in the 2000 release of the UMLS [10]. For example, the semantic type “Educational Activity” is defined as ‘an activity related to the organization and provision of education’. However, in practice, concepts such as “Hemodialysis training at home”, are assigned to this type. Concepts such as these are actually kinds of biomedical procedures and are, therefore, better clustered together with other procedures according to the semantic validity principle.
Semantic validity may also be measured by an analysis of the relationships in which the semantic groups participate. For example, a result that groups anatomical concepts together can be shown to have semantic validity when considering the relationships in which these concepts participate. We reviewed the stated relationships for each of the semantic types within a group and then evaluated the consistency and correctness of the set of relationships within and across each of the groups. For example, an anatomical structure can be connected to another anatomical structure, and it can be the location of a disorder. These facts help validate grouping all anatomical structures together and also separately grouping all disorders together.
The exclusivity principle implies that the domain is fully partitioned with no overlap between groups. A partitioning of the UMLS must provide not only disjoint groups of semantic types, but also disjoint groups of concepts. We tested compliance to the exclusivity principle on the full set of 730,000 concepts and analyzed those cases where concepts had been assigned to multiple semantic types. In many cases, multiple semantic typing did not result in a violation of the exclusivity principle, since the semantic types were classed together in the same group. For example, most chemicals are assigned both a structural and a functional semantic type. The former is related to the essential properties of chemicals, and the latter to the role they play. (See [2 and 11] for some discussion of this distinction.) Since structural and functional chemical types are classed together in the “Chemicals & Drugs” groups, this does not represent a violation of the exclusivity principle. In some cases, multiple typing of a concept did, however, result in that concept being assigned to more than one semantic group. A small number of these cases were not resolvable and are discussed below.
Results
The list of semantic groups is shown in Table 1, together with the number and percentage of UMLS concepts in each group.
Table 1.
Repartition of UMLS concepts using Semantic Groups (Percentage is greater than 100 because of some group overlap).
| Semantic Type Groups | No. Types | UMLS Concepts | |
|---|---|---|---|
| No. | % | ||
| Activities & Behaviors | 9 | 3224 | .4 % |
| Anatomy | 11 | 34,386 | 4.7 % |
| Chemicals & Drugs | 26 | 356,211 | 48.8 % |
| Concepts & Ideas | 12 | 17,639 | 2.4 % |
| Devices | 2 | 31,092 | 4.3 % |
| Disorders | 12 | 136,389 | 18.7 % |
| Genes & Molecular Sequences | 5 | 904 | .1 % |
| Geographic Areas | 1 | 949 | .1 % |
| Living Beings | 23 | 29,699 | 4.1 % |
| Objects | 5 | 6,857 | .9 % |
| Occupations | 2 | 890 | .1 % |
| Organizations | 4 | 2,124 | .3 % |
| Phenomena | 6 | 4,943 | .7 % |
| Physiology | 9 | 27,930 | 3.8 % |
| Procedures | 7 | 81,847 | 11.2 % |
| Totals | 134 | 735,084 | 100.6 % |
The number of concepts per group ranges from 904 for “Genes & Molecular Sequences” to 356,211 for “Chemicals & Drugs”. Because a small number of concepts fall into multiple semantic groups, the total number of concepts shown in the table exceeds the current number of concepts in the UMLS (730,155) and the total percentage slightly exceeds 100%.
The 15 groups almost realize a complete partition of the UMLS since 725,242 of the 730,155 concepts in the 2000 release of the UMLS are categorized into one and only one group. Of the remaining 4913 concepts, most are assigned to two groups, with only 16 concepts assigned to three groups. For example, “Chromatin”, is assigned 3 semantic types, “Cell Component”, which belongs to the group “Anatomy”, “Genetic Function”, which belongs to the group “Physiology”, and “Amino Acid, Peptide or Protein”, which belongs to the group “Chemicals & Drugs”. No concepts fall into more than 3 semantic groups.
Some of the relationships that obtain between the groups shown in Table 1 are listed in Table 2 below.
Table 2.
Some relationships between Semantic Groups
| Semantic Group | Relationship | Semantic Group |
|---|---|---|
| Anatomy | developmental_form_of | Anatomy |
| Chemicals & Drugs | treats | Disorders |
| Devices | treats | Disorders |
| Procedures | treats | Disorders |
| Living Beings | exhibits | Activities & Behaviors |
| Genes & Molecular Sequences | carries_out | Physiology |
| Genes & Molecular Sequences | property_of | Chemicals & Drugs |
To test the utility of the groupings, we compared the distribution of concepts in the UMLS as a whole with the distribution of concepts in a single vocabulary (PDQ – Physician Data Query Online System). Comparing Figure 2 with Figure 3, we can see at a glance that chemicals and drugs represent the largest class of concepts in the full UMLS, followed by disorders and procedures; while in PDQ, procedures represent the largest class, followed by chemicals and drugs, and then disorders. This indicates that the semantic groups can readily be used to create a high-level profile of a vocabulary, complementary to a more detailed analysis of the full semantics of that vocabulary.
Figure 2.
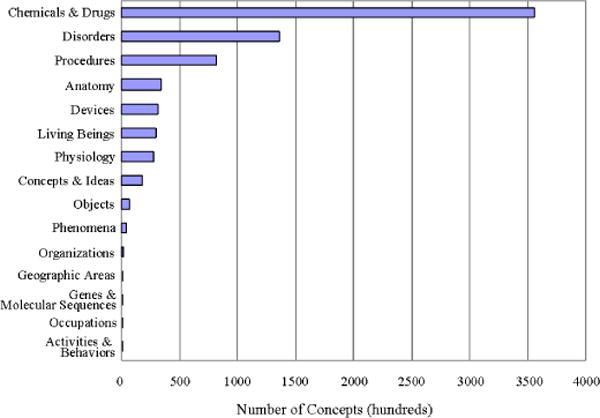
Distribution of concepts in the UMLS
Figure 3.
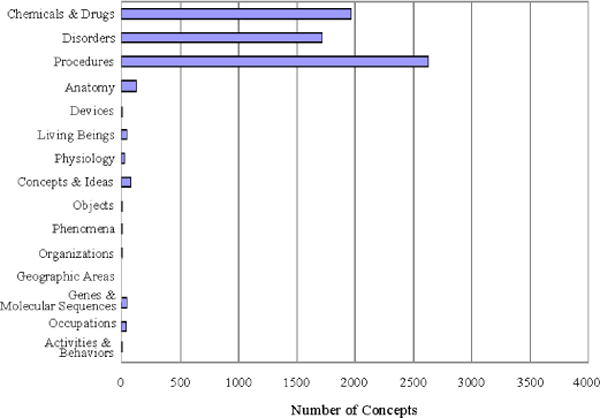
Distribution of concepts in PDQ
Conclusions
In some cases, it was not possible to resolve anomalies in our attempt to create a coherent and semantically valid set of groupings. This is partly because of the nature of meaning itself. Some concepts logically belong to multiple semantic groups. For example, an adenoma may be simultaneously thought of as an anatomical abnormality (that may have to be surgically removed) and as a disease (with a prognosis and with potential complications). In other cases, anomalies arise because of errors or inconsistencies in the assignment of semantic types to UMLS concepts. Examples of errors include concepts that wrongly refer to both a physiologic function and to a procedure analyzing this function. For example, “Glomerular Filtration Rate” is assigned to both the “Physiology” and “Procedures” semantic groups, since it has incorrectly been assigned two semantic types, “Organ or Tissue Function” and “Diagnostic Procedure”. In a few cases, stated relationships between semantic types also caused problems in grouping semantic types appropriately. The methods described here, therefore, afford another way to “audit” the correctness and consistency of the UMLS data. (See [12] for additional semantic auditing methods.)
There are many reasons why it is desirable to reduce conceptual complexity when dealing with a large domain. We have presented one method for doing so. The resulting semantic groups may be used for display purposes; they may provide a broad overview of a conceptual space, such as that represented in a terminology system; and they might be used to discover inconsistencies in the representation of that domain. Our future work will investigate some of these latter in the context of the UMLS.
Contributor Information
Alexa T. McCray, Email: mccray@nlm.nih.gov.
Anita Burgun, Email: burgun@nlm.nih.gov.
Olivier Bodenreider, Email: olivier@nlm.nih.gov.
References
- 1.Wickens CD, Gordon SE, Liu Y. An Introduction to Human Factors Engineering. New York: Longman; 1998. [Google Scholar]
- 2.McCray AT. High-Performance Medical Libraries: Advances in Information Management for the Virtual Era. Westport: Meckler Publishing; 1993. Representing biomedical knowledge in the UMLS Semantic Network; pp. 45–55. [Google Scholar]
- 3.Chandrasekaran R, Josephson JR, Benjamins VR. What are ontologies, and why do we need them? IEEE Intelligent Systems. 1999:20–26. [Google Scholar]
- 4.Pratt W. Dynamic organization of search results using the UMLS. Proc AMIA Annu Fall Symp. 1997:480–4. [PMC free article] [PubMed] [Google Scholar]
- 5.Pisanelli DM, Gangemi A, Steve G. An ontological analysis of the UMLS Metathesaurus. Proc AMIA Symp. 1998:810–4. [PMC free article] [PubMed] [Google Scholar]
- 6.Gu H, Perl Y, Geller J, Halper M, Liu LM, Cimino JJ. Representing the UMLS as an object-oriented database: modeling issues and advantages. J Am Med Inform Assoc. 2000 Jan-Feb;7(1):66–80. doi: 10.1136/jamia.2000.0070066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen M, Halper M, Geller J, Perl Y. A structural partition of the Unified Medical Language System’s Semantic Network. Proc IEEE Information Technology Applications in Biomedicine. 2000:296–301. [Google Scholar]
- 8.UMLS Knowledge Sources. Documentation. U.S National Library of Medicine; 1990. Experimental edition. [Google Scholar]
- 9.Tuttle MS, Cole WG, Sheretz DD, Nelson SJ. Navigating to knowledge. Methods Inf Med. 1995 Mar;34(1–2):214–31. [PubMed] [Google Scholar]
- 10.UMLS Knowledge Sources. Documentation. 11. U.S. National Library of Medicine; 2000. [Google Scholar]
- 11.Zweigenbaum P, Bachimont B, Bouaud J, Charlet J, Boisvieux JF. Issues in the structuring and acquisition of an ontology for medical language understanding. Methods Inf Med. 1995 Mar;34(1–2):15–24. [PubMed] [Google Scholar]
- 12.Cimino JJ. Auditing the Unified Medical Language System with semantic methods. J Am Med Inform Assoc. 1998 Jan-Feb;5(1):41–51. doi: 10.1136/jamia.1998.0050041. [DOI] [PMC free article] [PubMed] [Google Scholar]


