Abstract
Ryanodine receptors (RyRs) mediate rapid release of calcium (Ca2+) from intracellular stores into the cytosol, which is essential for numerous cellular functions including excitation-contraction coupling in muscle. Lack of sufficient structural detail has impeded understanding of RyR gating and regulation. Here, we report the closed-state structure of the 2.3 MDa complex of the rabbit skeletal muscle type 1 RyR (RyR1), solved by single-particle cryo-electron microscopy at an overall resolution of 4.8 Å. We fitted a polyalanine-level model to all 3939 ordered residues in each protomer, defining the transmembrane pore in unprecedented detail and placing all cytosolic domains as tertiary folds. The cytosolic assembly is built on an extended α-solenoid scaffold connecting key regulatory domains to the pore. The RyR1 pore architecture places it in the six-transmembrane (6TM) ion channel superfamily. A unique domain inserted between the second and third transmembrane helices interacts intimately with paired EF-hands originating from the α-solenoid scaffold, suggesting a mechanism for channel gating by Ca2+.
Ryanodine receptors (RyRs) are intracellular calcium (Ca2+) release channels on the sarcoplasmic and endoplasmic reticula (SR/ER) required for fundamental cellular functions in most tissues, including skeletal and cardiac muscle excitation-contraction (EC) coupling, synaptic transmission and pancreatic beta cell function 1. The type 1 ryanodine receptor (RyR1) mediates EC-coupling in skeletal muscle. It is a homotretramer of four ~565 kDa channel-forming protomers, as well as regulatory subunits, enzymes and their respective targeting/anchoring proteins, forming a macromolecular complex that exceeds three million daltons 2. In most tissues, RyRs are activated by the inward flow of Ca2+ via plasma-membrane Ca2+ channels, resulting in a massive and rapid release of Ca2+ from intracellular stores (a process known as Ca2+-induced Ca2+ release). In contrast, in skeletal muscle approximately 50% of RyR1 channels are mechanically activated by direct interaction with voltage-gated Ca2+ channels on the plasma membrane 3. RyR1 is also activated by Ca2+ in vitro, and hence those RyR1s not associated with DHPRs are presumably activated by Ca2+, possibly in concert with coupled gating between adjacent RyRs, amplifying Ca2+ release 4,5,6.
RyR channels are subject to stress-induced Ca2+ leak which is critically involved in heart failure 2, cardiac arrhythmias 7, post-traumatic stress disorder (PTSD) 8, age-dependent loss of muscle function 9, and muscular dystrophy 10. RyRs have recently become therapeutic targets for some of these disorders 11,12.
Prior single-particle cryo-electron microscopy (cryo-EM) studies revealed that RyR1 adopts a fourfold symmetric mushroom-like superstructure, with about 80% of the mass in the cytosol (the ‘cap’ of the mushroom) and a stalk embedded in the SR/ER membrane. The highest-resolution reconstruction obtained for intact RyR1 prior to this study was 9.6 Å 13, which, though giving a good picture of the overall dimensions of the molecule and approximate protomer boundaries, was insufficient to identify secondary structural elements, or locate key domains.
Architecture of RyR1 at 4.8 Å resolution
The calcium-free (low nM free Ca2+) RyR1-calstabin2 (FKBP12.6) complex was purified from rabbit (Oryctolagus cuniculus) skeletal muscle by calstabin-affinity chromatography (see online Methods and Extended Data Fig. 1 for details). Recent advances in cryo-EM detector technology and data processing 14 have allowed us to obtain 3D-reconstructions at overall resolutions as high as 4.8 Å (Extended Data Fig. 2).
3D-classification using RELION 1.2 15 identified distinct classes of particles (Extended Data Fig. 3) differing in the cytosolic assembly conformation but not in the transmembrane pore. The best-resolved class yielded a reconstruction with C4 symmetry and a resolution of 4.8 Å according to the FSC=0.143 gold standard criterion (Extended Data Fig. 4a) 15.
The quality of the density map was excellent in the transmembrane region, and lower in the cytoplasmic region. A reconstruction obtained from dephosphorylated (see Methods for details) RyR1 at 5.0 Å resolution had considerably improved density in regions that were poorly defined in the original sample (Fig. 1a and Extended Data Fig. 4b–e and 5). This map has been used for the majority of interpretation and model building (model-map correlation in Extended Data Fig. 6).
Figure 1. The architecture of RyR1 at 4.8 Å.
a, View from the plane of the SR membrane of a slab of density (blue mesh) coinciding with the channel axis b, Color coded schematic representation of the RyR1. c, view in the plane of the SR membrane d, from the cytosol e, and from the lumen of the electron density map of skeletal muscle RyR1 at 5.0 Å resolution, with one protomer segmented according to the domains assigned in the model, colored as follows: blue, N-terminal domain; cyan, SPRY1, SPRY2 and SPRY3; salmon, clamp region (RY12 repeats), and phosphorylation domain (RY34 repeats); yellow, calstabin; green, the bridge solenoid scaffold; red, the core solenoid; and orange, transmembrane and C-terminal domains; purple, putative Ca2+ binding domain (EF). Dashed lines represent major disordered segments.
The architecture of RyR1 consists of four protomers surrounding a central transmembrane pore coincident with the four-fold symmetry axis of the tetramer. Each protomer is built around an extended scaffold of α-solenoid repeats 16. This scaffold is extraordinary in scale, comprising 37 repeats in three contiguous segments totaling 2217 residues, or 56% of the ordered residues in the polypeptide. The α-solenoid scaffold is capped at the N-terminus by two beta-trefoil domains, NTD-A and NTD-B, which form a central cytosolic vestibule; and at the C-terminus by the transmembrane pore which adopts a fold placing it in the 6TM superfamily of ion channels that includes the voltage-gated sodium and potassium channels and the transient receptor potential (TRP) channels (Fig. 1b). The α-solenoid scaffold incorporates five major domains: three SPRY domains (SPRY1-3), and two pairs of RYR repeats (RY12 and RY34, the latter containing a regulatory PKA-phosphorylation site at S2843). Several smaller insertions were also identified, including most importantly a previously predicted EF-hand pair 17 that constitutes the presumed conserved Ca2+-binding domain (CBD) in RyRs (Fig. 1b). The RyR1 model exhibits well-defined protomer boundaries (Fig. 1c–e).
The flexible α-solenoid scaffold of RyR1
The α-solenoid scaffold of RyR1 can be divided into three major segments (Extended Data Fig. 7). The smallest of these is the N-terminal solenoid (NTD-C), consisting of four repeats, linking the central cytosolic vestibule of the channel to the three SPRY domains at the outer corner of the tetramer (Fig. 1b). The first two of these repeats were present in the crystal structure of the N-terminal fragment, in which they were assigned as ‘Domain C’ of this fragment 18.
The second and largest of the three major α-solenoid regions is the bridging solenoid, which curls around the periphery of the cytosolic assembly in a right-handed superhelix towards the SPRY domains of the adjacent protomer. The bridging solenoid may be further subdivided into N-terminal, central and C-terminal subdomains, denoted BrA, BrB and BrC (Extended Data Fig. 8).
The N-terminal subdomain, BrA, adopts a distinctly different architecture from the rest of the bridging solenoid, in which distorted, interlocking alpha-solenoid repeats wrap around a 72 Å-long central helix that projects towards the three SPRY domains. An additional feature of interest in BrA is the calstabin-binding helix which projects out from the central helix, with its N-terminus buried in calstabin2 (Extended Data Fig. 9).
Midway along BrB, the second RYR-repeat pair (RY34) projects away from the SR membrane into the cytosol. The connections between RY34 and BrB are not clearly resolved, leading to uncertainty in the numbering of this region. BrC, the C-terminal subdomain of the bridging solenoid, interacts closely with the SPRY3 domain of the neighboring subunit, before terminating in a disordered segment.
The last of the major solenoid regions, which we have termed the core solenoid (Fig. 1b), begins with a linker helix (starting approximately at residue 3679) embedded in BrA, and progresses through eight repeats towards the transmembrane pore (Extended Data Fig. 8b). An EF-hand pair, described in detail below, is inserted in the seventh repeat of the core solenoid (Extended Data Fig. 10). Between the seventh and eighth repeat, the core solenoid is interrupted by an extended loop (4180-4200), which likely adopts a beta-hairpin structure. The beta hairpin and the eighth repeat of the core solenoid form a ‘vice’ surrounding the C-terminal domain of RyR1, as described in detail below.
Inspection of the initial model suggested that the fold of three major regions of the polypeptide could be categorized into one of the many families of α-solenoid repeat proteins. We used the polyalanine model of each region to search the PDB (using the DALI web server 19). For all three major solenoid regions, the top hits (Z-scores > 15) were Armadillo repeat-containing proteins including APC and beta-catenin (Extended Data Fig. 7b–d).
Previously characterized Armadillo and HEAT repeats are flexible protein-protein interaction modules 20,21. We suggest that the flexible α-solenoid scaffold of RyR1 could facilitate assembly of regulatory proteins, and allow coupling of conformational changes of the scaffold, induced either by protein binding or post-translational modifications, to RyR1 activity.
RyR1 is a 6TM ion channel
The transmembrane domain is one of the best-resolved regions of the map (Fig. 2 a–b), allowing us to identify all secondary structural elements. The architecture of the pore clearly identifies it as a member of the six-transmembrane (6TM) ion channel superfamily22, with 6 transmembrane helices per protomer surrounding a central pore. An extended peptide (the P-segment), structurally analogous to the selectivity filter of potassium and sodium channels, delineates the luminal aperture (Fig. 2c). The transmembrane region contains two domains: the pore domain, formed by S5, S6, the pore helix and the P-segment; and a pseudo voltage-sensor domain (pVSD) formed by S1-S4 that interfaces with the pore domain of the adjacent subunit (Fig 2b). The RyR1 pore domain most closely resembles that of the voltage-gated sodium channel, NavAb (Fig. 2d), although the P-segment is more similar to the equivalent region of the TRPV1 cation channel (Fig. 3a) 23,24. Moreover, the luminal aperture is comparable to that of the highly Ca2+-permeable 25 TRPV1. The brevity of the constriction suggests it may accommodate only one Ca2+-binding site.
Figure 2. RyR1 transmembrane pore and C-terminal domain.
a, Electron density map of the pore region (black mesh) and the core solenoid (blue mesh), displayed with the transmembrane region of one protomer in spectral coloring (N-terminus is blue, C-terminus is red). b, A 90° slab through the plane of the membrane, with numbering indicating the positions of the six transmembrane helices. c, The transmembrane region of a protomer in spectral coloring, as labeled d, superposition of the RyR1 pore with the structure of a voltage-gated sodium channel (NavAB, PDB ID 3RVY). The two structures were aligned using SUPERPOSE 39, giving a core RMSD of 3.42 Å across 628 aligned Cα atoms.
Figure 3. The RyR1 conduction pathway.
a, superposition of the RyR1 pore domain (gray) with structures of the potassium channel KcsA (blue), the voltage-gated sodium channel NavAB (yellow), and TRPV1 (red) pores. Only two subunits are shown for clarity. The core RMSDs of the three alignments, calculated using SUPERPOSE are 3.44 Å (over 306 aligned Cα atoms), 3.42 Å (over 628 aligned Cα atoms) and 4.80 Å (over 635 aligned Cα atoms) respectively. b, scheme indicating the relative positioning of all the negatively charged residues in the ionic pathway (red dots) and the sequence of the P-loop and the cytosolic helix extending from S6 indicating in red the negatively charged residues. c, model of two opposing pore regions of RyR1 indicating the bundle crossing and the 24° kink in S6.
The P-segment contains many acidic residues that are expected to be anionic under physiological conditions, likely contributing to the high conductance of RyR1 (e.g. 110 pS for Ca2+, >700 pS for K+) 26. Seven additional negatively charged residues are located along the cytosolic extension of S6, presumably serving a similar function (Fig. 3b).
The transmembrane region includes three apparently amphipathic helices (JM1, the first helix of the S2-S3 insertion and the S4-S5 linker helix) that are expected to lie in the plane of the cytosolic surface of the membrane.
There is a single cytosolic constriction in the ion conduction pathway, at the S6 bundle crossing. The Cα-Cα distance at this point is 11 Å, similar to the minimal Cα-Cα distance observed in the structure of NavAb (10.41 Å. At Cys-217). Based on structural alignment with the closed NavAb pore (Fig. 2d), and the diameter of hydrated Ca2+ (8.2 Å 27) we conclude that the pore is in a non-conducting state.
The S6 helix has a 24° kink in the middle, centered on G4934 near the cytosolic face of the membrane, which orients the cytosolic extension of S6 roughly parallel to the channel axis (Fig. 3c). Conserved glycine residues in the pore-lining helices of other 6TM ion channels may operate as ‘glycine-hinges’, facilitating reorientation of the pore-lining helix and consequent alteration in the cytosolic aperture of the channel 28. G4934 is conserved in all RyR isoforms and in a closely related channel, the inositol 1,4,5-trisphosphate receptor.
The S6 helix extends ~30 Å into the cytosol, terminating in the C-terminal domain (CTD), a small alpha-helical domain that extends laterally from the channel axis into the core solenoid (Fig. 1b). The CTD is secured by an extended loop and a pair of helices that are reminiscent of a thumb and forefingers (Fig. 4).
Figure 4. The Ca2+ sensing machinery.
a, color coded representation of elements in the RyR1 structure that may participate in Ca2+ sensing and allosteric Ca2+ dependent force transduction to the channel gate. Green, calmodulin-like domain/EF hand pair; orange, C-terminal domain and S6; purple, core solenoid hairpin insertion; orange, S2-S3 helical bundle. b, two views of the “thumb and forefingers” motif originating from the core solenoid that engulfs the CTD.
An additional helix, in weaker density, may correspond to half of the predicted TM hairpin located between a.a. 4325-4370 (Extended Data Fig. 6).
A cytosolic Ca2+ sensor contacts the pore
Unlike other 6TM channels, RyR1 possesses a compact, folded cytosolic domain inserted within the pVSD, between helices S2 and S3 (residues 4675-4790). A curved, cytosolic extension of S2 leads into a bundle of four short helices, which are separated by an unresolved segment spanning 31 aa from an amphipathic helix leading into S3 (Fig. 2c). The S2-S3 helical bundle lies in close proximity to the putative Ca2+ binding domain and the C-terminal domain (Fig. 4) suggesting this domain may be involved in transmitting conformational changes in the cytosolic assembly to the pore.
RyR1 is activated by cytosolic [Ca2+]cyt 29. A calmodulin-like domain inserted within the core solenoid was previously suggested to be a Ca2+-sensor 17. This region is highly conserved in all RyRs, and shares 26% sequence identity with the C-lobe of human calmodulin (Extended Data Fig. 10a). Six of the eight residues that coordinate Ca2+ in calmodulin are conserved in the putative Ca2+-binding domain of RyR1.
The putative Ca2+-binding domain of RyR1 is adjacent to the S2-S3 insertion from a neighboring protomer and the C-terminal domain of the originating protomer (Fig. 4a), suggesting a mechanism for Ca2+-mediated gating whereby Ca2+-dependent changes in the conformation of the CBD are transmitted to the pore via contacts with the S2-S3 loop and/or the CTD, inducing a conformational change that alters the cytosolic aperture of the channel.
N-terminal domains form the cytosolic vestibule
A prominent feature of the RyR1 structure, already evident at lower resolution 30,31, is a large cytosolic vestibule around the four-fold axis above the pore in the cytosolic domain (Fig. 5). We docked the crystal structure of the three-domain N-terminal fragment of RyR1 18 into the density map in the same location as previously proposed. Several loops that were unresolved in the crystal structure are now clearly visible and were added into the model, as were residues 533-603 of NTD-C, which were not resolved in the previous structure. This addition completes the N-terminal solenoid, which connects to the beginning of SPRY1 through a short linker that is well resolved in the density map (Fig. 5a).
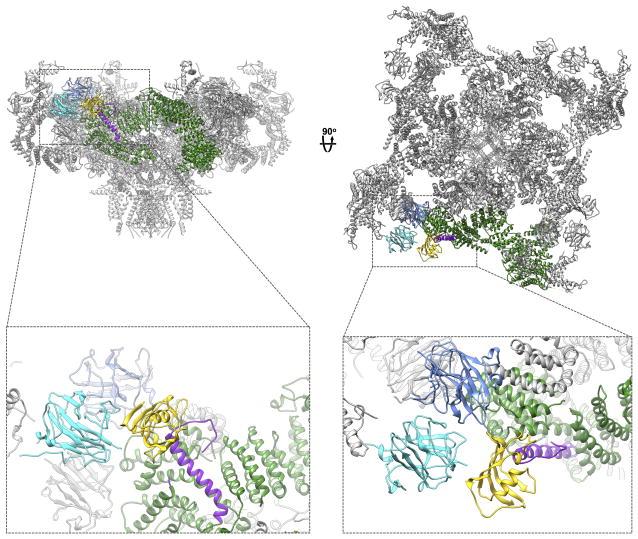
Figure 5. Intra- and inter-protomer interactions formed by cytosolic domains.
Views from the cytosol and in the membrane plane of RyR1, with regions colored as follows: the N-terminal domain (blue), core solenoid (red), and bridge solenoid (green). For clarity, one protomer is depicted in brighter coloring. In the view parallel to the membrane, most of the front subunit has been clipped for clarity. a, View from the cytosol, showing contacts between NTD-A and NTD-B of the neighboring protomer (1), NTD-B with the bridging solenoid of the adjacent protomer (2), and NTD-C forming a structurally contiguous solenoid with the bridging solenoid of the same protomer (3). b, view in the plane of the membrane showing interactions formed by NTD-C with the core solenoid of the same protomer (4), and of NTD-B with the core solenoid of the adjacent protomer (5). c, Views in the membrane plane and from the cytosol of RyR1 with the following domains colored as in Fig. 1b; core solenoid, the bridging solenoid scaffold, the RY34 repeats-phosphorylation domain, SPRY1, SPRY2, SPRY3, RY12 repeats, and the N-terminal domain. Position of the docked RY34 phosphorylation domain (salmon) abutting the bridge solenoid (dark green) within the density map (gray mesh). d, Locations of SPRY domains 1, 2 and 3 (cyan), calstabin2 (yellow), and RY12 (salmon) within the density map.
The N-terminal domains make key interactions stabilizing the cytosolic assembly, both with other domains in the same protomer, and with domains contributed by neighboring protomers. NTD-A contacts the fifth repeat in the core solenoid of the adjacent protomer via interactions mediated by R76 and E177 (Fig. 5b), and interacts with NTD-B of the neighboring protomer via Q156 (NTD-A) and D385 (NTD-B) (Fig. 5a). NTD-A interfaces with the sixth repeat of BrB in the bridging solenoid of the adjacent protomer, in an interaction involving the 121-134 loop on NTD-A, as well as F195, M196 and S175 (Fig. 5a). R392 and Q465 in NTD-C contribute to an interface with the core solenoid of the same protomer (Fig. 5b). NTD-C forms a structurally contiguous segment of α-solenoid repeats with the first set of repeats in the bridging solenoid region following SPRY3 (Fig. 5a). A major feature of the cytosolic domain are four prominent openings in the structure (one in each protomer) that are bounded by outer edges of the NTDs, the inner edges of the SPRY domains, and the bridging solenoid from the adjacent protomer (Fig. 5a).
RYR repeats, SPRY and calstabin interactions
RyR1 contains two pairs of RYR repeats, RY12 (residues 850-1055) and RY34 (residues 2734-2941). RY12 is located between SPRY1 and SPRY2, while RY34 is inserted in BrB (Fig. 1b). RY12 and RY34 are expected to adopt the same overall fold, as they share 29% sequence identity. The structure of RY34 (which contains a regulatory PKA-phosphorylation site at S2843) has been solved by two independent groups, and both placed it in the distinctly “U-shaped” density located at the extreme outer corners of the receptor, also referred to as the “clamp” region 32,33.
In our 5.0 Å map, we see two densities that share this distinctive shape. One is located in the previously identified location, abutting the SPRY domains, while another, less well-resolved density with the same overall shape protrudes upwards from the bridging solenoid. In the primary sequence, RY12 is located between SPRY1 and SPRY2. Therefore, we place RY12 at the corner of the channel, and RY34 (containing the PKA phosphorylation site) protrudes from BrB on top of the cytosolic assembly projecting into the cytosol (Fig. 5c–d). This assignment agrees with the observation that RY34, which appears to be the most exposed and flexible of the RYR repeat pairs, is also the most susceptible to proteolysis 34.
Interestingly, the location of RY12 (Fig. 5c), projecting laterally from the corners of the cytosolic assembly, places it in an appropriate position to serve as a contact site between neighboring channels in the paracrystalline RyR arrays that have been shown to assemble on the SR membrane 35.
RyR1 contains three SPRY domains, SPRY1 (residues 643-794), SPRY2 (residues 1073-1205) and SPRY3 (residues 1419-1567). Structures of SPRY domains from other proteins have shown that the domain adopts a distinctive β-sandwich structure consisting of two antiparallel 7- or 8-stranded β-sheets 36. In our 5.0 Å map of dephosphorylated RyR1, we identify the three SPRY domains based on the two curved sheets that form the β-sandwich (Fig. 5c–d). The plane of the triangle is oriented roughly perpendicular to the channel axis, and parallel to the edge of the cytoplasmic assembly.
High-affinity binding of calstabin1 and 2 plays a critical role in stabilizing the closed states of RyR1 and RyR2, respectively, and in preventing pathological intracellular calcium leak 2,37. Calstabin2 binds with high affinity to both RyR1 and RyR2 (but is not expressed in skeletal muscle), and calstabin1 binds preferentially to RyR1 38. Calstabin1 and calstabin2 differ at only 18 positions out of 108 residues. The calstabin-binding site is located at the mutual interface of the bridging solenoid, SPRY1 and SPRY2 (Extended Data Fig. 9). The N-terminal end of a helix originating from subdomain BrA is located in the hydrophobic binding pocket of calstabin2. Calstabin binding may stabilize the connection between the cytoplasmic regulatory domains and the pore by rigidifying the BrA/SPRY1/SPRY2 interface and possibly altering the relative orientation of these domains.
Concluding Remarks
The model and electron density maps presented here provide a first glimpse of the transmembrane architecture of ryanodine receptors. Our analysis confirms that RyR1 is a 6TM ion channel, reveals a flexible cytosolic alpha-solenoid scaffold of remarkable size, and places every previously known domain, including a putative calcium-binding domain. This structure is of RyR1 in the closed state, but its features provide a foundation for understanding the open state and the process of Ca2+-activation for all members of the RyR family.
ON LINE METHODS
RyR1 purification from rabbit skeletal muscle
Detergent-solubilized RyR1 was purified from rabbit (Oryctolagus cuniculus) skeletal muscle by calstabin-affinity chromatography using a procedure modified from Xin et. al. 41. The protein was eluted using calstabin2 (also known as FKBP12.6), rather than the physiological binding partner calstabin1 (also known as FKBP12), due to the higher affinity of the former for the RyR1 channel. Importantly, calstabin1 and calstabin2 share the same binding site on RyRs, and both have the same effects on RyR1 function. Addition of EGTA reduced the concentration of free Ca2+ in the sample to the low nM range such that the channel is closed according to functional studies using purified RyR1 reconstituted into planar lipid bilayers 42.
100 grams of rabbit skeletal muscle flash frozen in liquid nitrogen were blended for 90 seconds in 500 ml of ice-cold buffer containing 10 mM Tris-Maleate pH 6.8, 1 mM DTT, 1 mM EDTA, 150 μM PMSF and 1 mM benzamidine. The mixture was then centrifuged for 10 min at 8000g. The supernatant was centrifuged for 20 min at 40,000g. Pellets were solubilized in 50 mL buffer containing 10 mM HEPES pH 7.5, 1% (w/v) CHAPS, 1 M NaCl, 2 mM EGTA, 2 mM TCEP, and a protease inhibitor cocktail (Roche). The solubilized membranes were then diluted 1:1 in the same buffer without NaCl and centrifuged for 30 min at 100,000g. The supernatant was then filtered using a 0.2μm filter and allowed to bind overnight at 4 °C to a pre-equilibrated 5 mL GSTrap (GE healthcare) column with bound GST-calstabin1 (derived from 0.5 L BL-21 cells overexpressing GST-calstabin1). The column was then washed with 10 column volumes of cold solubilization buffer (with the following alterations: 0.5% (w/v) CHAPS and 0.5M NaCl) and RyR1 was eluted with two column volumes of 10 μM calstabin2 in the same buffer. The eluent was pre-cleared with 0.5 ml glutathione beads and treated with calf intestinal alkaline phosphatase (CIP, NEB, 100 U/ml) for 4 hr at room temperature. The CIP-treated RyR1 was concentrated in a 100kDa cut-off centrifugal filter unit (Millipore) to less than 500 μl, loaded on a size exclusion column (Tosoh G4SWXL) and eluted in solubilization buffer with the following alterations and substitutions: 5 mM EGTA, 0.25% (w/v) CHAPS and 0.001% (w/v) DOPC (Avanti). The fractions corresponding to the RyR1 peak (Extended Data Fig. 3b) were then combined and concentrated in a 100 kDa cut-off centrifugal filter unit (Millipore) to a concentration of ~10 mg/mL as determined using spectroscopy (A280nm at 1mg/mL=1.018, measured using Nanodrop spectrophotometer).
Grid preparation and data collection
3 μl of each sample was applied to holey carbon grids (C-flat CF-1.2/1.3-2C-T, Protochips Inc, NC) without prior plasma cleaning. After a 30s incubation time on the grids, sample was blotted with filter paper (Whatman; pre-treated with 1mM EGTA to remove trace Ca2+) for 0.5–2s at 100% humidity and vitrified by rapidly plunging into liquid ethane at −180°C with a Vitrobot (FEI) 43,44. Data acquisition was performed on a FEI Tecnai F30 Polara (FEI, Eindhoven) operating at 300 kV and at a nominal magnification of 31,000x. Datasets were collected with the automated data collection system Leginon 45 on a K2 Summit direct electron detector (Gatan, Pleasanton) with a physical pixel size of 5μm, corresponding to a calibrated pixel size of 1.255 Å per pixel at the specimen when operating in electron counting mode. Micrographs were recorded in dose-fractionation mode as a series of 23 frames, with an accumulation time of 0.2 s per frame. The dose rate was set to 8 counts per physical pixel per second (~10e−/pixel/s). The total exposure time was 4.6 s, leading to a total accumulated dose of approximately 25 e−/Å2 on the specimen.
Image processing, 3D classification and refinement
Dose-fractionated image stacks were aligned using dosefgpu_driftcorr 46 and a sum of frames 3 to 20 in each image stack was used for further processing. The first 2 frames were omitted as they contained motions greater than 4Å per frame on average. For the EGTA dataset, 3423 micrographs were collected in total. After visual assessment of the micrographs and their power spectra, 3190 micrographs were selected for subsequent processing. For the CIP treated dataset, 6194 micrographs were collected in total and 5243 micrographs were selected for further processing.
The data were then preprocessed using pySPIDER and Arachnid (www.arachnid.us; Langlois and Frank, unpublished). Arachnid is a Python-encapsulated version of SPIDER 47, replacing SPIDER batch files with Python scripts. It also contains novel procedures such as Autopicker 48, which was used for the reference-free automated particle selection (Extended Data Fig. 2a). 230,000 particles were extracted from a first dataset of RyR1 prepared in the presence of EGTA. Those particles were aligned and refined using the RELION 1.2 autorefine procedure 49. Map quality was good in the transmembrane region but lower in the most peripheral parts of the cytoplasmic region, suggesting the presence of conformational heterogeneity. The particles were therefore subjected to 3D-classification using RELION 50 without symmetry imposed and yielded different classes (Extended Data Fig. 3a) with the cytoplasmic assembly of RyR1 in different conformations. Class 1 (46860 particles) was the most homogeneous and yielded a reconstruction at 4.8 Å using the autorefine procedure implemented in RELION 1.2 with C4 symmetry imposed. The distribution of particle orientations in the ice was very homogeneous (Extended Data Fig. 2c) and all views could be recovered following reference-free 2D classification (Extended Data Fig. 3d). This map allowed us to determine the transmembrane topology of the channel, place the previously solved structures of the N-terminal fragment and the two RYR repeat pairs, and build a preliminary Cα trace for the protomer. A class of particles was identified in this dataset in which the entire cytosolic region of a single protomer was disordered (Extended Data Fig. 3a). A difference map calculated between this class and a class where all protomers were ordered supports the protomer boundaries assigned in our model (Extended Data Fig. 3c). Although the quality of the density was excellent in the transmembrane region in this dataset, with helices distinctly defined and some bulky side chains visible, the quality of the map in the cytoplasmic region was lower, impeding model completion.
RyR1 is regulated by phosphorylation 51, and we were concerned that heterogeneous phosphorylation could contribute to the observed structural heterogeneity. In an attempt to improve the conformational homogeneity of the sample, as well as the overall resolution, we treated purified RyR1 with calf intestinal alkaline phosphatase (CIP) to obtain a uniformly dephosphorylated sample (Extended Data Fig. 1c). From this sample, we collected a dataset from which 322,400 particles were extracted. 3D-classification of CIP-treated RyR1 performed following the same procedure as for the original sample gave several well-populated classes (Extended Data Fig. 3b), with different conformations of the cytoplasmic assembly, similar to those observed in the first dataset. The best resolved class had an overall resolution of 5.0 Å, refined as a C4-symmetric particle. Although the overall resolution did not improve, the local resolution improved substantially in some regions compared to the previous protein preparation (Extended Data Fig. 4 c, d). This map has been used for the majority of map interpretation and model building described herein. It is important to note that the observed improvements in the map may relate to dephosphorylation of the sample, but may also be due to the larger size of this dataset.
Importantly, the local resolution in the cytosolic assembly was greatly improved in reconstruction obtained from the CIP treated sample (Extended Data Fig. 4b,c), and this map was utilized for completion of the model, as described below. The local resolution, as measured using the program ResMap, varies between 4.5 and 6 Å across most of the map (Extended Data Fig. 4 b–d).
Reported resolutions are based on the gold-standard FSC=0.143 criterion, and were corrected for the effects of a soft mask on the FSC curve using high-resolution noise substitution 52. Prior to visualization, all density maps were sharpened by applying a negative B-factor that was estimated using automated procedures 53.
Density maps segmentation and display
Cryo-EM reconstructions were segmented using the SEGGER module 54 implemented in UCSF Chimera 55. Segments were manually combined to generate segmented densities corresponding to individual structural domains, as represented in Fig 1 c–e and Ext data Fig. 5. All other maps were left unsegmented. All density figures were rendered and displayed using UCSF Chimera.
Model building
The 5.0 Å map of dephosphorylated RyR1 (Fig 1a) has allowed us to place all domains in RyR1 (Fig. 1b), as well as to define the boundaries of the four identical protomers that comprise the channel (Fig. 1c–e), and to build a largely complete C-α model for each protomer, comprising 3939 out of the 5027 residues that constitute the full-length polypeptide chain, in addition to the 108 residues comprising the co-purified subunit, calstabin2. Most of the unassigned residues are located in regions of the sequence that are predicted to be disordered or to lack regular secondary structure. In addition, several of the regions that are either not present in the model or are less well ordered (for example RY34), have previously been shown to be highly accessible to proteolysis consistent with their surface-exposure 56.
Given the absence of homologous structures for most domains in RyR1 it is important to address the limitations of our model. In some regions, specifically NTD A-C, the RY12 and RY34 repeat pairs, and the EF-hand pair, we were able to place a crystal structure or high sequence identity homology model. For the SPRY1/2/3 domains, we placed homology models, and although we are confident of the domain orientation and connectivity, the numbering within the SPRY domains is uncertain because of limited sequence identity with template structures. In all other areas we built the model de novo as a polyalanine trace, and did not attempt to model side chains, even where visible. Helices are clearly defined in the density in most regions of the map; in the best areas, the pitch and curvature of the helices is clearly defined and bulky side chains are visible, though not identifiable (Extended Data Fig. 5). Features of the map correspond well to the local resolution measured using the program ResMap (Extended Data Fig. 4 b–d). The model is of good quality in most regions, as demonstrated by analyses of the local correlation with the map (Extended Data Fig. 6). The two main regions where the correlation of the model with the map is lower are the SPRY domains (which are subject to the difficulties described above), and an unmodelled rod of density on the periphery of the transmembrane region, which we discuss below. The final model has been presented as a C-alpha trace, with sequence assignments based on the distance from known landmarks in the sequence, such as the N-terminal domains, for which high-resolution crystal structures are available (Extended Data Fig. 5).
Model building was performed manually in Coot 57. First, the high-resolution structure of the N-terminal three-domain fragment of RyR1 (PDB ID 2XOA) was placed in the previously assigned location, and the orientation of the domain was determined using the “jiggle-fit” algorithm implemented in Coot, using 1000 random candidate orientations of the domain followed by rigid body fitting. Homology models for the three SPRY domains were generated using the PHYRE2 web server, and fit to the density using the same procedure as for the N-terminal fragment, assigned as SPRY1-3 according to the rationale described in the main text. The two pairs of RYR repeats (RY12 and RY34) were modeled based on the structure of RY34 58 and fit to the density in a similar fashion.
One of the SPRY domains is directly connected to the N-terminal solenoid via an ordered, alpha-helical linker, identifying it as SPRY1. Another of the SPRY domains is directly connected to RY12, placing it as SPRY2, and the remaining SPRY domain has been assigned as SPRY3. The three SPRY domains pack side-by-side in a triangular arrangement near the corner of the cytoplasmic assembly.
In the primary sequence, RY12 is located between SPRY1 and SPRY2, while RY34 is located ~1000 residues after SPRY3, inserted in the stretch of sequence we have assigned to the bridging solenoid. Two densities of an appropriate size and shape to accommodate RYR repeat pairs are present in the map; one is located at the corner of the tetramer, contacting the SPRY domains, while the other is directly adjacent to the bridging solenoid. Based on these observations we have revised the assignment of the position of RYR repeats such that RY12 is located at the extreme outer corner of the channel (directly adjacent to the SPRY domains), and RY34 (containing the PKA phosphorylation site) projects upward from the bridging solenoid (Fig. 6).
The putative Ca2+ binding domain was placed in a similar fashion to the other domains for which template structures were available, using the structure of the C-lobe of yeast calmodulin (PDB ID 1LKJ) as a starting model. The rest of the model was built manually in Coot, using the secondary structure recognition algorithms in Coot to place and extend helices and build the linkages between them. Directionality was determined based on extension of the structure from known fragments, most importantly the N-terminal fragment, and secondary structure predictions 59 were used to guide alignment of the sequence with the model. After an initial build, segments of the structure were flexibly fit to the density using the real-space refinement algorithms implemented in Coot, with ProSMART restraints (based on high-resolution reference structures where available, otherwise on the assigned secondary structure) applied to allow for deformation and curvature of helices without degrading local geometry. The final model is presented as a C-alpha trace, comprising in full 3939 ordered residues for each of the four protomers.
Extended Data
Extended data figure 1. Skeletal muscle RyR1 purification.
a, Coomassie blue staining of SDS-PAGE showing CHAPS-solubilized SR membrane, calstabin2 elution from glutathione-GST-calstabin1 affinity chromatography column (AC) and the eluted RyR1 from FPLC size exclusion chromatograph (SEC). b, FPLC plot showing the RyR1 peak at ~7 mL elution and the excess calstabin2 (Cs2) peak at ~12 mL elution. c, Immunoblot analysis of CIP-treated RyR1 probed at indicated timepoints with, from bottom, anti-RyR1 (34C) antibody, anti-Phosphotyrosine antibody (Abcam ab10321), anti-Phosphothreonine antibody (Abcam ab79851), anti-Phosphoserine antibody (Abcam ab9332) and anti-RyR phospho-specific antibody (P-Ser 2843) that recognizes the PKA phosphorylated site on RyR1.
Extended data figure 2. Particle picking and 2D class averages.
a, Sample micrograph of the RyR1-CIP-EGTA dataset after motion correction, with red boxes around the particles picked by Autopicker 40 (Scale bar=500 Å). b, Sample power spectrum of a twice-decimated micrograph after motion correction. c, Euler angle distribution before symmetry was imposed of the particles that went into the CIP-treated dataset final reconstruction. Latitude corresponds to θ from 0 to 90°. Longitude corresponds to ψ, from 0 to 360°. The dots color and area represent the number of particles in each view. d, 2D projections of the final CIP-treated map (left columns) compared to their respective reference-free 2D class averages (right columns).
Extended data figure 3. Classification and protomer boundaries.
a, Classification of the RyR1-EGTA dataset. First row: refined volume with all particles. Second row: primary classification with a number of classes K=10 giving rise to two major classes, one refined to 4.8 Å (blue box) and one refined to 5.0Å (green box). This class (green box) was subclassified with K=10 (third row) and yielded one class with a missing or disordered cytosolic portion of a protomer (red box). b, Classification of the RYR1-EGTA-CIP-treated dataset with K=8. Class8 (purple box) was refined to 5.0 Å. c, Views from the cytosol, membrane plane and lumen of the RyR1 model superimposed with a difference map between the full tetramer map (blue box) and the map with the cytosolic region of one protomer missing or disordered (red box).
Extended data figure 4. RyR1 cryo-EM local resolution map.
a, Gold-standard Fourier shell correlation curve for the 3D reconstructions, marked with resolutions corresponding to FSC=0.143. b, Cytosolic, membrane plane and luminal views of RyR1 (EGTA and CIP treated data set) local resolution distribution from 4 (blue) to 6 (red) Å resolution. c, local resolution distribution through a slab of density coincident with channel axis. d, Same slab as (c) for the EGTA treated without CIP. e, Slices through the volume of the CIP-EGTA dataset (top) and EGTA dataset (bottom). Slice direction and number are indicated on the images.
Extended data figure 5. Representative densities of RyR1 selected regions.
Representative electron density (gray mesh) in selected regions of the map. The protomers are represented as C-α traces, in different colors for clarity, with enlarged views of the following regions; a, calstabin2 b, the bridge solenoid c, N-terminal domain d, the pore region e, S6.
Extended data figure 6. RyR1 local model to map correlation.
a, Cytosolic, membrane plane and luminal views of the local correlation (calculated in a 5X5 voxel sliding window) between a map calculated from the model (filtered to 5 Å) and the electron density map of dephosphorylated RyR1, depicted in spectral coloring from 0.7 (red) to 1 (blue). b, local model/density map correlation within a slab of density through the plane of the membrane, highlighting the unmodelled rod of density on the periphery of the TM region and c, a slab coinciding with the channel axis.
Extended data figure 7. α-solenoid subdomains.
RyR1 electron density map (grey semitransparent surface) superimposed with the α-solenoid scaffold of RyR1. a, Core of the α-solenoid scaffold (insertions and elaborations are not shown). Green, bridging solenoid; blue, NTD solenoid; red, core solenoid. b, Alignment of NTD with an α-solenoid structure (PDB ID 3NMX). c, Alignment of core solenoid with an αsolenoid structure (PDB ID 1G3J). d, overlay of bridging solenoid with an α-solenoid structure (PDB ID 1WA5). In panels b through d, RyR1 α-solenoid repeats are depicted in spectral coloring from blue (N-terminus) to red (C-terminus), and the aligned α-solenoid protein is represented in dark grey.
Extended data figure 8. Architecture of bridging and core solenoids.
a, Electron density map of RyR1 in dark blue mesh superimposed with the bridging solenoid shown in detail on right, as labeled. b, two views of the interaction of the core solenoid (spectral coloring) containing the putative Ca2+-binding domain with CTD (grey) as labeled.
Extended data figure 9. Calstabin2 binding site.
Views in the membrane plane and cytosol of RyR1 with enlarged views of calstabin2 (yellow) bound to RyR1. SPRY1 is depicted in light blue, SPRY2 in cyan, the bridging solenoid in green and the calstabin binding helix in purple.
Extended data figure 10. Putative Ca2+ binding domain in RyR1.
a, sequence alignment of rabbit RyR1,2&3 with the C-lobe of human calmodulin. b, structural alignment of the C-lobe of yeast calmodulin with the model of RyR1.
Acknowledgments
We thank Richard Axel for helpful discussions and Melissa Thomas for assistance with the preparation of illustrations. This work was supported by grants from the National Institutes of Health (R01AR060037 and R01HL061503 to A.R.M, U54GM095315 to W.A.H., R01GM29169 to J.F.), by HHMI (to J.F.). R.Z. was a fellow of the American Heart Association (0625919T), and O.B.C. was supported by an overseas biomedical fellowship (NHMRC; Australia).
Footnotes
Author Contributions A.R.M. conceived the project, designed experiments, analyzed data and wrote the manuscript. W.A.H. conceived the project, designed experiments, analyzed data and wrote the manuscript. J.F. designed the cryo-EM experiments, analyzed data and wrote the manuscript. R.Z. designed experiments, isolated the RyR1 used in this work, analyzed data and wrote the manuscript. O.B.C. designed experiments, analyzed data, built the atomic model and wrote the manuscript. F.M. designed experiments. A.G. designed experiments and conducted the cryo-EM experiments, including data acquisition and processing and wrote the manuscript. R.A.G. conducted the cryo-EM experiments. S.R. conducted Phospho-RyR Western blots experiments.
Author Information Cryo-EM reconstructions of RyR1 have been deposited in the Electron Microscopy Data Bank under the accession number EMD-6106, EMD-6107 (RyR1)
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of the paper.
Online content Methods, Extended Data display items and Source Data are available in the online version of the paper; references unique to these sections appear only in the online paper.
References
- 1.Zalk R, Lehnart SE, Marks AR. Modulation of the ryanodine receptor and intracellular calcium. Annual review of biochemistry. 2007;76:367–385. doi: 10.1146/annurev.biochem.76.053105.094237. [DOI] [PubMed] [Google Scholar]
- 2.Marx SO, et al. PKA phosphorylation dissociates FKBP12.6 from the calcium release channel (ryanodine receptor): defective regulation in failing hearts. Cell. 2000;101:365–376. doi: 10.1016/s0092-8674(00)80847-8. [DOI] [PubMed] [Google Scholar]
- 3.Paolini C, Protasi F, Franzini-Armstrong C. The relative position of RyR feet and DHPR tetrads in skeletal muscle. Journal of molecular biology. 2004;342:145–153. doi: 10.1016/j.jmb.2004.07.035. [DOI] [PubMed] [Google Scholar]
- 4.Meissner G, Lu X. Dihydropyridine receptor-ryanodine receptor interactions in skeletal muscle excitation-contraction coupling. Bioscience reports. 1995;15:399–408. doi: 10.1007/BF01788371. [DOI] [PubMed] [Google Scholar]
- 5.Stern MD, Pizarro G, Rios E. Local control model of excitation-contraction coupling in skeletal muscle. The Journal of general physiology. 1997;110:415–440. doi: 10.1085/jgp.110.4.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marx SO, Ondrias K, Marks AR. Coupled gating between individual skeletal muscle Ca2+ release channels (ryanodine receptors) Science. 1998;281:818–821. doi: 10.1126/science.281.5378.818. [DOI] [PubMed] [Google Scholar]
- 7.Wehrens XH, et al. FKBP12.6 deficiency and defective calcium release channel (ryanodine receptor) function linked to exercise-induced sudden cardiac death. Cell. 2003;113:829–840. doi: 10.1016/s0092-8674(03)00434-3. [DOI] [PubMed] [Google Scholar]
- 8.Liu X, et al. Role of leaky neuronal ryanodine receptors in stress-induced cognitive dysfunction. Cell. 2012;150:1055–1067. doi: 10.1016/j.cell.2012.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andersson DC, et al. Ryanodine receptor oxidation causes intracellular calcium leak and muscle weakness in aging. Cell metabolism. 2011;14:196–207. doi: 10.1016/j.cmet.2011.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bellinger AM, et al. Hypernitrosylated ryanodine receptor calcium release channels are leaky in dystrophic muscle. Nature medicine. 2009;15:325–330. doi: 10.1038/nm.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marks AR. Calcium cycling proteins and heart failure: mechanisms and therapeutics. The Journal of clinical investigation. 2013;123:46–52. doi: 10.1172/JCI62834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andersson DC, Marks AR. Fixing ryanodine receptor Ca leak - a novel therapeutic strategy for contractile failure in heart and skeletal muscle. Drug discovery today. Disease mechanisms. 2010;7:e151–e157. doi: 10.1016/j.ddmec.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Serysheva, et al. Subnanometer-resolution electron cryomicroscopy-based domain models for the cytoplasmic region of skeletal muscle RyR channel. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9610–9615. doi: 10.1073/pnas.0803189105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernandez IS, et al. Molecular architecture of a eukaryotic translational initiation complex. Science. 2013;342:1240585. doi: 10.1126/science.1240585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. Journal of structural biology. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Groves MR, Barford D. Topological characteristics of helical repeat proteins. Current opinion in structural biology. 1999;9:383–389. doi: 10.1016/s0959-440x(99)80052-9. [DOI] [PubMed] [Google Scholar]
- 17.Xiong L, Zhang JZ, He R, Hamilton SL. A Ca2+-binding domain in RyR1 that interacts with the calmodulin binding site and modulates channel activity. Biophysical journal. 2006;90:173–182. doi: 10.1529/biophysj.105.066092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tung CC, Lobo PA, Kimlicka L, Van Petegem F. The amino-terminal disease hotspot of ryanodine receptors forms a cytoplasmic vestibule. Nature. 2010;468:585–588. doi: 10.1038/nature09471. nature09471 [pii] [DOI] [PubMed] [Google Scholar]
- 19.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic acids research. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Groves MR, Barford D. Current opinion in structural biology. 1999;9:383–389. doi: 10.1016/s0959-440x(99)80052-9. [DOI] [PubMed] [Google Scholar]
- 21.Huber AH, Nelson WJ, Weis WI. Cell. 1997;90:871–882. doi: 10.1016/s0092-8674(00)80352-9. [DOI] [PubMed] [Google Scholar]
- 22.Yu FH, Yarov-Yarovoy V, Gutman GA, Catterall WA. Overview of molecular relationships in the voltage-gated ion channel superfamily. Pharmacological reviews. 2005;57:387–395. doi: 10.1124/pr.57.4.13. [DOI] [PubMed] [Google Scholar]
- 23.Payandeh J, Scheuer T, Zheng N, Catterall WA. Nature. 2011;475:353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504:107–112. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Owsianik G, Talavera K, Voets T, Nilius B. Permeation and selectivity of TRP channels. Annual review of physiology. 2006;68:685–717. doi: 10.1146/annurev.physiol.68.040204.101406. [DOI] [PubMed] [Google Scholar]
- 26.Tripathy A, Xu L, Mann G, Meissner G. Calmodulin activation and inhibition of skeletal muscle Ca2+ release channel (ryanodine receptor) Biophys J. 1995;69:106–119. doi: 10.1016/S0006-3495(95)79880-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Volkov AG, Paula S, Deamer DW. Two mechanisms of permeation of small neutral molecules and hydrated ions across phospholipid bilayers. Bioelectroch Bioener. 1997;42:153–160. doi: 10.1016/S0302-4598(96)05097-0. [DOI] [Google Scholar]
- 28.Jiang Y, et al. Nature. 2003;423:33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- 29.Bezprozvanny I, Watras J, Ehrlich BE. Bell-shaped calcium-response curves of Ins(1,4,5)P3- and calcium-gated channels from endoplasmic reticulum of cerebellum. Nature. 1991;351:751–754. doi: 10.1038/351751a0. [DOI] [PubMed] [Google Scholar]
- 30.Ludtke SJ, Serysheva II, Hamilton SL, Chiu W. The pore structure of the closed RyR1 channel. Structure (Camb) 2005;13:1203–1211. doi: 10.1016/j.str.2005.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Samso M, Wagenknecht T, Allen PD. Internal structure and visualization of transmembrane domains of the RyR1 calcium release channel by cryo-EM. Nature structural & molecular biology. 2005;12:539–544. doi: 10.1038/nsmb938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuchi Z, Lau K, Van Petegem F. Disease mutations in the ryanodine receptor central region: crystal structures of a phosphorylation hot spot domain. Structure. 2012;20:1201–1211. doi: 10.1016/j.str.2012.04.015. [DOI] [PubMed] [Google Scholar]
- 33.Sharma P, et al. Structural determination of the phosphorylation domain of the ryanodine receptor. The FEBS journal. 2012;279:3952–3964. doi: 10.1111/j.1742-4658.2012.08755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marks AR, Fleischer S, Tempst P. Surface topography analysis of the ryanodine receptor/junctional channel complex based on proteolysis sensitivity mapping. The Journal of biological chemistry. 1990;265:13143–13149. [PubMed] [Google Scholar]
- 35.Yin CC, Han H, Wei R, Lai FA. Two-dimensional crystallization of the ryanodine receptor Ca2+ release channel on lipid membranes. Journal of structural biology. 2005;149:219–224. doi: 10.1016/j.jsb.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 36.Woo JS, et al. Structural and functional insights into the B30. 2/SPRY domain. The EMBO journal. 2006;25:1353–1363. doi: 10.1038/sj.emboj.7600994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brillantes AB, et al. Stabilization of calcium release channel (ryanodine receptor) function by FK506-binding protein. Cell. 1994;77:513–523. doi: 10.1016/0092-8674(94)90214-3. [DOI] [PubMed] [Google Scholar]
- 38.Gaburjakova M, et al. FKBP12 binding modulates ryanodine receptor channel gating. The Journal of biological chemistry. 2001;276:16931–16935. doi: 10.1074/jbc.M100856200. [DOI] [PubMed] [Google Scholar]
- 39.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta crystallographica. Section D, Biological crystallography. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 40.Langlois R, et al. Automated particle picking for low-contrast macromolecules in cryo-electron microscopy. Journal of structural biology. 2014;186:1–7. doi: 10.1016/j.jsb.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xin HB, Timerman AP, Onoue H, Wiederrecht GJ, Fleischer S. Affinity purification of the ryanodine receptor/calcium release channel from fast twitch skeletal muscle based on its tight association with FKBP12. Biochemical and biophysical research communications. 1995;214:263–270. doi: 10.1006/bbrc.1995.2283. [DOI] [PubMed] [Google Scholar]
- 42.Bezprozvanny I, Watras J, Ehrlich BE. Bell-shaped calcium-response curves of Ins(1,4,5)P3- and calcium-gated channels from endoplasmic reticulum of cerebellum. Nature. 1991;351:751–754. doi: 10.1038/351751a0. [DOI] [PubMed] [Google Scholar]
- 43.Dubochet J, et al. Cryo-electron microscopy of vitrified specimens. Quarterly reviews of biophysics. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- 44.Wagenknecht T, Frank J, Boublik M, Nurse K, Ofengand J. Direct localization of the tRNA--anticodon interaction site on the Escherichia coli 30 S ribosomal subunit by electron microscopy and computerized image averaging. Journal of molecular biology. 1988;203:753–760. doi: 10.1016/0022-2836(88)90207-0. [DOI] [PubMed] [Google Scholar]
- 45.Suloway C, et al. Automated molecular microscopy: the new Leginon system. Journal of structural biology. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 46.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nature methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frank J, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. Journal of structural biology. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 48.Langlois R, et al. Automated particle picking for low-contrast macromolecules in cryo-electron microscopy. Journal of structural biology. 2014;186:1–7. doi: 10.1016/j.jsb.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scheres SH. A Bayesian view on cryo-EM structure determination. Journal of molecular biology. 2012;415:406–418. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. Journal of structural biology. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reiken S, et al. PKA phosphorylation activates the calcium release channel (ryanodine receptor) in skeletal muscle: defective regulation in heart failure. The Journal of cell biology. 2003;160:919–928. doi: 10.1083/jcb.200211012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen S, et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy. 2013;135:24–35. doi: 10.1016/j.ultramic.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. Journal of molecular biology. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 54.Pintilie GD, Zhang J, Goddard TD, Chiu W, Gossard DC. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. Journal of structural biology. 2010;170:427–438. doi: 10.1016/j.jsb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 56.Marks AR, Fleischer S, Tempst P. Surface topography analysis of the ryanodine receptor/junctional channel complex based on proteolysis sensitivity mapping. The Journal of biological chemistry. 1990;265:13143–13149. [PubMed] [Google Scholar]
- 57.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica. Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tung CC, Lobo PA, Kimlicka L, Van Petegem F. The amino-terminal disease hotspot of ryanodine receptors forms a cytoplasmic vestibule. Nature. 2010;468:585–588. doi: 10.1038/nature09471. nature09471 [pii] [DOI] [PubMed] [Google Scholar]
- 59.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction 0server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]