Abstract
The use of targeted therapeutics directed against BRAFV600-mutant metastatic melanoma improves progression-free survival in many patients; however, acquired drug resistance remains a major medical challenge. By far, the most common clinical resistance mechanism involves reactivation of the MAPK (RAF/MEK/ERK) pathway by a variety of mechanisms. Thus, targeting ERK itself has emerged as an attractive therapeutic concept, and several ERK inhibitors have entered clinical trials. We sought to preemptively determine mutations in ERK1/2 that confer resistance to either ERK inhibitors or combined RAF/MEK inhibition in BRAFV600-mutant melanoma. Using a random mutagenesis screen, we identified multiple point mutations in ERK1 (MAPK3) and ERK2 (MAPK1) that could confer resistance to ERK or RAF/MEK inhibitors. ERK inhibitor–resistant alleles were sensitive to RAF/ MEK inhibitors and vice versa, suggesting that the future development of alternating RAF/MEK and ERK inhibitor regimens might help circumvent resistance to these agents.
Introduction
BRAF is mutated in approximately 50% of melanomas, resulting in constitutive activation of the MAPK pathway ((B)RAF–MEK–ERK; ref. 1). Inhibitors targeting RAF and MEK (a key downstream effector kinase in the pathway) improve the survival of patients with BRAF-mutant melanoma, and are thus used clinically for this indication (2–4). In particular, combined RAF/MEK inhibition appears to improve progression-free survival compared with RAF or MEK monotherapy (2, 5). Despite these therapeutic successes, nearly all patients develop progressive disease that shows resistance to these agents (2, 6).
Multiple studies describing mechanisms of resistance to RAF/MEK inhibition have been published. Reactivation of the MAPK pathway through a variety of means, including alternatively spliced BRAF, NRAS or MEK1/2 mutations, MAP3K8 upregulation, or receptor tyrosine kinase signaling, is the most common clinical drug resistance mechanism (7–11). Because all of these alterations converge on sustained activation of ERK, the clinical development of small-molecule ERK inhibitors is of considerable interest. More generally, ERK signaling represents a key downstream effector of RAS mutations in many cancer types, suggesting that ERK inhibitors might eventually have multiple indications in oncology.
ERK1 and ERK2 proteins are 84% identical and comprise the only known substrates of MEK. MEK activity on ERK results in dual phosphorylation on the Thr-Glu-Tyr motif of ERK1T202/Y204 and ERK2T185/Y187 that fully activates ERK kinase activity. Conversely, ERKs are negatively regulated by dephosphorylation, which is accomplished by dual specificity phosphatases (DUSP; refs. 12, 13). DUSPs also become induced by ERK signaling, creating a negative feedback loop (14). Other downstream ERK effectors include kinases such as RSK and MSK, cytoskeletal molecules, nucleoporins, and transcription factors (e.g., c-FOS, ELK-1, ETS-1, and MITF). Oncogenic dysregulation of this ERK program may profoundly impact cell proliferation and survival (15).
Several small-molecule ERK inhibitors have entered clinical trials (16, 17). Given the importance of secondary kinase mutations as resistance mechanisms in many oncogene-driven cancers, we wished to discover mutations in ERK that confer resistance to ERK inhibitors. We reasoned that at least some such ERK mutations might also confer resistance to RAF/MEK inhibitors. (In this regard, these studies might also identify constitutively active variants of ERK, which have remained elusive despite intensive study.) Random mutagenesis screens have successfully identified clinically relevant resistance alleles (18, 19) in multiple kinase oncogene-driven malignancies. Thus, we employed random mutagenesis in BRAF-mutant melanoma cells to identify mutations in ERK1 or ERK2 that could confer resistance to MAPK inhibitors.
Materials and Methods
Cell lines and reagents
A375, 293T, SKMEL-19, and WM266.4 cells were grown in DMEM with 10% FBS. A375 and 293T cells were acquired from the ATCC. WM266.4 cells were acquired through the Cancer Cell Line Encyclopedia (20). SKMEL-19 cells were a gift from N. Rosen (Memorial Sloan Kettering Cancer Center, New York, NY). GSK1120212, GSK2118436, VX-11e, and AZD6244 were obtained from Chemietek. PLX4720 was obtained from Selleck. SCH772984 was synthesized by J & W PharmLab. Lentiviral production and infection were performed as previously described (19).
Random mutagenesis
Mutagenesis screens were performed as described previously (19) with additional detail in Supplementary Materials and Methods.
Drug screens with mutagenized libraries
A375 cells (8 × 106) expressing tet-inducible GFP, wild-type ERK1/ERK2, or mutant library ERK1/ERK2 were plated in T150 flasks with VX-11e (2 μmol/L), trametinib (3 nmol/L), or trametinib + dabrafenib (1 nmol/L + 10 nmol/L) and doxycyline (DOX; 1 μg/mL) for 2 to 4 weeks until resistant cells emerged. At that time, genomic DNA (gDNA) was isolated (DNeasy kit; Qiagen). Exogenous ERK1/2 was amplified by the PCR using vector-specific primers (Supplementary Materials and Methods) and AccuPrime PFX supermix (Life Technologies). Individual colonies were collected and transferred into 96-well plates and expanded. Genomic DNA was isolated, and PCR was performed as above, then analyzed by Sanger sequencing.
Library generation and massively parallel sequencing
Massively parallel sequencing of PCR products from muta-genesis screens was performed as described previously (19). Additional detail is provided in Supplementary Materials and Methods.
Viability assays
A375, WM266.4, or SKMEL-19 cells were plated in 96-well plates, with six replicates for each drug. VX-11e (2 μmol/L), trametinib (3 nmol/L), dabrafenib (50 nmol/L), trametinib + dabrafenib (1 nmol/L + 10 nmol/L), or SCH772984 (1 μmol/L or 0.5 μmol/L) were added 24 hours later with or without DOX (1 μg/mL). Drugs were diluted 1:1,000 from stock concentrations in DMSO. Viability was analyzed 72 to 96 hours later by CellTiter 96 AQueous One Solution Cell Proliferation assay (Promega). The raw absorbance values were normalized to vehicle-treated controls after background subtraction and graphed as percent viability, with vehicle-treated cells at 100%. Heat maps were created from normalized viability values using GENE-E (21).
Western blotting
A375 cells were exposed to DOX with or without VX-11e (2 μmol/L), or trametinib (3 nmol/L), then harvested in RIPA buffer, and run on SDS-PAGE. Proteins were analyzed using the following antibodies: phosphoERK1T202/Y204/ERK2T185/Y187, ERK1/2, phosphoELK-1S383 (Santa Cruz Biotechnology), phosphoRSKS380, RSK, tubulin, DUSP6 (Cell Signaling Technology), cyclin D1 (Thermo Scientific), and V5 epitope (Abcam). Relative expression was calculated by dividing the relative densities of each phoshoELK-1 band by total ERK (or V5) using Image J. Values were normalized to untreated wild-type, which was set at 1.
Immunoprecipitation kinase assays
Described in detail in Supplementary Materials and Methods.
Site-directed mutagenesis
Primers were designed using the QuickChange Site-Directed Mutagenesis Protocol (Agilent) and are listed in Supplementary Materials and Methods. PCR was performed using iProof DNA polymerase and GC buffer using primers specific for each mutation (Bio-Rad). Mutations were confirmed by Sanger sequencing using vector-specific primers.
Aside from the mutagenesis screens, all experiments were repeated at least three times, with representative graphs or blots shown. Error bars represent SDs.
Results
ERK1/ERK2 random mutagenesis
To discover mutations in ERK1 or ERK2 that may confer resistance to MAPK inhibitors, a six-part random mutagenesis screen was performed. Here, we mutagenized ERK1 and ERK2 within the pDONR223 vector using a mutator strain of E. coli (22). The resulting mutagenized cDNA library was transferred into a DOX-inducible vector (pCW57.1), which was infected into A375 (BRAF-mutant) melanoma cells that are sensitive to MAP kinase pathway inhibitors. These cells were cultured in the presence of either the ERK inhibitor VX-11e (23), the MEK inhibitor trametinib (GSK1120212), or the combination of trametinib and the RAF inhibitor, dabrafenib (GSK2118436), in the presence of DOX. In all cases, drug-resistant A375 daughter populations emerged within 2 to 4 weeks. Genomic DNA was isolated, and ERK1 or ERK2 cDNA was amplified by the PCR followed by massively parallel sequencing (see Materials and Methods). In parallel experiments, a series of VX-11e– resistant ERK2 colonies were isolated, and ERK2 cDNA was analyzed by Sanger sequencing.
Overall, we identified 33 putative resistance variants spanning 28 amino acids in ERK1, and 24 substitutions affecting 20 amino acids in ERK2 (above a 5% massively parallel sequencing false discovery rate; see Supplementary Materials and Methods; Fig. 1; Supplementary Fig. S1A–F; and Supplementary Table S1). Sanger sequencing of 23 VX-11e–resistant colonies identified seven ERK2 mutations in five amino acids, two of which were also identified in the mutagenesis screens. We have no evidence that these mutagenesis screens were saturating; therefore, additional candidate ERK1/2 resistance mutations may remain to be identified. Nonetheless, our screening results suggested that a diverse range of ERK mutations might be associated with resistance to these inhibitors.
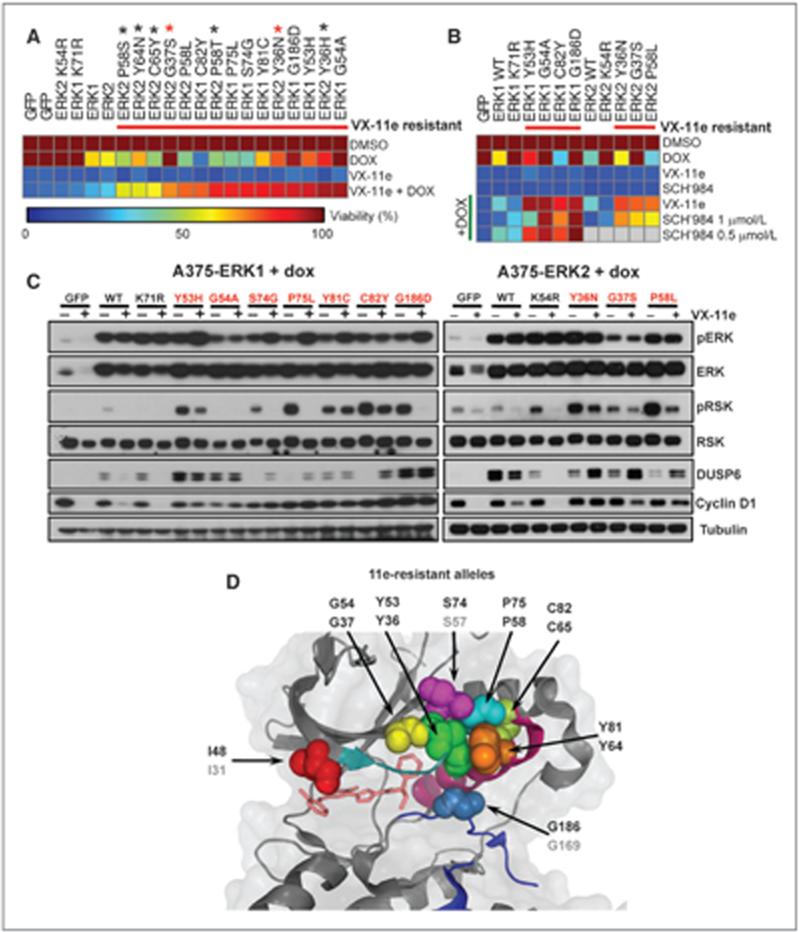
Figure 1.
The landscape of ERK1 and ERK2 resistance alleles following ERK or RAF/MEK inhibitor mutagenesis screens. A and B, the frequency of mutations from the ERK1 (A) and ERK2 (B) random mutagenesis screens is graphed at cDNA base pair resolution for VX-11e (red), trametinib (GSK′212; blue), and trametinib + dabrafenib (′212 + ′436; green). Amino acid substitutions for the most recurrently mutated nucleotides are highlighted in red (ERK inhibitor), blue (MEK inhibitor), and green (RAF + MEK inhibitor). Some amino acids were identified by more than one drug screen (black). Analogous alterations between ERK1 and ERK2 are underlined. C and D, Venn diagram depicting the overlap of all significantly mutated nucleotides in ERK1 or ERK2 from the ERK inhibitor screen (C) or the single-agent MEK and combined RAF/MEK inhibitor screens (D).
We observed five analogous resistance alleles between ERK1 and ERK2 in cells resistant to the ERK inhibitor in both the screen and in isolated colonies (ERK1Y53H/ERK2Y36H/N, ERK1G54A/ERK2G37S, ERK1P75L/ERK2P58L, ERK1Y81C/ ERK2Y64N, and ERK1C82Y/ERK2C65Y; Fig. 1C), and one analogous residue in MEK inhibitor–resistant cells (ERK1Y148H and ERK2Y131N/H/F/C/S; Fig. 1D), suggesting convergent resistance mechanisms between the ERK isoforms. There was relatively little overlap between the ERK inhibitor screens and the RAF/MEK inhibitor screens (three overlapping alleles of 67 total significant nucleotide alterations), raising the possibility that mutations identified through RAF/MEK inhibitor treatment might be resistant to one or both of these agents, but not to ERK inhibitors (and vice versa). Several ERK codons had multiple distinct amino acid substitutions (these include ERK1Y53C/H, ERK1G54A/S, ERK2Y36N/H, and ERK2P58L/S/T), suggesting the importance of these residues for inhibition by ATP-competitive ERK inhibitors.
In general, candidate ERK1/2 resistance mutations arising in the ERK inhibitor resistance screen clustered within the ATP/ drug binding pocket, suggesting that these variants might interfere with drug binding. In contrast, the RAF/MEK inhibitor resistance alleles were distributed throughout the ERK proteins, although some mutations clustered in the αC-helix and the common docking domain. The ERK αC-helix undergoes a conformational change when phosphorylated that is necessary for kinase activation, whereas the common docking domain represents one of the ERK substrate binding domains. These observations suggested that ERK resistance alleles that emerged under RAF/MEK inhibition might facilitate kinase activation. In addition, several of these alterations have been previously identified through DNA sequencing efforts. Alterations at ERK1R84, ERK1G186, ERK2D321, and ERK2E322 are listed in the COSMIC database (24), and ERK2E322K is recurrently mutated in cervical and head and neck cancers (25, 26); however, none of these variants have been linked to drug resistance.
ERK1 and ERK2 mutations confer resistance to ERK inhibitors by interfering with drug binding
We first characterized ERK1/2 mutations that arose in the setting of ERK inhibition. To verify their resistance effects, we engineered mutations into the DOX-inducible vector by site- directed mutagenesis. The resulting constructs were stably introduced into A375 cells and evaluated for growth in the presence of VX-11e. In total, we tested 20 candidate DOX-inducible ERK1/2 resistance alleles in cell growth inhibition assays, of which 16 conferred >4-fold increased cell viability compared with cells cultured without DOX (Fig. 2A; Supplementary Table S1; Supplementary Fig. S1G). In these experiments, wild-type ERK1 and ERK2 conferred a minor growth advantage (1.5-fold) in the presence of the ERK inhibitor, whereas kinase-dead ERK1 and ERK2 (27) and GFP had no effect (Fig. 2A).
Figure 2.
Pharmacologic and biochemical validation of ERK resistance alleles from ERK inhibitor screens. A, viability of A375 cells expressing tet-inducible ERK inhibitor resistance mutations exposed to VX-11e with or without DOX is shown as a heat map. GFP, wild-type ERK1/2, and kinase-dead ERK1K71R/ERK2K54R served as controls. Alleles are sorted by sensitivity to VX-11e + DOX. ERK2 alleles identified by sequencing drug-resistant colonies (black asterisks), or in both drug-resistant colonies and the ERK inhibitor random mutagenesis screen (red asterisks), are indicated. B, A375 cells expressing ERK inhibitor resistance mutations were treated as above except with SCH722984 (SCH'984). Viability was depicted as in A. Gray squares indicate mutations that were not tested. C, expression of phospho-ERK (pERK), phospho-RSK (pRSK), DUSP6, and cyclin D1 was examined in A375 cells expressing ERK resistance mutations or controls. Cells were treated with DOX in the presence or absence of VX-11e. D, structural localization of validated ERK1 and ERK2 mutations (spheres) mapped onto available ERK2 crystal structure data (cartoon) cocrystallized with VX-9a, a parent compound of VX-11e [PDB:3I60]. The glycine-rich loop (teal), the αC-helix (magenta), and the activation loop (blue) are indicated.
To determine whether these ERK1 and ERK2 resistance alleles might confer cross-resistance to other ERK inhibitors, we also performed cell growth inhibition assays using the ERK inhibitor SCH772984. For ERK1, we chose four alleles that conferred growth in the presence of the VX-11e, three of which conferred maximal growth (>90%) and one that conferred moderate resistance (>75%). We also included the three ERK2 alleles identified by the random mutagenesis screen. Indeed, these alleles were also resistant to SCH772984, as measured by >4-fold increases in viability following mutant allele expression. One allele, ERK1Y53H, showed only a 2.5-fold increase in viability at 1 μmol/L SCH772984, but conferred a 5-fold growth advantage at 0.5 μmol/L SCH772984, similar to the other ERK1 alleles (Fig. 2B). Conceivably, this difference may relate to compound-specific differences in ERK binding associated with this particular residue.
Next, we examined downstream ERK signaling from ten validated alleles upon DOX induction. As a control, we included wild-type ERK1 and ERK2, both of which produced robust RSK phosphorylation and elevated DUSP6 expression in this setting (Supplementary Fig. S2). In contrast, kinase-dead ERK1 and ERK2 showed no increased RSK phosphorylation, but did induce DUSP6 expression, suggesting a possible kinase-independent function (Supplementary Fig. S2). In the absence of ERK inhibitor, all validated ERK resistance alleles examined augmented RSK phosphorylation, DUSP6 expression, or both (Supplementary Fig. S2).
Although the ERK inhibitor–resistant ERK alleles conferred growth advantage in the presence of 11e, some alleles produced discordant effects on ERK signaling. For example, both ERK1G54A and the analogous variant ERK2G37S had minimal effects on RSK phosphorylation following DOX induction, but produced elevated DUSP6 expression (similar to the kinase-dead ERK variants). In contrast, ERK1S74G, ERK1P75L, and the orthologous allele ERK2P58L had modest effects on DUSP6 expression but robust upregulation of RSK phosphorylation. Thus, although some ERK resistance alleles may only partially augment downstream signaling, they may still confer a growth advantage in the setting of ERK inhibition.
Next, we analyzed ERK signaling from these resistance alleles upon addition of ERK inhibitors. All ten ERK inhibitor resistance alleles enabled sustained ERK signaling based on RSK phosphorylation, accumulation of DUSP6, or both (Fig. 2C). Cyclin D1, another downstream effector of ERK signaling, was also maintained in the presence of drug compared with controls. In addition, ERK signaling was maintained in the presence of SCH772984 (Supplementary Fig. S3A). In contrast to VX-11e, which does not alter phosphorylation of ERK (Fig. 2), SCH772984 decreased phosphorylation of wild-type ERK1/2, while having no effect on phosphorylation of the ERK2-resistant alleles (Supplementary Fig. S3A). Thus, these resistance alleles can maintain ERK signaling in the presence of an ERK inhibitor.
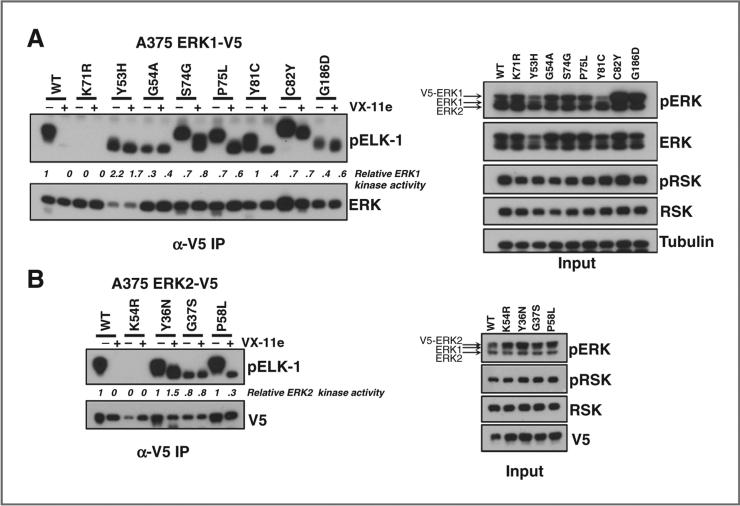
To determine whether the intrinsic kinase activity of the ERK1/2 alleles was maintained with inhibition of ERK, we examined the kinase activity of the ERK alleles via phosphor-ylation of an Elk-1 peptide using immunoprecipitation (IP) kinase assays and immunoblotting (see Materials and Methods). As expected, VX-11e blocked wild-type ERK1- and ERK2-mediated ELK-1 phosphorylation. In contrast, the ERK resistance alleles showed persistent ERK kinase activity in the presence of either VX-11e (Fig. 3A and B) or SCH772984 (Supplementary Fig. S3B). As expected, kinase-dead ERK1/2 did not phosphorylate the ELK-1 peptide (Fig. 3A and B). Thus, validated ERK resistance alleles maintained kinase activity even with concomitant ERK inhibitor exposure.
Figure 3.
Kinase activity of ERK mutants from ERK inhibitor resistance screens. A and B, the kinase activity of ERK1 (A) and ERK2 (B) was examined in lysates from A375 cells expressing validated ERK inhibitor resistance alleles or controls (wild-type and kinase-dead ERK1K71R/ERK2K54R) in the absence or presence of VX-11e using IP kinase assays. pERK and pRSK expression in the input was analyzed by immunoblotting.
Finally, we mapped the validated ERK1 and ERK2 alleles within the three-dimensional crystal structure of ERK2 bound to a structurally similar ERK inhibitor (VX-9a, PDB:3I60; ref. 23). The resistance alleles lie in close proximity to the ATP/drug binding pocket, albeit within distinct functional domains (Fig. 2D). In particular, they occupy the glycine-rich loop (ERK1I48N, ERK1Y53H, ERK1G54A, ERK2Y36N/H, and ERK2G37S), an interval between the b3-strand and αC-helix (ERK1S74G, ERK1P75L, and ERK2P58L), the αC-helix itself (ERK1Y81C, ERK1C82Y, ERK2Y64N, and ERK2C65Y), and the activation loop (ERK1G186D). Presumably, these substitutions interfere with engagement of the ATP binding cleft by ERK inhibitors.
ERK mutations that confer resistance to RAF/MEK inhibition preserve ERK activity despite MEK inhibition
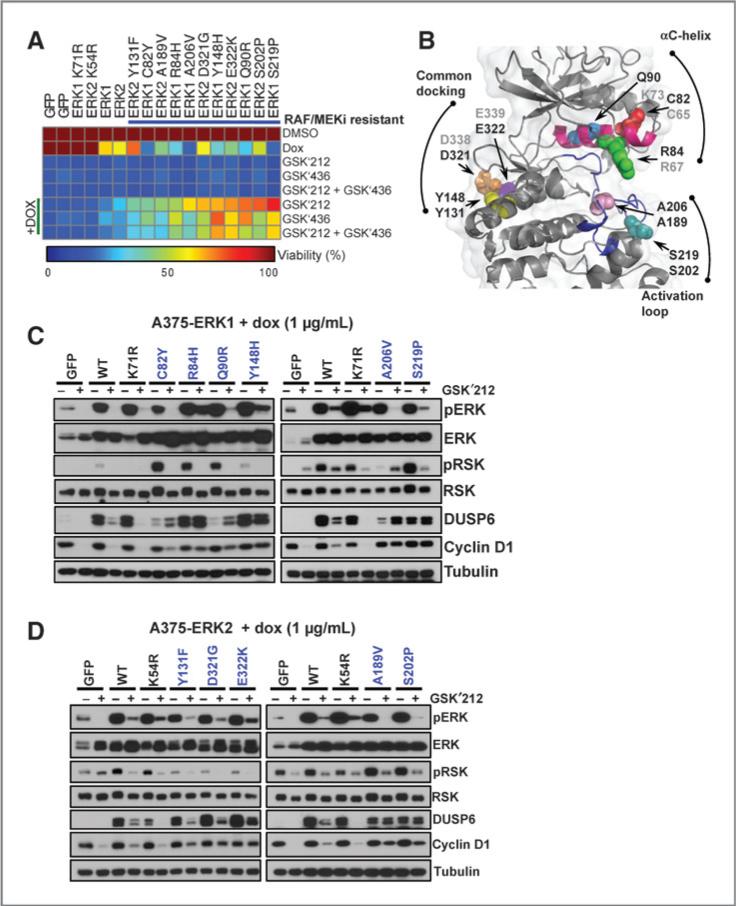
Next, we validated a series of candidate ERK resistance alleles that emerged from the RAF/MEK inhibitor mutagenesis screens. After cloning and expressing these mutations as described above, we performed cell growth inhibition assays using the MEK inhibitor trametinib and the RAF inhibitor dabrafenib, either alone or in combination. Of 17 mutated alleles examined, 10 were confirmed as resistant to each agent and the combination, conferring >2.5-fold growth advantage in the presence of the inhibitor(s) compared with controls (Fig. 4A; Supplementary Table S2). Of the 7 alleles that failed validation, at least two may have been passenger mutations that co-occurred with bona fide resistance alleles during the mutagenesis procedure (See Supplementary Materials and Methods and Supplementary Fig. S4). Because we observed robust growth advantage for two ERK1 alleles (ERK1A206V and ERK1S219P), we engineered the analogous mutations within ERK2 (because these were not observed in the primary screen). The resulting alleles (ERK2A189V and ERK2S202P) also conferred resistance to RAF/MEK inhibition (Fig. 4A).
Figure 4.
Pharmacologic and biochemical validation of ERK mutations that confer resistance to RAF/MEK inhibitors. A, A375 cells expressing tet-inducible RAF/MEK inhibitor resistance mutations were analyzed for viability in the presence of trametinib (GSK′212), dabrafenib (GSK′436), or trametinib + dabrafenib (GSK′212 + GSK′436), with or without DOX. Cell viability was normalized to DMSO, and values are depicted in the heat map. Alleles are sorted by sensitivity to trametinib + DOX. GFP, wild-type ERK1/2, and kinase-dead ERK1K71R and ERK2K54R served as controls. B, structural localization of validated RAF/MEK inhibitor–resistant alleles (spheres) mapped onto the ERK2 crystal structure (cartoon; PDB: 2ERK). The αC-helix (magenta) and the activation loop (blue) are labeled. The nonvalidating/ untested analogous ERK1/2 alleles are labeled in gray. C and D, cells expressing validated ERK1 (C) or ERK2 (D) RAF/MEK inhibitor resistance alleles were exposed to DOX with or without trametinib and pERK, pRSK, DUSP6, and cyclin D1 expression was analyzed by immunoblotting.
The observation that validated ERK1/2 alleles conferred cross-resistance to all RAF/MEK inhibitor conditions raised the possibility that in contrast to the ERK inhibitor resistance mutations (which may primarily interfere with inhibitor binding), these might represent activating events. This notion was buttressed by the observation that the ERK2E322K mutation—a known activating allele that occurs in cervical cancers (26)— also arose in this context. Moreover, ERK1/2A206V/A189V and ERK1/2S219P/S202P occur in the activation lip of ERK1/2 and in close proximity to the two phosphorylation residues that confer full ERK kinase activity when phosphorylated (ERK1T202/Y204 and ERK2T185/Y187; Fig. 4B and Supplementary Fig. S5A). Other validated RAF/MEK inhibitor resistance alleles map to the αC-helix (ERK1C82Y, ERK1R84H, and ERK1Q90R) and common docking domain (ERK1Y148H, ERK2Y131F, ERK2D321G, and ERK2E322K). Also known as the D-site recruitment site, this docking domain represents the means by which ERK1/2 binds to some substrates. Mutations in the common docking domains at ERK2D321N and ERK2E322K confer elevated activity in vivo due to reduced DUSP binding (28, 29). Alterations at ERK1/2Y148/Y131 have not been described, but may in principle impair phosphatase and substrate binding considering their three-dimensional localization near the common docking domain (Supplementary Fig. S5B).
Although our mutagenesis screens were unlikely to be saturating, we only observed αC-helix resistance mutations in ERK1, whereas common docking domain alterations (ERK2D321G and ERK2E322K) were only found in ERK2 (Fig. 4B). Interestingly, the analogous common docking domain mutations in ERK1 (ERK1D338N and ERK1E339K) did not confer resistance to RAF/MEK inhibition in our hands (Supplementary Fig. S6). This suggests that although ERK1 and ERK2 are highly homologous, “activating” variants of ERK1 and ERK2 may contain distinctive functional attributes, thus allowing dissection of isoform-specific functions in the future.
Next, we analyzed ERK signaling after expression of the validated alleles. As with the ERK inhibitor resistance mutations, DOX-induced expression of RAF/MEK inhibitor resistance alleles in both ERK1 and ERK2 activated ERK signaling, as evidenced by RSK or DUSP6 expression (Supplementary Fig. S2). The common docking domain mutations (e.g., ERK2D321G and ERK2E322K) did not induce RSK phosphorylation, consistent with published observations that such alterations may disrupt RSK binding (30). When these same ERK mutations were expressed in the presence of the MEK inhibitor, DUSP6 and cyclin D1 expression were maintained, whereas RSK phosphorylation was more variable (Fig. 4C and D). Thus, ERK mutations arising in the setting of RAF/MEK inhibition could maintain ERK signaling, even in the presence of upstream pathway inhibition.
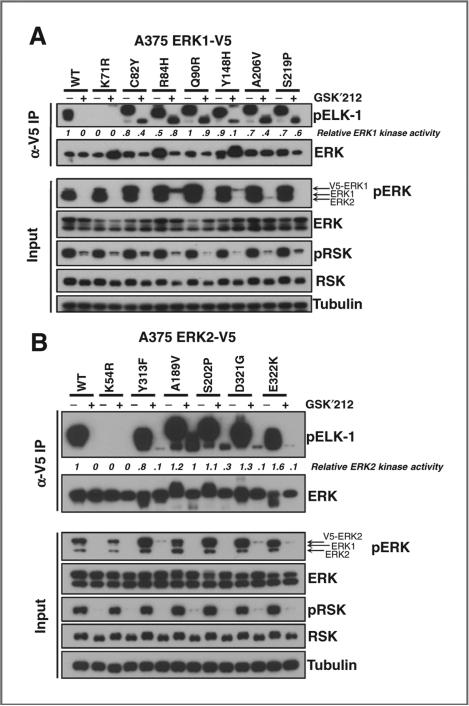
To determine whether the intrinsic kinase activity of the RAF/MEK inhibitor–resistant ERK alleles was maintained despite MEK inhibitor exposure, we analyzed ERK kinase activity by IP kinase assays. Here again, all ERK resistance alleles examined maintained ERK kinase activity (as measured by ELK-1 peptide phosphorylation) in the presence of MEK inhibitor, even though MEK-dependent (and activating) ERK phosphorylation was decreased (Fig. 5, input). ELK-1 binds to both the F-site recruitment motif and the common docking domain within ERK. Therefore, mutations confined to the common docking domain (ERK1Y148/ERK2Y131, ERK2D321G, ERK2E322K) should not be sufficient to abolish ERK-ELK-1 interactions (30, 31). However, we observed diminished residual kinase activity in the setting of trametinib exposure with the common docking domain mutants compared with other alleles (Fig. 5). Altogether, these results suggest that ERK resistance alleles arising in the setting of RAF/MEK inhibition maintain sufficient kinase activity to rescue BRAF-mutant melanoma cell growth—even in the presence of upstream pathway inhibition.
Figure 5.
Kinase activity of ERK mutants from RAF/MEK inhibitor resistance screens. A and B, the kinase activity of ERK1 (A) and ERK2 (B) was examined in lysates from A375 cells expressing validated RAF/MEK inhibitor resistance alleles or controls (wild-type and kinase-dead ERK1K71R/ERK2K54R) treated with trametinib (GSK′212). pERK and pRSK were monitored in the input cell lysates by immunoblotting.
RAF/MEK inhibitor–derived ERK resistance alleles are sensitive to ERK inhibition and vice versa
Previous studies have shown that several clinically validated genetic mechanisms of resistance to RAF and MEK inhibitors remain sensitive to ERK inhibition (Supplementary Fig. S7A; refs. 7, 8, 32). In our hands, RAF or MEK inhibitor–resistant BRAF-mutant melanoma cell populations generated by continuous exposure to tool compound inhibitors (PLX-4720 and AZD6244, respectively) following N-ethyl-N-nitrosourea (ENU) mutagenesis were also sensitive to ERK inhibition (Supplementary Fig. S7B). Because ERK resistance mutants arising from ERK inhibitor screens were largely distinct from those that emerged from the RAF/MEK inhibitor screens, we wished to determine whether either category of ERK mutations might confer cross-resistance to the alternative MAP kinase pathway inhibitor(s).
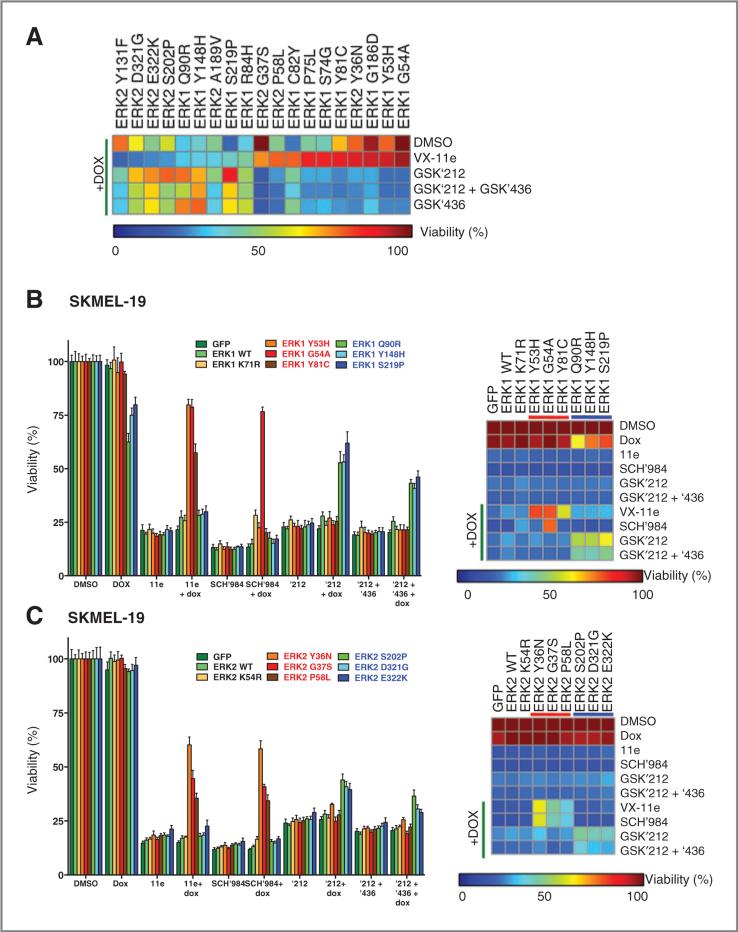
To investigate this question, we performed cell growth inhibition assays in the presence of RAF, MEK, or ERK inhibitors. In general, ERK resistance mutations that arose from RAF/MEK inhibitor screens were sensitive to ERK inhibition. Conversely, the ERK mutations from ERK inhibitor screens remained sensitive to RAF and MEK inhibition (or both; Fig. 6A). The same was true in two additional cell lines (SKMEL-19 and WM266.4); however, the overall magnitude of resistance was lower in these lines (Fig. 6B and C and Supplementary Fig. S8A). Interestingly, one ERK mutation (ERK1C82Y) conferred resistance to all MAP kinase inhibitors tested—though the magnitude of the resistance phenotype was more modest [45% (RAF/MEK inhibitors) and 75% (ERK inhibitor) viability compared with untreated controls]. Overall, ERK resistance mutations arising from ERK inhibition remained vulnerable to combined RAF/MEK inhibition, and the converse was also true.
Figure 6.
Drug-sensitivity studies of ERK resistance mutants using RAF, MEK, and ERK inhibitors. A, viability of A375 cells expressing tet-inducible ERK1/2 resistance alleles from Figs. 2 and 4 following exposure to VX-11e, trametinib (GSK′212), dabrafenib (GSK′436), or trametinib + dabrafenib (GSK′212+GSK ′436) in the presence or absence of DOX. Normalized viability is depicted by heat map. Alleles are sorted by their sensitivity to VX-11e in the presence of DOX. B and C, viability of SKMEL-19 cells expressing ERK1 (B) or ERK2 (C) resistance alleles after exposure to VX-11e, SCH772984 (SCH′984), trametinib (GSK′212), and trametinib + dabrafenib (GSK′212 + GSK′436) is graphed (left). Normalized viability is depicted by heat map (right). VX-11e–resistant alleles (red) and RAF/MEK inhibitor– resistant alleles (blue) are indicated.
ERK1/2 overexpression reduces the viability of some BRAF-mutant melanoma cells
While performing these studies, we noticed that ERK over-expression was lethal to A375 cells in the absence of MAP kinase pathway inhibition. Although this effect could certainly result from nonspecific toxicities associated with ectopic protein expression, it was also possible that “too much” MAP kinase pathway activation might be deleterious to BRAF-mutant melanoma cells—particularly in light of the “drug holiday” effect that has occasionally been observed after RAF inhibition in melanoma models (33). To test this possibility, we assessed cell proliferation following ectopic expression of validated ERK resistance mutations.
Overexpression of wild-type ERK1 and ERK2 indeed decreased the viability of A375 cells (Fig. 7A and Supplementary Fig. S2). Moreover, the growth-suppressive effect of ERK overexpression was generally linked to kinase activity, as kinase-dead ERK1/2 (ERK1K71R/ERK2K54R) had no effect. Overexpression of ERK1G54A and ERK2G37S, which exhibit reduced basal kinase activity compared with wild-type ERK (Fig. 3), also had no effect on viability (Fig. 7A). In contrast, ERK mutations arising in the setting of RAF/MEK inhibition produced the greatest reductions in viability (Fig. 7B, highlighted in blue), consistent with the above observations that these mutations enhance intrinsic or in vivo ERK kinase activity. We observed similar viability defects in a second BRAF-mutant melanoma cell line (WM266.4 cells; Supplementary Fig. S8). However, in a third cell line (SKMEL-19 cells), only the most active ERK1 resistance alleles (e.g., those with augmented kinase activity) reduced cell viability (ERK1Q90R, ERK1Y148H, and ERK1S219P; Fig. 6B and C). These data raise the possibility that excessive ERK signaling may be detrimental to the growth of some BRAF-mutant melanoma cells, as observed by others (33, 34).
Figure 7.
Lethality of ERK resistance mutants in BRAF-mutant melanoma cells. A, viability of A375 cells expressing tetinducible ERK1/2 resistance alleles after exposure to DOX. Controls (GFP, wild-type ERK1/2, and kinase-dead ERK1K71R/ERK2K54R; green), ERK inhibitor resistance mutations (red), and RAF/MEK inhibitor resistance mutations (blue) are shown. ERK1C82Y is resistant to RAF, MEK, and ERK inhibitors (purple). B, phase contrast micrographs of A375 cells expressing tet-inducible RAF/MEK inhibitor–resistant alleles (ERK1A206V and ERK1S219P) or controls treated with DOX (top), or DOX + trametinib (GSK′212; bottom) for 24 hours (magnification, ×10).
Discussion
In melanoma, clinical resistance to RAF/MEK inhibition often involves reactivation of the MAPK pathway. Although multiple distinct mechanisms to achieve this effect have been described, ERK activation stands as their chief point of convergence. Thus, ERK represents an attractive therapeutic target in MAP kinase–driven cancers, and several ERK inhibitors have entered clinical trials. However, even if ERK inhibitors are successful clinically, resistance to these agents will likely emerge, as has been seen with most other kinase-based anticancer therapeutics.
Secondary mutations within the kinase target represent a common resistance mechanism for this class of therapeutics. The present study used random mutagenesis screens to identify mutations in ERK1 and ERK2 that confer resistance to tool compounds that bear structural similarity to ERK inhibitors in clinical trials. Multiple resistance mutations were identified in both ERK isoforms, and additional mutations could emerge in the future if such screens are carried out to saturation. ERK mutations we identified using the VX-11e compound were cross-resistant to SCH772984, suggesting that these resistance alleles might represent generalizable resistance mechanisms to ATP-competitive ERK inhibitors.
In addition to identifying mutations in ERK that confer resistance to ERK inhibitors, we identified several mutations in both ERK1 and ERK2 that retain robust ERK signaling despite pharmacologic RAF/MEK inhibition. Although we have not formally proved that these represent bona fide constitutive active ERK isoforms, the mutations may nonetheless prove useful for future biochemical studies.
One of the RAF/MEK inhibitor–resistant alleles detected in our screen, MAPK1E322K (ERK2), was identified in cervical and head and neck carcinoma (25, 26). Although ERK2E322K does not exhibit increased basal kinase activity in vitro, our data are consistent with the notion that this variant may remain activated in vivo because of reduced DUSP binding and hence loss of negative regulation (35), thereby conferring resistance to RAF/MEK inhibitors. Conceivably, treatment of patients containing this residue with RAF and/or MEK inhibitors may prove ineffective.
Our results raise the possibility that tumors with somatic ERK mutations arising in the setting of ERK inhibition might still be sensitive to RAF/MEK inhibition. Reciprocally, ERK mutations that confer resistance to RAF/MEK inhibition could remain sensitive to ERK inhibitors. If such mutations were to arise clinically, one could speculate that an alternative RAF/MEK and ERK dosing regimen might be attempted to achieve more prolonged disease control. Indeed, anecdotal reports of successful alternating targeted therapeutic regimens have emerged for other oncogene-driven cancers, such as EGFR-driven lung cancer. Of course, such speculation must be tempered by the fact that somatic ERK mutations have not yet been described as a clinical resistance mechanism to RAF/MEK inhibition in melanoma or other cancers.
This work also implies that BRAF-mutant melanoma cells must maintain tight control over ERK signaling to enable optimal growth. On the one hand, if flux through the MAPK pathway diminishes too much, growth arrest or apoptosis ensues. On the other hand, our results suggest that excessive MAPK signaling (over and above that conferred by BRAFV600E) can also reduce cell viability, at least in some cases. This result is concordant with recent observations that “acute” withdrawal of vemurafenib from a patient-derived xenograft tumor model that had progressed to drug resistance produced a temporary tumor regression (34). This has also been termed the “drug holiday” effect (33). Presumably, the flare of MAP kinase signaling that ensues upon drug discontinuation is deleterious to melanoma cells (of note, BRAF amplification was a resistance mechanism in this study, making an acute signaling flare even more likely upon drug removal).
If this general phenomenon (e.g., lethality of acute drug withdrawal) were broadly relevant at some point during the treatment course, one could envision intermittent therapeutic strategies for RAF/MEK inhibition in melanoma. Indeed, discontinuous vemurafenib allowed for disease stabilization in the aforementioned study by Das Thakur, Stuart and colleagues (34). There have also been reports of successful rechallenge with vemurafenib in patients that initially relapsed on the drug (36–38). Moreover, recent clinical trial results, in which vemurafenib was administered chronically, while the MEK inhibitor was given intermittently, suggest that intermittent MEK inhibition was no worse than chronic administration of both drugs, but MEK inhibitor toxicity may have been reduced by intermittent dosing (39). The ERK resistance alleles identified herein may provide a useful set of tools for detailed preclinical evaluation of intermittent ERK inhibition alternating with “acute” ERK hyperactivation as a novel therapeutic approach.
In summary, this study has identified a set of mutations in ERK1 and ERK2 that confer resistance to MAP kinase pathway inhibition at the level of RAF/MEK and ERK. These findings may anticipate future resistance mechanisms, enable new biochemical studies of ERK isoforms, and support preclinical studies of novel therapeutic strategies. They could also inform the development of “next-generation” ERK inhibitors that could be developed for melanoma and many other MAP kinase–driven cancers.
Supplementary Material
Acknowledgments
Grant Support
E.M. Goetz was supported by the Novartis Institutes for BioMedical Research. L.A. Garraway was supported by the Novartis Institutes for BioMedical Research, Melanoma Research Alliance, Starr Cancer Consortium, Dr. Miriam and Sheldon Adelson Medical Research Foundation, and NCI.
Footnotes
Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors’ Contributions
Conception and design: E.M. Goetz, L.A. Garraway
Development of methodology: E.M. Goetz, N. Wagle, L.A. Garraway
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): E.M. Goetz, D.J. Treacy
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): E.M. Goetz, M. Ghandi, N. Wagle, L.A. Garraway
Writing, review, and/or revision of the manuscript: E.M. Goetz, M. Ghandi, N. Wagle, L.A. Garraway
Study supervision: L.A. Garraway
Disclosure of Potential Conflicts of Interest
L.A. Garraway received a commercial research grant from Novartis. He is also a consultant/advisory board member for Novartis and Foundation Medicine. No potential conflicts of interest were disclosed by the other authors.
References
- 1.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 2.Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012;367:1694–703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–19. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flaherty KT, Robert C, Hersey P, Nathan P, Garbe C, Milhem M, et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med. 2012;367:107–14. doi: 10.1056/NEJMoa1203421. [DOI] [PubMed] [Google Scholar]
- 6.Trunzer K, Pavlick AC, Schuchter L, Gonzalez R, McArthur GA, Hutson TE, et al. Pharmacodynamic effects and mechanisms of resistance to vemurafenib in patients with metastatic melanoma. J Clin Oncol. 2013;31:1767–74. doi: 10.1200/JCO.2012.44.7888. [DOI] [PubMed] [Google Scholar]
- 7.Van Allen EM, Wagle N, Sucker A, Treacy DJ, Johannessen CM, Goetz EM, et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 2014;4:94–109. doi: 10.1158/2159-8290.CD-13-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagle N, Van Allen EM, Treacy DJ, Frederick DT, Cooper ZA, Taylor-Weiner A, et al. MAP kinase pathway alterations in BRAF-mutant melanoma patients with acquired resistance to combined RAF/MEK inhibition. Cancer Discov. 2014;4:61–8. doi: 10.1158/2159-8290.CD-13-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–7. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature. 2011;480:387–90. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–72. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mandl M, Slack DN, Keyse SM. Specific inactivation and nuclear anchoring of extracellular signal-regulated kinase 2 by the inducible dual-specificity protein phosphatase DUSP5. Mol Cell Biol. 2005;25:1830–45. doi: 10.1128/MCB.25.5.1830-1845.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muda M, Theodosiou A, Rodrigues N, Boschert U, Camps M, Gillieron C, et al. The dual specificity phosphatases M3/6 and MKP-3 are highly selective for inactivation of distinct mitogen-activated protein kinases. J Biol Chem. 1996;271:27205–8. doi: 10.1074/jbc.271.44.27205. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Z, Kobayashi S, Borczuk AC, Leidner RS, Laframboise T, Levine AD, et al. Dual specificity phosphatase 6 (DUSP6) is an ETS-regulated negative feedback mediator of oncogenic ERK signaling in lung cancer cells. Carcinogenesis. 2010;31:577–86. doi: 10.1093/carcin/bgq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roskoski R., Jr. ERK1/2 MAP kinases: structure, function, and regulation. Pharmacol Res. 2012;66:105–43. doi: 10.1016/j.phrs.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 16.BioMed Valley Discoveries I. In ClinicalTrialsgov [Internet] National Library of Medicine; Bethesda, MD: 2013. [June 2014]. Phase I dose-escalation, safety, pharmacokinetic and pharmacodynamic study of BVD-523 in patients with advanced malignancies. Available from: < https://clinical-trials.gov/ct2/show/NCT01781429?term=bvd+523&rank=1>. [Google Scholar]
- 17.Genentech . In ClincialTrialsgov [Internet] National Library of Medicine; Bethesda, MD: 2013. [June 14, 2014]. A dose-escalation study of GDC-0994 in patients with locally advanced or metastatic solid tumors. Available from: < https://clinicaltrials.gov/ct2/show/NCT01875705?term=gdc+0994&rank=1>. [Google Scholar]
- 18.Ray A, Cowan-Jacob SW, Manley PW, Mestan J, Griffin JD. Identification of BCR-ABL point mutations conferring resistance to the Abl kinase inhibitor AMN107 (nilotinib) by a random mutagenesis study. Blood. 2007;109:5011–5. doi: 10.1182/blood-2006-01-015347. [DOI] [PubMed] [Google Scholar]
- 19.Emery CM, Vijayendran KG, Zipser MC, Sawyer AM, Niu L, Kim JJ, et al. MEK1 mutations confer resistance to MEK and B-RAF inhibition. Proc Natl Acad Sci U S A. 2009;106:20411–6. doi: 10.1073/pnas.0905833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Broad-Novartis Cancer Cell Line Encyclopedia (CCLE) Version 2.17. Broad Institute; Cambridge, MA: 2014. [September 5, 2014]. Available from: < http://broad-institute.org/ccle/>. [Google Scholar]
- 21.GENE-E [Internet] Broad Institute; Cambridge, MA: 2013. [September 5, 2014]. Available from: < http://www.broadinstitute.org/cancer/software/GENE-E/download.html>. [Google Scholar]
- 22.Muteeb G, Sen R. Random mutagenesis using a mutator strain. Methods Mol Biol. 2010;634:411–9. doi: 10.1007/978-1-60761-652-8_29. [DOI] [PubMed] [Google Scholar]
- 23.Aronov AM, Tang Q, Martinez-Botella G, Bemis GW, Cao J, Chen G, et al. Structure-guided design of potent and selective pyrimidylpyrrole inhibitors of extracellular signal-regulated kinase (ERK) using conformational control. J Med Chem. 2009;52:6362–8. doi: 10.1021/jm900630q. [DOI] [PubMed] [Google Scholar]
- 24.COSMIC: Catalogue of somatic mutations in cancer [Internet]. v70. Wellcome Trust Sanger Institute; Hinxton, Cambridgeshire, UK: 2014. [September 5, 2014]. Available from: < http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/>. [Google Scholar]
- 25.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ojesina AI, Lichtenstein L, Freeman SS, Pedamallu CS, Imaz-Rosshandler I, Pugh TJ, et al. Landscape of genomic alterations in cervical carcinomas. Nature. 2014;506:371–5. doi: 10.1038/nature12881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson MJ, Harkins PC, Zhang J, Baer R, Haycock JW, Cobb MH, et al. Mutation of position 52 in ERK2 creates a nonproductive binding mode for adenosine 50-triphosphate. Biochemistry. 1996;35:5641–6. doi: 10.1021/bi952723e. [DOI] [PubMed] [Google Scholar]
- 28.Bott CM, Thorneycroft SG, Marshall CJ. The sevenmaker gain-of-function mutation in p42 MAP kinase leads to enhanced signalling and reduced sensitivity to dual specificity phosphatase action. FEBS Lett. 1994;352:201–5. doi: 10.1016/0014-5793(94)00958-9. [DOI] [PubMed] [Google Scholar]
- 29.Chu Y, Solski PA, Khosravi-Far R, Der CJ, Kelly K. The mitogen-activated protein kinase phosphatases PAC1, MKP-1, and MKP-2 have unique substrate specificities and reduced activity in vivo toward the ERK2 sevenmaker mutation. J Biol Chem. 1996;271:6497–501. doi: 10.1074/jbc.271.11.6497. [DOI] [PubMed] [Google Scholar]
- 30.Burkhard KA, Chen F, Shapiro P. Quantitative analysis of ERK2 interactions with substrate proteins: roles for kinase docking domains and activity in determining binding affinity. J Biol Chem. 2011;286:2477–85. doi: 10.1074/jbc.M110.177899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee T, Hoofnagle AN, Kabuyama Y, Stroud J, Min X, Goldsmith EJ, et al. Docking motif interactions in MAP kinases revealed by hydrogen exchange mass spectrometry. Mol Cell. 2004;14:43–55. doi: 10.1016/s1097-2765(04)00161-3. [DOI] [PubMed] [Google Scholar]
- 32.Hatzivassiliou G, Liu B, O'Brien C, Spoerke JM, Hoeflich KP, Haverty PM, et al. ERK inhibition overcomes acquired resistance to MEK inhibitors. Mol Cancer Ther. 2012;11:1143–54. doi: 10.1158/1535-7163.MCT-11-1010. [DOI] [PubMed] [Google Scholar]
- 33.Sun C, Wang L, Huang S, Heynen GJ, Prahallad A, Robert C, et al. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature. 2014;508:118–22. doi: 10.1038/nature13121. [DOI] [PubMed] [Google Scholar]
- 34.Das Thakur M, Salangsang F, Landman AS, Sellers WR, Pryer NK, Levesque MP, et al. Modelling vemurafenib resistance in melanoma reveals a strategy to forestall drug resistance. Nature. 2013;494:251–5. doi: 10.1038/nature11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arvind R, Shimamoto H, Momose F, Amagasa T, Omura K, Tsuchida N. A mutation in the common docking domain of ERK2 in a human cancer cell line, which was associated with its constitutive phosphorylation. Int J Oncol. 2005;27:1499–504. [PubMed] [Google Scholar]
- 36.Guerreschi P, Scalbert C, Qassemyar A, Kluza J, Ravasi L, Huglo D, et al. Patient-derived tumor xenograft model to guide the use of BRAF inhibitors in metastatic melanoma. Melanoma Res. 2013 Jul 10; doi: 10.1097/CMR.0b013e328363ed92. [DOI] [PubMed] [Google Scholar]
- 37.Romano E, Pradervand S, Paillusson A, Weber J, Harshman K, Muehlethaler K, et al. Identification of multiple mechanisms of resistance to vemurafenib in a patient with BRAFV600E-mutated cutaneous melanoma successfully rechallenged after progression. Clin Cancer Res. 2013;19:5749–57. doi: 10.1158/1078-0432.CCR-13-0661. [DOI] [PubMed] [Google Scholar]
- 38.Seghers AC, Wilgenhof S, Lebbe C, Neyns B. Successful rechallenge in two patients with BRAF-V600-mutant melanoma who experienced previous progression during treatment with a selective BRAF inhibitor. Melanoma Res. 2012;22:466–72. doi: 10.1097/CMR.0b013e3283541541. [DOI] [PubMed] [Google Scholar]
- 39.McArthur GA, Callahan J, Ribas A, Gonzalez R, Pavlick AC, Hamid O, et al. Metabolic tumor burden for prediction of overall survival following combined BRAF/MEK inhibition in patients with advanced BRAF mutant melanoma. J Clin Oncol. 2014;32:5s. (suppl; abstr 9006^) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.