Abstract
It is thought that Retinal Determination gene products define the response made to cell-cell signals within the eye developmental field by binding to enhancers of genes that are also regulated by cell-cell signaling pathways. In Drosophila, Retinal Determination genes including Eyeless, teashirt, eyes absent, dachsous and sine oculis, are required for normal eye development and can induce ectopic eyes when mis-expressed. Characterization of the enhancers responsible for eye expression of the hedgehog, shaven, and atonal genes, as well as the dynamics of Retinal Determination gene expression themselves, now suggest a multilayered network whereby transcriptional regulation by either Retinal Determination genes or cell-cell signaling pathways can sometimes be indirect and mediated by other transcription factor intermediates. In this updated view of the interaction between extracellular information and cell intrinsic programs during development, regulation of individual genes might sometimes be several steps removed from either the Retinal Determination genes or cell-cell signaling pathways that nevertheless govern their expression.
Master Regulatory genes and their function in development
The concept of ‘master regulatory genes’ has attracted much interest since the remarkable findings that the eyeless (ey) gene from Drosophila and the pax6 gene that is mutated in Aniridia in humans and in small eye in rodents were homologs, and that either gene was capable of inducing ectopic eyes in many locations in the Drosophila body when ectopically expressed[1]. Without attempting to define ‘master regulatory gene’ precisely, since the term is nowadays applied in a range of situations, we think that a remarkable feature exemplified by Eyeless is to specify development of an organ that comprises multiple cell types. In whatever manner such master regulators work, it cannot be so simple as to initiate a program of gene expression, because one program of gene expression could not define multiple cell types. Instead, genes such as eyeless must define a program that interacts with extracellular information to allow different gene-expression programs to occur in the eye cell types that develop at different locations. In this regard they fulfill a function that may be necessary in many or all organs.
The current hypothesis is that combinatorial interactions at the enhancers of target genes occur between retinal determination gene products and transcription factors dedicated to cell-cell signaling pathways, a model first developed for other classes of master regulatory genes that act in the Drosophila thorax [2, 3]. This hypothesis makes a clear prediction that genes whose spatial expression is regulated during eye development should have regulatory sequences that combine the two classes of input. Transcriptional regulation of several genes expressed during eye development has now been characterized, including hedgehog (hh), shaven (sv) (also known as sparkling (spa) or D-Pax2), and atonal (ato), and we discuss these examples with regard to this prevailing hypothesis.
Our conclusion is that the current model can accommodate what has been discovered about regulation of the hh, sv, and ato genes, if recent studies of the expression of the retinal determination genes themselves are incorporated, along with the notions that both cell intrinsic and extracellular information can act indirectly through intermediaries, and that Retinal Determination genes themselves can play some roles in relaying extracellular information in addition to providing eye field identity. When these modifications are incorporated, it remains possible to understand how retinal determination genes define an eye field in terms of an interactions between cell intrinsic programs and cell-cell signaling pathways, as has proven useful for understanding much of development in the past.
Cell intrinsic programs and extracellular information contribute together to regulate cells within developmental fields
Cells within developmental fields share a common determination state but can still take distinct individual cell fates according to the distinct signals that cells encounter at their respective locations within the field. Cell fate specification, as well as other cell properties such as morphology and the control of cell proliferation, therefore depends on the interaction between extracellular information and the cell intrinsic programs that define each developmental field.
One common source of extracellular information is cell-cell signaling pathways, and this is certainly true of the developing Drosophila eye[4]. Certain cell-cell signaling pathways, including the Wnt, Hh, Notch (N), TGF-beta and Ras pathways, are used many times during development with distinct, context-dependent outcomes each time[5,6]. Because of the re-use of pathways these cell-cell signaling pathways are not unique to any particular field, and field-specific interpretation of cell-cell signaling pathways is one of many indications that cell intrinsic information is also essential for defining cellular responses.
This combinatorial view of developmental fields harkens back to the ‘positional information’ concept popularized by Wolpert, who posited that individual cells translate ‘positional information’ (ie extracellular information that corresponds to location within a tissue) into specific differentiation programs only in the context of pre-existing specification to a particular developmental field[7]. This makes it possible for the same cell-cell signals to encode extracellular information with diverse effects in different fields, while the master regulators define how each field responds to the signals. The same dichotomy has recently been recognized by Davidson, who classified some regulatory genes including pax6 into ‘kernels’ that control stable features of the body plan, eg ‘eye’; other regulatory genes, such as the cell-cell signaling pathways that act in multiple body regions, were classified as ‘plug-ins’, because they are used in many times and places[8].
Cell-cell signals can be integrated with intrinsic determination states at combinatorial enhancers
Each of the Wnt, Hh, Notch (N), TGF-beta and Ras pathways cell-cell signaling pathways affect transcription through one (or a few) dedicated transcription factor(s) that are generally used regardless of the context [5,6]. The Notch signaling pathway, for example, exerts most or all of its effects through the transcription factor Su(H), whose vertebrate homologs are also called CBF or RBP-Jk[9]. The specific responses these signaling pathways elicit are governed by the developmental history and specification of each developmental field.
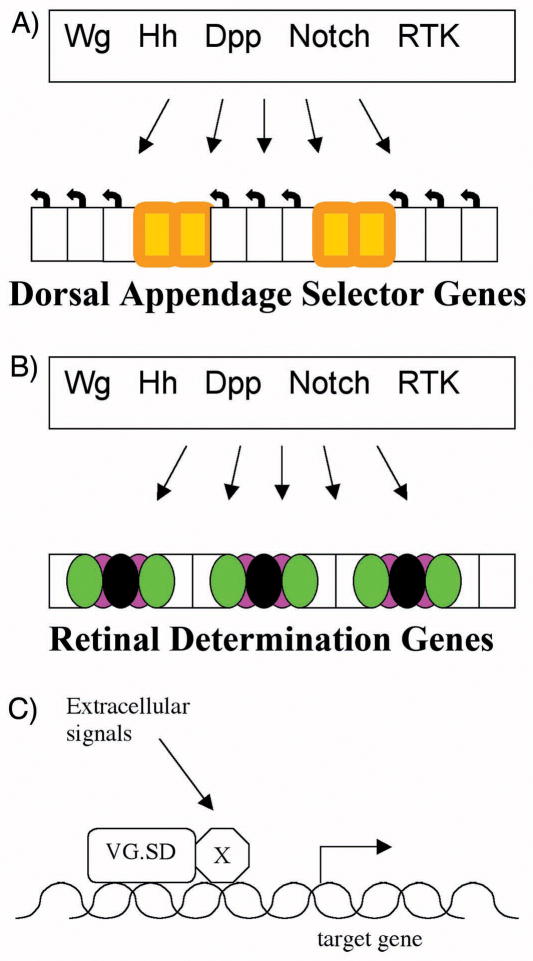
One way that cell-cell signals interact with cell determination states is by means of combinatorial interactions on the enhancers of target genes. Such interactions would involve the transcription factors that are regulated by cell-cell signaling pathways and the master regulatory proteins that define the developmental field (Figure 1). This paradigm has emerged from study of master regulatory genes that define developmental fields in the Drosophila thorax, and it has been proposed that it also applies to Drosophila eye development [2, 3]. For example, the dorsal body appendages in the Drosophila thorax (ie wing and haltere) are defined by expression of the Vestigial (Vg) protein, along with its cofactor Scalloped (Sd). As its name suggests, mutations in vg lead to minimal wing development. Genes are activated in particular wing or haltere cells by enhancers that integrate Vg/Sd binding with cell-cell signaling pathways. An enhancer that is active along the dorsal-ventral margin of the wing, for example, binds both Vg/Sd and Su(H), the transcription factor that mediates Notch signaling, since Notch signaling is active at this location. The combinatorial requirement for both factors confers activity to the enhancer specifically in wing cells experiencing Notch signaling. Exchanging Su(H) binding sites for binding by Ci, the Gli-family transcription factor that mediates Hh signaling, produces an enhancer active along the boundary between the anterior and posterior compartments of the wing instead, ie wing cells experiencing Hh signaling[10].
Figure 1. Integration of developmental identity and extracellular signals in developmental patterning.
Cell-cell signaling pathways that are used widely to confer spatio-temporal information are interpreted differently in each developmental field. The Drosophila wing and eye are each epithelia that are patterned by many of the same signals.
A) Typical wing cell fates include wing vein (orange), and wing-hair producing intervein cells (uncolored). All these fates depend on Dorsal Appendage selector genes Scalloped and vestigial, as well as other selector genes such as the Hox gene Antennapedia, but the fates of individual cells within the wing differ due to positional information from cell-cell signaling pathways.
B) Typical eye cell fates include clusters of photoreceptor neurons and various retinal support cells (black, green and magenta). These fates all depend on the Retinal Determination genes including Eyeless and its paralog twin of eyeless. Individual eye cells take different fates in response to positional information from the same cell-cell signaling pathways that are active in the wing.
C) The prevailing model is that positional information is integrated with developmental identity at combinatorial enhancers[62]. In the example of a wing target gene, enhancer requiring binding by the Vg/Sd complex proteins and by transducers of cell-cell signaling pathways could confer spatio-temporal regulation[12].
Another example is provided by the homeotic selector genes, homeodomain-containing transcription factors encoded in Hox gene complexes and which define trunk segments in insects and anterior-posterior body subdivisions in vertebrates. The name ‘selector genes’ reflects the notion that Hox genes select among alternative possible developmental pathways appropriate for one or other segment. Like Vestigial, Hox proteins are also thought to interact with gene promoters in synergy with the transcription factors that transduce cell-cell signaling pathways, providing segment-specific context to responses to the positional information[11][12]. In fact, it has been suggested that the transcription factors that mediate extracellular signals may generally be insufficient for transcription without cooperation from a master regulator partner[13,14]. If this hypothesis is correct, cell-cell signaling pathways would be unable to influence cells in the developing eye were there no master regulators for the eye.
It has been proposed that the eye developmental field in Drosophila develops in similar manner to the thoracic organs, with Eyeless (Ey) and other Retinal Determination genes (RD genes) defining the eye field and providing a context for eye cells to respond to cell-cell signaling pathways. This would be achieved through enhancer elements that require combinatorial input from RD gene products as well as from the transcription factors that transduce cell-cell signaling pathways[2, 3] (Figure 1). Since the transcriptional regulation of some eye target genes has now been reported, it is possible to evaluate the combinatorial hypothesis with regard to RD genes and cell-cell signaling pathways.
One difference between RD genes and vg, sd and the homeotic selector genes is that expression of RD genes is more dynamic. A related difference concerns the greater degree to which RD gene expression itself is more continuously regulated by cell-cell signaling pathways. At least in part, this is related to the progressive nature of eye differentiation[15]. A third difference from classic selector genes is that, while Eyeless is required for eye development, its absence leads to loss of the eye primordium rather than transformation to another structure, so that Eyeless does not select among alternative fates in the same simple manner as homeotic selector genes[10,16].
Several distinct patterns of Retinal Determination gene expression occur in the Drosophila eye field
One feature of RD gene expression is that it is dynamic during critical stages of eye patterning. This is different from vg and the Homeotic Selector Genes, which are expressed more stably in their developmental fields while patterning occurs. The eye develops in the eye imaginal disc, an epithelial sheet whose progenitor cells proliferate in the larval stages, then pattern and differentiate as metamorphosis approaches (Figure 2A)[17,18]. When the proliferative phase draws to a close, a morphogenetic furrow sweeps across the epithelium. As the morphogenetic furrow crosses each portion of the eye disc, proliferation ceases, the first retinal cell fates are specified, and photoreceptor differentiation begins within a few short hours (Figure 2A). Cell-cell signaling pathways coordinate these events (Figure 2B)[4]. Hh and Dpp, secreted from cells within and posterior to the morphogenetic furrow, continue to push the morphogenetic furrow forwards (Figure 2B). The advance of the furrow is antagonized by the Wnt gene wingless (wg) expressed at the anterior disc margins, so that the rate of progression depends on the balance between these signals (Figure 2B). Posterior to the morphogenetic furrow, photoreceptor and other cell types are specified by reiterative use of receptor tyrosine kinase and Notch signaling, and proliferation is limited to the progenitors of certain cell types (Figure 2B). Thus, the major cell-cell signaling pathways are each activated by ligands produced at particular locations within the eye disc epithelium itself (Figure 2B)[4,18].
Figure 2. Outline of retinal differentiation.
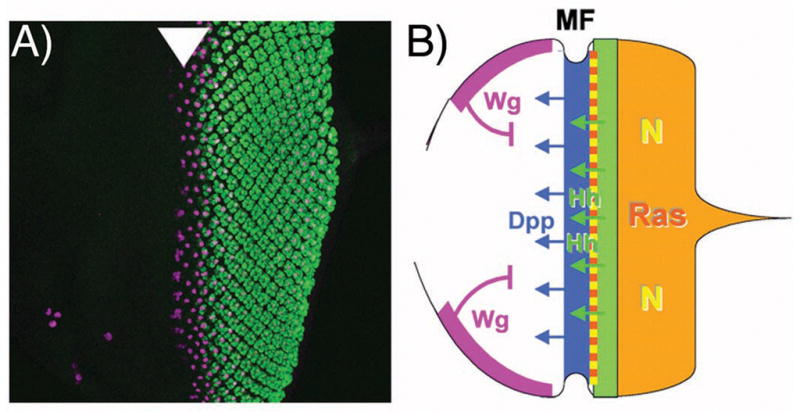
A) Photoreceptor differentiation begins at the posterior of the Drosophila eye imaginal disc, the larval primordium from which the retinal epithlium differentiates. Anterior is shown to the left, and photoreceptor differentiation labeled in green. The first founder R8 photoreceptor cell, specified slightly before the others, is labelled in magenta with an antibody against Senseless, a target of the proneural gene Atonal[63]. Atonal expression begins just before the morphogenetic furrow (arrowhead) where R8 cells are specified[51].
B) Cartoon diagraming the main cell-cell signals acting in the developing eye. The morphogenetic furrow is driven anteriorly by Hh and Dpp diffusing from differentiating photoreceptor cells and from the morphogenetic furrow respectively. These positive signals are antagonized by Wg, emanating from the anterior dorsal and ventral boundaries. Posterior to the morphogenetic furrow, episodes of signaling through the Ras and Notch pathways regulates the proliferation, recruitment and differentiation of many retinal cell types[4,17,18].
In addition to Ey and its paralog Twin of Eyeless, the tsh, dac, so, and eya genes are RD genes by the criterion that they can induce eyes when expressed ectopically (or synergize strongly with ectopic eye induction by other RD genes), and are required for normal eye development. All are transcription factors: Eya and Dac proteins interact with the DNA-binding protein So[19–21], and Dac can also bind DNA[22,23]. Although not able to induce ectopic eyes, Homothorax (Hth) is a another transcription factor that binds DNA and also interacts directly with the Ey and Tsh proteins[24]. These genes form a regulatory network whose homologs also regulate eye development in vertebrates. Many of these genes also play roles, alone and in combinations, in the development of other tissues[25–30]. There is an extensive literature on the cross-regulatory interactions among these genes which we do not review here[24,31–35]. The highly-connected nature of the network probably creates a robust program of eye development, and provides a hypothesis to explain how ectopic expression of one gene can be sufficient for ectopic eye development, if the whole network can be induced quite readily by just one member.
As the multipotent, proliferative stage of eye development ahead of the morphogenetic furrow comes to an end, the expression of Hth, Ey and Tsh proteins declines as the morphogenetic furrow approaches, and is replaced by expression of Dac, So and Eya (Figure 3A, B). The RD genes thus divide into two subsets that have a bistable relationship reinforced by negative regulatory interactions between them[24]. This makes the hypothesis that RD genes define the context to make eye-specific responses to cell-cell signaling pathways a complex one. For example, cell-cycle arrest occurs in response to Hh and Dpp[36], but RD gene expression is itself switching at the same time, making the contribution of RD genes to this process uncertain (Figure 3B). Perhaps the reality is that the eye comprises two successive, temporally distinct fields, one corresponding approximately to Ey/Tsh/Hth expression ahead of the morphogenetic furrow, where cells are not yet specified to individual fates, transitioning to Dac/Eya/So expression posterior to the morphogenetic furrow, where the cell cycle is arrested and terminal differentiation underway[24](Figure 3B). This has to be a simplification, however, because the transition between two RD gene expression ‘states’ is not sharp but gradual, because the transitions of the individual genes are not in complete synchrony, and because it is also possible that members of each set of genes might act individually, as well as together.
Figure 3. Complex changes in RD gene expression accompanies eye differentiation.
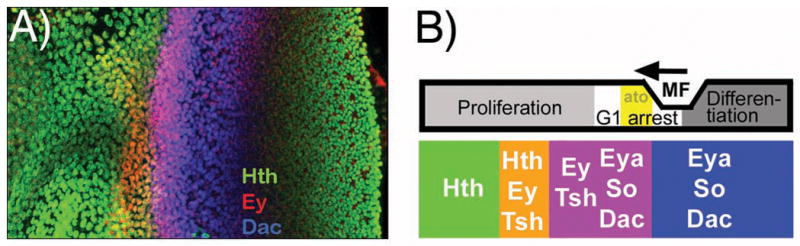
A) Examples of RD gene expression in the Drosophila eye imaginal disc as the wave of differentiation crosses the retinal epithelium. This eye disc has been immune-labelled to reveal overlapping, dynamic expression of three proteins, Homothorax (Hth: green), Eyeless (Ey: red), and Dachsous (Dac: blue).
B) Summary diagram correlating dynamic RD gene expression patterns with developmental processes in the eye imaginal disc. Cells mature from an undifferentiated, proliferative state through a cell cycle arrest that accompanies expression of the first fate specification genes such as the proneural gene atonal (ato) and is followed by progressive differentiation of the various retinal cell types. During this period, cells evolve from expression of the antennal selector gene homothorax and RD genes ey and tsh to expression of the retinal determination genes eya, so, and dac. Figure adapted from[40].
The detailed roles of the two states defined by the alternative RD gene expression remain surprisingly little known. The Hth gene is important in maintaining proliferation by the multipotent eye cells ahead of the furrow[37–39]. Perhaps Dac/Eya/So could be responsible for terminal eye differentiation, while the Ey/Tsh/Hth combination plays an earlier role either setting up this state, or preventing the Dac/Eya/So ‘differentiation field’ becoming active precociously, or perhaps simply regulates an unrelated set of earlier eye functions.
Retinal Determination gene expression is regulated dynamically by cell-cell signaling pathways in the eye
The transition in RD gene expression sweeps across the eye disc over time largely but not entirely as a response to Dpp signaling[24](see below). Dpp signaling also contributes to many other changes in gene expression, cell cycle regulation, and cellular morphology as the eye begins differentiation. Because these are precisely the eye field-specific transcriptional responses predicted to depend on the master regulatory gene context, it complicates the combinatorial concept if Dpp contributes to both RD gene expression and its outputs at the same time.
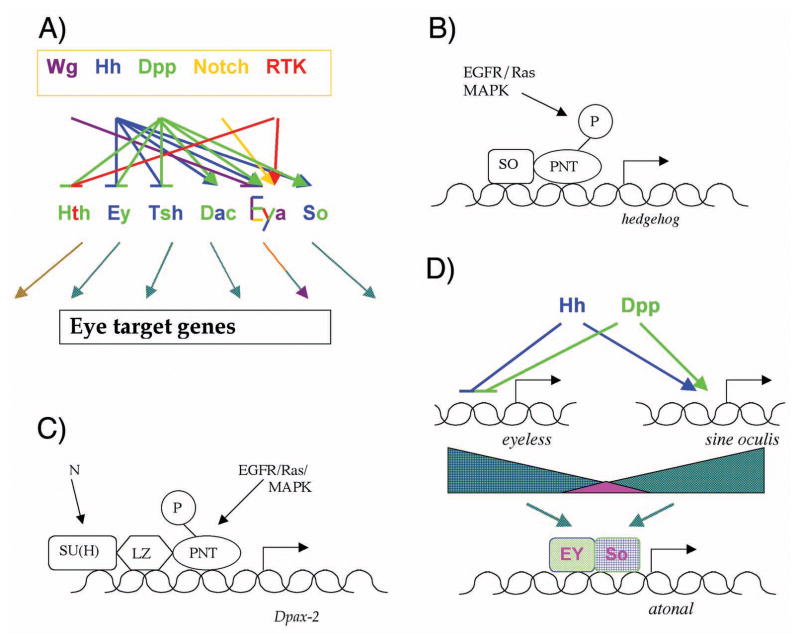
Dpp is not the only cell-cell signal regulating RD gene expression. The other major extracellular signaling pathways have roles too, contributing to dynamically shifting RD gene expression patterns during retinal development[40]. For example, Dpp signaling is not sufficient to eliminate all Ey expression, and loss of Dpp signaling only delays the loss of Ey. The additional, Dpp-independent regulation of Ey is due to Hh. Hh and Dpp together are required for completely normal regulation of Ey expression, and this combination of signals also controls induction of Dac. N, Wg, and the EGF Receptor pathway that signals through Ras also contribute to the spatial and temporal regulation of RD gene expression, and it is necessary to consider the effects of all these pathways to provide an account of the spatial and temporal regulation of RD gene expression[40]. A glance at the complexity of the summary of how cell-cell signaling pathways affect RD gene expression that is shown in Figure 4A suffices to appreciate multiple, complicated inputs.
Figure 4.
A) Recent studies describe a complex network of cell-cell signaling that controls the spatiotemporal pattern of RD gene expression during eye development. The point of the color-coded summary shown here is not to follow the individual interactions in detail, but simply to note that all of these five cell-cell signaling pathways contribute to the spatiotemporal regulation of the RD genes that was summarized in Figure 3. This summary is probably incomplete, for example Dpp and Wg signaling pathways often act antagonistically, but most of the targets of Dpp have not been tested for regulation by Wg[40]. The figure also vertically compresses regulation by extracellular signals into a single layer, although in reality regulation is rendered multi-layered by regulatory relationships between the RD genes themselves. For example, Dac is necessary and sufficient for tsh repression, so it is possible that Hh and Dpp may regulate tsh indirectly as a secondary target of dac, which requires Hh and Dpp signaling for its expression[40]. As a result of the extensive regulation, the dynamic RD gene expression patterns contain information about the extracellular signaling pathway activities, and which could be conferred on further target genes in turn.
B) Regulation of hedgehog. The enhancer responsible for eye expression is bound directly by Pnt, an Ets-domain transcription factor regulated by the EGFR/Ras/MAPK pathway of extracellular signals, and the RD protein SO[44]. This combinatorial scheme resembles the paradigm established for dorsal appendages (see Figure 1).
C) Regulation of shaven (dPax-2). The sparkling (spa) enhancer responsible for eye expression is bound directly by Pnt and by Su(H), the transcription factor regulated by Notch signaling, as well as the Runx-family transcription factor Lozenge{[47]. Lz is not an RD gene, but its expression in the eye is itself regulated by So, bringing spa under RD control indirectly[49].
D) Regulation of atonal. Enhancer sequences that would be direct targets of Hh and Dpp signaling have not been found[54,55]; the observed dependence of ato expression on Hh and Dpp may instead be explained by the Hh- and Dpp-dependence of ey and so expression. Hh and Dpp regulate both ey and so expression, repressing ey and turning so on [40]. This regulation is itself likely to be multi-layered, since Ey appears to be a direct regulator of so expression (not diagrammed)[31,33], and since it is not known whether Mad (or Brinker) and Ci proteins bind directly to enhancers of the ey and so genes, or act indirectly through intermediate transcription factors. The Ey and So proteins are coexpressed ahead of the morphogenetic furrow, Ey declining as So rises. Transcription of ato initiates within the overlap. It is interesting that ato is activated by transcription factors that are co-expressed only transiently; in principle this provides a mechanism for a transient stripe of ato activation. The ato enhancer is active in only a subset of the cells coexpressing Ey and So, however, and other data also suggest that additional factors are also likely to limit ato transcription[40].
In order to understand the role of RD genes it is useful to examine the regulation of some individual eye target genes. We will not dwell on the many known examples of cell-autonomous cross-regulation of one RD gene by another [24,31–35], focussing on the more downstream target genes hh, sv, and ato that are actual effectors of eye development and differentiation (Figure 4).
Direct, combinatorial regulation of hh transcription in the eye
Hh expression is the primary driver of morphogenetic furrow movement[41,42]. Hh is expressed in a subset of differentiating photoreceptor neurons. As differentiation advances across the eye, new regions of the retina begin differentiating and secrete Hh, promoting further advance of the furrow [41,42]. Although hh is also expressed in many other tissues[43], the expression of hh in photoreceptor cells serves as an example of an eye gene regulation.
Expression of hh in the eye is directed by a specific enhancer, deletion of which produces mutant alleles defective only for the eye function of Hh[44]. This expression, and activity of the enhancer, occurs in developing photoreceptor cells whose specification depends on EGF receptor signaling. Accordingly, the enhancer is bound by Pnt, the ETS-domain transcription factor that mediates positive responses to receptor tyrosine kinase signals. The enhancer also binds the RD protein So, which is required for enhancer activity[44]. Thus, this hh enhancer is consistent with the simple hypothesis that RD genes act in combination with cell-cell signals to confer eye expression, as Vg and the Hox genes are thought to act in other body regions. In this case, So, likely with its binding partners Eya and Dac, provides the cell intrinsic information that makes the hh gene responsive to EGF receptor signaling posterior to the morphogenetic furrow of the eye, but not elsewhere in the body.
The shaven gene is directly regulated by cell-cell signaling pathways
The shaven (sv) gene encodes the D-Pax2 transcription factor. In the eye, D-Pax2 is required to specify cone cells, a non-neuronal cell type[45]. A subset of sv alleles that specifically affect sv expression in the eye were originally described as sparkling (spa) mutations, before allelism with sv was determined[46]. Cone cell specification depends on the combination of EGF receptor and Notch signaling, and accordingly the sparkling enhancer contains binding sites for Pnt, the ETS domain transcription factor downstream of receptor tyrosine kinases, and Su(H), the transcription factor downstream of Notch. It is thought that sv is activated by a combinatorial code comprising Pnt, Su(H), and Lozenge[47]. Lozenge is a Runx-family transcription factor. Lz is not an RD gene, is not capable of inducing ectopic eyes when overexpressed, and functions in the Drosophila hematopoietic system as well as the eye[48]. In this sv enhancer, then, the input of the EGFR and N signals that induce cone cell specification is likely direct, but the source of the eye specificity, and if and how it depends on RD genes, is not immediately obvious.
One possibility is that the eye-specificity of this enhancer depends on Lz. Within the eye, lz is expressed in all retinal cells in the posterior, differentiating part of the eye disc, after proliferation ceases, and is not specific for cone cell precursors. This lz expression has been shown to depend on the earlier expression of So. Thus, So may regulate sv indirectly, using Lz as an intermediary[49]. The reason that sv is not expressed in the hematopoietic system requires more study. It may be that receptor tyrosine kinase and N signaling are never both active in lz-expressing cells of the hematopoietic system, that Lz is not the only factor required to confer eye specificity on the spa enhancer, or that another protein expressed in the hematopoietic system blocks spa expression there. The sparkling enhancer of sv has recently been examined in much greater detail, with the conclusion that activity and patterning depend on interaction between many regulatory sites in addition to those binding Pnt, Su(H), and Lz, whose cognate binding partners are not yet known[50]. This is consistent with the idea that another protein might collaborate with Lz to confer eye specificity on the spa enhancer.
The sv gene suggests that direct binding by RD proteins is not always essential for enhancer activity in the eye. Such enhancers could depend on RD proteins indirectly, through a chain of other transcription factors initiated by RD genes [49].
The atonal gene is directly regulated by RD gene products
The atonal (ato) gene encodes the proneural bHLH gene that initiates neurogenesis in the morphogenetic furrow and sets terminal retinal differentiation in motion (Figure 3B)[51]. Expression of Ato in the eye depends on two enhancers. One initiates expression as the morphogenetic furrow approaches; the second is required for ato-dependent autoregulation and stable maintenance of R8 cell fate[52]. Ato also functions as a proneural gene in other tissues, but although it was once suggested that the eye might follow a core neurogenic expression pathway also common to other tissues[53], the data that led to this conclusion have not been reproduced by other groups, who find a distinct enhancer that initiates ato expression in the eye[54,55].
The initiation of ato in the eye depends on Hh and Dpp signaling[56,57]. The familiar combinatorial paradigm predicts binding sites for the transcription factors that mediate Hh and Dpp signaling, and for RD proteins. The ato 3′ enhancer that is responsible for initiating ato expression does indeed contain binding sites for the RD proteins Ey and So, and both of these sites are essential for activity[54](Figure 4D). Surprisingly, the 3′ ato enhancer does not contain canonical binding sites for any of the transcription factors that normally mediate Hh and Dpp signaling (the Zn-finger containing Gli protein Ci for Hh, and the Mad or Brinker (Brk) proteins for Dpp). Does this imply that Hh and Dpp have other mechanisms of signal transduction in the eye, by-passing Ci and Mad? This is not likely, because ci and mad genes are required for ato expression and to regulate eye development[57,58]. Although it cannot yet be excluded that Mad, Brk or Ci bind to this enhancer via sequences that differ from their previously-identified binding sites, it seems likely that the input of Hh and Dpp is mediated by other proteins.
By analogy with spa, where the input of RD genes could be mediated indirectly through a chain of other factors, for ato it seems that the input of extracellular signals might be mediated indirectly. It is possible that the factor(s) responsible are not known, because the ato enhancer contains conserved sequences whose cognate binding factors are unidentified[54]. It is not necessary to propose further regulatory factors, however, if the RD proteins themselves are responsible. It turns out that Hh and Dpp, the two main extracellular signals that activate ato, also define the Ey and So expression patterns in the eye disc[40]. Hh and Dpp together are required to repress Ey as the morphogenetic furrow approaches, and Hh and Dpp together are required to turn So on. We suggest that because the Ey and So expression patterns are dependent on Hh and Dpp signaling, Ey and So may relay Hh-dependence and Dpp-dependence onto ato expression (Figure 4D).
This dual requirement for Ey and So in ato regulation is also interesting because Ey and So expression only overlap transiently (Figure 3B, 4D). Ey is expressed in the anterior eye, and repressed by Hh and Dpp signaling as the morphogenetic furrow approaches. By contrast, So is turned on by Hh and Dpp signaling[40]. Because the Ey and So binding sites are adjacent on the ato promoter, we first wondered whether Ey might repress ato, and So activate its expression, providing a sensitive Hh/Dpp-dependent switch that initiates ato expression close to the morphogenetic furrow. Data from Zhang et al refute this model unequivocally. Although adjacent, both the So and Ey binding sites are essential for the activity of the Ato enhancer[54].
Since Ey expression is repressed by the same signals that initiate expression of So, both proteins can only be present together transiently, and it is during this time that ato expression begins (Figures 3B, 4D). This may provide part of the explanation for why the ato enhancer is active in a stripe that does not resemble the broad, graded distribution of the parent Hh and Dpp signaling activities. Ey and So do not seem to be the sole factors activating ato expression because they overlap before ato expression is first detected and because they are not sufficient for ato expression in the absence of Hh and Dpp signaling[40], however, so additional activators or repressors may also participate in limiting ato enhancer activity to a transient stripe (Figure 4D).
Conclusions: A modified model for eye gene expression
It is thought that each developmental field may be characterized by one or more ‘master regulatory genes’ that render cells within it competent to make appropriate responses to the extracellular signals they encounter. It has even been suggested that cell-cell signaling pathways may be unable to regulate gene expression unless a master regulator protein is also present[13,14]. Master regulators such as Vestigial and the Hox genes have achieved this through combinatorial interactions at target gene promoters, enabling developmental field identity to be integrated with cell-cell signaling pathways at the level of transcription of target genes. It has been suggested that the RD gene master regulators for the eye act similarly[2,3]. We argue that this model remains adequate in the face of more recent data, with some modifications. Some eye enhancers respond only indirectly to RD genes or to cell-cell signaling pathways. Eye specificity can be conferred by a secondary transcription factor that is not itself an RD gene, but which is a primary target of RD genes. On the other hand, it looks as though the overlapping and dynamic expression patterns of the RD genes may confer spatiotemporal specificity on some enhancers, without a direct input from the extracellular signaling pathways themselves.
These mechanisms supplement the known combinatorial mechanism to expand the repertoire of eye-specific gene expression, providing control by extracellular signals and RD genes in an indirect and eye-specific manner. In this view, RD gene expression can best be described as a multi-layered network. Deep in the network, many eye genes will be secondary targets of RD genes and extracellular signals, directly regulated by transcription factors that are direct, primary targets. It will be interesting to consider what are the particular features or advantages of this multilayer network, and whether different types of network are prone to particular defects when the system breaks down, such as in cancers that overexpress RD genes[29,30].
A topic for the future concerns the molecular basis for combinatorial gene regulation between these classes of transcription factors, which has only been discussed at a formal level in this article. Physical interactions between protein complexes on the enhancer DNA could be responsible, collaborating to provide enhancer function, or providing complementary co-activator components to activate transcription. There has not been much direct study of these mechanisms yet reported with regard to gene targets of RD genes. A further possibility is that while some proteins bind enhancers to interact with the transcriptional machinery, others define open or closed chromatin states[59]. Chromatin states might differ more between determination states (such as eye and thorax) than in response to cell-cell signaling pathways. Recent studies of the eye enhancer of the prospero gene suggested that the RD gene So makes the enhancer accessible, while the extracellular signals EGFR and Notch more directly affect transcription initiation[60]. There is also evidence that Pax6 may regulate lens fibre differentiation in mammals via chromatin state[61]. If master regulators as a rule define accessible chromatin regions for each developmental field, then it will be interesting to determine whether proteins of the RD gene network have additional properties that facilitate a multilayered control network.
Acknowledgments
Research in our laboratory was supported by NIH grant GM47892. The Department of Ophthalmology and Visual Sciences is the recipient of an Unrestricted Grant from Research to Prevent Blindness. We thank Ales Cvekl, Claude Desplan, Jing-Dong Han and Jessica Treisman and anonymous reviewers for comments on an earlier version of the manuscript.
Abbreviations
- RD genes
Retinal Determination genes
Bibliography
- 1.Halder G, Callaerts P, Gehring WJ. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science. 1995;267:1788–92. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- 2.Curtiss J, Halder G, Mlodzik M. Selector and signalling molecules cooperate in organ patterning. Nat Cell Biol. 2002;4:E48–51. doi: 10.1038/ncb0302-e48. [DOI] [PubMed] [Google Scholar]
- 3.Mann RS, Carroll SB. Molecular mechanisms of selector gene function and evolution. Curr Opin Genet Dev. 2002;12:592–600. doi: 10.1016/s0959-437x(02)00344-1. [DOI] [PubMed] [Google Scholar]
- 4.Roignant J-Y, Treisman JE. Pattern formation in the Drosophila eye disc. Int J Dev Biol. 2009;53:795–804. doi: 10.1387/ijdb.072483jr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerhart J. Warkany lecture: signaling pathways in development. Teratology. 1999;60:226–39. doi: 10.1002/(SICI)1096-9926(199910)60:4<226::AID-TERA7>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 6.Pires-daSiva A, Sommer RJ. The evolution of signalling pathways in animal development. Nature Reviews Genetics. 2002;4:39–49. doi: 10.1038/nrg977. [DOI] [PubMed] [Google Scholar]
- 7.Wolpert L. Pattern formation in biological development. Scientific American. 1978;239:154–64. doi: 10.1038/scientificamerican1078-154. [DOI] [PubMed] [Google Scholar]
- 8.Davidson EH, Erwin DH. Gene regulatory networks and the evolution of animal body plans. Science. 2006;311:796–800. doi: 10.1126/science.1113832. [DOI] [PubMed] [Google Scholar]
- 9.Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–89. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 10.Halder G, Polaczyk P, Kraus ME, Hudson A, et al. The Vestigial and Scalloped proteins act together to directly regulate wing-specific gene expression in Drosophila. Genes Dev. 1998;12:3900–9. doi: 10.1101/gad.12.24.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mann RS, Morata G. The developmental and molecular biology of genes that subdivide the body of Drosophila. Annu Rev Cell Dev Biol. 2000;16:243–71. doi: 10.1146/annurev.cellbio.16.1.243. [DOI] [PubMed] [Google Scholar]
- 12.Affolter M, Mann R. Development. Legs, eyes, or wings--selectors and signals make the difference. Science. 2001;292:1080–1. doi: 10.1126/science.1060856. [DOI] [PubMed] [Google Scholar]
- 13.Furriols M, Bray S. A model Notch response element detects Suppressor of Hairless-dependent molecular switch. Current Biology. 2001;11:60–4. doi: 10.1016/s0960-9822(00)00044-0. [DOI] [PubMed] [Google Scholar]
- 14.Barolo S, Posakony JW. Three habits of highly effective signaling pathways: principles of transcriptional control by developmental cell signaling. Genes and Development. 2002;16:1169–81. doi: 10.1101/gad.976502. [DOI] [PubMed] [Google Scholar]
- 15.Burke R, Basler K. Hedgehog signalling in Drosophila eye and limb development - conserved machinery, divergent roles? Current Opinions in Neurobiology. 1997;7:55–61. doi: 10.1016/s0959-4388(97)80120-1. [DOI] [PubMed] [Google Scholar]
- 16.Benassayag C, Plaza S, Callaerts P, Clements J, et al. Evidence for a direct functional antagonism of the selector genes proboscipedia and eyeless in Drosophila head development. Development. 2003;130:575–86. doi: 10.1242/dev.00226. [DOI] [PubMed] [Google Scholar]
- 17.Wolff T, Ready DF. Pattern formation in the Drosophila retina. In: Bate M, Martinez Arias A, editors. The Development of Drosophila melanogaster. Cold Spring Harbor Laboratory Press; 1993. pp. 1277–325. [Google Scholar]
- 18.Nagaraj R, Canon J, Banerjee U. Cell fate specification in the Drosophila eye. Results and Problems in Cell Differentiation. 2002;37:73–88. doi: 10.1007/978-3-540-45398-7_6. [DOI] [PubMed] [Google Scholar]
- 19.Bonini NM, Bui QT, Gray-Board GL, Warrick JM. The Drosophila eyes absent gene directs ectopic eye formation in a pathway conserved between flies and vertebrates. Development. 1997;124:4819–26. doi: 10.1242/dev.124.23.4819. [DOI] [PubMed] [Google Scholar]
- 20.Pignoni F, Hu B, Zavitz KH, Xiao J, et al. The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell. 1997;91:881–91. doi: 10.1016/s0092-8674(00)80480-8. [DOI] [PubMed] [Google Scholar]
- 21.Chen R, Amoui M, Zhang Z, Mardon G. Dachshund and eyes absent proteins form a complex and function synergistically to induce ectopic eye development in Drosophila. Cell. 1997;91:893–903. doi: 10.1016/s0092-8674(00)80481-x. [DOI] [PubMed] [Google Scholar]
- 22.Ikeda K, Watanabe Y, Ohto H, Kawakami K. Molecular interaction and synergistic activation of a promoter by Six, Eya, and Dach proteins mediated through CREB binding protein. Mol Cell Biol. 2002;22:6759–66. doi: 10.1128/MCB.22.19.6759-6766.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim S-S, Zhang R, Braunstein SE, Joachlmlak A, et al. Structure of the retinal determination protein Dachshund reveals a DNA binding motif. Structure. 2002;10:787–95. doi: 10.1016/s0969-2126(02)00769-4. [DOI] [PubMed] [Google Scholar]
- 24.Bessa J, Gebelein B, Pichaud F, Casares F, et al. Combinatorial control of Drosophila eye development by Eyeless, Homothorax, and Teashirt. Genes and Development. 2002;16:2415–27. doi: 10.1101/gad.1009002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fabrizio JJ, Boyle M, DiNardo S. A somatic role for eyes absent (eya) and sine oculis (so) in Drosophila spermatocyte development. Dev Biol. 2003;258:117–28. doi: 10.1016/s0012-1606(03)00127-1. [DOI] [PubMed] [Google Scholar]
- 26.Sewell W, Williams T, Cooley J, Terry M, et al. Evidence for a novel role for dachshund in patterning the proximal arthropod leg. Dev Genes Evol. 2008;218:293–305. doi: 10.1007/s00427-008-0220-5. [DOI] [PubMed] [Google Scholar]
- 27.Estella C, Mann RS. Logic of Wg and Dpp induction of distal and medial fates in the Drosophila leg. Development. 2008;135:627–36. doi: 10.1242/dev.014670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cvekl A, Duncan MK. Genetic and epigenetic mechanisms of gene regulation during lens development. Progress in Retinal and Eye Research. 2007;26:555–97. doi: 10.1016/j.preteyeres.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Christensen KL, Patrick AN, McCoy EL, Ford HL. The six family of homeobox genes in development and cancer. Adv Cancer Res. 2008;101:93–126. doi: 10.1016/S0065-230X(08)00405-3. [DOI] [PubMed] [Google Scholar]
- 30.Popov VM, Wu K, Zhou J, Powell MJ, et al. The Dachshund gene in development and hormone-responsive tumorigenesis. Trends Endocrinol Metab. 2010;21:41–9. doi: 10.1016/j.tem.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Halder G, Callaerts P, Flister S, Walldorf U, et al. Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development. 1998;125:2181–91. doi: 10.1242/dev.125.12.2181. [DOI] [PubMed] [Google Scholar]
- 32.Czerny T, Halder G, Kloter U, Souabni A, et al. twin of eyeless, a second Pax-6 gene of Drosophila, acts upstream of eyeless in the control of eye development. Mol Cell. 1999;3:297–307. doi: 10.1016/s1097-2765(00)80457-8. [DOI] [PubMed] [Google Scholar]
- 33.Niimi T, Seimiya M, Kloter U, Flister S, et al. Direct regulatory interaction of the eyeless protein with an eye-specific enhancer in the sine oculis gene during eye induction in Drosophila. Development. 1999;126:2253–60. doi: 10.1242/dev.126.10.2253. [DOI] [PubMed] [Google Scholar]
- 34.Pauli T, Seimiya M, Blanco J, Gehring WJ. Identification of functional sine oculis motifs in the autoregulatory element of its own gene, in the eyeless enhancer, and in the signalling gene hedgehog. Development. 2005;132:2771–82. doi: 10.1242/dev.01841. [DOI] [PubMed] [Google Scholar]
- 35.Ostrin EJ, Li Y, Hoffman K, Liu J, et al. Genome-wide identification of direct targets of the Drosophila retinal determination protein Eyeless. Genome Res. 2006;16:466–76. doi: 10.1101/gr.4673006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Firth LC, Baker NE. Extracellular signals responsible for spatially regulated proliferation in the differentiating Drosophila eye. Dev Cell. 2005;8:541–51. doi: 10.1016/j.devcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 37.Peng HW, Slattery M, Mann RS. Transcription factor choice in the Hippo signaling pathway: homothorax and yorkie regulation of the microRNA bantam in the progenitor domain of the Drosophila eye imaginal disc. Genes Dev. 2009;23:2307–19. doi: 10.1101/gad.1820009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lopes CS, Casares F. hth maintains the pool of eye progenitors and its downregulation by Dpp and Hh couples retinal fate acquisition with cell cycle exit. Dev Biol. 2010;339:78–88. doi: 10.1016/j.ydbio.2009.12.020. [DOI] [PubMed] [Google Scholar]
- 39.Firth LC, Bhattacharya A, Baker NE. Cell cycle arrest by a gradient of Dpp signaling during Drosophila eye development. BMC Dev Biol. 2010;10:28. doi: 10.1186/1471-213X-10-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Firth LC, Baker NE. Retinal determination genes as targets and possible effectors of extracellular signals. Developmental Biology. 2009;327:366–75. doi: 10.1016/j.ydbio.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heberlein U, Wolff T, Rubin GM. The TGF-β homolog dpp and the segment polarity gene hedgehog are required for propagation of a morphogenetic wave in the Drosophila retina. Cell. 1993;75:913–26. doi: 10.1016/0092-8674(93)90535-x. [DOI] [PubMed] [Google Scholar]
- 42.Ma C, Zhou Y, Beachy PA, Moses K. The segment gene hedgehog is required for progression of the morphogenetic furrow in the developing Drosophila eye. Cell. 1993;75:927–38. doi: 10.1016/0092-8674(93)90536-y. [DOI] [PubMed] [Google Scholar]
- 43.Lee JJ, von Kessler DP, Parks S, Beachy PA. Secretion and localized transcription suggest a role in positional signalling for products of the segmentation gene hedgehog. Cell. 1992;71:33–50. doi: 10.1016/0092-8674(92)90264-d. [DOI] [PubMed] [Google Scholar]
- 44.Rogers EM, Brennan CA, Mortimer NT, Cook S, et al. Pointed regulates an eye-specific transcriptional enhancer in the Drosophila hedgehog gene, which is required for the movement of the morphogenetic furrow. Development. 2005;132:4833–43. doi: 10.1242/dev.02061. [DOI] [PubMed] [Google Scholar]
- 45.Fu W, Noll M. The Pax2 homolog sparkling is required for development of cone and pigment cells in the Drosophila eye. Genes Dev. 1997;11:2066–78. doi: 10.1101/gad.11.16.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fu W, Duan H, Frei E, Noll M. shaven and sparkling are mutations in separate enhancers of the Drosophila Pax2 homolog. Development. 1998;125:2943–50. doi: 10.1242/dev.125.15.2943. [DOI] [PubMed] [Google Scholar]
- 47.Flores G, Duan H, Yan H, Nagaraj R, et al. Combinatorial signaling in the specification of unique cell fates. Cell. 2000;103:75–85. doi: 10.1016/s0092-8674(00)00106-9. [DOI] [PubMed] [Google Scholar]
- 48.Daga A, Karlovich CA, Dumstrei K, Banerjee U. Patterning of cells in the Drosophila eye by Lozenge, which shares homologous domains with AML1. Genes Dev. 1996;10:1194–205. doi: 10.1101/gad.10.10.1194. [DOI] [PubMed] [Google Scholar]
- 49.Yan H, Canon J, Banerjee U. A transcriptional chain linking eye specification to terminal determination of cone cells in the Drosophila eye. Dev Biol. 2003;263:323–9. doi: 10.1016/j.ydbio.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 50.Swanson CI, Evans NC, Barolo S. Structural rules and complex regulatory circuitry constrain expression of a Notch- and EGFR-regulated eye enhancer. Dev Cell. 2010;18:359–70. doi: 10.1016/j.devcel.2009.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jarman AP, Grell EH, Ackerman L, Jan LY, et al. atonal is the proneural gene for Drosophila photoreceptors. Nature. 1994;369:398–400. doi: 10.1038/369398a0. [DOI] [PubMed] [Google Scholar]
- 52.Sun Y, Jan LY, Jan YN. Transcriptional regulation of atonal during development of the Drosophila peripheral nervous system. Development. 1998;125:3731–40. doi: 10.1242/dev.125.18.3731. [DOI] [PubMed] [Google Scholar]
- 53.Niwa N, Hiromi Y, Okabe M. A conserved developmental program for sensory organ formation in Drosophila melanogaster. Nat Genet. 2004;36:293–7. doi: 10.1038/ng1308. [DOI] [PubMed] [Google Scholar]
- 54.Zhang T, Ranade S, Cai CQ, Clouser C, et al. Direct control of neurogenesis by selector factors in the fly eye: regulation of atonal by Ey and So. Development. 2006;133:4881–9. doi: 10.1242/dev.02669. [DOI] [PubMed] [Google Scholar]
- 55.Tanaka-Matakatsu M, Du W. Direct control of the proneural gene atonal by retinal determination factors during Drosophila eye development. Dev Biol. 2008;313:787–801. doi: 10.1016/j.ydbio.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Greenwood S, Struhl G. Progression of the morphogenetic furrow in the Drosophila eye: the roles of Hedgehog, Decapentaplegic and the Raf pathway. Development. 1999;126:5795–808. doi: 10.1242/dev.126.24.5795. [DOI] [PubMed] [Google Scholar]
- 57.Curtiss J, Mlodzik M. Morphogenetic furrow initiation and progression during eye development in Drosophila: the roles of decapentaplegic, hedgehog and eyes absent. Development. 2000;127:1325–36. doi: 10.1242/dev.127.6.1325. [DOI] [PubMed] [Google Scholar]
- 58.Fu W, Baker NE. Deciphering synergistic and redundant roles of Hedgehog, Decapentaplegic and Delta that drive the wave of differentiation in Drosophila eye development. Development. 2003;130:5229–39. doi: 10.1242/dev.00764. [DOI] [PubMed] [Google Scholar]
- 59.Zaret KS, Watts J, Xu J, Wandzioch E, et al. Pioneer factors, genetic competence, and inductive signaling: programming liver and pancreas progenitors from the endoderm. Cold Spring Harb Symp Quant Biol. 2008;73:119–26. doi: 10.1101/sqb.2008.73.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hayashi T, Xu C, Carthew RW. Cell-type-specific transcription of prospero is controlled by combinatorial signaling in the Drosophila eye. Development. 2008;135:2787–96. doi: 10.1242/dev.006189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cvekl A, Mitton KP. Epigenetic regulatory mechanisms in vertebrate eye development and disease. Heredity. 2010;105:135–51. doi: 10.1038/hdy.2010.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Simon MA. Receptor tyrosine kinases: specific outcomes from general signals. Cell. 2000;103:13–5. doi: 10.1016/s0092-8674(00)00100-8. [DOI] [PubMed] [Google Scholar]
- 63.Nolo R, Abbot LA, Bellen HJ. Senseless, a Zn finger transcription factor, is necessary and sufficient for sensory organ development in Drosophila. Cell. 2000;102:349–62. doi: 10.1016/s0092-8674(00)00040-4. [DOI] [PubMed] [Google Scholar]