Abstract
How DNA demethylation is achieved in mammals is still under extensive investigation. One proposed mechanism is deamination of 5-hydroxymethylcytosine to form 5-hydroxymethyluracil (5hmU), followed by base excision repair to replace the mismatched 5hmU with cytosine. In this process, 5hmU:G mispair serves as a key intermediate and its localization and distribution in mammalian genome could be important information to investigate the proposed pathway. Here we describe a selective labeling method to map mismatched 5hmU. After converting other cytosine modifications to 5-carboxylcytosines, a biotin tag is installed onto mismatched 5hmU through β-glucosyltransferase-catalyzed glucosylation and click chemistry. The enriched 5hmU-containing DNA fragments can be subject to subsequent sequencing to reveal the distribution of 5hmU:G mispair with base-resolution information acquired.
Keywords: 5-Hydroxymethyluracil, Deamination, Demethylation, Tet proteins
1. Introduction
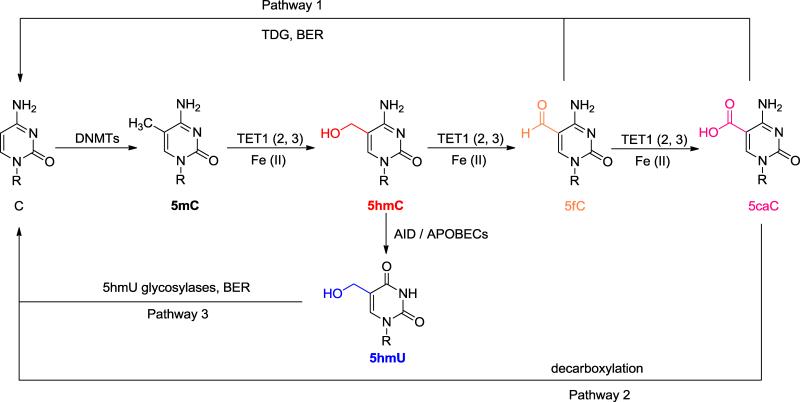
5-methylcytosine (5mC) is an important epigenetic mark in mammalian cells and is regarded as the fifth base besides A, T, C and G. 5mC has been known to impact various biological functions such as genomic imprinting, X chromosome inactivation and cancer development [1, 2]. Although 5mC appears to be a relatively stable modification, there are clear observations of demethylation in both zygotes and somatic cells, ranging from genome-wide to a few loci [3]. However, it is still obscure how demethylation is initiated with multiple pathways proposed [3]. Recently, the discovery of oxidative derivatives of 5mC in mammalian cells opens up new possibilities for demethylation mechanisms (Fig. 1) [4-7]. Pathway 1 is thymine DNA glycosylase (TDG)-mediated base excision repair (BER). Both 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC), oxidized products of 5mC, can be recognized by TDG and converted back to unmodified cytosine in high efficiency [6, 8, 9]. Pathway 2 is direct decarboxylation of 5caC. Although no decarboxylase has been identified, decarboxylation activity in mouse embryonic stem (mES) cell lysate has been suggested [10]. Pathway 3 is deamination of 5hmC followed by BER. AID (activation-induced deaminase) /APOBEC (apolipoprotein B mRNA-editing enzyme complex) family proteins, which catalyze the deamination of cytosine to uracil, may also work on 5hmC to produce 5hmU:G mispair that can be excised by glycosylases and repaired through BER [11, 12]. It has been reported that the amount of 5hmU is positively correlated with the expression level of Tet1 but negatively correlated with the expression level of AID in vivo, suggesting AID/APOBEC may contribute to demethylation [12]. However, recent biochemistry study showed that purified AID/APOBECs exhibit very low deamination activity on 5hmC, raising questions about the feasibility of this pathway [13].
Fig. 1.
Proposed 5mC demethylation pathways in mammals. Pathway 1, cleavage of 5fC and 5caC by TDG followed by BER; Pathway 2, direct decarboxylation of 5caC; Pathway 3, deamination of 5hmC followed by BER.
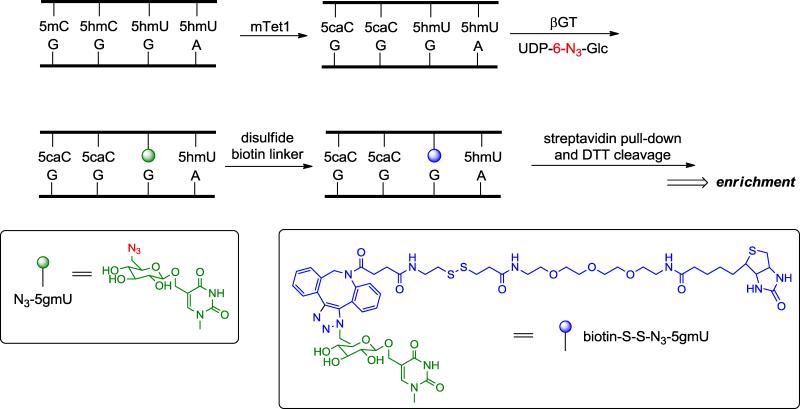
To further explore the plausibility of deamination-involved demethylation pathway, it will be important to gain the knowledge about the localization of mismatched 5hmU, derived directly from 5hmC deamination. Thererfore, an efficient and robust labeling approach is required. The structural similarities between 5hmC and 5hmU make it possible to apply certain 5hmC labeling and profiling methods to 5hmU. β-glucosyltransferase (βGT) from T4 bacteriophage is known to catalyze the glucosylation reaction on the hydroxyl group of 5hmC. We found that βGT can also work on mismatched 5hmU:G but not matched 5hmU:A, which prompted us to design the following strategy for chemical labeling of mismatched 5hmU (Fig. 2). First, recombinant mouse Tet1 is utilized to oxidize all 5mC and 5hmC in genomic DNA to 5caC. Then βGT is applied to install a modified N3-glucose onto the hydroxyl group of mismatched 5hmU followed by incorporation of disulfide biotin linker through click chemistry. After capture of the mismatched-5hmU-containing fragments with streptavidin-coupled beads, the bound DNA fragments can be readily released by simple DTT cleavage of the disulfide bond. The enriched fragments can be applied to deep sequencing to map the distribution of mismatched 5hmU. The precise position of the mismatched 5hmU may be determined by analyzing C-to-T mutation around the identified peaks.
Fig. 2.
Overview of the chemical labeling and enrichment strategy for mismatched 5hmU. Genomic DNA is first treated with recombinant mTet1 to convert all 5mC and 5hmC to 5caC. Then βGT is utilized to selectively label mismatched 5hmU with N3-glucose. After adding the biotin tag through click chemistry, the mismatched 5hmU-containing DNA fragments are enriched by streptavidin-coupled beads.
1.1 Mismatched 5hmU chemical labeling with N3-glucose and biotin
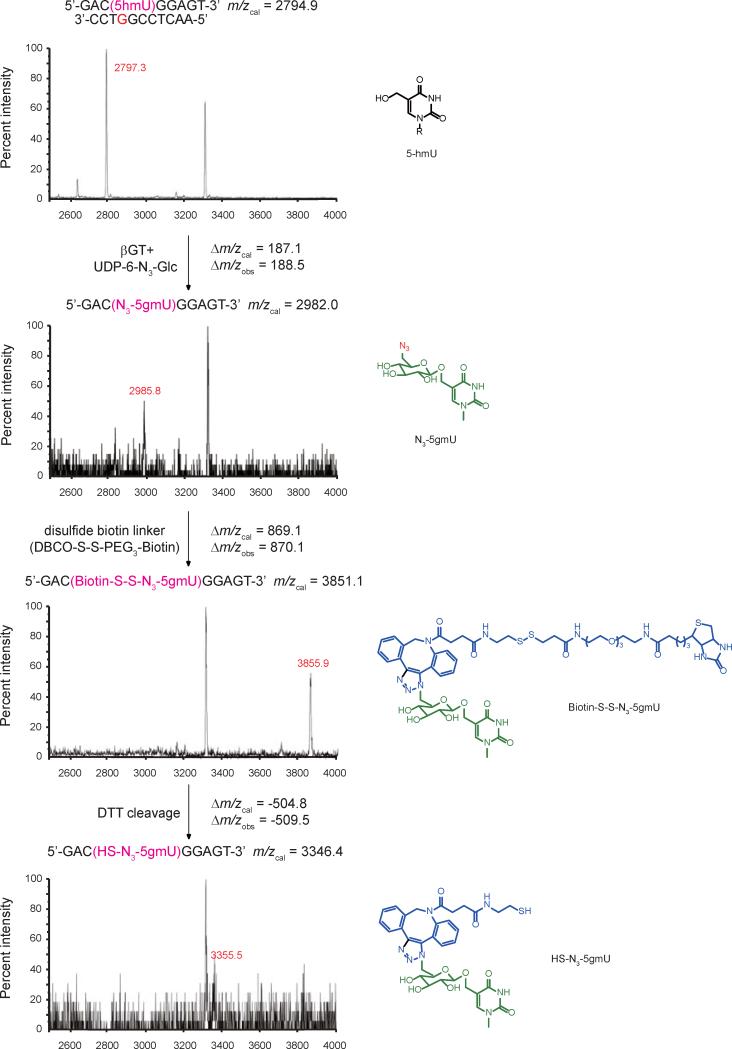
βGT transfers the glucose moiety from uridine diphosphoglucose (UDP-Glc) to 5hmC on double-stranded DNA [14, 15]. Previously, we have demonstrated that βGT can utilize modified UDP-6-N3-Glc as a cofactor with only slight decrease in reaction rate for 5hmC substrate [16]. However, there is not much study about the activity of βGT on 5hmU. To test whether βGT works on 5hmU in a similar way, we attempted the glucosylation reaction on a model 9mer-11mer duplex DNA containing one mismatched 5hmU (5hmU:G) or matched 5hmU (5hmU:A) site, and monitored the products by MALDI-TOF/TOF. In the presence of 2 μM βGT and 200 μM UDP-6-N3-Glc, only mismatched 5hmU could be glucosylated to form N3-5gmU while matched 5hmU stayed untouched (Fig. 3 and Fig S1A). The yield of glucosylation reaction on mismatched 5hmU is over 90%, which was confirmed by HPLC (Fig. S2). The resulting N3-5gmU could be further modified to add a biotin tag by reacting with the disulfide-containing biotin linker through copper-free click chemistry to yield biotin-S-S-N3-5gmU. The disulfide bond on the biotin linker can be readily cleaved under DTT treatment to recover the enriched DNA fragments after pull-down. The reaction products of each step were confirmed using the model 9mer-11mer duplex DNA containing one mismatched 5hmU with MALDI-TOF/TOF (Fig. 3).
Fig. 3.
Mass spectrometry characterization of the products in 5hmU-labeling reactions with model DNA. The duplex DNA with 5hmU on the 9-mer strand was annealed to the mismatched (opposite to 5hmU site) 11-mer complementary strand. The DNA was subject to the treatment described in Fig. 2 and monitored by MALDI-TOF/TOF with the calculated and observed molecular weight indicated.
1.2 Behavior of 5mC, 5hmC and mismatched 5hmU upon recombinant mTet1 treatment
The TET gene was first identified as the fusion partner of Mixed Lineage Leukemia (MLL) gene in acute myeloid leukemia (AML) with unknown biological function [17, 18]. In 2009, the TET family proteins were found to be responsible for converting 5mC to 5hmC through FeII- and 2-oxoglutarate-dependent oxidation [4]. Further investigation showed that TET proteins can further oxidize 5hmC to 5fC and 5caC [6, 7].
For efficient enrichment of mismatched 5hmU, it is critical to avoid the N3-glucose and biotin labeling on 5hmC (intrinsic ones in genomic DNA or derived from incomplete 5mC oxidation). Hence, the high conversion rate of 5mC/5hmC to 5caC by mTet1 is extremely important before specific labeling of 5hmU:G. On the other hand, mismatched 5hmU has to survive the mTet1 oxidation for the subsequent labeling. To test that, we performed the oxidation reaction on the 9mer-11mer duplex DNA containing one 5mC, 5hmC or mismatched 5hmU site with excess recombinant mTet1. Both 5mC and 5hmC were oxidized to 5caC almost completely while the mismatched 5hmU was not the substrate of mTet1-mediated oxidation (Fig. S1B).
1.3 Selectivity and efficiency of mismatched 5hmU labeling
To examine whether this method can discriminate between mismatched 5hmU and other modifications, we treated 76mer double-stranded DNA containing 5mC, 5hmC, 5hmU:G or 5hmU:A as described in Fig. 2, and checked the final pull-down yields after DTT cleavage and purification. As shown in Table 1, 26% of 5hmU:G-containing DNA was recovered while the recovery rate for all other DNAs were only ~1%, indicating that the labeling of 5hmU:G is both selective and efficient.
Table 1.
The pull-down yield of 76mer model DNA with different modifications.
| Modification | Pull-down yield (% of total input) |
|---|---|
| 5hmU:G (mismatch) | 26.3% |
| 5mC | 1.1% |
| 5hmC | 1.3% |
| 5hmU:A | 1.0% |
2. Method
2.1 mTet1 oxidation
The genomic DNA was sheared to desired size range and purified with QIAquick PCR purification kit (Qiagen) and elute in Milli-Q water. The oxidation reactions were performed in multiple 50-μl solution containing 50 mM HEPES (pH 8.0), 100 μM ammonium iron (II) sulfate, 1 mM α-ketoglutarate, 2 mM ascorbic acid, 2.5 mM DTT, 100 mM NaCl, 1.2 mM ATP, 10 ng/μl sheared genomic DNA and 3 μM recombinant mTet1. After incubating the reaction at 37 °C for 1.5 h, 1 μl proteinase K (20 mg/ml) was added, followed by another 1 h incubation at 50 °C. The oxidized genomic DNA was cleaned up with Micro Bio-Spin 30 Columns (Bio-Rad) first, then applied to QIAquick PCR purification kit (Qiagen). The purified DNA is eluted in Milli-Q water.
2.2 βGT labeling
The oxidized DNA was glucosylated in 50 mM HEPES (pH 8.0), 25 mM MgCl2, 2 μM βGT and 200 μM UDP-6-N3-Glc at 37 °C for 1.5 h. The glucosylated DNA was purified with QIAquick PCR purification Kit (Qiagen) and eluted in Milli-Q water. A stock solution (1 mM) of disulfide biotin linker (DBCO-S-S-PEG3-Biotin, Click Chemistry Tools) in DMSO was prepared. The disulfide biotin linker (150 μM) was mixed with glucosylated DNA and incubated at 37 °C for 2 h. The product was cleaned up using QIAquick PCR purification Kit (Qiagen) and eluted in 50 μl Milli-Q water. The purified labeled DNA was ready for the following pull-down procedures.
2.3 Enrichment of mismatched 5hmU-containing fragments
2.3.1 Solutions and reagents
Dynabeads MyOne Streptavidin C1 (Invitrogen); 1×Binding & Washing (B&W) buffer: 5 mM Tris-HCl (pH 7.5), 0.5 mM EDTA, 1 M NaCl, 0.01% Tween-20; 2×Binding & Washing (B&W) buffer: 10 mM Tris-HCl (pH 7.5), 1 mM EDTA, 2 M NaCl, 0.02% Tween-20; 100 mM DTT solution (freshly prepared).
2.3.2 Procedure
Resuspend the beads thoroughly and transfer 25 μl to a clean tube. Place the tube on magnet and remove the supernatant by careful aspiration. Remove the tube from the magnet and wash with 25 μl 1×B&W buffer four times. After final wash, resuspend the beads in 50 μl 2×B&W buffer and add the labeled DNA from previous step (50 μl in Milli-Q water). Incubate for 15 minutes at room temperature with gentle rotation. After incubation, separate the DNA coated beads with a magnet and discard the supernatant. Wash the beads six times with 100 μl 1×B&W buffer each time. To release the enriched DNA fragments from the beads, add 50 μl freshly prepared 100 mM DTT solution to the coated beads and incubate for 2 h at room temperature with gentle rotation. Separate the beads on magnet and purify the supernatant containing DNA with the Micro Bio-Spin 6 Columns (Bio-Rad) and QIAquick PCR purification kit (Qiagen). The eluate now contains the mismatched 5hmU-enriched DNA and may be applied to the library construction and sequencing to detect the 5hmU:G mispair.
3. Results
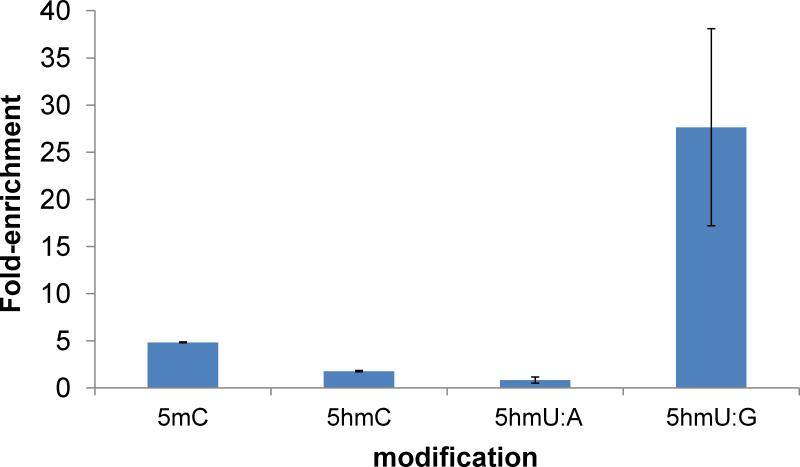
To measure the enrichment efficiency in the real sample, we added a pool of spike-ins containing C, 5mC, 5hmC, 5hmU:A or 5hmU:G to the mES genomic DNA and performed the mismatched 5hmU pull-down assay. The enrichment folds of each modification were analyzed by quantitative PCR and normalized to the ‘C’-containing control (Fig. 4). A significant enrichment of mismatched 5hmU control (~27 folds) was observed with slight enrichment of 5mC control (~5 folds), probably due to the incomplete oxidation. No enrichment was observed for 5hmC or matched 5hmU control.
Fig. 4.
Enrichment test of mismatched 5hmU pull-down assay. DNA containing C, 5mC, 5hmC, 5hmU:A, or 5hmU:G was added into mES genomic DNA as spike-in controls. Values shown are fold-enrichment over input, normalized to ‘C’-containing DNA.
In summary, taking advantage of the TET-mediated oxidation [19, 20], we provided a profiling method for mismatched 5hmU to examine the deamination product of 5hmC. The limitation of this method is that the level of 5hmU is much lower than that of 5mC and 5hmC in most genomic DNA, which may make it challenging to differentiate the mismatched 5hmU peaks from the background (non-oxidized 5hmC or partially oxidized 5mC). The presented method is able to identify the relatively abundant, mismatched 5hmU peaks with normal sequencing depth and the C-to-T mutation sites around the peak could mark the exact location of 5hmU site; however, for low abundance 5hmU sites, more sequencing depths are required to distinguish these peaks from the background. The screening of C-to-T mutations around the peak region can also help validate the identified 5hmU peaks.
Supplementary Material
Highlights.
Mismatched 5hmU can be selectively labeled with biotin through βGT-catalyzed glucosylation followed by click chemistry.
After Tet-mediated 5mC and 5hmC oxidation mismatched 5hmU can be efficiently labeled and enriched.
This selective labeling and pull-down assay can be applied to profile the mismatched 5hmU in genomic DNA.
Acknowledgments
The study described in this paper was supported by National Institutes of Health HG006827 and Howard Hughes Medical Institute (C.H.).
Abbreviations
- 5mC
5-methylcytosine
- 5hmC
5-hydroxymethylcytosine
- 5fC
5-formylcytosine
- 5caC
5-carboxylcytosine
- 5hmU
5-hydroxymethyluracil
- BER
base excision repair
- TET
Ten-eleven translocation
- TDG
thymine DNA glycosylase
- βGT
β-glucosyltransferase
- AID
activation-induced deaminase
- APOBEC
apolipoprotein B mRNA-editing enzyme complex
- UDP-Glc
uridine diphosphoglucose
- mES
mouse embryonic stem
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Gal-Yam EN, Saito Y, Egger G, Jones PA. Annu. Rev. Med. 2008;59:267–280. doi: 10.1146/annurev.med.59.061606.095816. [DOI] [PubMed] [Google Scholar]
- 2.Jones PA. Nature reviews. Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 3.Wu SC, Zhang Y. Nat. Rev. Mol. Cell. Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kriaucionis S, Heintz N. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, Sun Y, Li X, Dai Q, Song CX, Zhang K, He C, Xu GL. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maiti A, Drohat AC. J. Biol. Chem. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang L, Lu X, Lu J, Liang H, Dai Q, Xu GL, Luo C, Jiang H, He C. Nat. Chem. Biol. 2012;8:328–330. doi: 10.1038/nchembio.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schiesser S, Hackner B, Pfaffeneder T, Muller M, Hagemeier C, Truss M, Carell T. Angew. Chem. Int. Ed. Engl. 2012;51:6516–6520. doi: 10.1002/anie.201202583. [DOI] [PubMed] [Google Scholar]
- 11.Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, Abramowitz LK, Bartolomei MS, Rambow F, Bassi MR, Bruno T, Fanciulli M, Renner C, Klein-Szanto AJ, Matsumoto Y, Kobi D, Davidson I, Alberti C, Larue L, Bellacosa A. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo JU, Su Y, Zhong C, Ming GL, Song H. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nabel CS, Jia H, Ye Y, Shen L, Goldschmidt HL, Stivers JT, Zhang Y, Kohli RM. Nat. Chem. Biol. 2012;8:751–758. doi: 10.1038/nchembio.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Josse J, Kornberg A. The Journal of biological chemistry. 1962;237:1968–1976. [PubMed] [Google Scholar]
- 15.Lariviere L, Morera S. J. Biol. Chem. 2004;279:34715–34720. doi: 10.1074/jbc.M404394200. [DOI] [PubMed] [Google Scholar]
- 16.Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, Wang J, Zhang L, Looney TJ, Zhang B, Godley LA, Hicks LM, Lahn BT, Jin P, He C. Nat. Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ono R, Taki T, Taketani T, Taniwaki M, Kobayashi H, Hayashi Y. Cancer. Res. 2002;62:4075–4080. [PubMed] [Google Scholar]
- 18.Lorsbach RB, Moore J, Mathew S, Raimondi SC, Mukatira ST, Downing JR. Leukemia. 2003;17:637–641. doi: 10.1038/sj.leu.2402834. [DOI] [PubMed] [Google Scholar]
- 19.Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, Min JH, Jin P, Ren B, He C. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L, Szulwach KE, Hon GC, Song CX, Park B, Yu M, Lu X, Dai Q, Wang X, Street CR, Tan H, Min JH, Ren B, Jin P, He C. Nat. Commun. 2013;4:1517. doi: 10.1038/ncomms2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.