Abstract
Although amyloid imaging with PiB-PET, and now with F-18-labelled tracers, has produced remarkably consistent qualitative findings across a large number of centers, there has been considerable variability in the exact numbers reported as quantitative outcome measures of tracer retention. In some cases this is as trivial as the choice of units, in some cases it is scanner dependent, and of course, different tracers yield different numbers. Our working group was formed to standardize quantitative amyloid imaging measures by scaling the outcome of each particular analysis method or tracer to a 0 to 100 scale, anchored by young controls (≤45 years) and typical Alzheimer’s disease patients. The units of this scale have been named “Centiloids.” Basically, we describe a “standard” method of analyzing PiB PET data and then a method for scaling any “non-standard” method of PiB PET analysis (or any other tracer) to the Centiloid scale.
INTRODUCTION
As biomarkers have been incorporated with increasing frequency into multicenter research collaborations and clinical trials, the need for standardization of: 1) specimen or data collection; 2) biomarker assay; 3) analysis of data; and 4) reporting of results has become apparent. A lack of comparable methods across laboratories impedes the combination of data across sites within a single study and limits meta-analyses across studies. Lack of standardization prevents the application of universal cutoffs between normal and abnormal ranges. It is also difficult to compare longitudinal changes in quantitative terms without standardized units. The sources of variability vary with the particular biomarker are a cause for concern in all biomarker studies. Biomarker researchers working with cerebrospinal fluid (CSF) analytes and brain volumetric measurements by magnetic resonance imaging (MRI) have recognized this and have already begun collaborative efforts to standardize methods and outcomes across laboratories [1-6].
The need for standardization is equally important in amyloid positron emission tomography (PET). In amyloid PET, causes of variability include the particular amyloid tracer used, acquisition time duration, method of analysis, target and reference regions employed and partial volume correction (of lack thereof). Instrumentation issues such as scanner model, reconstruction algorithm and method of attenuation correction also challenge efforts towards standardization. The recent proliferation of amyloid PET tracers, each with somewhat different properties, has added to the variability in quantitatively expressed outcome data. The result of this lack of standardization in amyloid PET has led to: 1) a fairly wide range of “typical” values in amyloid-negative subjects (i.e., the normal range); 2) lack of a clear definition of amyloid loads typically associated with clinical dementia vs. levels that are only just outside of the amyloid-negative range but are seldom associated with dementia (i.e., a dementia cutoff); 3) difficulty comparing data across studies in both natural history and treatment studies; and 4) difficulty comparing longitudinal changes across sites.
For these reasons, our working group was convened after a presentation at the 2012 Alzheimer’s Imaging Consortium pre-meeting of the Alzheimer’s Association International Conference. That presentation of a general standardization approach by one of the co-authors of this report (MM) evolved into the specific approach that is presented here in detail. This relatively simple approach hypothesizes that comparable results can be achieved across analysis techniques and tracers by linearly scaling the outcome data of any amyloid PET method to an average value of zero in “high-certainty” amyloid-negative subjects and to an average value of 100 in “typical” Alzheimer’s disease (AD) patients. The unit of this 100-point scale has been termed the “Centiloid” (CL).
In this report, we outline a standard approach that is tailored to assessment of a large cortical area that represents the typical regions of high amyloid load in Alzheimer’s disease (AD). We have gathered cases we believe can adequately define average “high-certainty” amyloid-negative subjects and typical AD patients. To be included in our analysis, subjects had to have dynamic PET datasets available to increase the generalizability of their use. Methods are presented to take this “standard” approach and adapt it to most approaches currently used in the field so that only a simple scaling of data is required and no significant change in locally-preferred practice is necessary. The approach is meant to be broadly applicable and, as such, some shortcomings were accepted in order to improve simplicity and accessibility by most groups. The approach is based on the most widely applied method up to this point: [C-11]Pittsburgh Compound-B (PiB) tissue ratios gathered 50-70 min post-injection. Whenever choices were made based on optimization of outcomes, the data used was PiB data and no consideration was given to optimization of any of the F-18-labeled tracers. However, we recognize that many sites will not have access to carbon-11, so we also describe how scaling can be accomplished using only fluorine-18 tracers when necessary.
A key component of the optimal use of the Centiloid method will be free access to all necessary data on a public database, and all of the scan data used in this initial report has been deposited on the Global Alzheimer’s Association Information Network (GAAIN; http://www.gaain.org) for free public access. This initial description is intended to be a serviceable first iteration. We assume that further research will be necessary to fully examine the assumptions made and to fine-tune the process and fully understand the strengths and limitations.
APPROACH and RESULTS
There are three “levels” to the Centiloid process. The first level is described in this report and need not be repeated by other sites. The purpose of this level-1 process is to set the “typical” 0-anchor and 100-anchor points for all future scaling operations. The second level of the Centiloid process is the method suggested for individual sites to scale their unique method of PiB-PET or any method using a tracer other than PiB to the Centiloid scale. The third level is to be used when an individual site simply wants to exactly reproduce a method that has previously been scaled to Centiloids and is basically a check of the processing pipeline to ensure that errors are eliminated before the processing and scaling of site-collected data is begun.
1. Level-1: The Standard Method and Anchor Points (see Supplemental Flowchart 1)
Level-1 is the main component of this study. A diagram of the general Level-1 process is given in Supplemental Flowchart 1. It is the process of choosing subjects to define the 0- and 100-anchor points of the CL scale and prescribing the method for normalization. It should not be necessary for any group to repeat this step of the process to employ the Centiloid scale. An effort was made to give these “anchor points” biological relevance. The 0-anchor was intended to represent a definitively amyloid(−) brain. The 100-anchor was intended to represent the amount of global amyloid deposition found in a typical mild-moderate AD subject. Since both are mean values, some amyloid(−) scans will have slightly negative CL values. Likewise, there will be a range of “typical AD values” around 100 CL and about half of all AD subjects will fall above, sometimes significantly above a value of 100 CL.
1.1 Subjects
None of the subjects described below should be considered unique to this study as all have been included in a variety of previously published analyses.
1.1.1 The Young Control 0-Anchor (YC-0) Subject Set
PiB PET data that included data from at least 50-70 min after injection of PiB was collected from 34 subjects under the age of 45 (31.5 ± 6.3 years; range 22-43) who were judged to be cognitively normal after a standard neuropsychological and clinical evaluation [7-9] This age range was used because it lends great certainty that the subject will be truly amyloid-negative [10; 11]. Twenty subjects were studied at Washington University in St. Louis [18 on Siemens BioGraph TruePoint TrueV (Model 1094) and two on Siemens ECAT Exact HR+) and 14 subjects were assessed at the University of California at Berkeley/Lawrence Berkeley National Laboratory (UCB/LBNL) (five on Siemens BioGraph TruePoint TrueV (Model 1094) and nine on Siemens ECAT Exact HR). Of the 32 younger subjects tested for APOE genotype (2 younger subjects refused testing), 8 (25%) were APOE ε4 carriers and 24 (75%) were ε4 non-carriers. These young control 0-anchor (YC-0) subjects were used to define the 0-anchor point by determining the mean of the group.
1.1.2 The AD 100-Anchor (AD-100) Subject Set
PiB PET data that included data from at least 50-70 min after injection of PiB was collected from 47 subjects diagnosed with AD using the 1984 NINCDS-ADRDA criteria and assessed at the University of California at San Francisco (UCSF) and LBNL (n=22: all from Siemens ECAT Exact HR PET scanner), the University of Pittsburgh (Pitt; n=18; all from Siemens ECAT Exact HR+ PET scanners) or through the Australian Imaging, Biomarkers and Lifestyle study (AIBL; n=7; all from Phillips Allegro PET scanner) according to previously described procedures [12-15]. All AD 100-anchor subjects (AD-100) had a Clinical Dementia Rating (CDR) Scale Global Score of 0.5 or 1 [16].
Since the intention of this cohort was to define the average PiB retention of a “typical” AD cohort, we excluded outlier subjects that were suspected to be clinically misdiagnosed. A “mild” outlier was defined as PiB retention that exceeded the third quartile by >1.5 times the interquartile range (Q3+1.5×IQR) or was beneath the first quartile by an amount <1.5 times the interquartile range (Q1-1.5×IQR), that is, any observation outside the “inner fences” of the boxand-whisker plot [17]. This approach has previously been used in the “Iterative Outlier” method of defining PiB-positive cutoffs [13; 18]. As described below, this resulted in the exclusion of two AD subjects with low PiB retention (no AD subjects were outliers due to high PiB retention) leaving 45 AD-100 subjects for further analysis. The average age of these 45 subjects was 67.5 ± 10.5 yrs (range 50 to 89 yrs). Of these 45, 44 AD subjects were tested for APOE genotype (1 AD subject refused testing) and 28 (64%) were APOE ε4 carriers.
1.1.3 The Global Cortical Target (CTX) Region Subject Set
In order to avoid using the same subjects for definition of the CTX volume-of-interest (VOI) (see below) as were used to define the 0-Anchor and 100-Anchor points, a separate set of 19 “AD-CTX” subjects (CDR =0.5-1; 72.1 ± 11.8 yrs; range 53-94 yrs) and 25 older control (OC-CTX) subjects (71.4 ± 9.8 yrs; range 45-88 yrs) were used. The OC-CTX subjects were clearly amyloid-negative by previously published quantitative criteria and by visual assessment [13; 18]. All were adjudicated as AD or cognitively normal according to previously published criteria [12; 13]. The AD-100, AD-CTX and OC-CTX groups were not significantly different from each other in age. Thirteen of the 19 AD-CTX subjects had an [F-18]fluorodeoxyglucose (FDG) scan on the same day as their PiB scan. These 13 subjects were used to generate the Cerebeller Gray (CG) VOI as described below.
Both of the AD-CTX and OC-CTX groups were evaluated and scanned at the University of Pittsburgh using the same methods described above [13; 19]. All subjects were injected with 10-15 mCi of PiB at the start of the experiment and imaged according to previously published methods [20]. Reconstruction algorithms varied by site and scanner-type and included filtered back-projection, FORE and OSEM (Fourier rebinding, ordered-subsets expectation maximization; BioGraph PET/CT) and 3D-Ramla (row-action maximum likelihood algorithm; Philips Allegro). In addition, FDG data were acquired for the AD-CTX subjects using ~7 mCi FDG. FDG PET data were acquired over 25 min (five 5 min frames) after a 35 min uptake period as the subjects rested quietly in a dimly lit room with their eyes open. The FDG-PET data were analyzed as previously described in detail [19]. Magnetic resonance imaging (MRI) was performed at either 1.5T or 3T using previously published methods [7; 14; 15; 20].
1.2 The Standard VOIs
1.2.1 The Normalization Process
The normalization process proved to be a potential source of error and strict adherence to the process described here appears to be important for exact replication of the standard method. For example, the Statistical Parametric Mapping, version 8, (SPM8), revision 4290 (http://www.fil.ion.ucl.ac.uk/spm/software/spm8/) unified method proved superior to the SPM5 segmentation method [21] or DARTEL [22]. An area of particular concern was the brainstem, that was handled less well than cortical areas by all methods. The PiB and FDG PET data were averaged over frames corresponding to the 50-70 minute and 40-60 minute post-injection intervals, respectively. SPM8 was used for all subsequent registration and normalization processes. The MRI and PET scans were first manually reoriented to match the orientation of the MNI-152 T1-weighted template provided with the SPM8 software (2 mm resolution). The subject MRIs were then individually registered to the MNI-152 template using the “Coregister: Estimate” module (Reference Image: MNI-152 template; Source Image: MRI) with default parameters. In turn, each averaged PET image was registered to its MRI (and, thus, coregistered to MNI-152) also using the “Coregister: Estimate” module (Reference Image: MRI; Source Image: PET) with default parameters. The unified segmentation method [23] was subsequently applied to all coregistered MRI scans. This method combines segmentation, bias correction, and spatial normalization into a single unified model. Within SPM8, this was performed using the “Segment” module with default parameters, utilizing the provided tissue probability maps at 1 mm resolution (note that 2 mm resolution used throughout this process produced equivalent results). The “Segment” module produces two MATLAB formatted binary files containing the forward (*_seg_sn.mat) and inverse (*_seg_inv_sn.mat) normalization parameters. The forward parameters were applied to the coregistered MRI and PET scans for each individual using the “Normalise: Write” module in SPM8 (Parameter File: forward parameters; Images to Write: coregistered MR and PET). Default parameters were used in this process, with the exception of the Bounding Box, which was modified to [−90 −126 −72; 91 91 109] to reflect MNI-152 space.
1.2.2 Reference VOIs
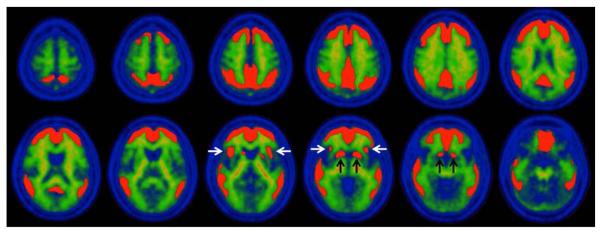
Four reference VOIs were assessed in the development of the “standard method.” These included: 1) Cerebellar Gray (CG); 2) Whole Cerebellum (WC); 3) Whole Cerebellum plus Brainstem (WC+B); and 4) Pons (Figure 1). The CG VOI was designed in a data-driven fashion to maximize the contribution of gray matter while minimizing the contribution of white matter areas that non-specifically accumulate PiB [24]. To do this, we identified cerebellar gray matter voxels by first applying a WC VOI mask in MNI-152 space to the average of the normalized AD-CTX FDG PET scans. This average FDG image was computed using the image calculator within SPM8. To minimize inclusion of cerebellar white matter nonspecific PiB retention, we first averaged the normalized PiB PET scans of these same AD-CTX subjects, and then masked this average with the WC+B VOI in MNI-152 space. The average masked FDG image was then normalized to its maximum value, and the average masked PiB image was normalized to its maximum white matter value, which was assessed in the pons. The resulting PiB image (i.e., mainly white matter) was subtracted from the resulting FDG image (i.e., mainly gray matter). Negative voxels in this difference image were thresholded to zero, and the result was binarized, providing a CG VOI that had minimal white matter contamination.
Figure 1.
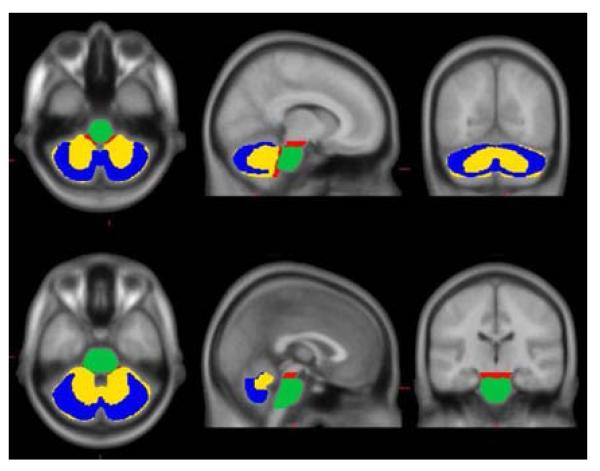
The WC and Pons VOIs were modified from those previously defined in the International Consortium for Brain Mapping (ICBM) Single Subject MRI Anatomical Template [25], last revised August 22, 2012. (http://www.loni.ucla.edu/ICBM/Downloads/Downloads_ICBMtemplate.shtml). The WC+B VOI was defined using a threshold on the ICBM 152 MR atlas. All four reference VOIs were then edited by manually “cleaning up” the regions both by removing spurious voxels was well as filling in any small “holes” in the region. After this cleaning process, the CG was a subset of WC (missing the cerebellar white matter) and WC was a subset of WC+B (missing the brainstem). The Pons VOI is a subset of the brainstem portion of WC+B. In order to apply these ICBM VOIs in MNI-152 space, the ICBM Single Subject MRI template was normalized to MNI-152 space using the registration and normalization procedures described above. The forward normalization parameters were then applied to the WC, Pons, and WC+B VOIs.
The superior limit of the CG, WC and WC+B VOIs was set at z = −15 mm of the SPM8 MNI-152 template to avoid spillover from specific signal in the occipital cortex of amyloid-positive cases. The superior limit of the Pons VOI was set at z = −20 mm of the SPM8 MNI-152 template according to the ICBM template anatomical boundaries. The inferior limit of all four reference VOIs was truncated at z = −52 mm in order to compensate for two common potential sources of error: 1) the most inferior portions of the cerebellum may be outside of the field-of-view as a result of poor positioning, particularly in older PET-only scanners having a smaller axial field-of-view and 2) the spatial normalization procedure is less optimized for and may not provide a good match with voxels in the lower brainstem region. This allows the Centiloid processing method to be applied to the vast majority of amyloid imaging scans.
1.2.3 The CTX VOI
The CTX VOI (Figure 2) was data-driven and determined by averaging the PiB PET 50-70 min standardized uptake volume ratio (SUVr; equivalent to tissue ratio) parametric images (using the WC reference VOI) from a set of subjects unique from those used below for the Centiloid analysis. These included 19 typical AD subjects (AD-CTX) and 25 age-matched controls (OC-CTX). Each image was normalized into MNI-152 space using the parameters obtained for the corresponding MRIs obtained as described above. An average AD-CTX and average OC-CTX image was generated in MNI-152 space using SPM8 and the OC-CTX image was subtracted from the AD-CTX image and smoothed with a 3D-Gaussian filter with full width at half maximum (FWHM) of 5.0 mm. After exploring several thresholds, the resultant difference-image was thresholded at 1.05 SUVr units. This threshold produced a large VOI representing areas of the brain with the greatest amyloid load while avoiding areas that are primarily white matter and minimizing discontinuity in the VOI (Figure 2). This mask was then edited manually both to remove spurious voxels in the mask as well as filling in small holes in the mask. The resultant data-driven CTX VOI included the typical brain regions with high amyloid load in AD including the frontal, temporal and parietal cortices and precuneus. It also included the anterior striatum and insular cortex.
Figure 2.
1.3 The Standard Analysis Method
1.3.1 Choice of the Standard Reference Region
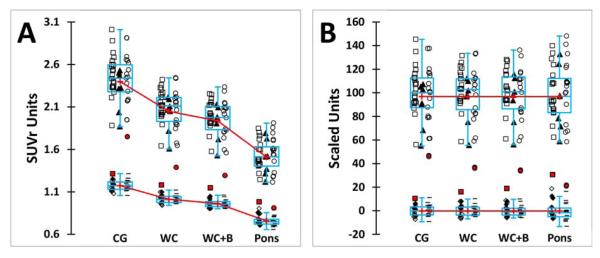
After normalization into MNI-152 space, a 50-70 min post-injection CTX SUVr was calculated using each of the four reference VOIs for the 34 YC-0 and the 45 AD-100 subjects. Choice of the standard reference was based on the variance observed in the data as well as on the effect size of the difference between the AD-100 and YC-0 groups (Table 1). The Pons gave the largest (worst) variance and smallest effect size, the CG performed better, but was consistently outperformed by the WC and WC+B which produced predictably similar results. While the WC and WC+B were nearly equivalent, greater weight was given to the lower variance obtained using the WC reference VOI in the YC-0 group as this group represents noise in the method and minimization of this noise was considered a critical parameter. Other considerations included: 1) the variability inherent in accurately normalizing (and thus fixing the VOI) using the CG due to the proximity of the non-specific signal from the cerebellar peduncles and 2) the poorer performance of many normalization routines (including those used here) in brainstem regions such as the pons. Use of the WC without pons minimizes both of these difficulties. Therefore, the WC was chosen as the reference VOI for the “standard PiB method.”
Table 1.
Summary Statistics of SUVr and Scaled Data.
| SUVr | Scaled Units | |||||||
|---|---|---|---|---|---|---|---|---|
| CG | WC | WC+B | Pons | CG | WC | WC+B | Pons | |
| AD 100 | ||||||||
| Mean | 2.428 | 2.076 | 1.962 | 1.535 | 100.0 | 100.0 | 100.0 | 100.0 |
| SD | 0.246 | 0.191 | 0.180 | 0.175 | 19.6 | 17.9 | 18.0 | 22.6 |
| COV | 10.1% | 9.2% | 9.2% | 11.4% | --- | --- | --- | --- |
| YC 0 | ||||||||
| Mean | 1.170 | 1.009 | 0.959 | 0.761 | 0.0 | 0.0 | 0.0 | 0.0 |
| SD | 0.057 | 0.046 | 0.046 | 0.058 | 4.52 | 4.34 * | 4.56 | 7.44 |
| COV | 4.9% | 4.6% | 4.8% | 7.6% | --- | --- | --- | --- |
| effect size | 6.55 | 7.14 | 7.11 | 5.57 | 6.55 | 7.14 | 7.11 | 5.57 |
The variability in the YC-0 cohort is an important variable as it reflects mainly the noise in each method.
In this report, we refer to “scaled units” to generally refer to the outcomes from all reference regions. By definition, Centiloid units have a very specific meaning and should be reserved only for data derived in one of four ways: 1) from the level-1 analysis presented here specifically using PiB 50-70 SUVr data and the CTX and WC VOIs (i.e., the “standard PiB Method”); 2) from a level-2 analysis (see below) after calibrating to the standard PiB method; 3) from a level-2 analysis after calibrating to a “surrogate” F-18 method, previously, directly calibrated to the standard PiB method; and 4) from a level-3 analysis (see below).
1.3.2 Calculation of Centiloid Values
After co-registration of each subject’s PiB PET to their MRI, the MRI images of all 34 YC-0 and the 48 original AD-100 subjects were normalized into MNI-152 space using the SPM8 unified segmentation method. Coordinate transformation of the PiB PET into MNI-152 space was via subject’s MRI transformation parameters. The CTX VOI and all four reference VOIs were sampled in each subject and the tissue ratio of the CTX:reference VOI yielded the SUVr value (Supplemental Table A). The mean and SD values and effect sizes for each reference VOI are shown in Table 1 and presented graphically in Figure 3.
Figure 3.
Since one goal was to define the mean value of “typical” AD patients as 100 CL, the AD data was screened for outliers as defined above and two subjects were excluded for being low outliers, leaving 45 AD-100 subjects for further analysis (Figure 3). These subjects were low outliers with all four reference VOIs. There were no high AD outliers and no outliers were identified in the YC-0 data. AD-100 and YC-0 data is shown in Figure 3 according to the site acquiring the data and it is apparent that there is no significant difference in the range of either the AD-100 or YC-0 data. Also noticeable in the YC-0 data in Figure 3, is that the E4 carriers from UCSF/UCB had PiB retention that was equivalent to the non-carriers (Wash U ApoE data was available only as aggregate data, not identifiable with individual subjects). Finally, Figure 3 shows graphically the wide difference in average values of AD-100 and YC-0 SUVr data depending on the reference VOI (Figure 3A) and how these data can be scaled to the same 0-100 scale facilitating direct numeric comparisons across different measures (Figure 3B).
After calculation of the SUVr values (referenced to WC) for all 34 YC-0 and the 45 AD-100 subjects (Supplemental Table A), the mean value of the YC-0 subjects (1.009 SUVr; Table 1) was set to 0 CL and the mean of the AD-100 subjects (2.076 SUVr; Table 1) was set to 100 CL.
The standard Centiloid value (CL) for each individual subject was then defined as:
| (Eq.1.3a) |
Where: PiBSUVrIND is an individual’s SUVr value PiBSUVrAD-100 is the mean SUVr of the 45 AD-100 subjects PiBSUVrYC-0 is the mean SUVr of the 34 YC-0 subjects all determined using PiB 50-70 min data and the standard CTX target and WC reference VOIs.
Substituting the values in Table 1, Eq. 1.3a simplifies to:
| (Eq.1.3b) |
CL values were then calculated for each subject (Supplemental Table A). For completeness, Supplemental Table A also includes the scaled values that were derived using the mean SUVr values for alternate reference regions as shown in Table 1.
1.4 Level-1 Data Made Available for Unrestricted Use
All of the de-identified PET and MRI scans used in the derivatization of the Centiloid scaling and all of the reference and CTX VOIs have been made available at http://www.gaain.org.
2. Level-2: Calibrating a new method to the Centiloid Scale
Level-2 is the process independent groups should use to calibrate their specific method to CL units. This process is described in detail beginning in section 2.2 below and in Supplemental Flowchart 2, but we first discuss some general principles. Level-2 callibration is a process that will need to be performed whenever a procedure other than the “Standard PiB Method” (i.e., PiB 50-70 min SUVr data using the standard CTX and WC VOIs described above) is to be calibrated to the CL scale. For example, non-standard methods would include the use of: 1) any method of PiB analysis other than 50-70 min SUVr using the standard CTX and WC VOIs (including atrophy-corrected methods, use of different CTX or reference regions and direct PiB-to-MNI normalization without the use of MRIs) and 2) any other tracer by any method. A diagram of the general Level-2 process is given in Supplemental Flowchart 2 and explained below in section 2.2.
Because some sites may wish to use the Centiloid scale, but do not have local access to [C-11]PiB, an F-18 amyloid tracer that has previously been calibrated directly to the standard PiB method by this Level-2 process can be substituted for PiB as a “surrogate reference” tracer. A surrogate reference tracer should not be more than one step removed from a PiB reference. That is, if Tracer-A is calibrated directly to PiB by the process below (standard VOIs, etc.) it can be a surrogate reference tracer. If Tracer-B is then calibrated to Tracer-A (because [C-11]PiB is not available), Tracer-B results can be expressed in CL units, but Tracer-B cannot be used as a surrogate reference tracer for the calibration of Tracer-C. Either PiB or Tracer-A should be used to calibrate Tracer-C.
2.1 New Data Necessary for Level-2 Centiloid Calibration
2.1.1 Standard-Method PiB Data as a Scaling reference
A calibrating site should acquire PiB PET 50-70 SUVr and structural MRI data (for normalization/co-registration) on at least 25 subjects. PiB is preferable but another F-18 tracer/method would be an acceptable surrogate if it has been previously and directly calibrated against the standard PiB method. Of these 25 (or more) subjects, at least 10 should be cognitively normal subjects ≤45 yrs of age (and thus extremely likely to be amyloid-negative). In addition, at least 15 subjects should have a high likelihood of being amyloid-positive - with ~5 typical AD patients and ~10 subjects likely to have intermediate values of PiB retention. Although there is no absolute way to ensure acquisition of this intermediate group, we recommend inclusion of at least some cognitively normal ApoE4 carriers above age 80 and/or MCI subjects. The rationale is to spread the points along the range of the correlation in as continuous a manner as possible to increase the validity of the correlation outcome measures (i.e., slope, intercept and correlation coefficient) in contrast to the result that might be obtained from a group of YC subjects at one extreme and a group of high-amyloid AD subjects at the other. Note that the spread of the data is the important factor, not the clinical diagnosis.
2.1.2 Avoidance of Pre-Informed Selection of Data
In order to avoid any appearance of pre-informed selection of data, we recommend that: 1) all subjects be studied prospectively and no subject be excluded except for carefully justified technical reasons made explicit in the first manuscript that is published using these results; 2) previously collected data (with or without supplementation with new, prospective data) is acceptable only if the entire dataset with both PiB and the new tracer is used (i.e., no selection of a subset of subjects for the analyses below). This can be insured by inclusion of the entire cohort of a previously published study. If the study has not been published (i.e., a clinical trial), a description of the original study should be given that includes the total number of subjects in that study cross-referenced to public documents (e.g., FDA submissions or clinicaltrials.gov registries).
2.1.3 Non-Standard Method Data
2.1.3.1 Tracers Other Than PiB
If a new tracer is being used, it will be necessary to acquire the appropriate tracer retention data from the same 25 (or more) subjects described above within 3 months of the PiB scan used for scaling.
2.1.3.2 Non-Standard PiB Methods
If a non-standard method of PiB acquisition or analysis is being calibrated, appropriate non-standard data must be collected on the same 25 (or more) subjects studied with the standard PiB method. This should be simultaneous with the PiB 50-70 min data collection, if possible. However, since 40 of the 45 AD-100 datasets and all 34 YC-0 datasets available for downloading as described in section 1.4 contains dynamic data collected from 0-70 min post-injection of PiB, it may not be necessary to collect any new data and the calibration can be done completely with the downloaded data by performing the standard analysis, followed by the non-standard analysis to be calibrated. This section would also apply if an F-18 tracer that had previously been calibrated to PiB using one method was being used by a method different than the one originally calibrated to PiB.
2.2 Level-2 Analysis (see Supplemental Flowchart 2)
2.2.1 Replication of the Level-1 Analysis
Since there are potential errors that can be introduced into a new analysis pipeline - particularly in the MRI normalization step - the calibrating site should first demonstrate that it can accurately express the Level-1 PiB data on the Centiloid scale as follows. The site should download all 34 YC-0 and all 45 AD-100 scans from the Level-1 study, normalize into MNI-152 space and calculate standard SUVr values for each scan (i.e., using the standard CTX and WC VOIs). The mean Level-2 PiBSUVr of the 34 YC-0 subjects and 45 AD-100 subjects should fall within 2% of the mean PiBSUVr determined in the Level-1 analysis and reported above in Table 1 using the WC as the reference VOI (i.e., 2.05-2.13 SUVr for AD-100 and 1.05-1.09 SUVr for YC-100).
The mean of the 34 YC-0 PiB SUVr values determined in a Level-2 analysis (SUVrYC-0*) will be defined as 0 CL and the 100 CL point will be defined as the mean of the 45 AD-100 SUVr values (SUVrAD-100*). The following general equation will be used to convert the downloaded PiB data into Centiloids:
| (Eq.2.2.1) |
Where PiBSUVrS-IND* is the 50-70 min SUVr value determined for an individual subject of the 34 YC-0 or 45 AD-100 subjects by downloading and re-calculating data at the calibrating site. Eq. 2.2.1 differs from Eq. 1.3a only by the fact that the values for all variables are calculated by the calibrating site using the downloaded data for a Level-2 analysis. This is indicated by a single asterisk here only to clarify the source of the data (i.e., downloaded) for the purpose of this description. These asterisks are not intended to be used by the independent sites when expressing their CL data.
The calibrating site will then perform a linear correlation of their downloaded/re-calculated PiB Centiloid values vs. the PiB Centiloid values reported here. This should be included as supplemental data in the first publication with the slope, intercept and R2 reported in the manuscript. The expectation is that the slope will be between 0.98 and 1.02, the intercept will be between −2 and 2 CL and the R2 will be > 0.98.
2.2.2 Example of a Replication Analysis
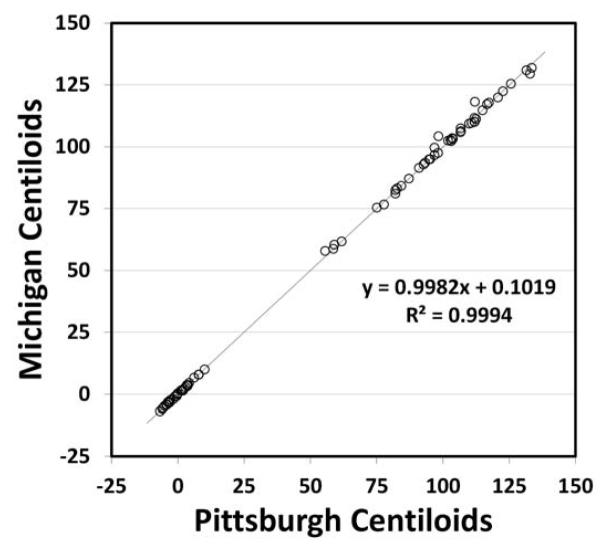
Since a key component of the Centiloid process is initial verification of the independent sites analysis pipeline, after the initial analysis method was developed and performed at the University of Michigan, the data and VOIs were used in an independent, but identical analysis performed at the University of Pittsburgh (except the normalized subject MRI scan data were resliced to 2 × 2 × 2 mm voxel size, rather than to 1 × 1 × 1 mm as done at Michigan site). Figure 4 shows the correlation of the two independent analyses.
Figure 4.
The agreement was well within the limits set above for all reference VOIs (Table 2).
Table 2.
Agreement of Independent Analyses by Michigan and Pittsburgh
| Reference VOI | Slope (0.98 to 1.02) |
Intercept (−2 to 2 CL) |
R2 (>0.98) |
% Difference YC (−2 to 2%) with SD ± 2% |
|---|---|---|---|---|
| CG | 0.9973 | 0.15 | 0.9992 | 0.0 ± 0.30 |
| WC* | 0.9982 | 0.10 | 0.9994 | 0.0 ± 0.26 |
| WC+B | 0.9985 | 0.08 | 0.9995 | 0.0 ± 0.25 |
| Pons | 0.9987 | 0.07 | 0.9998 | 0.0 ± 0.50 |
See Figure 4.
If the calibrating site is using a previously calibrated F-18 tracer in place of the standard PiB method, they should follow the steps described above, except substituting the downloaded F-18 tracer data (calculated using the standard CTX and WC VOIs) for the downloaded PiB data. The same is true of the steps described below for analysis of site-acquired F-18 tracer data.
2.2.3 Level-2 Calibration of Other Tracers or Methods
After the site’s analysis pipeline has been validated by the replication of the Level-1 analysis, the site should next calculate 50-70 min PiBSUVrIND** values for the site-acquired PiB scans using the standard CTX and WC VOIs (i.e., the ~10 YC and ~15 subjects likely to be amyloid-positive). The double asterisk refers to data both acquired and analyzed in a Level-2 analysis. This data will be used to scale the new tracer or method to the CL scale as described below (see Supplemental Flowchart 2).
If desired, a site may then convert these values to Centiloids with the following equation:
| (Eq.2.2.3) |
2.2.3.1 Calibration of another Tracer for Future Use as a Surrogate Reference
As mentioned above, it may be important to have one or more F-18 tracers available as a surrogate for PiB for calibration of site-specific methods into Centiloid units (e.g., when C-11 is not accessible). In this case, the calibration of the surrogate tracer to the standard PiB method should be done using the standard CTX and WC VOIs and calculation of a SUVr. This would not necessarily include the 50-70 min time window used for PiB in the standard method since this may not be optimal for the surrogate tracer. Section 2.2.3.2 describes a more general calibration of a tracer/method not intended for use as a surrogate reference and allows for unrestricted variation in cortical target and reference VOIs and method of analysis.
Using the standard CTX and WC VOIs, the site should calculate PiB 50-70 min SUVr values for the site-acquired 25 (or more) PiB scans (designated as PiBSUVrIND**, where the two asterisks refer to data both collected at and calculated at the calibrating site). Likewise, the calibrating site should use the CTX and WC VOIs to calculate the appropriate surrogate tracer SUVr value. After plotting the PiBSUVrIND** values on the x-axis and the TracerSUVrIND values on the y-axis a slope (TracermStd) and intercept (TracerbStd) is calculated, where the “Std” subscript designates that the standard CTX and WC VOIs were employed:
| (Eq.2.2.3.1a) |
A TracermStd of 1.0 means the surrogate has the same specific signal (or dynamic range) as PiB. A slope of 0.5, half the signal of PiB, a slope of 2, twice the signal of PiB. Thus, the numerical value of this slope is informative regarding the relative signals of PiB and the surrogate tracer. The conversion to Centiloid units would then be accomplished by first converting the TracerSUVrIND values into “PiB calculated” SUVr values (PiB-CalcSUVrIND):
| (Eq.2.2.3.1b) |
| (Eq.2.2.3.1c) |
| (Eq.2.2.3.1c) |
The slope (TracermStd) and intercept (TracerbStd) by any other site in a Level-2 or Level-3 (see below) analysis to generate PiB-CalcSUVrIND values that, in turn, can then be converted to CL units with equation 2.2.3.1c.
2.2.3.2 Calibration of another Tracer or Method for General Use
As stated above, this section allows for calibration of tracers and methods without restriction on the cortical target and reference VOIs and method of analysis. However, it should be recognized that there is likely a degree of departure from the standard CTX and WC VOIs at which the calibration is no longer valid as considered below in the discussion.
Similar to the process in section 2.2.3.1, using the standard CTX and WC VOIs, the site should calculate PiBSUVrIND** values for the site-acquired 25 (or more) PiB scans. Next, the calibrating site should use their preferred, non-standard target and/or reference VOIs to calculate the appropriate surrogate tracer value (in any preferred unit depending on the preferred analysis method). After plotting the PiBSUVrIND** values on the x-axis and the TracerUNITIND values on the y-axis a slope (TracermNS) and intercept (TracerbNS) is calculated, where the “NS” subscript designates that non-standard target and/or reference VOIs were employed:
| (Eq.2.2.3.2a) |
As stated above, the TracermS value informs on relative differences in tracer signal with the “method” being held constant. In this case, a comparison of the TracermNS and TracermS values informs on the additional differences in signal due to the difference between the standard PiB method and the non-standard method (with the tracer held constant). As in section 2.2.3.1, the conversion to Centiloid units would then be accomplished by first converting the TracerUNITIND values into “PiB calculated” SUVr values (PiB-CalcSUVrIND):
| (Eq.2.2.3.2b) |
Both the PiBSUVrIND** and the PiB-CalcSUVrIND are then converted to PiBCLStd** and TracerCLNS, respectively, using Eq. 2.2.3. The slope (TracermNS) and intercept (TracerbNS) can then be used by any other site in a Level-2 or Level-3 analysis.
2.2.3.3 Use of Small VOIs
Direct calibration of small regional VOIs (e.g., precuneus) to the standard (i.e., global) PiB method - if done individually – would force the mean CL value of AD patients for each region to be 100. This is likely to lead to a given tracer retention measure (e.g., 1.70 SUVr units) equating to different CL values in different brain regions. To avoid this undesirable situation, we suggest that if an independent site is working with multiple small VOIs, they first generate the conversion equation in section 2.2.3.1 or 2.2.3.2 based on the standard CTX and WC VOIs and then apply the conversion factors to the regional data either by: 1) converting the entire dataset to Centiloids on a voxel basis (i.e., create a Centiloid parametric image) using Eq. 2.2.3.1b or 2.2.3.2b and then sample the Centiloid parametric image with the non-standard VOIs or 2) calculate the regional tracer retention value in the same units (and by the same method) used for TracerUNITN in section 2.2.3.2 and then directly convert to Centiloids using Eq. 2.2.3.2b. Small-region Centiloid values such as these should be distinguished by a superscript. For example, Centiloid values from only a precuneus VOI could be noted as CLPRC.
2.3 Quality Control
The final aspect of implementing the Level-2 Centiloid process is evaluating the quality of the values produced by the new tracer method and the transformation process. Two basic aspects of quality can be evaluated: reliability across subjects and relative variance.
2.3.1 Reliability
The reliability of a new-method relative to the standard PiB method should be evaluated by calculating a correlation coefficient (R2) for the linear regression between the site-acquired standard PiB SUVr data and the non-standard method data (e.g., the scattergraph of 2.2.3.1a or 2.2.3.2a). This correlation coefficient (R2), should be reported in the first publication using the non-standard method. The expectation is that the R2 will be > 0.7 for a well-correlated tracer/method.
2.3.2 Relative Variance
Because the Centiloid process will collapse/expand all methods and all tracers to approximately the same dynamic range after conversion to Centiloid units, it is important that information not be lost about the true precision of the various tracers and methods. While TracermStd and TracermNS give some indication of dynamic range, information on precision is best found in the relative variance of the different methods. Since each tracer/method will have been scaled to PiB in the same set of subjects, the variance of each non-standard tracer/method relative to PiB should be calculated as follows. For the 10 cognitively normal subjects ≤45 yrs of age, the variance of the PiBCLStd** data and the non-standard data (i.e., either PiBCLNS, TracerCLStd or TracerCLNS) should be calculated and the “relative variance” be reported as a ratio. For example, if the mean ± SD of the PiBCLStd** data from the 10 cognitively normal subjects ≤45 yrs of age was 0.0 ± 5.0 and the mean ± SD of the TracerCLNS data in these same subjects was 0.0 ± 7.0 this would be reported and it would be concluded that the new tracer had a relative variance of 1.4 compared to PiB (SD of 7 divided by SD of 5). Both the SD (in CL) and the relative variance should be reported in the first publication calibrating a tracer to the Centiloid scale. However, it should be kept in mind that the relative variance of the different tracers or methods contains information from both the dynamic range and the noise in the tracer method. For example, if PiB and another tracer have the same relative variance, it is not necessarily because the dynamic range is the same. The dynamic range for of the non-standard tracer could be lower, but if the absolute variance of the non-standard tracer also is lower by an equivalent factor, then the relative variance will be approximately the same.
2.3.3 Test/Re-test Variation
Although it is beyond the scope of this paper, it is recommended that if a completely new tracer is being characterized, test-retest data should be acquired in some manner and published for this new tracer in Centiloid units. Calculation of test-retest parameters by comparing the mean ± SD of the difference in test-retest values expressed in Centiloid units may prove very useful in cross-tracer comparisons.
2.4 Level-2 Data made available for unrestricted use
The validating site will deposit (into a freely accessible database such as http://www.gaain.org) de-identified PET and MRI data from all 25 (or more) scans for both the non-standard tracer/method and PiB. Also, the calculated values of TracermStd, TracermNS, TracerbStd, and TracerbNS should be given along with a table of all individual subject values of TracerUNITIND, PiBSUVrIND**, PiB-CalcSUVrIND, PiBCLStd**, and TracerCLNS. Finally, a statement should be included in the first publication as to whether these data were collected according to the recommendations in section 2.1.2 about pre-selection of data.
2.5 Examples of Level-2 Analyses
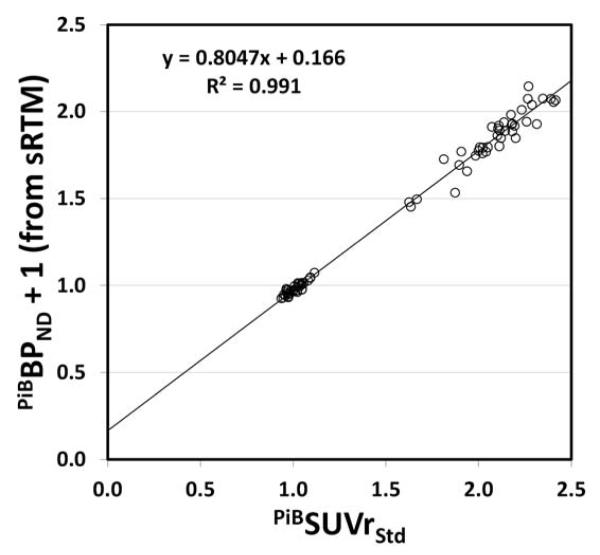
As an example of a Level-2 analysis we performed a Centiloid calibration of non-displaceable binding potential (BPND) data determined using the simplified reference tissue method (sRTM) from the 40 of 45 AD-100 and 34 of 34 YC-0 subjects who had 0-70 min of dynamic data available. In this case, we used the standard CTX and WC VOIs. The standard 50-70 min PiBSUVrStd and PiBBPND+1 values calculated using the same 40 AD-100 and 34 YC-0 subjects are shown in Supplemental Table B. Figure 5 shows the correlation between PiBBPND+1 values obtained from the sRTM method and the PiBSUVrStd values. The slope (PiBmNS) for this correlation is 0.805 suggesting that the dynamic range of the sRTM method is less than that of the PiBSUVr method, although this difference is caused mostly by the fact that the PiBSUVr method overestimates specific binding as tissue curves are continuing to clear and equilibrium is never reached [26; 27]. The intercept (PiBbNS) is 0.166. The conversion to Centiloid units was then accomplished by first converting the PiBBPND + 1 values into PiB-CalcSUVr values using Eq. 2.2.3.2b as follows:
Figure 5.
These values also are shown in Supplemental Table B. The PiBSUVr and the PiB-CalcSUVr were then converted to PiBCLStd and PiBCLNS, respectively, using Eq. 1.3b and these are shown in Supplemental Table B as well.
By way of evaluating the quality of the PiBCLNS values derived through this sRTM analysis method, we assessed the reliability by calculating a R2 value for the correlation in Figure 5, finding it to be 0.991. This is well above the suggested threshold of 0.7. However, given the fact that this was simply a recalculation of a single data set and thus would not have the variability induced by the use of a second tracer or movement of the subject to perform a second scan, it is not surprising that this sRTM method of analysis correlated very well with the standard PiB SUVr analysis. The relative variance of the sRTM method - determined by the ratio of the SD of the YC-0 PIBCLNS values (4.24 CL) to the SD of the YC-0 PIBCLStd values (4.34 CL) – was 0.98. As was the case with the R2, this is not surprising and suggests the two methods have very similar variance.
As another example, the Pons VOI was used as the reference region in a non-standard analysis (along with the standard CTX VOI). The slope (PiBmNS) for this correlation was 0.724 and the intercept (PiBbNS) was 0.0315. The R2 value for the correlation between the PiBSUVr and PiBSUVrPons values was 0.955; less than that for the correlation between PiBSUVr and PiBBPND+1 values, but still well above the suggested threshold of 0.7. The relative variance of the Pons reference method was 1.65, reflecting the greater variance of the Pons method as shown in Table 1.
3. Level-3: Exact Reproduction of a Previously Calibrated Method
This section would apply if a site simply wants to express their data in Centiloid units obtained using the standard PiB method or another previously calibrated method without modification.
3.1 New Data Necessary for Level-3 Centiloid Calibration
No new data need be acquired. The independent site downloads the previously calibrated dataset for the tracer and method they wish to reproduce. For example, to simply use the standard PiB SUVr 50-70 min method, they would download the 34 YC-0 and the 45 AD-100 subjects. To reproduce another previously validated tracer method, they would download the 25 (or more) scans available for that tracer/method.
3.2 Level-3 Analysis
In the same manner as done with the standard PiB data in section 2.2.1, the independent site should show that their analysis pipeline does not introduce errors into the data. The site should calculate the outcome data for the method to be reproduced for each downloaded scan (e.g., standard PiB 50-70 min SUVr values for the 34 YC-0 and the 45 AD-100 subjects). They will then convert that data into Centiloids using the equation provided for that tracer method (i.e., Eq. 2.2.1 for PiB data and a combination of Eq. 2.2.3.1b and or Eq. 2.2.3.2b). For the standard PiB method, Eq. 1.3b should be used along with the AD100 and YC0 mean SUVr value listed above.
The site will then produce a scattergraph and calculate a trendline and R2 value from their calculated Centiloid values and the published Centiloid values for the method of interest. This should be included as supplemental data in the first publication from that site using this validated method. The expectation is that the slope will be between 0.98 and 1.02, the intercept will be between −2 and 2 CL and the R2 will be > 0.98.
3.3 Level-3 Data made available for unrestricted use
No new data is acquired and thus there is no need for new publically available data.
DISCUSSION
In the sections above we outline a process to scale global cortical 50-70 min PiB PET SUVr data to a scale anchored at 0 to represent relatively young “high certainty” amyloid negative subjects and at 100 to represent typical AD patients. The data used to complete this process has been deposited on the publically accessible GAAIN website. This data can be downloaded so an interested site can verify that their data-analysis pipeline gives results essentially identical to those reported here. This pipeline can then be used to generate a site-acquired PiB 50-70 min dataset from ~25 subjects who have also been scanned with another tracer or had data analyzed by a method different from the “standard” PiB method described here. This “non-standard” data can then be scaled to the Centiloid scale to facilitate comparison to other appropriate data. We strongly encourage sites that perform what we term a “level-2” analysis of this nature to deposit the scan data they used to do that analysis on the GAAIN website. This data could serve two purposes. First, if performed in the appropriate manner, data from an F-18 tracer could allow sites who do not have C-11 tracers available, to use this F-18 data to scale other non-standard methods to the Centiloid scale. Second, it could serve as data to verify the pipeline of a site that simply wants to reproduce that exact method using the Centiloid scale (i.e., a “level-3” analysis).
It is important to stress that the Centiloid process is not intended to replace any method that a site has determined is optimal for its specific purposes. It is intended to be a simple means to translate those outcomes into units that are more easily compared across sites. In practice, this could take the form of presenting results only in Centiloid units, presenting results in both original and Centiloid units (or providing an equation for that purpose), making a Centiloid “translation” available as supplemental online data, or simply being prepared to provide results in Centiloid units if requested. It should be noted that while this process was developed with amyloid imaging data in mind, an analogous approach could be used for most any class of tracers, including emerging tau tracers [28-30].
Several aspects of the Centiloid process deserve further discussion. First, this approach is based on PiB since this tracer has been the most widely employed and thoroughly studied tracer to-date. The choice of the SUVr 50-70 min method of analysis was based on this being a widely accessible measure. While some may argue that this approach is inferior to a variety of dynamic analysis methods for PiB data, our example above shows that the 50-70 SUVr method correlates well with an sRTM dynamic method in the same subjects. The requirement for sites to acquire 0-90 min dynamic data for a level-2 analysis was felt to be an unnecessary burden. Partial volume correction (PVC) was not included in this “standard” approach since both the practice of applying PVC at all and the process by which PVC is accomplished is highly variable across centers. PVC has value in certain situations and can be applied as a non-standard approach and calibrated to the Centiloid scale.
Second, we chose the MNI-152 template and the specified SPM8 normalization approach on the basis that these were widely available approaches that have been validated in a large variety of studies. While these were found to function well for the purpose of this study, and out-performed several other methods tested, we do not claim that these are the only, or even the best, approaches for this purpose.
Third, the four reference regions tested encompass the vast majority of those used in published studies. While there is an appeal to using a purely gray matter reference region like the data-driven CG VOI, the WC and WC+B VOIs clearly outperformed the CG (and Pons) region. The larger size of WC and WC+B is likely to be the major factor, but the variability present in the exclusion of white matter from the cerebellar peduncles when normalizing the CG VOI is likely another important factor. The poor performance of the Pons VOI was surprising, but also likely relates to the smaller size of this VOI and the fact that the SPM8 normalization algorithm seems to handle brainstem structures less well than cortical structures. Normalization of brainstem structures was even worse for other algorithms tested (i.e., SPM5 and DARTEL). While the WC and WC+B VOIs produced essentially equivalent results, we believed it was important to define a single “standard” reference VOI. Based on the variance in the YC-0 group, we judged the WC VOI to be slightly better. We acknowledge that the choice of reference region was guided solely by between-group comparisons using cross-sectional data. The performance of various reference regions across time in longitudinal studies could be different and is an area for further investigation. Nothing in the Centiloid process would prohibit the use of a reference region other than WC for longitudinal (or cross-sectional) studies after calibrating the non-standard method to the standard PiB method or another previously calibrated method.
Fourth, the standard Centiloid process is designed to produce a measure of “global” cortical amyloid deposition (including signal from the striatum). Early in the Centiloid development process, the decision was made that it would be preferential to have the CTX VOI completely defined by the amyloid imaging data itself. This was based on the fact that Aβ deposition does not follow any atlas-based VOI in an exact manner and the deposition is blurred by PET resolution at gray-white borders making MR-based segmentation imperfect for this purpose. We reasoned that this process would include the majority of the amyloid signal without the loss that segmentation or restrictive atlas VOIs might cause while avoiding additional non-amyloid areas that overly-inclusive atlas areas might add. A similar approach has been used by others, most frequently in the use of “signature regions” for FDG data in AD [31-33]. The use of smaller regional measures, while acceptable, will produce CL values that may not carry the same connotation as the global measures. For example, a group of AD subjects with a mean of 100 PiBCLStd may have a PiBCLNS of significantly greater than 100 in precuneus and significantly less than 100 in occipital cortex. This is true even if the regional measures are generated from parametric Centiloid images as suggested. Also, care must be taken to understand that cutoff values will be tied to the region from which they were generated. For example, if a cutoff of 25 PiBCLStd – determined using the CTX and WC VOIs – was found to effectively define early amyloid positivity, then a regional PiBCLNS value of 25 from a small brain region does not necessarily represent an amyloid positive subject. In addition, much more care must be taken if a small target VOI is employed in a non-standard analysis in place of the standard CTX target VOI. While this may be acceptable, we expect that, at some point, a target VOI may be too small to use in a valid manner. Precise definition of the limits of valid target VOIs will have to await future explorations of the nuances of the Centiloid process.
Fifth, there are two separate scaling processes for tracers other than PiB. The first is to be used if that F-18 tracer is to be used as a surrogate for PiB in future scaling procedures when a site does not have access to [C-11]PiB. This use comes with the restrictions of using the standard CTX and WC VOIs. The second process is not limited in this way and is meant solely to scale the new tracer to PiB without the intention of using that tracer method as a PiB-surrogate. Of course, the two processes can often be performed on a single dataset.
The ultimate purpose, of course, is for this Centiloid scaling process to enhance the comparison of data obtained at different sites and even with different tracers. An indication that this could occur can be seen by consideration of the SUVr data in Figure 3. If expressed in SUVr units determined using CG as a reference, the typical range AD values would be about 1.90-3.00 SUVr units. If pons was used as a reference region, as has been done in several published studies [34; 35], the typical AD range would be about 1.20-1.90. The confusion about the interpretation of a value of 1.80 SUVr units becomes clear here – and this is without other sources of confusion such as the use of atrophy-correction and different target VOIs. Furthermore, this does not even consider differences caused by the use of different tracers. In contrast, when the typical AD range using CG as reference is expressed on the Centiloid scale it is about 55-150 CL and when pons is used as reference, the typical AD range is about 60-155 CL. The same phenomenon occurs in the YC range. One possible benefit of this consistency is that we may be able to consistently define three ranges of amyloid deposition: 1) the amyloid-negative range [36]; 2) the “just-positive” range [37]; and 3) the “AD-like” range [38]. The lack of a clearly defined AD-like range, separate from that of amyloid-positive controls, may have contributed to limitations regarding the value of a positive scan in the FDA-approved labels for the two currently-approved amyloid imaging PET tracers (Amyvid™ and Vizamyl™). Indeed, the labels state that a positive scan indicates moderate to frequent amyloid neuritic plaques such as is typically present in patients with AD, but may also be present in patients with other types of neurologic conditions as well as older people with normal cognition. If further study makes it possible to subdivide the amyloid-positive range into two ranges: 1) a range rarely seen in AD patients and 2) a range typical of clinical AD, then expression of this AD range in a quantitative and consistent manner using the Centiloid scale may increase the value of amyloid imaging for a positive diagnosis of AD, rather than just excluding this diagnosis.
There are several limitations that became apparent in the development of the Centiloid process. One critical assumption when translating the online level-1 data into site-acquired Centiloid data is that, when analyzed by the standard method presented here, any given subject would yield essentially the same PiB SUVr 50-70 min value if scanned at two different sites several days apart. It is not currently clear how differences in scanners, reconstruction algorithms, instrument resolution or methods of attenuation correction might affect this assumption, but we are aware that this assumption is incorrect at least to some degree. Subjects in this study were scanned on similar instruments with comparable resolution, however we have not investigated effects of substantial differences in scanner design and software nor can we account for future developments in instrumentation that might render our assumptions about the similarities invalid. We have attempted to minimize site-specific nuances in the level-1 data by including data from three separate sites – although two of these sites used the same scanner. It was reassuring to see that the range of AD-100 and YC-0 values was very similar across the three sites.
We emphasize that when scaling other tracers to the standard method PiB data reported here, the R2 and SD values determined are solely to test the overall validity of the scaling process and are not valid comparisons of the performance of different tracers. This is even more true of slopes and intercepts derived from these correlations. This is because different groups of subjects will be used to scale different tracers to PiB data and thus, uncontrollable subject variables will impact these R2 and SD values. Therefore, it is not valid, for example, to make inferences about the relative similarities of two different F-18-labelled tracers to PiB by simply comparing the R2 and SD values. Direct comparison between F-18 tracers will require studies performed in the same group of subjects. However, since it is desirable to not lose measures of the relative dynamic range (or signal-to-noise ratio) of different tracers or methods as they are all expanded or compressed to the same Centiloid scale, the “relative SD” compared to PiB in the same set of subjects is an important tracer-specific parameter. The higher this relative SD, the less likely that method or tracer will be able to reliably distinguish the earliest evidence of amyloid deposition or small changes in amyloid load over time or in response to anti-amyloid therapy.
In summary, it is hoped that widespread use of the Centiloid standardization method will facilitate: 1) direct comparison of results across labs even when different analysis methods or tracers are employed; 2) clear definition of cutoffs for the earliest signs of amyloid-positivity in cognitively normal controls; 3) further definition of the range of amyloid positivity characteristic of AD (AD-like levels vs. earliest evidence of positivity in controls); 4) more consistent representation of longitudinal change in standard units (rather than as percent change); 5) direct comparison of the characteristics of different tracers. Facile combination of results across studies (including studies that employ more than one tracer) would make the combination of difficult-to-perform studies possible. Thus, conversion to Centiloid units may allow combination of results across ADNI (PiB and AV-45), Japan-ADNI (PiB and BF-227) and AIBL (PiB, flutemetamol, AV-45). These cross-center analyses could include combination of results across difficult-to-perform studies such as postmortem pathology to in vivo data correlations and data pooling in therapeutic trials. If these goals are realized, the Centiloid standardization process will be a valuable addition to the field.
Supplementary Material
Supplemental Table A. SUVr and Centiloid Values for Each Reference VOI.
Supplemental Table B. Scaling sRTM BPND + 1 Data to Centiloid Units
Systematic Review: We used PubMed to search all articles under the search terms “amyloid imaging, Pittsburgh Compound B, PiB, AV45, florbetapir, Amyvid, flutemetamol, Vizamyl, florbetaben, AZ4694 and NAV4694.” We found no papers dealing with standardization of quantitative outcome measures of tracer retention.
Interpretation: The current report presents a starting point for the field to standardize the expression of quantitative amyloid imaging results. This could lead to: 1) facilitation of cross-center comparison of results; 2) clear definition of cutoffs; 3) facilitation of longitudinal studies; 4) direct comparison of the different tracers and 5) combination of results from multi-center studies.
Future Directions: The value of this proposed “Centiloid” method will rely on whether it is widely accepted and used by the field. With widespread use, the limitations and capabilities of this initial proposed standardization method will become clearer and refinements to overcome the limitations will then need to be developed.
Acknowledgements
This work was supported in-part by grants from the National Institute on Aging: R37 AG025516 (WEK), P01 AG025204 (WEK), P50 AG005133 (WEK), R01 AG034570 (WJ), P01 AG026276 (TB), U19 AG032438 (TB). We would like to thank Marybeth Howlett for her organizational and administrative support.
Footnotes
Disclosures:
1) GE Healthcare holds a license agreement with the University of Pittsburgh based on the PiB technology described in this manuscript. Drs. Klunk and Mathis are co-inventors of PiB and, as such, have a financial interest in this license agreement. GE Healthcare provided no grant support for this study and had no role in the design or interpretation of results or preparation of this manuscript. All other authors have no conflicts of interest with PiB-related technology and had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
2) Dr. Jagust is a consultant to Genentech, Synarc, and F. Hoffman LaRoche
3) Dr. Rowe has received research grants for imaging in dementia from Bayer-Schering Pharma, Avid Radiopharmaceuticals, GE Healthcare, Piramal and Astra Zeneca. He has been a consultant and conference speaker for Bayer Schering Pharma and GE Healthcare.
4) Dr. Benzinger has received research grants for imaging in dementia from Avid Radiopharmaceuticals, a wholly owned subsidiary of Eli Lilly.
5) Dr. Devous has received grants from Avid Radiopharmaceuticals, and has served as a consultant for Eli Lilly, Piramal, Navidea, and GE Healthcare.
6) Drs. Pontecorvo, Skovronsky and Mintun are employees of Avid Radiopharmaceuticals, a wholly owned subsidiary of Eli Lilly and Company.
All other authors have no conflicts with this work. All authors had full access to the data used in this analysis and approved the final manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Frisoni GB, Jack CR. Harmonization of magnetic resonance-based manual hippocampal segmentation: a mandatory step for wide clinical use. Alzheimers Dement. 2011;7:171–174. doi: 10.1016/j.jalz.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 2.Jack CR, Jr., Barkhof F, Bernstein MA, Cantillon M, Cole PE, Decarli C, Dubois B, Duchesne S, Fox NC, Frisoni GB, Hampel H, Hill DL, Johnson K, Mangin JF, Scheltens P, Schwarz AJ, Sperling R, Suhy J, Thompson PM, Weiner M, Foster NL. Steps to standardization and validation of hippocampal volumetry as a biomarker in clinical trials and diagnostic criterion for Alzheimer’s disease. Alzheimers Dement. 2011;7:474–485. e4741. doi: 10.1016/j.jalz.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boccardi M, Bocchetta M, Ganzola R, Robitaille N, Redolfi A, Duchesne S, Jack CR, Jr, Frisoni GB. Operationalizing protocol differences for EADC-ADNI manual hippocampal segmentation. Alzheimers Dement. 2013 doi: 10.1016/j.jalz.2013.03.001. (in press): DOI 10.1016/j.jalz.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Frisoni GB, Bocchetta M, Chetelat G, Rabinovici GD, de Leon MJ, Kaye J, Reiman EM, Scheltens P, Barkhof F, Black SE, Brooks DJ, Carrillo MC, Fox NC, Herholz K, Nordberg A, Jack CR, Jr., Jagust WJ, Johnson KA, Rowe CC, Sperling RA, Thies W, Wahlund LO, Weiner MW, Pasqualetti P, Decarli C. Imaging markers for Alzheimer disease: Which vs how. Neurology. 2013;81:487–500. doi: 10.1212/WNL.0b013e31829d86e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vanderstichele H, Bibl M, Engelborghs S, Le Bastard N, Lewczuk P, Molinuevo JL, Parnetti L, Perret-Liaudet A, Shaw LM, Teunissen C, Wouters D, Blennow K. Standardization of preanalytical aspects of cerebrospinal fluid biomarker testing for Alzheimer’s disease diagnosis: a consensus paper from the Alzheimer’s Biomarkers Standardization Initiative. Alzheimers Dement. 2012;8:65–73. doi: 10.1016/j.jalz.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 6.Carrillo MC, Blennow K, Soares H, Lewczuk P, Mattsson N, Oberoi P, Umek R, Vandijck M, Salamone S, Bittner T, Shaw LM, Stephenson D, Bain L, Zetterberg H. Global standardization measurement of cerebral spinal fluid for Alzheimer’s disease: an update from the Alzheimer’s Association Global Biomarkers Consortium. Alzheimers Dement. 2013;9:137–140. doi: 10.1016/j.jalz.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 7.Mintun MA, Larossa GN, Sheline YI, Dence CS, Lee SY, Mach RH, Klunk WE, Mathis CA, DeKosky ST, Morris JC. [11C]PIB in a nondemented population: potential antecedent marker of Alzheimer disease. Neurology. 2006;67:446–452. doi: 10.1212/01.wnl.0000228230.26044.a4. [DOI] [PubMed] [Google Scholar]
- 8.Morris JC, Aisen PS, Bateman RJ, Benzinger TL, Cairns NJ, Fagan AM, Ghetti B, Goate AM, Holtzman DM, Klunk WE, McDade E, Marcus DS, Martins RN, Masters CL, Mayeux R, Oliver A, Quaid K, Ringman JM, Rossor MN, Salloway S, Schofield PR, Selsor NJ, Sperling RA, Weiner MW, Xiong C, Moulder KL, Buckles VD. Developing an international network for Alzheimer research: The Dominantly Inherited Alzheimer Network. Clin Investig (Lond) 2012;2:975–984. doi: 10.4155/cli.12.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oh H, Madison C, Haight TJ, Markley C, Jagust WJ. Effects of age and beta-amyloid on cognitive changes in normal elderly people. Neurobiol Aging. 2012;33:2746–2755. doi: 10.1016/j.neurobiolaging.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rowe CC, Ellis KA, Rimajova M, Bourgeat P, Pike KE, Jones G, Fripp J, Tochon-Danguy H, Morandeau L, O’Keefe G, Price R, Raniga P, Robins P, Acosta O, Lenzo N, Szoeke C, Salvado O, Head R, Martins R, Masters CL, Ames D, Villemagne VL. Amyloid imaging results from the Australian Imaging, Biomarkers and Lifestyle (AIBL) study of aging. Neurobiol Aging. 2010;31:1275–1283. doi: 10.1016/j.neurobiolaging.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 11.Morris JC, Roe CM, Xiong C, Fagan AM, Goate AM, Holtzman DM, Mintun MA. APOE predicts amyloid-beta but not tau Alzheimer pathology in cognitively normal aging. Ann Neurol. 2010;67:122–131. doi: 10.1002/ana.21843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 13.Aizenstein HJ, Nebes RD, Saxton JA, Price JC, Mathis CA, Tsopelas ND, Ziolko SK, James JA, Snitz BE, Houck PR, Bi W, Cohen AD, Lopresti BJ, DeKosky ST, Halligan EM, Klunk WE. Frequent amyloid deposition without significant cognitive impairment among the elderly. Arch Neurol. 2008;65:1509–1517. doi: 10.1001/archneur.65.11.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rowe CC, Ng S, Ackermann U, Gong SJ, Pike K, Savage G, Cowie TF, Dickinson KL, Maruff P, Darby D, Smith C, Woodward M, Merory J, Tochon-Danguy H, O’Keefe G, Klunk WE, Mathis CA, Price JC, Masters CL, Villemagne VL. Imaging beta-amyloid burden in aging and dementia. Neurology. 2007;68:1718–1725. doi: 10.1212/01.wnl.0000261919.22630.ea. [DOI] [PubMed] [Google Scholar]
- 15.Rabinovici GD, Furst AJ, O’Neil JP, Racine CA, Mormino EC, Baker SL, Chetty S, Patel P, Pagliaro TA, Klunk WE, Mathis CA, Rosen HJ, Miller BL, Jagust WJ. 11C-PIB PET imaging in Alzheimer disease and frontotemporal lobar degeneration. Neurology. 2007;68:1205–1212. doi: 10.1212/01.wnl.0000259035.98480.ed. [DOI] [PubMed] [Google Scholar]
- 16.Morris JC. The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology. 1993;43:2412–2414. doi: 10.1212/wnl.43.11.2412-a. [DOI] [PubMed] [Google Scholar]
- 17.Schwertman NC, de Silva R. Identifying outliers with sequential fences. Computational Statistics & Data Analysis. 2007;51:3800–3810. [Google Scholar]
- 18.Cohen AD, Mowrey W, Weissfeld LA, Aizenstein HJ, McDade E, Mountz JM, Nebes RD, Saxton JA, Snitz B, Dekosky S, Williamson J, Lopez OL, Price JC, Mathis CA, Klunk WE. Classification of amyloid-positivity in controls: Comparison of visual read and quantitative approaches. Neuroimage. 2013;71:207–215. doi: 10.1016/j.neuroimage.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen AD, Price JC, Weissfeld LA, James J, Rosario BL, Bi W, Nebes RD, Saxton JA, Snitz BE, Aizenstein HA, Wolk DA, Dekosky ST, Mathis CA, Klunk WE. Basal Cerebral Metabolism May Modulate the Cognitive Effects of Aβ in Mild Cognitive Impairment: An Example of Brain Reserve. J Neurosci. 2009;29:14770–14778. doi: 10.1523/JNEUROSCI.3669-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Price JC, Klunk WE, Lopresti BJ, Lu X, Hoge JA, Ziolko SK, Holt DP, Meltzer CC, DeKosky ST, Mathis CA. Kinetic modeling of amyloid binding in humans using PET imaging and Pittsburgh Compound-B. J Cereb Blood Flow Metab. 2005;25:1528–1547. doi: 10.1038/sj.jcbfm.9600146. [DOI] [PubMed] [Google Scholar]
- 21.Ashburner J, Friston KJ. Nonlinear spatial normalization using basis functions. Hum Brain Mapp. 1999;7:254–266. doi: 10.1002/(SICI)1097-0193(1999)7:4<254::AID-HBM4>3.0.CO;2-G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashburner J. A fast diffeomorphic image registration algorithm. Neuroimage. 2007;38:95–113. doi: 10.1016/j.neuroimage.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 23.Ashburner J, Friston KJ. Unified segmentation. Neuroimage. 2005;26:839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 24.Klunk WE, Engler H, Nordberg A, Wang Y, Blomqvist G, Holt DP, Bergström M, Savitcheva I, Huang GF, Estrada S, Ausén B, Debnath ML, Barletta J, Price JC, Sandell J, Lopresti BJ, Wall A, Koivisto P, Antoni G, Mathis CA, Långström B. Imaging brain amyloid in Alzheimer’s disease with Pittsburgh Compound-B. Ann Neurol. 2004;55:306–319. doi: 10.1002/ana.20009. [DOI] [PubMed] [Google Scholar]
- 25.Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, Woods R, Paus T, Simpson G, Pike B, Holmes C, Collins L, Thompson P, MacDonald D, Iacoboni M, Schormann T, Amunts K, Palomero-Gallagher N, Geyer S, Parsons L, Narr K, Kabani N, Le Goualher G, Boomsma D, Cannon T, Kawashima R, Mazoyer B. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM) Philos Trans R Soc Lond B Biol Sci. 2001;356:1293–1322. doi: 10.1098/rstb.2001.0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tolboom N, Yaqub M, Boellaard R, Luurtsema G, Windhorst AD, Scheltens P, Lammertsma AA, van Berckel BN. Test-retest variability of quantitative [11C]PIB studies in Alzheimer’s disease. Eur J Nucl Med Mol Imaging. 2009;36:1629–1638. doi: 10.1007/s00259-009-1129-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yaqub M, Tolboom N, Boellaard R, van Berckel BNM, van Tilburg EW, Luurtsema G, Scheltens P, Lammertsma AA. Simplified parametric methods for [11C]PIB studies. NeuroImage. 2008;42:76–86. doi: 10.1016/j.neuroimage.2008.04.251. [DOI] [PubMed] [Google Scholar]
- 28.Chien DT, Bahri S, Szardenings AK, Walsh JC, Mu F, Su MY, Shankle WR, Elizarov A, Kolb HC. Early Clinical PET Imaging Results with the Novel PHF-Tau Radioligand [F-18]-T807. J Alzheimers Dis. 2012 doi: 10.3233/JAD-122059. (in press): DOI 10.3233/JAD-122059. [DOI] [PubMed] [Google Scholar]
- 29.Maruyama M, Shimada H, Suhara T, Shinotoh H, Ji B, Maeda J, Zhang MR, Trojanowski JQ, Lee VM, Ono M, Masamoto K, Takano H, Sahara N, Iwata N, Okamura N, Furumoto S, Kudo Y, Chang Q, Saido TC, Takashima A, Lewis J, Jang MK, Aoki I, Ito H, Higuchi M. Imaging of tau pathology in a tauopathy mouse model and in Alzheimer patients compared to normal controls. Neuron. 2013;79:1094–1108. doi: 10.1016/j.neuron.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okamura N, Furumoto S, Harada R, Tago T, Yoshikawa T, Fodero-Tavoletti M, Mulligan RS, Villemagne VL, Akatsu H, Yamamoto T, Arai H, Iwata R, Yanai K, Kudo Y. Novel 18F-labeled arylquinoline derivatives for noninvasive imaging of tau pathology in Alzheimer disease. J Nucl Med. 2013;54:1420–1427. doi: 10.2967/jnumed.112.117341. [DOI] [PubMed] [Google Scholar]
- 31.Herholz K, Salmon E, Perani D, Baron JC, Holthoff V, Frolich L, Schonknecht P, Ito K, Mielke R, Kalbe E, Zundorf G, Delbeuck X, Pelati O, Anchisi D, Fazio F, Kerrouche N, Desgranges B, Eustache F, Beuthien-Baumann B, Menzel C, Schroder J, Kato T, Arahata Y, Henze M, Heiss WD. Discrimination between Alzheimer dementia and controls by automated analysis of multicenter FDG PET. Neuroimage. 2002;17:302–316. doi: 10.1006/nimg.2002.1208. [DOI] [PubMed] [Google Scholar]
- 32.Yamane T, Ikari Y, Nishio T, Ishii K, Kato T, Ito K, Silverman DH, Senda M, Asada T, Arai H, Sugishita M, Iwatsubo T. Visual-Statistical Interpretation of 18F-FDG-PET Images for Characteristic Alzheimer Patterns in a Multicenter Study: Inter-Rater Concordance and Relationship to Automated Quantitative Evaluation. AJNR Am J Neuroradiol. 2013 doi: 10.3174/ajnr.A3665. (in press): DOI 10.3174/ajnr.A3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jagust WJ, Bandy D, Chen K, Foster NL, Landau SM, Mathis CA, Price JC, Reiman EM, Skovronsky D, Koeppe RA. The Alzheimer’s Disease Neuroimaging Initiative positron emission tomography core. Alzheimers Dement. 2010;6:221–229. doi: 10.1016/j.jalz.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klunk WE, Price JC, Mathis CA, Tsopelas ND, Lopresti BJ, Ziolko SK, Bi W, Hoge JA, Cohen AD, Ikonomovic MD, Saxton JA, Snitz BE, Pollen DA, Moonis M, Lippa CF, Swearer JM, Johnson KA, Rentz DM, Fischman AJ, Aizenstein HJ, DeKosky ST. Amyloid deposition begins in the striatum of presenilin-1 mutation carriers from two unrelated pedigrees. J Neurosci. 2007;27:6174–6184. doi: 10.1523/JNEUROSCI.0730-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Villemagne VL, Ataka S, Mizuno T, Brooks WS, Wada Y, Kondo M, Jones G, Watanabe Y, Mulligan R, Nakagawa M, Miki T, Shimada H, O’Keefe GJ, Masters CL, Mori H, Rowe CC. High striatal amyloid beta-peptide deposition across different autosomal Alzheimer disease mutation types. Arch Neurol. 2009;66:1537–1544. doi: 10.1001/archneurol.2009.285. [DOI] [PubMed] [Google Scholar]
- 36.Cohen AD, Bi W, Weissfeld LA, Aizenstein HA, McDade E, Mountz JM, Nebes RD, Saxton JA, Snitz BE, Lopez OL, Price JC, Mathis CA, Klunk WE. Classification of amyloid-positivity in controls: Comparison of visual read and quantitative approaches. Neuroimage. 2012;71:207–215. doi: 10.1016/j.neuroimage.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mormino EC, Brandel MG, Madison CM, Rabinovici GD, Marks S, Baker SL, Jagust WJ. Not quite PIB-positive, not quite PIB-negative: slight PIB elevations in elderly normal control subjects are biologically relevant. Neuroimage. 2012;59:1152–1160. doi: 10.1016/j.neuroimage.2011.07.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klunk W, Cohen A, Bi W, Weissfeld L, Aizenstein H, McDade E, Mountz J, Nebes R, Saxton J, Snitz B, Lopez O, Price J, Mathis C. Why we need two cutoffs for amyloid imaging: Early versus Alzheimer’s-like amyloid-positivity. Alzheimer’s & dementia : the journal of the Alzheimer’s Association. 2012;8:P453–P454. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table A. SUVr and Centiloid Values for Each Reference VOI.
Supplemental Table B. Scaling sRTM BPND + 1 Data to Centiloid Units