Abstract
Imaging biomarkers derived from magnetic resonance imaging (MRI) data are used to quantify normal development, disease, and the effects of disease-modifying therapies. However, motion during image acquisition introduces image artifacts that, in turn, affect derived markers. A systematic effect can be problematic since factors of interest like age, disease, and treatment are often correlated with both a structural change and the amount of head motion in the scanner, confounding the ability to distinguish biology from artifact. Here we evaluate the effect of head motion during image acquisition on morphometric estimates of structures in the human brain using several popular image analysis software packages (FreeSurfer 5.3, VBM8 SPM, and FSL Siena 5.0.7). Within-session repeated T1-weighted MRIs were collected on 12 healthy volunteers while performing different motion tasks, including two still scans. We show that volume and thickness estimates of the cortical gray matter are biased by head motion with an average apparent volume loss of roughly 0.7%/mm/min of subject motion. Effects vary across regions and remain significant after excluding scans that fail a rigorous quality check. In view of these results, the interpretation of reported morphometric effects of movement disorders or other conditions with increased motion tendency may need to be revisited: effects may be overestimated when not controlling for head motion. Furthermore, drug studies with hypnotic, sedative, tranquillizing, or neuromuscular-blocking substances may contain spurious “effects” of reduced atrophy or brain growth simply because they affect motion distinct from true effects of the disease or therapeutic process.
Keywords: Head Motion, MRI, Cortical Gray Matter Estimates, Spurious Effect, Bias, Volume, Thickness, Quality Control
1. Introduction
In neuroimaging, structural MRI is frequently acquired to study a wide variety of diseases such as Alzheimer’s, Huntington’s disease, schizophrenia, cancer, and stroke. Furthermore, the analysis of within-subject longitudinal changes allows the assessment of the response to drug treatment, or the quantification of progression in neurodegeneration or brain development. A large array of imaging biomarkers are derived from MRI, most often with automatic processing methods to reduce noise caused by within- or cross-rater variability and to facilitate the analysis of large data sets.
Despite the intuitive appeal that, within short time intervals, brain structure should be constant and derived measures stable, there are confounds. For example, hydration levels affect brain and ventricular volume1–3 making it difficult, for example, to attribute causes of brain recovery after alcohol abuse4 to rehydration versus actual regrowth. The increase in ventricular volume, as well as gray and white matter losses, reported in aging, Alzheimer’s, Huntington’s or other degenerative diseases, may also be confounded by subjects’ hydration states. Hydration is not the only confounder that should be considered in morphometric studies of the brain.
There is a general awareness that image quality is affected by head motion during the acquisition, which can result in image artifacts. Motion can cause structured artifacts, shading, and blurring in structural MR images that are best appreciated via qualitative assessment5,6. Reduced image quality can affect derived volume or cortical thickness estimates and reduce reliability. However, as of yet, it remains unclear if motion artifacts produce a directional (systematic) bias or simply an increase in the variance of the measures. This is an important distinction.
While increased variability can reduce power to detect group differences or longitudinal changes, a bias may induce spurious effects that are not directly caused by disease or treatment, but rather by the amount of head motion. This is particularly problematic when studying movement disorders such as Huntington’s disease, or even normal aging, where the amount of head motion correlates with the variable of interest. Spurious effects of head motion have recently been reported for resting-state functional connectivity MRI7–10 and diffusion MRI11. Here we explore whether similar, systematic effects are present in structural MRI measures.
We adopt a prospective within-subject design: Healthy volunteers were scanned repeatedly both as they remained still and as they performed different motion tasks. Knowing a priori that there should be no changes in the subjects’ brain structure during a single scan session, we explored the effect of head motion on volume and thickness estimates of cortical and subcortical regions produced by a variety of automated tools. Our findings demonstrate a systematic bias in all tested software packages, resulting in the spurious detection of apparent cortical atrophy due entirely to increased motion.
2. Materials
Twelve healthy adult volunteers (having given informed consent) were scanned on a 3T TIM Trio MRI system (Siemens Healthcare, Erlangen) using the vendor-supplied 12-channel head matrix coil supplied. Each subject’s visit was broken into two “blocks”, between which the subject was removed from the scanner and given a short break. At the start of each block, subjects were positioned such that the juncture between the forehead and the bridge of the nose was at isocenter.
Five multiecho MPRAGE (MEMPRAGE)12,13 scans were collected with 256 mm × 256 mm × 176 mm FOV, 1 mm isotropic resolution, 4 echoes with bandwidth of 650 Hz/pixel, and 2 × GRAPPA acceleration (the 4 echoes were combined via RMS to give one output volume for analysis). At the start of each MEMPRAGE, the Autoalign system14 was used to automatically detect the current position of the subject and align the MEMPRAGE field of view.
The order of the scans within each block was randomized. For one scan in each block the subjects were directed to remain still. Three different task-scans were then randomly assigned to the first or the second block. For these three scans, subjects were asked to perform a task when a visual cue appeared on a projected screen viewed via a mirror. The three tasks were nod (superior-inferior head rotation), shake (left-right head rotation), and free motion that the subjects were asked to invent and repeat for the duration of the screen display (each subject was given the suggestion of “for example, draw a figure eight with your nose”). Subjects were randomized into two even-sized groups: those whose action prompts lasted 15 sec out of every min during scans, and those whose prompts lasted 5 sec per min. By directing subjects to perform varied types of motion and additionally varying the motion duration, we aimed to ensure that measurements were made over a range of motion amplitudes, durations, and trajectories, beyond what would occur due to natural inter-subject variation. This, in turn, gives greater confidence that our results are neither limited to a specific type of motion, nor to specific durations or amplitudes.
Volumetric navigator images (vNavs)15 were collected during each scan to provide real-time estimates of subject motion. Neither prospective nor retrospective motion correction was applied. Navigator images were, however, used for the analysis of subject motion between TR’s during each scan. To keep motion levels in a realistic range, scans were immediately stopped and repeated if a subject’s motion was estimated to have exceeded 8 degrees rotation or 20 mm translation in one TR. This limit is enforced by Siemens’ PACE motion-tracking system16, which the vNavs system is based upon.
All MEMPRAGE images were visually evaluated by an expert for motion-related artifacts such as blurring, ghosting and striping5,6, as well as general criteria that can affect image quality, including: head coverage, wrapping artifact, radiofrequency noise, signal inhomogeneity, susceptibility artifact, and ringing artifact. An ordinal score was given to each criterion (none, mild, moderate, severe), and an overall qualitative score was given to each image (pass, warn, fail) using standardized methodology (Harvard Center for Brain Science17).
Note that, while we considered several types of motion in this work, we did not include continuous tremor motion for two reasons: first, we did not expect our healthy volunteers to reliably maintain a consistent tremor motion for the duration of the scans; and second, the vNavs tracking system only estimated subject motion every ~2.53 seconds, which is quite slow relative to the frequency of a tremor. As such, we cannot be sure if the results shown here extend to tremor-induced motion. Tremor is likely better studied with a high-frequency optical18 or similar tracking system, although simulating tremors in healthy subjects may still be challenging.
3. Methods
We analyzed the association of the motion severity on the anatomical markers using all scans in a linear mixed effects model. We further explored the effect of quality control and exclusion of individual low quality scans on our results to mimic common practice in the field.
During image acquisition, navigator images were collected at each TR during the scan and can be used to quantify the amount of motion during each scan and provide a measure (RMSpm) for the average displacement per minute. The sequential rigid transformations Ti,i+1 from navigator image i to navigator (i + 1) were estimated via rigid registrations to the baseline navigator image (index 0) and composition of the transforms1. Then the root mean square (RMS) deviation19 was computed for each incremental motion update Ti,i+1 and averaged across the whole sequence. The RMS deviation quantifies the average voxel displacement (in mm) inside a spherical volume for a given affine transformation T = (M, t⃗), where M is a 3 × 3 linear transformation matrix (in our case a rotation) and t⃗ the corresponding 3 × 1 translation vector. The RMS deviation for a spherical volume with radius r is described by:
where tr is the trace and Id the identity matrix. In this work we use a sphere centered at the isocenter with a radius of 64 mm to represent the full brain. Since navigator images were acquired at each TR (TR = 2.53 seconds), we can estimate the average motion in mm/min (the RMS displacement per minute RMSpm) via:
MEMPRAGE volumes were analyzed using the following popular and freely available processing software: Percent brain volume change between two scans was directly estimated with FSL Siena20 5.0.7; Gray matter (GM) volumes were estimated using voxel based morphometry VBM8 toolbox21 of the SPM8 package22; Cortical thickness and gray matter volume were estimated using both the independent23,24 and longitudinal image-processing stream25,26 of FreeSurfer 5.3 (FS). In FSL Siena standard-space masking was used as well as BET (-m option). Furthermore, the lower part of MNI152/Talairach space (–b -50) was ignored and the approximate center of the head passed to BET (–B “ –c 135 100 90”). In VBM8 images were corrected for bias-field inhomogeneity and tissue-classified into GM, white matter and cerebrospinal fluid (with partial volume estimates). Gray matter volumes were estimated from the reported GM segmentation. For the voxel-based analysis the modulated images were smoothed at 8 mm full width half maximum (FWHM), and a mixed-effects analysis was performed for voxels with a minimal modulated GM volume of 0.2 (across all images). For FreeSurfer analysis, the default settings for both the independent and longitudinal pipelines were employed. In the longitudinal processing, surfaces were estimated first on a robust within-subject template, with a subsequent fine-tuning step on each time point. This approach reduces variability and prevents completely incorrect placement of surfaces in cases with severe motion. Surface analysis was performed on the pre-existing FreeSurfer fsaverage template after smoothing (with 15 mm FWHM) inside the cortical regions of both hemispheres.
Analysis of the repeated measure data was performed with a linear mixed effects model for gray matter volume estimates27 as well as via a spatio-temporal approach for FreeSurfer’s cortical thickness estimates28. To analyze dependency of morphometric estimates on motion severity, the following linear mixed effects model was fitted to all the data across the different motion types (subject i, scan j):
| (1) |
with intercept bi + β1 as the random effect and eij the measurement error. Here the dependent variable Yij is the gray matter volume estimate from SPM/VBM or FreeSurfer, mij is the motion measure (RMSpm). For FreeSurfer analysis the estimates were taken from both the independently processed images (regular stream) and from the longitudinal stream using all five time points to construct the within-subject template. For cortical surface analysis, false discovery rate (FDR) was controlled at the level 0.05 using an adaptive two-stage linear step-up procedure29.
4. Results
4.1. Motion Estimates
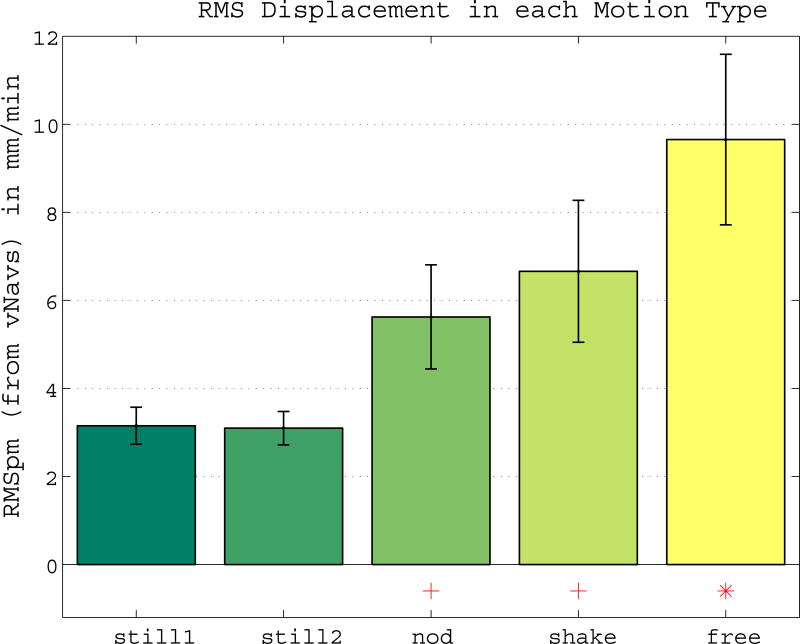
First we analyzed if motion levels differed across the different motion types. Figure 1 shows the average RMSpm displacement for each type, with increasing motion in nod, shake and free compared to still. Note that even in still scans, we estimated an average of 3 mm/min of accumulated motion (1.5 to 5.7 mm/min across subjects). Since repeated measures were available for each subject with differently severe motion, a mixed effects analysis was performed with all acquired data to estimate the effect of motion on morphometric measurements.
Figure 1. Different Motion Levels across Motion Types.
Mean RMSpm (RMS displacement per minute) for each motion type with ± standard error. Compared to the still scans, motion increases in nod, shake and free. Significance of the paired Wilcoxon signed rank test is indicated by a red + (p < 0.01) and * (p < 0.001).
4.2. Gray Matter Volume and Thickness Estimates
The following results emerged for both SPM/VBM gray matter and FS cortical gray matter volume as dependent variables in the linear mixed effects model (Eq. 1):
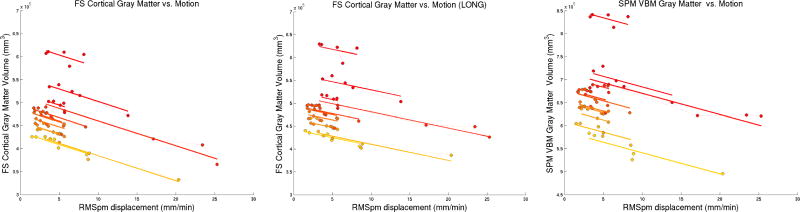
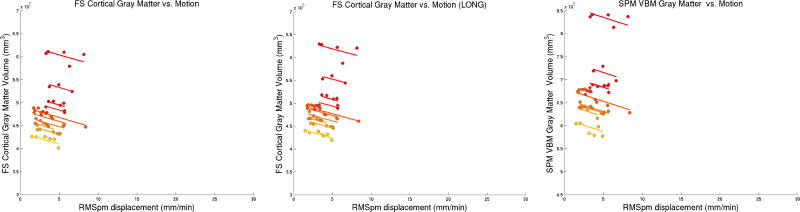
Significant association of SPM/VBM gray matter and FS cortical gray matter volume with motion (p < 10−10) of approximately 5400 mm3 (1%, FS independent), 3500 mm3 (0.7%, FS longitudinal), and 4600 mm3 (0.7%, SPM/VBM) volume loss associated with 1 mm/min RMSpm increase in motion. Figure 2 shows these results and highlights the linear dependence of gray matter volume estimates on motion severity.
The general dependence of volume loss on motion (at slightly different slopes) remains significant when analyzing images from only a specific motion type (i.e. nod-still, shake-still, free-still), but not for still-still. This can be expected given the good fit in Figure 2: dropping points does not substantially affect slopes, indicating that motion severity seems to be the driving factor. Further disentangling motion type and severity was not possible in this dataset, probably due to the strong correlation of both variables (see Fig. 1), with severity increasing from still, nod, shake to free motion.
When testing for a quadratic effect (adding a quadratic fixed effect term to Eq. 1) we find that FS (longitudinal) and SPM/VBM measurements demonstrate a small, but significant quadratic dependence on motion. This effect, however, is no longer significant when the three measurements with severe motion (RMSpm > 20 mm/min) are dropped from the model, indicating a potential floor effect.
Figure 2. Cortical Gray Matter Volume Estimates are explained by Motion.
FS and SPM cortical GM volume change is accurately explained by motion in the 12 different subjects via a linear mixed effects model. The slopes are approximately 1% (FS independent, left), 0.7% (FS longitudinal, middle) and 0.7% (SPM, right) volume loss per 1 mm/min RMSpm increase (p < 10−10). Different colors indicate different individuals, sorted with respect to baseline GM volume from smallest (yellow) to largest (red).
Some analysis methods, such as FSL Siena20, are designed to directly quantify change between two images. Using Siena, we computed percent brain volume change (PBVC) comparing the first still scan with the four motion types (still, nod, shake and free) and find apparent volume loss in all motion conditions, except for the still test-retest (see Table 1).
Table 1.
Percent brain volume change (PBVC) with respect to the first still scan, computed with FSL Siena for the four different motion types.
| still-still | nod-still | shake-still | free-still | |
|---|---|---|---|---|
| Mean PBVC | −0.1% | −1.1% * | −1.0% ** | −2.4% ** |
Significance of the one-sided Wilcoxon signed rank test for median below zero is reached in nod (* p<0.05), shake and free motion (** p<0.001).
To analyze localized dependence of GM volume loss on motion severity, we perform a mass-univariate linear mixed effects analysis (Eq. 1) with the modulated VBM GM volume images on a voxel-by-voxel basis (using the FreeSurfer mixed effects tools). Results are shown in Fig. 3 (p-value maps after FDR thresholding), showing volume loss at the GM/CSF boundary (pial) and some volume gain at the GM/WM boundary. Effect sizes (not shown) are mostly between 1% and 3% local volume loss for each 1mm/sec RMSpm increase.
Figure 3. VBM GM Volume Estimates Correlate with Motion.
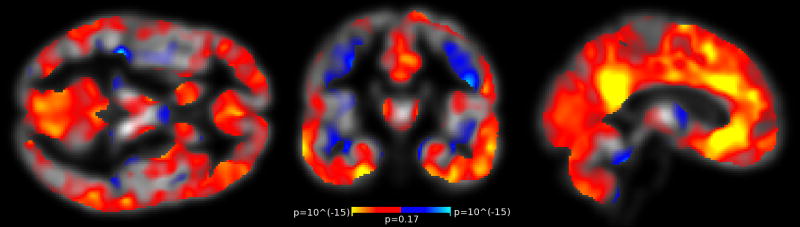
Regions of significant VBM GM volume change associated with increased motion. The maps show p-values testing the association (β2 in Eq. 1) and are FDR thresholded at level 0.05. Red/yellow indicate GM volume loss, blue indicates GM volume gain with increased motion.
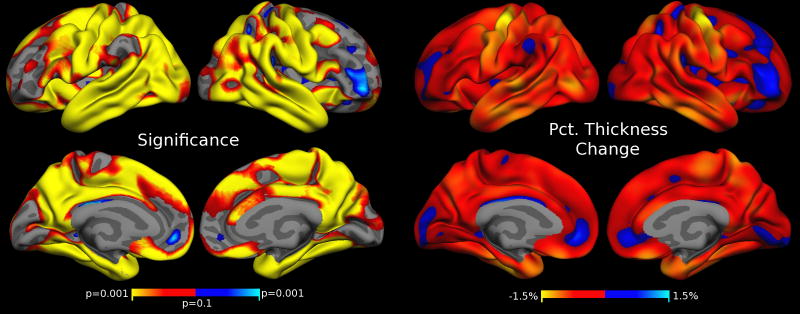
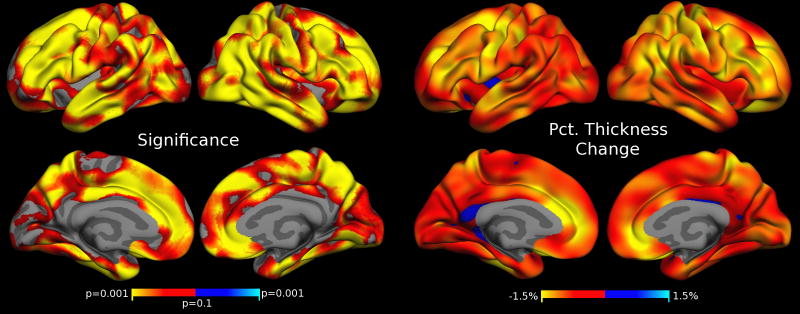
Similarly, to analyze localized dependence of cortical thickness on the motion severity, we employ FreeSurfer’s spatial-temporal linear mixed effects modeling approach again using the model in Eq. 1. We find that increased motion is correlated with thickness reduction in large parts of the cortex. Fig. 4 shows the FDR thresholded p-values and percent thickness changes associated with 1 mm/min increase in RMSpm motion. The most severe thickness reduction can be seen in the pre- and post-central cortex, in the temporal lobes and pole, as well as enthorhinal and parahippocampal regions. Some frontal regions and deep sulci demonstrate thickness gains with increased motion (e.g., medial orbital frontal, lateral frontal), indicating that results are regionally specific. The results shown in Fig. 4 were obtained using FreeSurfer’s longitudinal pipeline including all five time points. We also analyzed the association of thickness and motion on the cortex (not shown) using independent processing, i.e. the regular stream in FS, and found similar results with generally larger effect sizes. Independent processing is more susceptible to outlier measurements on images with strong motion.
Figure 4. Cortical Thickness Estimates Correlate with Motion.
Regions of significant cortical thickness change associated with increased motion. Left: FDR thresholded (at level 0.05) p-values testing the association (β2 in Eq. 1) with increased motion. Red/yellow indicate thickness loss, blue indicates thickness gain with increased motion. Right: Effects as percent thickness change for a 1 mm/min increase in RMSpm motion. Yellow regions of approx. 1.5% thinning correspond to a decrease in thickness of approx. 0.04 mm (per 1 mm/min RMSpm motion increase).
4.4. Quality Control
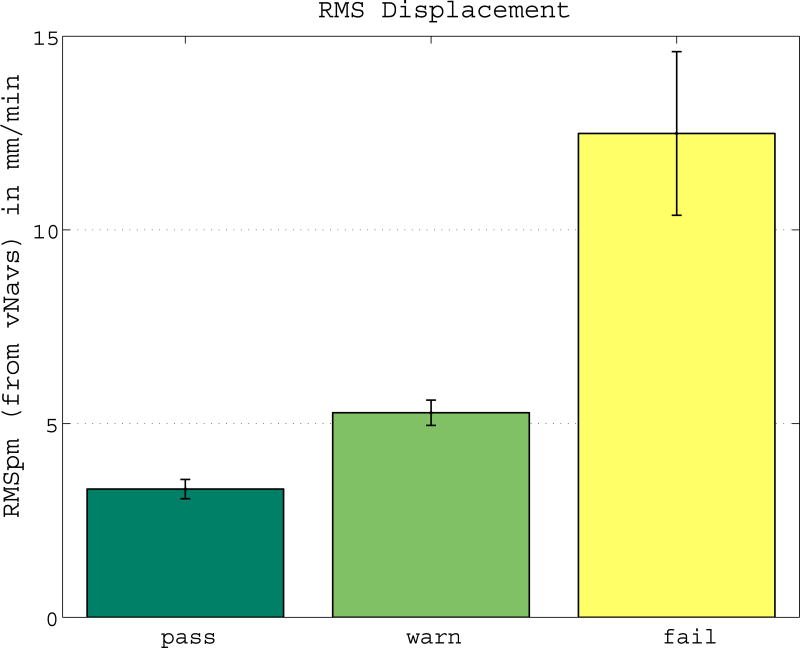
Researchers frequently perform quality-control (QC) on their data with regards to motion and sometimes remove low quality scans from their study. An expert manually assessed the quality of each scan. Of the 60 scans (12 subjects, each 5 scans), 12 failed the test (4 nod, 2 shake, 6 free), 15 had a warning (3 still, 1 nod, 5 shake, 6 free) and the remaining 33 scans passed the test. Figure 5 shows the average RMSpm for each quality score (pass, warn, fail). The quality score “fail” accurately identifies the cases with the most severe motion. Images that fail the thorough quality check are sometimes excluded from a study, while images with a warning are usually not excluded, but processing is checked carefully.
Figure 5. Manual Quality Check Identifies Increased Motion.
Different RMSpm motion levels for “pass”, “warn”, and “fail” indicate that the manual quality check correctly identifies cases with motion.
Here, in order to analyze whether exclusion of low quality scans would be sufficient to remove the directional measurement bias (more precisely: our ability to detect it), all scans that failed QC were dropped, and the linear mixed effects model was rerun (Eq. 1) with RMSpm to explain VBM and FS gray matter volume on the remaining scans that had pass or warn QC quality. A similar association was found for this subset of scans as in the previous section: a gray matter volume loss of approximately 5500 mm3 (0.8%) and 4600 mm3 (0.9%) for VBM and for FS longitudinal processing respectively, and 4900 mm3 (1%) for FS independent processing, associated with a 1 mm/min RMSpm motion increase (p < 0.0001) (see Fig. 6 and compare to Fig. 2).
Figure 6. Cortical Gray Matter Volume Estimates after regular QC are explained by Motion.
FS and SPM/VBM cortical GM volume change is accurately explained by motion in the 12 different subjects via a linear mixed effects model, even after removing scans that fail QC. The slopes are approximately 1% (FS independent, left), 0.9% (FS longitudinal, middle) and 0.8% (SPM, right) volume loss per 1 mm/min RMSpm increase (p < 10−10). Compare to Figure 2.
Similarly, Figure 7 (VBM analysis) can be compared to Figure 3, and Figure 8 (FS thickness analysis) to Figure 4, showing the same local morphometric analyses again after removing scans that fail QC. These results indicate that motion is a confounder even after typical quality control procedures are applied to remove scans with artifacts.
Figure 7. VBM GM Volume Estimates Correlate with Motion after regular QC.
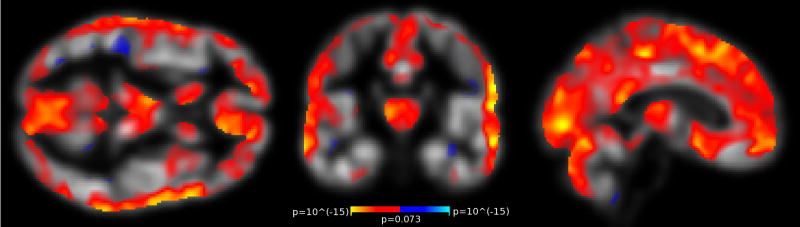
Regions of significant GM volume change associated with increased motion after removing scans that fail QC. Compare to Fig. 3 and see description for details.
Figure 8. Cortical Thickness Estimates Correlate with Motion after regular QC.
Regions of significant cortical thickness change associated with increased motion after removing scans that fail QC. Compare to Fig. 4 and see description for details.
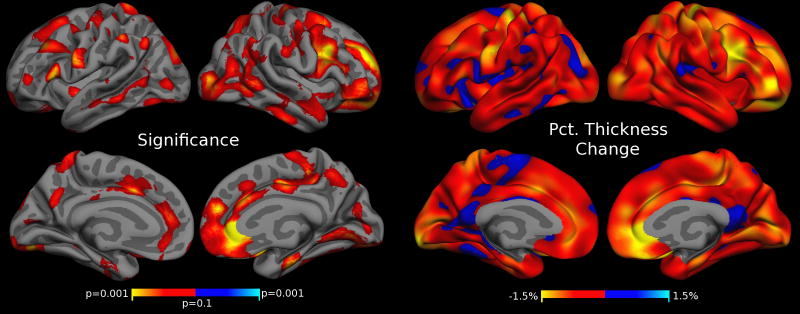
When additionally removing scans deemed as ‘warn’ in our manual QC procedure, we no longer detect an association of motion severity and volume measurements in the GM volume and thickness analysis after FDR correction. Figure 9 shows the FS thickness analysis after removing scans with a warning and failed QC. As opposed to Figs. 4 and 8, the p-value map in Fig. 9 (left) is not FDR thresholded (FDR correction removes all effects). Fig. 9 shows a trend towards thinning with increasing motion and similar trends in a VBM analysis are seen on the high quality images (not shown). The general impression is that stringent removal of images with motion artifacts mitigates misestimation of quantitative structural data measures but residual effects are still present. Whether they are weaker versions of the bias observed with more profoundly affected data or whether a loss in statistical power limits their significance will have to be investigated with a larger dataset.
Figure 9. Cortical Thickness Estimates Correlate with Motion after extreme QC.
Regions of significant cortical thickness change associated with increased motion after removing scans that fail QC and scans with a warning. The p-value map (left) is not FDR thresholded (effects disappear after FDR). Compare to Figs. 8 and 4 and see description for details.
5. Discussion
These results demonstrate spurious, systematic effects of motion in morphometric estimates. Even small amounts of motion are sufficient to bias results enough to potentially overshadow real effects. For example, for a small increase in motion of approx. 2mm/min RMSpm the apparent ~1.4–2% GM volume loss detected by VMB8, FS and Siena is comparable to yearly atrophy rates in (early stages or mild) neurodegenerative diseases30–33. The cortical thickness analysis indicates that spurious atrophy is not necessarily global but rather varies across regions. Longitudinal image processing methods, such as the method available in FreeSurfer, are capable of reducing variability but cannot remove the motion-induced effects, nor can they be used in cross-sectional studies. It is important to stress that our results imply that the spurious effects do not reflect a processing failure of the analyzed morphometry tools. Rather, the images themselves contain consistent changes, such as motion-induced blurring, that appear similar to gray matter atrophy and cause systematic bias in morphometric estimates across many analysis approaches.
Critically, the exclusion of scans that fail a thorough quality check is not sufficient to account for motion as a confounding variable, as significant measurement bias can still be detected after removing these scans. Furthermore, when keeping only the highest quality scans (removing all scans that received a data quality warning), we still detect a similar trend, i.e. that motion causes spurious thinning, but the data exclusion mitigates the biased estimates. These findings imply that great care needs to be taken when studying movement disorders or any disease/condition that affects motion directly or indirectly. In particular, cross-sectional studies that do not quantify and control for motion may overestimate disease effects, developmental effects, or effects of advanced aging. Even in longitudinal studies, motion levels likely increase concurrent with disease severity, inducing increased spurious atrophy rates. Furthermore, similar to drugs that primarily increase hydration levels, drugs with hypnotic, sedative, tranquillizing or neuromuscular-blocking substances may provide the desired “effect” of reduced atrophy rates or even apparent neural augmentation, simply because they inhibit motion rather than providing a true anti-disease effect on brain structure.
Solutions to these problems are limited and not always applicable. Obviously, restricting motion in the scanner in the first place will improve image quality, reliability, and reduce motion-induced bias. Devices such as facemasks, bite bridges and special pillows have been designed to immobilize the subject’s head during acquisition. Furthermore, in some types of studies patients can be sedated, to promote stillness, but sedation or head immobilization of volunteers may not be feasible in most situations. Head motion can be reduced by selecting a pillow with good support and by padding the head inside the coil (at the sides). This procedure is unobtrusive and, while it will not prevent motion, nor necessarily restrict nodding movements, it can help to reduce motion artifacts.
Alternatively, methods that prospectively correct for motion during image acquisition by continuously localizing and following head position throughout the scan show promise in improving image quality and mitigating motion artifacts15,34. However, the impact of these technologies on different patient groups and resultant measurement reliability has yet to be explored.
Independent of immobilization or on-line motion correction during image acquisition, it is highly recommended to track motion during the scan (or estimate the motion retrospectively) and then control for motion in the statistical model. We only find quadratic motion effects when including cases with severe motion, indicating that a linear motion co-variate could be a reasonable approximation to remove most of the motion bias, especially after manual QC is performed.
However, working with corrupted images will limit reliability and statistical power even if the amount of motion is known. Outliers with severe motion artifacts may have unpredictable effects on the statistical analysis. Non-parametric statistical approaches may be most appropriate in such instances. Also, motion can affect images differently, depending on when (in k-space) it occurs. Regional results may change, depending on the type of image processing (longitudinal vs. cross-sectional, multi-time point vs. paired analysis) and on the type of motion. In any case, if motion estimates are available, a correlation analysis among motion and other predictors should always be performed and any co-linearity should be reported. High co-linearity between predictors makes it difficult to disentangle their effects and is symptomatic of insufficient information, which cannot be rectified by simple data manipulation. In this case, inclusion of a motion co-variate into the statistical model can shadow any real effects. This is problematic, as in many settings motion can be expected to be correlated with the variable of interest (disease severity, age, drug dose etc.).
Collecting several structural scans and manually selecting one without motion artifacts for the structural analysis seems to be a commonly used option even in the presence of a costly increase in scan time. This procedure can reduce the spurious motion effect, but does not completely eliminate it. Furthermore, it is difficult in many study groups to obtain even a single scan without visible motion artifacts. While a visual inspection of automatically generated results is always recommended, it is often up to the individual expert to decide whether to exclude or repeat a scan or not. Since even small and visually inconspicuous, yet consistent, motion artifacts might bias the results, we believe that reducing motion during the scan is currently the best option. Controlling for motion in the statistical analysis is a second alternative that ideally should go hand in hand with a correlation analysis between motion and the predictors of interest.
Processed results of this study including quality scores and motion estimates are attached in a spreadsheet as supplementary material for further analysis.
Supplementary Material
Highlights.
MRI head motion induces a consistent bias in morphometric analysis.
Increased motion generally causes smaller gray matter volume and cortical thickness estimates.
Effects of movement disorders may be severely overestimated when not controlling for head motion.
Drugs that inhibit motion likely provide a spurious effect of reduced atrophy rates.
Exclusion of scans that fail a visual quality check is not sufficient to remove this bias.
Acknowledgments
Support for this research was provided in part by the National Cancer Institute (1K25-CA181632-01), the NIH Eunice Kennedy Shriver National Institute of Child Health and Human Development (R01-HD071664, K99-HD074649), the National Institute of Mental Health (R21-MH096559), the National Center for Research Resources (P41-RR014075, U24-RR021382, 1UL1-RR025758-01, 1S10-RR023401, 1S10-RR019307, 1S10-RR023043), the National Institute for Biomedical Imaging and Bioengineering (8P41-EB015896-15), the National Institute on Aging (5R01-AG008122-23, 2R01-AG016495, R21-AG046657), the National Institute for Neurological Disorders and Stroke (5R21-NS072652-02, 5R01-NS070963-03, 1R01-NS083534). In addition, BF has a financial interest in CorticoMetrics, a company whose medical pursuits focus on brain imaging and measurement technologies.
Footnotes
BF’s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies.
MR and MDT had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Martin Reuter, Massachusetts General Hospital, Department of Neurology, 55 Fruit Street, Boston, MA 02114, USA. Massachusetts General Hospital, Department of Radiology, A.A. Martinos Center for Biomedical Imaging, 149 Thirteenth Street, Suite 2301, Charlestown, MA 02129, USA. Massachusetts Institute of Technology, Computer Science and Artificial Intelligence Laboratory, Cambridge, MA 02139, USA. Harvard Medical School, 25 Shattuck St., Boston, MA 02115, USA.
M. Dylan Tisdall, Massachusetts General Hospital, Department of Radiology, A.A. Martinos Center for Biomedical Imaging, 149 Thirteenth Street, Suite 2301, Charlestown, MA 02129, USA. Harvard Medical School, 25 Shattuck St., Boston, MA 02115, USA.
Abid Qureshi, Massachusetts General Hospital, Department of Neurology, 55 Fruit Street, Boston, MA 02114, USA. Harvard Medical School, 25 Shattuck St., Boston, MA 02115, USA.
Randy L. Buckner, Massachusetts General Hospital, Department of Radiology, A.A. Martinos Center for Biomedical Imaging, 149 Thirteenth Street, Suite 2301, Charlestown, MA 02129, USA. Harvard Medical School, 25 Shattuck St., Boston, MA 02115, USA
André J. W. van der Kouwe, Massachusetts General Hospital, Department of Radiology, A.A. Martinos Center for Biomedical Imaging, 149 Thirteenth Street, Suite 2301, Charlestown, MA 02129, USA. Harvard Medical School, 25 Shattuck St., Boston, MA 02115, USA
Bruce Fischl, Massachusetts General Hospital, Department of Radiology, A.A. Martinos Center for Biomedical Imaging, 149 Thirteenth Street, Suite 2301, Charlestown, MA 02129, USA. Massachusetts Institute of Technology, Computer Science and Artificial Intelligence Laboratory, Cambridge, MA 02139, USA. Harvard Medical School, 25 Shattuck St., Boston, MA 02115, USA.
References
- 1.Duning T, Kloska S, Steinstrater O, Kugel H, Heindel W, Knecht S. Dehydration confounds the assessment of brain atrophy. Neurology. 2005 Feb 8;64(3):548–550. doi: 10.1212/01.WNL.0000150542.16969.CC. [DOI] [PubMed] [Google Scholar]
- 2.Dickson JM, Weavers HM, Mitchell N, et al. The effects of dehydration on brain volume -- preliminary results. Int J Sports Med. 2005 Jul-Aug;26(6):481–485. doi: 10.1055/s-2004-821318. [DOI] [PubMed] [Google Scholar]
- 3.Kempton MJ, Ettinger U, Schmechtig A, et al. Effects of acute dehydration on brain morphology in healthy humans. Hum Brain Mapp. 2009 Jan;30(1):291–298. doi: 10.1002/hbm.20500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartsch AJ, Homola G, Biller A, et al. Manifestations of early brain recovery associated with abstinence from alcoholism. Brain. 2007 Jan;130(Pt 1):36–47. doi: 10.1093/brain/awl303. [DOI] [PubMed] [Google Scholar]
- 5.Bellon EM, Haacke EM, Coleman PE, Sacco DC, Steiger DA, Gangarosa RE. MR artifacts: a review. AJR Am J Roentgenol. 1986 Dec;147(6):1271–1281. doi: 10.2214/ajr.147.6.1271. [DOI] [PubMed] [Google Scholar]
- 6.Smith TB, Nayak KS. MRI Artifacts and Correction Strategies. Imaging in Medicine. 2010;2(4):445–457. [Google Scholar]
- 7.Van Dijk KR, Sabuncu MR, Buckner RL. The influence of head motion on intrinsic functional connectivity MRI. Neuroimage. 2012 Jan 2;59(1):431–438. doi: 10.1016/j.neuroimage.2011.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage. 2012 Feb 1;59(3):2142–2154. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Satterthwaite TD, Wolf DH, Loughead J, et al. Impact of in-scanner head motion on multiple measures of functional connectivity: relevance for studies of neurodevelopment in youth. Neuroimage. 2012 Mar;60(1):623–632. doi: 10.1016/j.neuroimage.2011.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeng LL, Wang D, Fox MD, et al. Neurobiological basis of head motion in brain imaging. Proc Natl Acad Sci U S A. 2014 Apr 22;111(16):6058–6062. doi: 10.1073/pnas.1317424111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yendiki A, Koldewyn K, Kakunoori S, Kanwisher N, Fischl B. Spurious group differences due to head motion in a diffusion MRI study. Neuroimage. 2014 Mar;88:79–90. doi: 10.1016/j.neuroimage.2013.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Kouwe AJ, Benner T, Salat DH, Fischl B. Brain morphometry with multiecho MPRAGE. Neuroimage. 2008 Apr 1;40(2):559–569. doi: 10.1016/j.neuroimage.2007.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mugler JP, 3rd, Bao S, Mulkern RV, et al. Optimized single-slab three-dimensional spin-echo MR imaging of the brain. Radiology. 2000 Sep;216(3):891–899. doi: 10.1148/radiology.216.3.r00au46891. [DOI] [PubMed] [Google Scholar]
- 14.van der Kouwe AJ, Benner T, Fischl B, et al. On-line automatic slice positioning for brain MR imaging. Neuroimage. 2005 Aug 1;27(1):222–230. doi: 10.1016/j.neuroimage.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 15.Tisdall MD, Hess AT, Reuter M, Meintjes EM, Fischl B, van der Kouwe AJ. Volumetric navigators for prospective motion correction and selective reacquisition in neuroanatomical MRI. Magn Reson Med. 2012 Aug;68(2):389–399. doi: 10.1002/mrm.23228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thesen S, Heid O, Mueller E, Schad LR. Prospective acquisition correction for head motion with image-based tracking for real-time fMRI. Magn Reson Med. 2000 Sep;44(3):457–465. doi: 10.1002/1522-2594(200009)44:3<457::aid-mrm17>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 17.Harvard Center for Brain Science. Quality Control. 2014 http://cbs.fas.harvard.edu/science/core-facilities/neuroimaging/information-investigators/qc.
- 18.Zaitsev M, Dold C, Sakas G, Hennig J, Speck O. Magnetic resonance imaging of freely moving objects: prospective real-time motion correction using an external optical motion tracking system. Neuroimage. 2006 Jul 1;31(3):1038–1050. doi: 10.1016/j.neuroimage.2006.01.039. [DOI] [PubMed] [Google Scholar]
- 19.Jenkinson M. Measuring Transformation Error by RMS Deviation. Oxford Center for Functional Magnetic Resonance Imaging of the Brain (FMRIB); 1999. [Google Scholar]
- 20.Smith SM, De Stefano N, Jenkinson M, Matthews PM. Normalized accurate measurement of longitudinal brain change. J Comput Assist Tomogr. 2001 May-Jun;25(3):466–475. doi: 10.1097/00004728-200105000-00022. [DOI] [PubMed] [Google Scholar]
- 21.Gaser C. Voxel-based Morphometry Extension to SPM8. 2014 http://dbm.neuro.uni-jena.de/vbm/
- 22.Ashburner J, Friston KJ. Unified segmentation. Neuroimage. 2005 Jul 1;26(3):839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 23.Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis II: Inflation, flattening, and a surface-based coordinate system. Neuroimage. 1999;9(2):195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- 24.Fischl B, Sereno MI, Tootell RB, Dale AM. High-resolution intersubject averaging and a coordinate system for the cortical surface. Hum Brain Mapp. 1999;8(4):272–284. doi: 10.1002/(SICI)1097-0193(1999)8:4<272::AID-HBM10>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reuter M, Rosas HD, Fischl B. Highly accurate inverse consistent registration: a robust approach. Neuroimage. 2010 Dec;53(4):1181–1196. doi: 10.1016/j.neuroimage.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reuter M, Schmansky NJ, Rosas HD, Fischl B. Within-subject template estimation for unbiased longitudinal image analysis. Neuroimage. 2012 Jul 16;61(4):1402–1418. doi: 10.1016/j.neuroimage.2012.02.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernal-Rusiel JL, Greve DN, Reuter M, Fischl B, Sabuncu MR Alzheimer’s Disease Neuroimaging Initiative. Statistical analysis of longitudinal neuroimage data with Linear Mixed Effects models. Neuroimage. 2013 Feb 1;66:249–260. doi: 10.1016/j.neuroimage.2012.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bernal-Rusiel JL, Reuter M, Greve DN, Fischl B, Sabuncu MR Alzheimer’s Disease Neuroimaging Initiative. Spatiotemporal linear mixed effects modeling for the mass-univariate analysis of longitudinal neuroimage data. Neuroimage. 2013 Nov 1;81:358–370. doi: 10.1016/j.neuroimage.2013.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benjamini Y, Krieger AM, Yekutieli D. Adaptive linear step-up procedures that control the false discovery rate. Biometrika. 2006 Sep;93(3):491–507. [Google Scholar]
- 30.Resnick SM, Pham DL, Kraut MA, Zonderman AB, Davatzikos C. Longitudinal magnetic resonance imaging studies of older adults: a shrinking brain. J Neurosci. 2003 Apr 15;23(8):3295–3301. doi: 10.1523/JNEUROSCI.23-08-03295.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jack CR, Jr, Petersen RC, Xu Y, et al. Rate of medial temporal lobe atrophy in typical aging and Alzheimer’s disease. Neurology. 1998 Oct;51(4):993–999. doi: 10.1212/wnl.51.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderson VM, Schott JM, Bartlett JW, Leung KK, Miller DH, Fox NC. Gray matter atrophy rate as a marker of disease progression in AD. Neurobiol Aging. 2012 Jul;33(7):1194–1202. doi: 10.1016/j.neurobiolaging.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosas HD, Reuter M, Doros G, et al. A tale of two factors: what determines the rate of progression in Huntington’s disease? A longitudinal MRI study. Mov Disord. 2011 Aug 1;26(9):1691–1697. doi: 10.1002/mds.23762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.White N, Roddey C, Shankaranarayanan A, et al. PROMO: Real-time prospective motion correction in MRI using image-based tracking. Magn Reson Med. 2010 Jan;63(1):91–105. doi: 10.1002/mrm.22176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.