Abstract
Atlantic killifish (Fundulus heteroclitus) thrive in New Bedford Harbor (NBH), MA, highly contaminated with polychlorinated biphenyls (PCBs). Resident killifish have evolved tolerance to dioxin-like (DL) PCBs, whose toxic effects through the aryl hydrocarbon receptor (AhR) are well studied. In NBH, non-dioxin like PCBs (NDL PCBs), which lack activity toward the AhR, vastly exceed levels of DL congeners yet how killifish counter NDL toxic effects has not been explored. In mammals and fish, NDL PCBs are potent activators of ryanodine receptors (RyR), Ca2+ release channels necessary for a vast array of physiological processes. In the current study we compared the expression and function of RyR related pathways in NBH killifish with killifish from the reference site at Scorton Creek (SC, MA). Relative to the SC fish, adults from NBH displayed increased levels of skeletal muscle RyR1 protein, and increased levels of FK506-binding protein 12 kDa (FKBP12), an accessory protein essential for NDL PCB-triggered changes in RyR channel function. In accordance with increased RyR1 levels, NBH killifish displayed increased maximal ligand binding, increased maximal response to Ca2+ activation and increased maximal response to activation by the NDL PCB congener PCB 95. Compared to SC, NBH embryos and larvae had increased levels of mtor and ryr2 transcripts at multiple stages of development, and generations, while levels of serca2 were decreased at 9 days post-fertilization in the F1 and F2 generations. These findings suggest that there are compensatory and heritable changes in RyR mediated Ca2+ signaling proteins or potential signaling partners in NBH killifish.
Keywords: Non-dioxin like PCBs, ryanodine receptor, Fundulus heteroclitus, PCB Tolerance
1.0 Introduction
The Atlantic killifish (Fundulus heteroclitus) is a non-migratory fish species, with small genetically distinct subpopulations (Nacci et al., 2010) inhabiting coastal and inland regions of the Atlantic coast of North America. Several of these subpopulations have developed a heritable resistance to the effects of various contaminants, leading to the fish’s success in heavily polluted U.S. EPA Superfund locations (Wirgin and Waldman, 2004). Here, we use the terms resistance and tolerance that both have been used in other studies to indicate a killifish population that has reduced sensitivity to a chemical as demonstrated through a reduction in overt toxicity or induction of molecular pathways involved in a toxic outcome. At the Superfund National Priority Site found in New Bedford Harbor (NBH), on the Acushnet River, MA, USA, killifish have evolved tolerance to extreme levels of polychlorinated biphenyls (PCBs). PCB concentrations in NBH killifish have been recorded as high as 1370 µg g−1 (Lake et al., 1995), levels that far exceed those known to cause adverse biological effects in sensitive populations or other fish species (Wirgin and Waldman, 2004).
Several studies have addressed the mechanistic basis of this resistance to PCB toxicity in killifish and other resistant fish species inhabiting severely polluted sites (Wirgin and Waldman, 2004; Burnett et al., 2007; Nacci et al., 2010). These studies have focused on PCB congeners that lack ortho-substitution and that mimic dioxins by activating the aryl hydrocarbon receptor (AhR) and related pathways. Tolerant killifish populations, as compared to susceptible reference populations (Nacci et al., 2002), exhibit reduced embryo toxicity when exposed to the potent AhR agonist PCB 126, reduced induction of AhR target genes and pathways (Bello et al., 2001), and decreased sensitivity to AhR ligands in the F1 and F2 generations (Wirgin and Waldman, 2004; Burnett et al., 2007), indicating that resistance to toxicity of dioxin-like (DL) PCBs in the killifish is heritable (Nacci et al., 2002; Nacci et al., 2010). While studies to date have focused primarily on AhR-related pathways, they have not fully explained the basis for tolerance to the pathophysiological potential of PCBs. Genetic studies show multiple loci are likely under selection in polluted versus non-polluted killifish populations (Williams and Oleksiak, 2008), suggesting multiple contributing mechanisms leading to acquired tolerance to extreme pollutant concentrations.
Those PCBs that have one or more ortho-chlorine substitution are considered non-dioxin like (NDL) because they show little to no interaction with the AhR in mammalian and teleost species (Giesy and Kannan, 1998). Mechanisms of tolerance to NDL PBCs have not been addressed even though these congeners account for the majority of the total tissue load in NBH killifish (Lake et al., 1995), and other organismal samples including mammals, and have several modes of toxicity that are independent of the AhR pathway (Simon et al., 2007; Pessah et al., 2010). Of interest to the current study is the fact that NDL PCBs are potent and direct sensitizers of ryanodine-sensitive Ca2+ channels, termed ryanodine receptors (RyRs), in both mammalian (Pessah et al., 2006; Samsó et al., 2009) and fish species (Fritsch and Pessah, 2013).
RyRs are integral membrane Ca2+ channels anchored within the sarcoplasmic reticulum (SR) of muscle and endoplasmic reticulum (ER) of non-muscle cells. In addition to their well understood contribution to excitation-contraction (EC) coupling in striated muscle, they are important for the normal development of neuronal networks, endocrine health and neurodegenerative disorders (Pessah et al., 2010). The ramifications of RyR channel sensitization by NDL PCBs observed in vitro on muscle development and long-term health are not fully understood, but exposure to nanomolar NDL PCBs and their hydroxylated metabolites can alter important aspects of skeletal muscle EC coupling, and this action is highly dependent on chemical structure (Niknam et al., 2013). Moreover, RyR isoforms are broadly expressed in neurons where they contribute to the developmental neurotoxicity induced by NDL PCBs (Kenet et al., 2007; Yang et al., 2009; Wayman et al., 2012a; Wayman et al., 2012b).
The importance of the RyR to diverse physiological processes together with the extreme body burdens of NDL PCBs (Lake et al., 1995), suggests that NBH killifish may have developed altered RyR mediated Ca2+ signaling dynamics and differential sensitivity to NDL PCB induced disruption of the RyR. To begin addressing these hypotheses, we examined RyR related expression and functional differences between NBH killifish and fish from the reference population from Scorton Creek (SC), MA, USA. The goals of the current study were three fold: (1) obtain an up-to-date measure of NDL PCB concentrations, relative to DL PCBs, in NBH and SC killifish, (2) assess potential changes in the expression of RyR related pathways (Figure 1) in adult killifish collected in the field and across three generations of embryos and larvae reared in a laboratory setting and (3) assess potential RyR function or NDL PCB sensitivity differences between NBH and SC killifish populations.
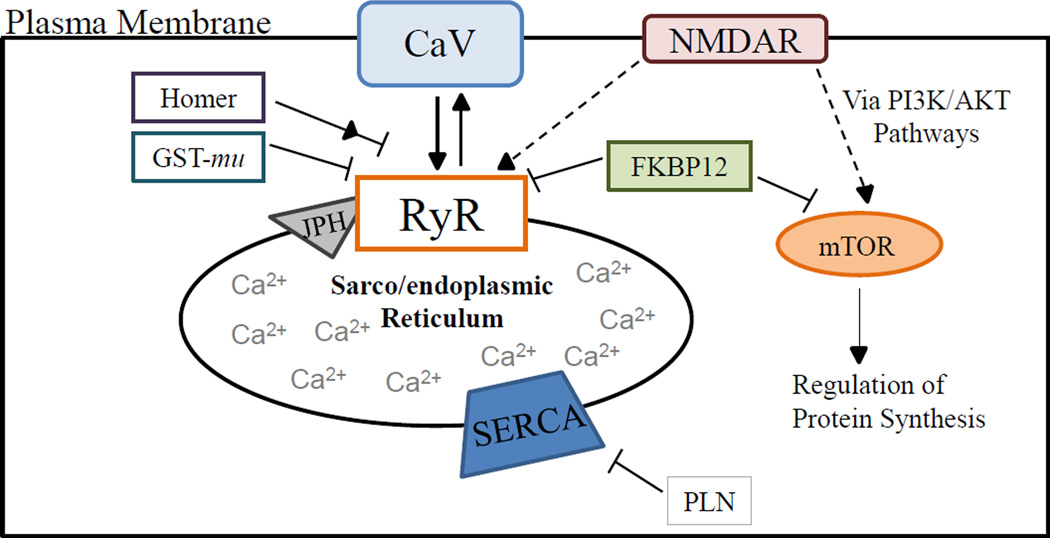
Figure 1.
Proteins involved in RyR Ca2+ signaling dynamics and potential RyR related signaling partners. Arrow heads and blunt ended lines represent activating or inhibiting regulation respectively. Full versus dashed lines represent an established versus a proposed relationship, respectively. Note that the diagram is not isoform or tissue specific but rather proteins displayed relate to Ca2+ signaling in skeletal muscle, cardiac muscle or brain tissue. Also, this is not an exhaustive list of RyR regulatory proteins or signaling partners. The Ca2+ channel interacts with or is regulated by numerous other plasma membrane bound signaling receptors, cytoplasmic accessory proteins and integral and luminal sarcoplasmic reticulum proteins not measured in the current study. Abbreviations see Table 1. Information gathered from (Pouliquin and Dulhunty, 2009; Van Dongen et al, 2009; Hoeffer and Klann, 2010; Pessah et al., 2010; Martín-Cano et al., 2013).
2.0 Materials and Methods
2.1 Animal Collection and Maintained Killifish Cultures
Adult killifish used for PCB detection, tissue dissections, and adult breeding stock were collected from NBH and SC (Latitude and longitude; 41.6676 by 70.9150; 41.7649 by 70.4800, respectively) utilizing baited aluminum semi-collapsible traps that were deployed from the shoreline and collected after 1 to 3 hours (Nacci et al., 1999; Nacci et al., 2002). Fish for chemical detection or tissue dissection were collected in August and September of 2009 or 2010, respectively, under permits from the Massachusetts Division of Marine Fisheries, and protocols approved by Animal Care and Use Committees at WHOI and USEPA. Upon capture fish from SC and NBH were returned from the field and maintained in uncontaminated laboratory conditions consisting of flow through aquaria receiving 5 µM filtered sea water. Fish used for tissue and PCB data were dissected or frozen within 48 h of collection. Here, fish were euthanized using an overdose of MS-222 at which time fish were measured for length and sex was noted. The average standard length of sampled fish was 63.5 mm or 57.6 mm for NBH and SC, respectively, and samples consisted of an approximate 50:50 male to female ratio. It should be noted; however, that fish were selected to achieve an approximate match in size and sex ratio for NBH and SC and therefore this information should not be used to represent potential differences or similarities between the killifish populations. Once dissected, skeletal muscle, cardiac muscle and brain tissue were immediately flash frozen in liquid nitrogen and stored at −80 °C until molecular analysis. For chemical analysis euthanized fish were stored at −20 °C until chemical extraction procedures were conducted.
Embryos and larvae utilized for qPCR were obtained from the breeding stocks maintained in uncontaminated conditions at the US EPA National Health and Environmental Effects Laboratory (Atlantic Ecology Division, Narragansett RI), as described previously (Nacci et al., 2002; Nacci et al., 2010). Briefly, adult fish collected by trapping were kept in flow through tanks receiving 5 µM filtered sea water for at least 6 months and no longer than 2 years. Adults were feed Tetramin flakes and minced krill and were held in tanks at ambient temperatures and photoperiod, such as summertime conditions of 23°C with a 14:10 light:dark-cycle. Under these conditions killifish were reproductively active from spring through fall, producing embryos on an approximately semi-lunar cycle. For several days surrounding the full and new moons, spawning chambers were introduced into breeding tanks and fertilized eggs were removed from the aquaria daily after unstimulated spawning events. Embryos were then maintained at 23°C in fresh seawater provided daily. Once hatched some larvae were reared to adulthood (~1 year) in flowing uncontaminated seawater to maintain multiple generations of breeding stocks.
For the current study, we were interested in developmental and generational expression differences and therefore fish were collected at 3 or 9 days post fertilization (dpf; referred to as embryos) or 1 day post hatch (dph; referred to as larvae) from each of the F1, F2 and F3 generations of NBH killifish and compared to the same developmental stage of the F1 generation from SC. This treatment structure was repeated for both the 2012 and 2013 breeding seasons. At collection fish were placed in pools of 10 and immediately flash froze in liquid nitrogen and stored at −80°C.
2.2 PCB Measurements
Three individual adult fish per site were measured for PCBs. Here, a sagittal half of an entire fish was utilized for chemical detection. This included half of all organs contained in the body cavity and half of the brain. We detected concentrations of the 18 PCB congeners commonly measured under the NOAA National Status and Trends Program (nsandt.noaa.gov), which were determined using microwave extraction following methods described and utilized elsewhere (Jayaraman et al., 2001; Nacci et al., 2002). Detection of AhR potent DL PCBs (77, 81,126 and 169; Van den Berg et al. 2006) were detected following methods described by Gutjahr-Gobbel and colleagues (1999), which has previously been utilized to detect DL PCBs in killifish tissue (Gutjahr- Gobell et al., 1999).
2.3 Differences in RyR-Related Gene Transcription
Total RNA from adult killifish skeletal and cardiac muscle, brain tissue or pools of embryos or larvae was extracted using TRIzol Reagent (Invitrogen) following manufacturer’s instructions. RNA concentrations were determined on a Nanodrop 1000 (Thermo Scientific) and quality of each sample assessed using 260/280 ratios and through visualization on an agarose gel stained with ethidium bromide. Complementary DNA was synthesized with 1 µg of total RNA using Random Primers and SuperScript Reverse Transcriptase III (Invitrogen).
Genes of interest (GOI) along with suitable reference genes (Table 1) were measured utilizing quantitative Polymerase Chain Reaction (qPCR) as described elsewhere (Connon et al., 2012). Sequences for the ryr1, ryr3, fkbp12 and mtor were graciously supplied by Dr. Sibel Karchner (Woods Hole Oceanographic Institution) and sequences for cav1.1, jph1, homer1 and nmdar1 were acquired using degenerate primers designed and implemented elsewhere (Fritsch et al., 2013). In the present study sequences were obtained directly from degenerate PCR amplicons after purification with a QIAquick PCR Purification Kit (Qiagen). Purification products were sequenced at the UC Davis DNA Sequencing Facility at the College of Biological Sciences and submitted to the NCBI databank.
Table 1.
Genes utilized to assess NDL PCB mediated differences in New Bedford Harbor and Scorton Creek Atlantic killifish.
| Gene Name | Gene Symbol |
Accession Number |
Forward/Reverse Primers | Probe |
|---|---|---|---|---|
| Ryanodine Receptor 1 | ryr1 | a | gccgttcggtaagcagtagt cctctgaagctgctcacaaa |
120 |
| Ryanodine Receptor 2 | ryr2 | KJ955487 | agatgctgatcgcttccttca gcactgagaggagataggggaga |
71 |
| Ryanodine Receptor 3 | ryr3 | a | tacctgggtcgcattgaaa tcgctgatctcgaagtagacc |
44 |
| L-type Voltage Gated Ca2+ Channel, subunit 1αS |
cav1.1 | KJ955488 | cctcaaacgttgaaacggtga gaaacagttctggctagaagagagtg |
89 |
| FK506-Binding Protein 1A, 12kDa | fkbp12 | a | aaagtcttcgactcgtcgaggt ggccgacactcatctgagcta |
52 |
| Junctophilin 1 | jph1 | KJ955489 | ctcgctgtctgagtggaagttg cagagcaacggcgcagtt |
52 |
| Homer 1 | homer1 | KJ955490 | acgacatctgcgactccacc cccagtgctttgtcatggc |
106 |
| GST mu | gst-mu | AY725220 | ggacgaaaaggtcagagtgg acatcctcacaaagccgttt |
22 |
| Mechanistic Target of Rampamycin | mtor | a | acctccatagcgttggtcag gaagttccccgaaaagatcc |
153 |
| Sacrco/endoplasmic Reticulum Calcium ATPase, 2 |
serca2 | U58326 | cacgctgaccaccaatca atcccgctccgctttatc |
112 |
| Cardiac Phospholamban | pln | CV817790 | cgtcgagcgtctcagatcg gcagatgaggatgagcgtgaa |
128 |
| N-methyl-D-aspartate Glutamate Receptor 1 |
nmdar1 | KJ955491 | cgcaccttctcctattccag agatgccctcaccttgtcat |
2 |
| Ribosomal Protein L8 | rpl8 | DQ066926 | gcccagctgaacattggta caggcagcagatgatggtt |
96 |
| Ribosomal Protein S20 | rps20 | CN984003 | cgcatgccaaccaagact gaagcgatcccaggttttg |
9 |
| β-actin | β-actin | DR046767 | ggccagaaggacagctatgt tggggtacttcagggtcaag |
165 |
| Elongation Factor 1 alpha | ef1α | AY430091 | tcaccatcgacatcgctct gcatcgatgatggtcacgta |
54 |
Gene sequences provided by S. Karchner; WHOI
2.4 Crude Microsomal Protein Preparations
Skeletal muscle tissue of adult killifish was utilized to obtain crude microsomal protein preparations enriched in RyR isoform 1 (RyR1) following protocols described previously (Franck et al., 1998; Pessah et al., 2006). Briefly, approximately 1 g of skeletal muscle tissue, which represented tissue combined from at least four fish collected from either NBH or SC, were minced with scissors in 5 volumes ice cold homogenization buffer consisting of (in mM) 300 sucrose, 20 HEPES, 1 PMSF, 10 NaF, 2 β-Glycerol, 5 Na4P2O7 and 0.5 Na3VO4 and 2µg ml−1 leupeptin (pH 7.2). Tissue was further homogenized with a Polytron E2000 with 2 bursts of 20s at half speed, with 2 min on ice between bursts. Following centrifugation at 6,500 g (4°C for 15min) the supernatant was collected, the pellet re-suspended and homogenization and centrifugation steps repeated. The combined supernatants underwent ultracentrifugation at 100,000 g (4°C for 1h), the supernatant was discarded and the pellet re-suspended in 300 mM sucrose containing 20 mM HEPES using a glass Dounce homogenizer. Protein concentrations were determined in triplicate using a BCA Protein Assay (Fisher Scientific, Pittsburgh, PA).
2.5 Protein Quantification Using Western Blot Analysis
Protein, 10 µg of each homogenate, were resolved on a 4–12% gradient Bis-Tris gel (Life Technologies) and blotted onto a PVDF membrane (Millipore). Membranes were probed for anti-mouse RyR1 (1:1000, 34C, DSHB), anti-mouse CACNA1S (1:500, AbCam, here after referred to as CaV1.1), anti-rabbit FKBP12 (1:500, AbCam) and reference protein anti-rabbit GAPDH (1:2000, Cell Signaling). Bands were detected with goat anti-mouse or goat anti-rabbit secondary antibodies conjugated to fluorescent dyes (DyLight™ 680 or 800, respectively), which were visualized and quantified using the Odyssey Infrared Imaging System (LI-COR).
2.6 Saturation Radioligand Binding for RyR1 and CaV1.1
Radioligand-receptor binding assays were utilized to determine potential changes in receptor ligand interactions. For both RyR1 and CaV1.1, saturation binding was completed with 100 µg ml−1 of the crude SR preparation incubated in the presence of 0.75–50 µM [3H]-Ryanodine ([3H]Ry, 57 Ci mmol−1; Perkin Elmer) or 0.1–10 nM [3H]-PN200-110 ([3H]PN, 82.2 Ci mmol−1; Perkin Elmer) in a final volume of 300 µl. RyR1 binding did not reach saturation at 50nM; therefore to address binding at low-affinity sites; protein samples were incubated with 10nM [3H]Ry with increasing amounts of unlabeled Ry (Total ligand at 50–300 nm) and corrected for differences in specific activity for saturation calculations. RyR binding buffer consisted of (mM) 250 KCl, 15 NaCl, 20 Hepes, and 0.05 CaCl2 (pH=7.1) and samples were incubated at 25°C for 16h. DHPR binding buffer contained (mM) 140 NaCL, 15 KCl, 20 Hepes (pH=7.1) and samples were incubated for 2h at 25°C in the dark. Non-specific binding was determined as above with the addition of 1000-fold unlabeled ryanodine and 200 µM EGTA or 4000-fold unlabeled PN200-100. After incubation, reactions were quenched with rapid filtration through Whatman GF/B filters and washed with 15 ml ice cold buffer containing (in mM) 140 KCl, 0.1 CaCl2, 10 Hepes (pH=7.3). Filters were extracted in 5 ml of scintillation cocktail (Fisher Scientific),stored overnight and radioligand concentrations determined using liquid scintillation counting (Beckman LS6500) Specific binding was determined as the total ligand bound minus non- specific binding for a given radio-labeled compound. Specific binding of [3H]Ry or [3H]PN represented 94.5 and 39.3 average percent of the total ligand bound, respectively.
2.7 RyR1 Sensitivity to Ca2+ Regulation or Disruption by NDL PCBs
The plant alkaloid ryanodine, for which the RyR is named, binds selectively to the open channel state of the RyR (Pessah et al., 1985; Pessah et al., 1987) and thus increased [3H]Ry binding represents enhanced receptor activity. Impacts of variable Ca2+ concentrations on receptor activity were assessed under the RyR1 saturation binding conditions described above but were completed in the presence of 10 nM [3H]Ry and free Ca2+ adjusted between 0.25–10,000 µM using titrations of CaCl2 buffered with variable amounts of EGTA as determined by the Bound And Determined software (Brooks and Storey, 1992). Differences in RyR1 NDL PCB sensitivity between killifish from the two populations was assessed under the same conditions listed under the saturation binding methods but was completed with 40 µg ml−1 of the crude protein homogenates incubated in the presence of 8 nM [3H]Ry, 50 µM CaCl2 and a 0.5% DMSO solvent control or 0.01–20 µM PCB or PCB 95 dissolved in 0.5% DMSO.
2.8 Replication and Statistical Analysis
2.8.1 Replication
For each population, three fish were used to conduct PCB measurements and data are presented as an average of the PCB congener levels detected in the three fish (n=3). For protein based assays, four different skeletal muscle homogenates were created from each population and each homogenate was representative of the skeletal muscle of at least four fish. All protein homogenates were run on five different gels. For each homogenate, proteins of interest were normalized to GAPDH by gel and the normalized intensity was then averaged across gels to determine the level expressed in each population (n=4). Radioligand binding assays were each completed 2–4 times (i.e 2–4 biological replicates) and each time an assay was run it was done so in triplicate or quadruplicate (i.e. 3–4 technical replicates). Once corrected for non-specific binding,, values were averaged to determine specific binding at variable radioligand, Ca2+ or NDL PCB concentrations. Tissue specific expression of the genes of interest were measured in the skeletal muscle, cardiac muscle and brain tissue of six adult killifish from each population (n=6). Finally, quantification of genes of interest was completed in embryos and larvae collected at 3 dpf, 9dpf and 1 dph from the F1, F2, and F3 generations of NBH or the F1 generation of SC. At each developmental stage per generation and population 6–8 biological replicates were assessed for levels of gene transcription and each biological replicate consisted of pools of ten fish (n=6–8).
2.8.2 Statistical Analysis
Parameters for radio-ligand binding assessments were determined using non-linear regression models (Prism 5.0; Graphpad Software). For saturation binding, one-site and two-site ligand binding models were compared and utilized to determine the affinity (Kd) and maximal binding (Bmax) for radioligands. Scatchard plots were created using the ratio of bound to free ligand with the x-intercept or y-intercept represented by Bmax or Bmax/Kd, respectively, as determined through non-linear regression. The ability of the endogenous regulator Ca2+ or the potent RyR activator PCB 95 to alter RyR1 in skeletal muscle was assessed using a bell shaped or sigmoidal-dose response curve, respectively.
For qPCR, the suitability of the chosen reference genes was determined using the geNorm® algorithm (Vandesompele et al., 2002). Genes l8, rps20, β-actin and ef1α were determined to be stable (M) across all tissue and age classes assessed and it was determined that the pair-wise variability (V) was improved by using four rather than three reference genes for normalization. Therefore each gene of interest was normalized to the geNorm® normalization factor calculated from all four reference genes. Normalized mRNA values were placed on a log-2-scale for parametric statistical testing. Differences between adult transcript levels and protein were assessed using a student’s t-test while differences in mRNA transcripts in embryos and larvae were compared using multifactorial ANOVAs or one-way ANOVAs (Minitab 16.0;State College, PA).
3.0 Results and Discussion
3.1 Adult killifish NDL PCB Concentrations
The total PCB concentration detected in NBH killifish was 118,746 ng g−1 (Table 2). These levels were much greater than the total PCB concentration detected in SC killifish, 174 ng g−1 (Table 2), confirming that the SC subpopulation is a suitable reference site for comparison against highly contaminated locations. The high level recorded in NBH killifish in the current study, which represented PCB burdens in numerous tissues including skeletal muscle, brain and other body cavity organs, were consistent with previously published findings. Specifically, PCB levels have been documented at 22,666 ng g−1 or 100,000 ng g−1 (dry weight) in NBH sediment or adult killifish livers, respectively (Nacci et al., 2002; Nacci et al., 2010). The PCB concentrations found in NBH far exceed PCB concentrations detected in other fish species in the United States. For example, in a national study, that measured the same 20 PCB congeners assessed in the current research, the maximum PCB concentration detected in estuarine fish was 1160 ng g−1 (Harvey et al., 2008). These levels were detected in estuarine catfish populations, not killifish, from the Northeastern Atlantic but the differences exemplify the extreme pollution levels found in NBH.
Table 2.
PCB concentrations detected in SC and NBH killifish
| Population |
|||
|---|---|---|---|
| Scorton Creek | New Bedford Harbor | ||
| PCB Congeners | Chlorine Substitution | Concentration (ng g−1 dry) (SEM) | |
| Non ortho | |||
| PCB 77b | 3,3’,4,4’ | 3.11 (1.78) | 1358.72 (432.15) |
| PCB81b | 3,4,4’,5 | 0.40 (0.30) | 82.036 (28.15) |
| PCB126b | 3,3’,4,4’,5 | 0.39 (0.22) | 109.82 (31.15) |
| PCB169b | 3,3’,4,4’,5,5’ | ND | 1.56 (0.35) |
| Mono ortho | |||
| PCB008 | 2,4’ | <MDL | 7819.19 (1410.46) |
| PCB028 | 2,4,4′ | 23.96 (13.69) | 16468.85 (3065.18) |
| PCB066 | 2,3′,4,4′ | 8.86 (3.57) | 3425.62 (1469.43) |
| PCB105b | 2,3,3’,4,4 | 3.75 (0.43) | 3160.61 (405.66) |
| PCB118b | 2,3’,4,4’,5 | 20.27 (3.44) | 9206.99 (1437.15) |
| Ortho | |||
| PCB018a | 2,2’,5 | 9.19 (8.80) | 14909.95 (2795.34) |
| PCB044a | 2,2’,3,5’ | 6.12 (3.38) | 8531.69 (1423.56) |
| PCB052a | 2,2′,5,5′ | 26.40 (13.91) | 22578.74 (4376.61) |
| PCB101a | 2,2′,4,5,5′ | 18.56 (4.63) | 11233.93 (1830.98) |
| PCB128 | 2,2’,3,3’,4,4’ | 1.27 (0.43) | 1856.53 (293.19) |
| PCB138a | 2,2’,3,4,4’,5’ | 18.73 (2.16) | 5748.89 (752.310 |
| PCB153a | 2,2′,4,4′,5,5′ | 27.19 (3.63) | 7895.77 (1301.28) |
| PCB170a | 2,2′,3,3′,4,4′,5 | <MDL | 927.48 (144.10) |
| PCB180a | 2,2′,3,4,4′,5,5′ | 1.95 (0.69) | 1467.24 (220.22) |
| PCB187a | 2,2′,3,4′,5,5′,6 | 4.28 (1.88) | 1819.53 (374.70) |
| PCB195 | 2,2’,3,3’,4,4’,5,6 | <MDL | 81.76 (15.41) |
| PCB206 | 2,2’,3,3’,4,4’,5,5’,6 | <MDL | 56.31 (10.12) |
| PCB209 | 2,2’,3,3’,4,4’,5,5’,6,6 | <MDL | 5.14 (2.45) |
| ΣPCBs | 174.43 | 118746.36 | |
Congener has a demonstrated effect on the RyR1 in mammals or fish (Pessah et al. 2006) or (Fritsch and Pessah, 2013)
Congener has a demonstrated effect on the AhR, CB105 and CB118 are minor AhR active congeners (Van den Berg et al., 2006)
Method Detection Limit (MDL); Not Detected (ND)
In NBH and SC killifish, the PCB congeners known to activate the RyR1 greatly exceeded the relative concentration of AhR active PCBs found in killifish tissue, as reported previously (Lake et al., 1995). These trends are consistent with relative levels of NDL and DL PCBs detected in other studies (Hwang et al., 2001; Stahl et al., 2009). The ortho-PCB congeners detected in the NBH killifish greatly exceed the levels that activate RyRs in mammals (Pessah et al., 2006) and fish (Fritsch and Pessah, 2013). For example, the concentration of di-ortho congener PCB 52, a potent RyR activator, was found in NBH killifish at a 77 µM equivalent (Mass normalized to 291.99 MW). This concentration is ~25 times or ~150 times greater than the concentration known to significantly increase RyR1 activity in the skeletal muscle of rainbow trout (Fritsch and Pessah, 2013) or mammals (Pessah et al., 2006), respectively. Such extreme NDL PCB concentrations found in the killifish strongly suggest that there could be NDL specific adaptations involving the RyRs.
3.2 Compensatory and Heritable Difference in RyR Related mRNA Expression
3.2.1 Tissue specific differences in gene expression in NBH versus SC adult killifish
Adult killifish from both populations displayed tissue specific expression of the 12 genes assessed supporting isoform specific primer design (data not shown). To aid visualization of population differences, genes with low tissue specific expression levels (detected at >31 CT for qPCR; Figure 2) were excluded for a given tissue (e.g. ryr1, ryr3, and cav1.1 in cardiac muscle).
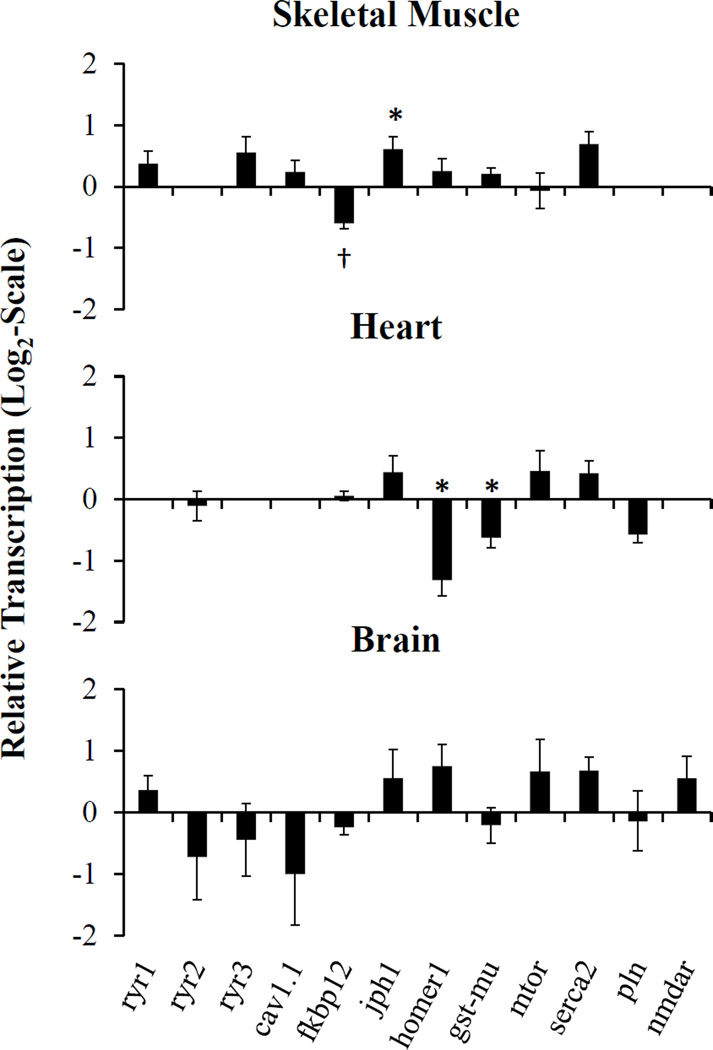
Figure 2.
Tissue specific transcript levels in adult killifish from SC and NBH. Data shown is relative to the transcript levels observed for a given tissue collected for SC (0 on the y-axis). Abbreviations see Table 1. Means ± SEM, n=6, *p ≤ 0.05, †p ≤ 0.01.
Expression of the skeletal muscle RyR isoform 1 (RyR1) in adult killifish collected from NBH appeared to be slightly increased, compared to the mRNA transcript levels measured in the reference killifish from SC; however, these levels were not significantly different (Figure 2; p = 0.13). The so called brain RyR isoform, RyR3, which has been demonstrated to occur in skeletal muscle of fish (O'Brien et al., 1995; Darbandi and Franck, 2009), displayed high variability in NBH killifish skeletal muscle, and the expression levels were not different from levels found in SC killifish. Levels of cav1.1, the L-type Ca2+ channel, α subunit of isoform 1, which mechanically interacts with the RyR1 to drive excitation contraction (EC) coupling in skeletal muscle, also were consistent between killifish from the two populations. Of the other genes assessed in skeletal muscle, RyR accessory proteins fk506 binding protein 12 kDa (fkbp12) and junctophilin 1 (jph1) were significantly decreased and increased, respectively, in NBH as compared to SC killifish (fkbp12 p<0.01; jph1 p<0.05). FKBP12 is an accessory protein that stabilizes the RyR conformational state in skeletal and cardiac muscle (Pessah et al., 2010). NDL PCB induced activity of RyR is dependent on FKBP12 and the resulting toxicity can be reduced when FKBP12 is inhibited by the immunosuppressants FK506 or rapamycin (Wong and Pessah, 1997). The accessory protein JPH1 is essential for maintaining the SR/ER in close opposition to plasma membranes in skeletal muscle and neuron cells. In skeletal muscle specifically, JPH1 permits the close association of RyR1 with CaV1.1 in the plasma membrane and is necessary for coordinated EC-coupling and muscle contraction. Finally, in NBH killifish skeletal muscle levels of the SR/ER Ca2+ ATPase, slow isoform 2 (serca2), responsible for refilling SR/ER Ca2+ levels after muscle contraction, were increased (p<0.05) as compared to killifish from SC.
Similar to skeletal muscle the major RyR isoform found in the heart, expression of the so called cardiac isoform, RyR2, was not significantly different between NBH and SC killifish hearts. Likewise, expression of fkbp12, known to interact with RyR2 in fish (Jeyakumar et al., 2001), was not different between NBH and SC killifish hearts even though it was significantly altered in the skeletal muscle of NBH killifish. This tissue specific expression in the PCB tolerant killifish may be due to differential interactions of FKBP12, versus FKBP12.6, with RyR1 and RyR2 channels, respectively, which currently is not fully understood (Jeyakumar et al., 2001) especially in regard to PCB toxicity. It should be noted however, that NDL PCBs are capable of interacting with both RyR1 and RyR2 in a stereospecific manner (Pessah et al., 2008). There was also a significant decrease in the accessory protein homer1 and the redox sensitive glutathione-s-transferase, isoform mu (gst-mu), in NBH versus SC killifish hearts. Homer1 is known to regulate signal transduction, synaptogenesis and receptor trafficking in neurons. Homer1, is also present in skeletal and cardiac muscle, at levels relative to that found in brain (Soloviev et al., 2000), and the accessory protein has been shown to have a biphasic modulatory effect on RyR1 and RyR2 channel activity (Feng et al., 2002; Huang et al., 2007; Pouliquin and Dulhunty, 2009). The redox-sensitive GSTmu is found in both skeletal and cardiac muscle, where it is believed to have an isoform specific regulatory role toward either RyR1 or RyR2. In cardiac tissue specifically, GSTmu has an inhibitory effect on RyR2 where it is suggested to help conserve SR Ca2+ levels (Abdellatif et al., 2007).
Finally, in the brain of SC and NBH killifish, the expression of RyR related genes was highly variable between individuals, such that there were no obvious differences between the two populations. Examination of the tissue specific expression of RyR related proteins did suggest a potential difference in Ca2+ signaling dynamics between PCB tolerant and sensitive populations. Interestingly, the most significant differences were seen in accessory or modulator proteins, whereas main Ca2+ channel expression did not vary widely. Currently, the role of RyR accessory proteins in NDL PCB toxicity has not been addressed outside of FKB12. Research regarding the implication of the observed differences in expression seen here may further elucidate mechanisms of NDL PCB effects on the function of RyR.
3.2.2 Developmental and generational mRNA transcript levels in killifish embryos and larvae
In order to address developmental and transgenerational effects of NDL PCBs, we examined expression of the various genes in developmental stages and multiple generations derived from PCB tolerant NBH killifish, as compared to PCB sensitive SC killifish. The F1-F3 generations were progeny of a parental generation (P) obtained in the wild that were maintained and embryos and larvae were reared under identical uncontaminated conditions in the laboratory. RyR-related mRNA expression was measured in 3 or 9 dpf embryos or 1 dph larvae collected from each of the F1, F2 and F3 generations of NBH killifish and compared to the mRNA levels found at the same developmental stage of the F1 generation from SC. We completed a multifactorial ANOVA with two factors, namely developmental stage and population, and determined that for most genes, mRNA expression differences between NBH and SC occurred in a developmental stage specific manner. Differences between populations were thus determined using one-way ANOVAs comparing NBH and SC embryos and larval expression at each developmental stage for a given generation (Figure 3).
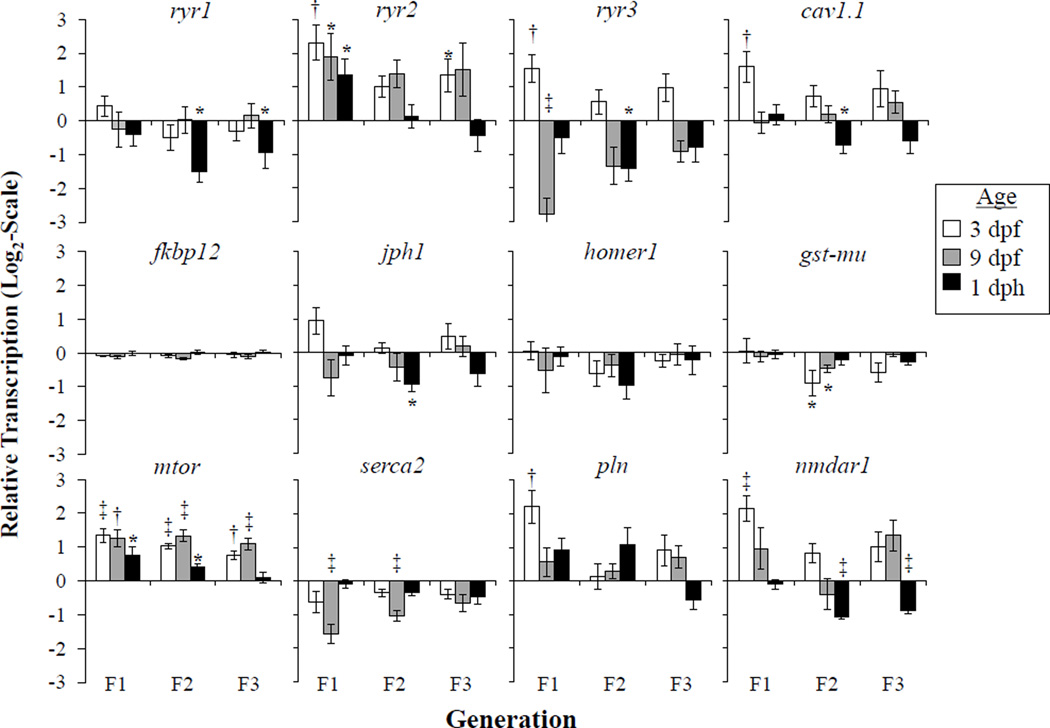
Figure 3.
RyR related mRNA differences between SC and NBH embryos and larvae from various developmental stages across multiple generations. Data shown is relative to the transcript levels observed at the same age class for SC (0 on the y-axis). Means ± SEM; n≥6; * p ≤ 0.05, † p ≤ 0.01, ‡ p ≤ 0.001. Abbreviations see Table 1.
Of the twelve genes assessed, ryr2, ryr3, mtor and serca2 displayed the most consistent age related patterns across the F1, F2 and F3 generations. Levels of ryr2 were consistently increased in NBH killifish embryos, as compared to embryos from SC, mainly at 3 and 9 dpf developmental stages. This was observed for the F1, F2 and F3 generations of NBH fish, although the most significant increases relative to the SC fish were detected in the F1 generation only. Population differences in ryr3 were at 3 dpf, and there was a significant decrease in transcription at 9 dpf, and while this trend was observed in all generations of NBH killifish embryos it was only significant in the F1 generation. Transcripts of serca2 showed a trend to decrease in all NBH embryos, as compared to SC embryos, however the levels of expression were significantly decreased only at 9 dpf in the NBH F1 and F2 generations.
Of the genes assessed, NBH embryos consistently demonstrated increased transcript levels of the Ser/Thr kinase known as the mechanistic target of rapamycin (mtor). The increased expression was evident in all developmental stages for the three generations of embryos and larvae reared in the laboratory. To our knowledge, this is the first study to demonstrate a potential impact of PCBs on mtor in an organismal exposure scenario. However, there have been recent findings, gathered from exposed adipocytes (Kim et al., 2012) and liver carcinoma cells (An et al., 2014), which identify mTOR as a potential target of PCBs. In those studies, alterations in mTOR expression were observed in cells exposed to the NDL PCB congener 153 or Aroclor 1254 (predominantly comprised of ortho rich NDL PCBs), but these effects were not seen in cells exposed to TCDD. Thus, while the DL PCBs present in NBH cannot be ruled out as potential contributors to the observed generational patterns in mtor we hypothesize that the changes may be related to NDL PCBs specifically.
Supporting this hypothesis are studies demonstrating a relationship between RyR related proteins and mTOR protein complexes. The protein now known as mTOR was previously termed FKBP12 rapamycin associated protein 1 (FRAP1; genecard), where it is known that in the presence of rapamycin FKBP12 has an inhibitory effect on the formation of mTOR complex 1 and to a lesser extent mTOR complex 2 (mTORC1 and mTORC2), thereby blocking mTOR signaling (Hoeffer and Klann, 2010). As discussed above (see mRNA results in adult killifish skeletal muscle), FKBP12 is a key accessory protein toward RyR channel function and is a necessary component in NDL PCB-induced RyR-dependent toxicity. To date, however, the direct protein-protein or protein-chemical interactions driving NDL PCB perturbation of the FKBP12/RyR complex have not been described. Potentially, NDL PCBs may act directly on FKBP12 altering its regulatory interaction with a number of proteins leading to leaky RyR channels (Pessah et al., 2010), or altered formation of mTOR complexes. Apart from the FKBP12-mTOR interaction, there is a recent study demonstrating that altered mTOR activity leads to altered RyR mediated Ca2+ release, which may represent a direct connection between mTOR and RyR signaling. To understand these relations further investigation is needed, where future research regarding the cause or the implications of altered mTOR expression in PCB exposed organisms such as the PCB tolerant killifish population, will provide important information regarding potential disease states induced by persistent exposure to PCBs. This is especially true as mTOR is well known for its role in translation initiation, cellular growth and division and is a potential culprit in diseases ranging from cancer (Zoncu et al., 2010) to neurodevelopmental disorders (Santini and Klann, 2011)
3.3 Changes in Protein Levels and Channel Function in Skeletal Muscle of Adult Killifish
3.3.1 Protein Levels in Adult Skeletal Muscle
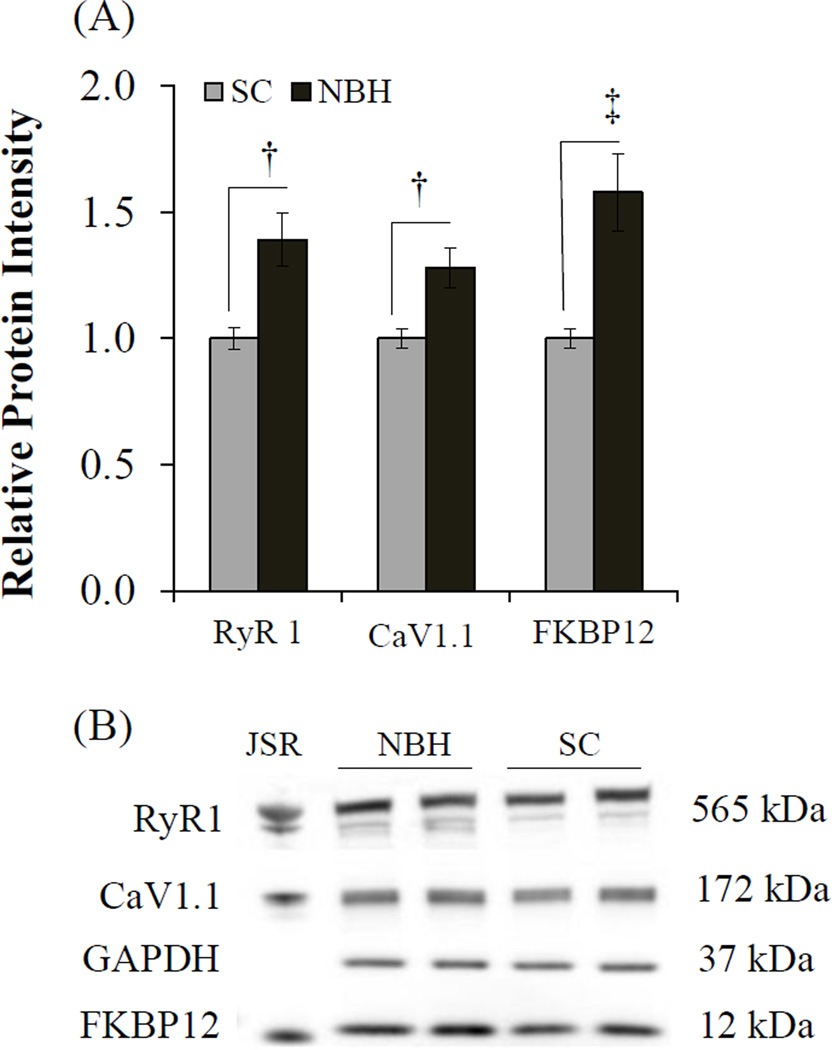
In skeletal muscle protein homogenates from killifish, antibody 34C recognized one distinct band at approximately 565 kDa, the region of where mammalian RyR1 migrates (Figure 4). There was a faint second band recognized suggesting the presence of two isoforms, which would be consistent with what has been observed elsewhere in teleost white muscle (O'Brien et al., 1995; Franck et al., 1998). The slight variations in isoform size (Franck et al., 1998) suggests that these bands represent RyR1 (RyR1b found in white muscle) and RyR3, respectively. In contrast, antibodies for CaV1.1 and FKBP12 identified single bands at approximately 172 kDa and 12 kDa, respectively, which is consistent with results published elsewhere (Qi et al., 1998; Schredelseker et al., 2005). Unlike transcript expression, protein levels of RyR1, CaV1.1, and FKBP12 all were found to be significantly increased in the skeletal muscle of NBH killifish as compared to that from SC fish. This discrepancy, especially the inflection in FKBP12 mRNA versus protein, may be due to variation in tissue preparation, where mRNA was measured in whole skeletal muscle while protein levels were measured in microsomal fractions. Therefore, while the constitutive expression of fkbp12 in whole tissue may be decreased, that found in the microsomal fraction may represent an increase in FKBP12 protein bound to increased levels of RyR1 channels.
Figure 4.
Differences in RyR1, CaV1.1 and FKBP12 protein levels in the skeletal muscle of killifish from NBH and SC. (A) Average protein band intensity shown as a ratio to SC after normalization to GAPDH. (B) Representative protein separation visualized on 5 gels; JSR, junctional sarcoplasmic reticulum preparation from white skeletal muscle of a New Zealand White Rabbit. Means ± SEM; (n = 4); †p ≤ 0.01,‡ p ≤ 0.001.
Increased levels of RyR1 and FKBP12 in adult killifish represent a potential compensatory response that could maintain Ca2+ signaling homeostasis in the face of extreme NDL-PCB burdens. This is supported by studies demonstrating that increased RyR number and function is linked to increased muscle contractility and swimming performance in fish and mammals (Anttila et al., 2006; Anttila et al., 2008; James et al., 2011; Seebacher et al., 2012). Increased levels of FKBP12 may confer tighter regulation of channel gating because: 1) the number of FKBP12 proteins bound to the RyR homotetramer varies by physiological and pathophysiological state (Zalk et al., 2007), 2) pharmacologically inhibited binding or genetically induced deficiencies in FKBP12 increases RyR channel open probability and mean open time; and 3) RyR conformational control can be regained when recombinant FKBP12 is added to channels exposed to Bastidin 10, a marine sponge extract known to disrupt the FKBP12/RyR complex (Pessah et al., 2010). These current findings are in agreement with chronic RyR1 leak and SR Ca2+ depletion as NDL-PCB induced consequences
3.3.2 Receptor Ligand Interactions
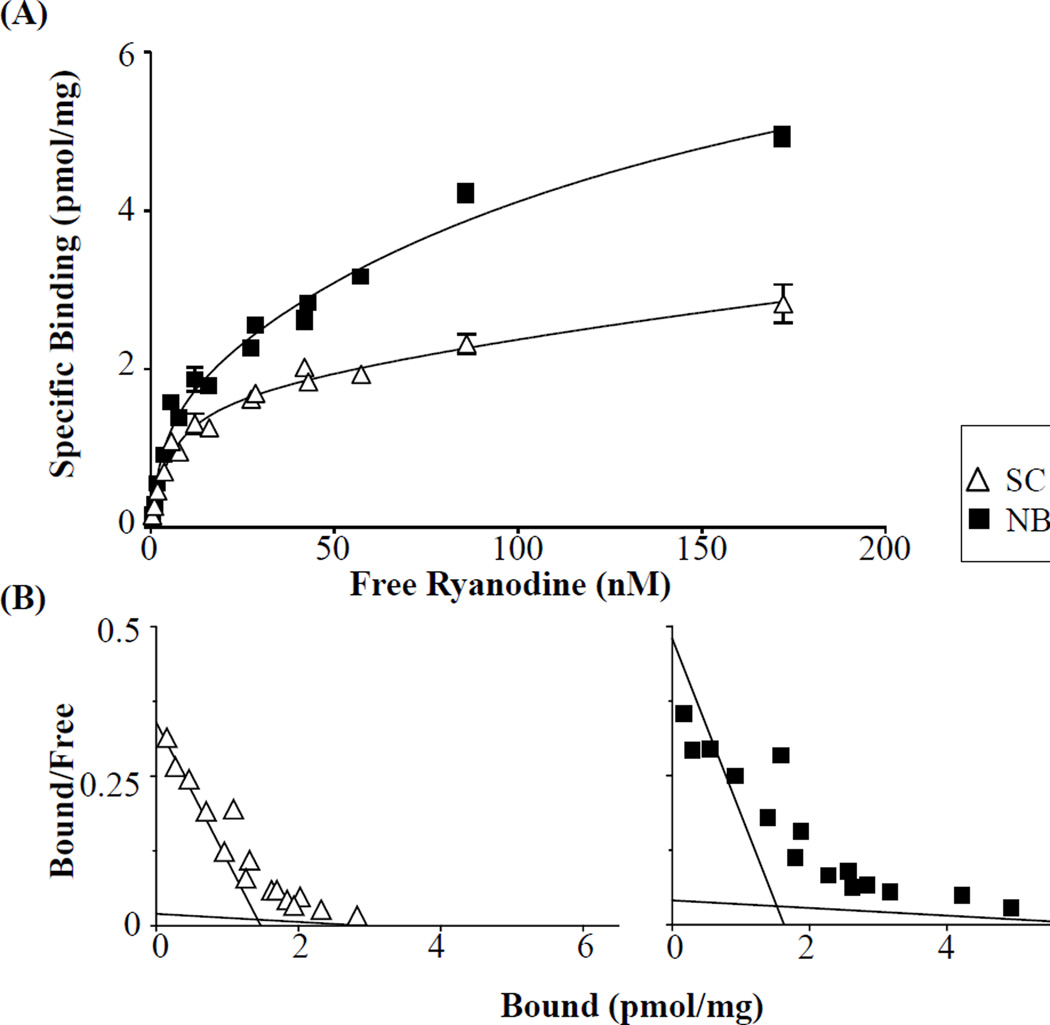
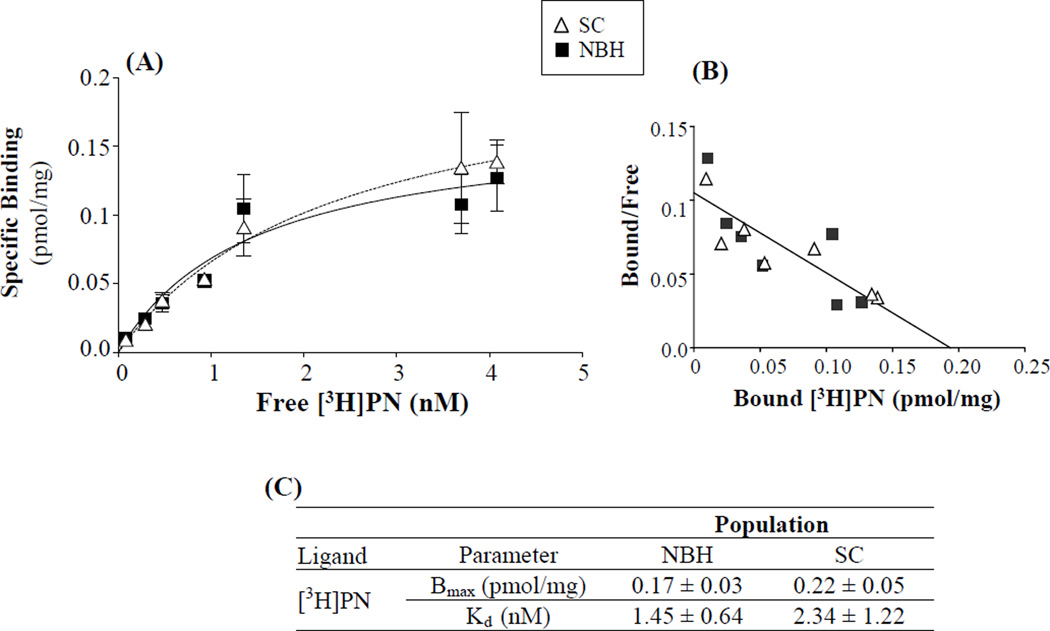
Binding of ryanodine to RyR1 of skeletal muscle from killifish fit a two-site model (Figure 5), rather than a one-site model (p<0.001), which is similar to that demonstrated in mammals (Pessah and Zimanyi, 1991). The high affinity site’s maximal receptor-ligand binding (Bmax) and ligand affinity (Kd) did not differ between killifish from NBH and SC (Table 3). The low affinity sites of NBH killifish did display a much higher Bmax compared to SC killifish, consistent with the observed increase in RyR1 protein level differences. However, it should be noted that the low affinity binding parameters were not significantly different between the populations due to a wide 95% confidence interval observed in SC killifish (Bmax 0.81 to 5.05 pmol/mg; Kd 0.0 to 556.9). [3H]PN binding to the L-type Ca2+ channel in killifish skeletal muscle followed a one-site model but neither Bmax nor Kd differed significantly between NBH and SC (Figure 6), suggesting that the observed difference in protein expression was not detected with the receptor-radioligand binding conditions used.
Figure 5.
Saturation binding of [3H]Ry in the skeletal muscle of adult killifish from SC and NBH. (A) Specific [3H]Ry binding or (B) Scatchard analysis with predicted high and low affinity binding sites by population. Means ± SEM; n=2; with 4 subsamples.
Table 3.
Binding of [3H]Ry in the skeletal muscle of Atlantic killifish from SC versus NBH.
| Site | |||
|---|---|---|---|
| Binding Site | Parameter | SC | NBH |
| High Affinity | Bmax (pmol/mg) |
1.47 ± 0.31 | 1.63 ± 0.32 |
| Kd (nM) | 4.31 ± 1.57 | 3.41 ± 1.25 | |
| Low Affinity | Bmax (pmol/mg) |
2.93 ± 1.07 | 6.69 ± 0.92 |
| Kd (nM) | 191.9 ± 184.3 | 163.2 ± 63.71 | |
Parameters represent estimates defined by non-linear regression ± SE Bmax represents the maximum amount of ligand bound per binding site Equilibrium dissociation constant ( Kd) represents binding site affinity
Figure 6.
Saturation binding curves for [3H]PN to in skeletal muscle of adult killifish from SC or NBH. (A) Specific [3H]PN binding or (B) Scatchard analysis; and (C) Population radioligand binding parameters. Means ± SEM; n=2; with 4 subsamples.
3.3.3 RyR-Channel Sensitivity to Regulation by Ca2+ or by NDL PCBs
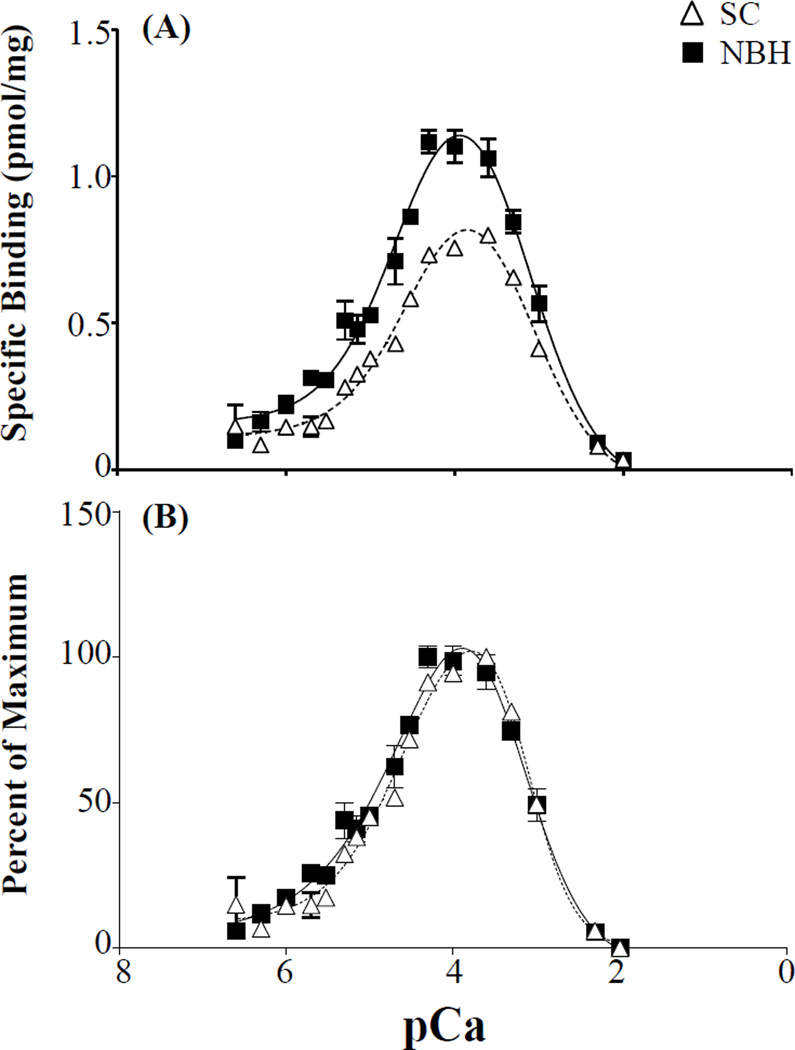
The binding of [3H]Ry to the open channel state of the RyR1 in Atlantic killifish displayed a typical bell shaped Ca2+ sensitivity curve, where Ca2+ levels below 250 µM lead to receptor activation and levels above 500 µM lead to receptor inactivation (Figure 7). Maximum receptor activation occurred at a pCa of 4 (100 µM of Ca2+), which was consistent in the crude microsomal preparations from both NBH and SC killifish skeletal muscle. While there was no shift in Ca2+ sensitivity between the two populations there was a significant difference (p < 0.01; Student t-test) in the maximal binding achieved at a pCa of 4.
Figure 7.
Ca2+ induced [3H]Ry binding in the skeletal muscle of adult killfish from SC and NBH. (A) Specific [3H]Ry bound per mg of protein; (B) Data normalized to the maximum response observed per population. Means ± SEM; n=3, with 4 subsamples.
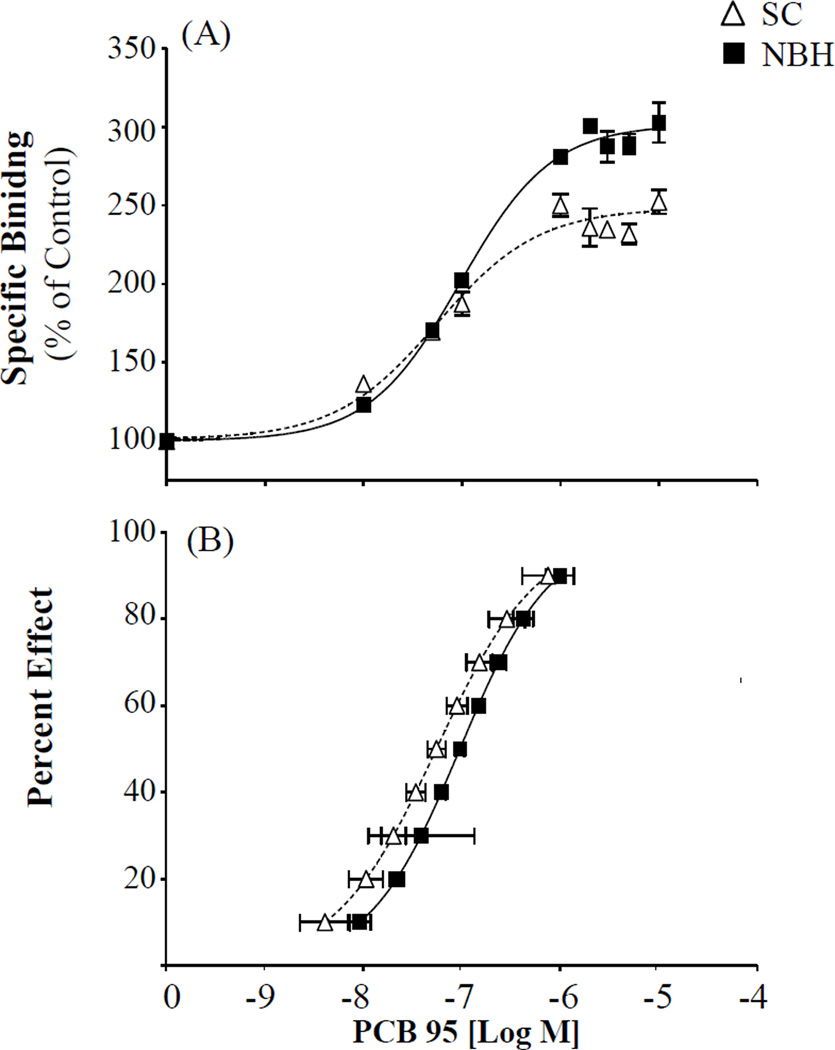
There was a characteristic rise in RyR1 activity in the presence of the potent NDL congener PCB 95 (Figure 8). There was a significant difference (p < 0.0001; F-test) in the maximum response achieved between the two populations. The maximum response achieved was 301% or 248% of the DMSO control measured for either NBH or SC, respectively. PCB 95 enhanced receptor activity at levels below 10 nM (EC10; NBH=9.23 nM; SC=4.11 nM) with a half-maximal effect concentration (EC50) at 96.2 nM and 55.6 nM for NBH and SC, respectively (95% CI; 77.7–119.1 NBH; 36.6–84.6 nm SC). Even though there was no obvious shift in the effect concentration curve, the EC50 values were predicted to vary significantly between NBH and SC (p < 0.016). Normalizing the concentration-response curves by predicted percent effect concentrations, ranging from EC10 to EC90, did show a slight shift in NBH sensitivity as compared to SC and it was determined that the concentration response curves varied significantly by population (p < 0.001).
Figure 8.
PCB 95 induced [3H] Ry binding in the skeletal muscle of adult killfish from SC and NBH. (A) Specific binding shown as a percent of the DMSO solvent control for a given population; (B) Percent effect concentrations ranging from 10–90% of predicted PCB 95 induced RyR activity in the two populations. Means ± SEM; n=4, with 3 subsamples.
Together these findings demonstrate increased RyR1 protein levels in NBH versus SC killifish but suggest that there are only minor differences in ligand interactions or endogenous and chemical regulation of the RyR between the tolerant and sensitive killifish populations. In the case of NDL PCB activation of the RyR1, there was only a minor shift in chemical sensitivity. Perhaps the lack of an obvious chemical response difference in NBH versus SC killifish reflects the abundance of NDL PCBs already present in NBH killifish tissue (Table 2); tissues for RyR assays were sampled from fish within 48 h of field capture. Thus, future research in thoroughly depurated fish or those from generations reared in a laboratory may better explain potential phenotypic adaptations to NDL PCBs rather than compensatory changes due to exposure in the field. Additionally, studies that focus more on receptor function in light of changing FKBP12 levels may better explain population differences in NDL PCB tolerance, especially as this accessory protein appears to be necessary for RyR toxicity. Such mechanisms (i.e. altered receptor/accessory protein interactions) would be in accordance with that described for DL PCB specific tolerance in the PCB resistant tomcod (Microgadus tomcod) found in the Hudson River (Wirgin et al., 2011). Notably, AhR variants found in sensitive versus tolerant tomcod populations display differential TCDD binding, which was explained by a six base pair deletion occurring in the AhR region essential for binding the AhR interacting protein (AIP). We find it intriguing that AIP’s N-terminal sequence contains a peptidyl-prolyl cis/trans isomerase (PPIase) domain characteristic of FK506 binding proteins, including FKBP12 (Linnert et al., 2013). Therefore PPIase regulatory domains may represent a convergent target of DL and NDL PCBs where future studies examining RyR1or FKBP12 population variants, and the kinetic interaction thereof, may elucidate changes in receptor functioning in tolerant versus sensitive killifish populations.
CONCLUSIONS
The Atlantic killifish is tolerant to a wide range of both natural and anthropogenic stressors acting as a useful model organism (Burnett et al., 2007). Regarding acquired tolerance to the pollutants present in NBH, research has mainly focused on select PCB congeners, namely DL PCBs, thereby considering NDL congeners as insignificant contributors to directional selection. NDL PCBs comprise the majority of the PCB burden found in NBH sediment and NBH killifish and likely represent pathway specific selective pressures. Here we focused on altered RyR related pathways in the PCB tolerant killifish from NBH versus the sensitive population found at SC. Our findings demonstrate that adult NBH killifish display increased RyR1 and FKBP12 protein levels and increased mRNA levels of key RyR accessory or modulatory proteins. It is postulated that these findings represent compensatory responses in NBH killifish because tighter control of more RyR channels, especially by FKBP12, would likely mitigate chronic RyR leak and SR Ca2+ depletion, which are believed to result from prolonged NDL PCB action at the Ca2+ release channel. Additionally, we identify potential heritable difference in the RyR-related signaling partner mTOR (Hoeffer and Klann, 2010; Santini and Klann, 2011; Martín-Cano et al., 2013). These findings likely relate to NDL PCB ability to alter FKBP12’s interaction with its modulatory proteins and the increased levels in NBH embryos and larvae suggests that mtor may contribute to long-term (generational) effects of NDL PCBsAs such, the current study identifies RyR related pathways as a candidate that may contribute to the NBH killifish’s ability to thrive in extreme NDL PCB concentrations. Moving forward, research aimed at understanding potential phenotypic (or inferred genetic) mechanisms of developed NDL PCB tolerance in NBH killifish will provide information regarding potential costs associated with extended PCB exposure. In general, however, there is a need to establish NDL PCB mediated toxic effects in non-mammalian vertebrates, where furthering this understanding will provide a more comprehensive appreciation of the exposure risks associated with total PCB burdens, adding to the extensive research currently available for DL congeners. Addressing these issues, specifically in the killifish population, will not only provide information about long-term multigenerational PCB impacts but may also identify genetic variation as determinants of NDL PCB sensitivity in vertebrates.
Highlights.
Non dioxin-like PCBs predominate in New Bedford Harbor (NBH) killifish tissue
Several congeners are >20X that known to activate the ryanodine receptor (RyR)
NBH killifish have elevated RyR and RyR-regulator FKBP12 protein levels
Laboratory reared NBH embryos have elevated mechanistic target of rapamycin
Supports NDL PCB compensatory and heritable responses in the PCB tolerant killifish
Acknowledgements
Funding was provided through the NIEHS Superfund Research Program UC Davis (INP and EBF; P42-ES004699) and Boston University (JJS and JVG; P42-ES007381). Support was supplied via the UC Davis NHLBI (EBF; T32-HL086350). Additional support came from NIEHS 1R01-ES014901, 1R01-ES017425, the UC Davis Center for Children’s Environmental Health (1P01-ES011269, U.S. Environmental Protection Agency Grant 8354320), and an unrestricted JB Johnson Foundation gift grant. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH or USEPA. Appreciation is extended to Sibel Karchner, Woods Hole Oceanographic Institution, for supplying gene sequences important to this study and Ashley Bertrand and Ian Kirby, USEPA Office of Research and Development Narragansett, RI, for help with embryos and larvae sampling.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abdellatif Y, Liu D, Gallant EM, Gage PW, Board PG, Dulhunty AF. The Mu class glutathione transferase is abundant in striated muscle and is an isoform-specific regulator of ryanodine receptor calcium channels. Cell Calcium. 2007;41:429–440. doi: 10.1016/j.ceca.2006.08.004. [DOI] [PubMed] [Google Scholar]
- An J, Wang X, Guo P, Zhong Y, Zhang X, Yu Z. Hexabromocyclododecane and polychlorinated biphenyls increase resistance of hepatocellular carcinoma cells to cisplatin through the phosphatidylinositol 3-kinase/protein kinase B pathway. Toxicology Letters. 2014;229:265–272. doi: 10.1016/j.toxlet.2014.06.025. [DOI] [PubMed] [Google Scholar]
- Anttila K, Järvilehto M, Mänttäri S. Ca2+ handling and oxidative capacity are greatly impaired in swimming muscles of hatchery-reared versus wild Atlantic salmon (Salmo salar) Canadian Journal of Fisheries and Aquatic Sciences. 2008;65:10–16. [Google Scholar]
- Anttila K, Mänttäri S, Järvilehto M. Effects of different training protocols on Ca2+ handling and oxidative capacity in skeletal muscle of Atlantic salmon (Salmo salar L.) Journal of Experimental Biology. 2006;209:2971–2978. doi: 10.1242/jeb.02341. [DOI] [PubMed] [Google Scholar]
- Bello SM, Franks DG, Stegeman JJ, Hahn ME. Acquired Resistance to Ah Receptor Agonists in a Population of Atlantic Killifish (Fundulus heteroclitus) Inhabiting a Marine Superfund Site: In Vivo and in Vitro Studies on the Inducibility of Xenobiotic Metabolizing Enzymes. Toxicological Sciences. 2001;60:77–91. doi: 10.1093/toxsci/60.1.77. [DOI] [PubMed] [Google Scholar]
- Brooks SPJ, Storey KB. Bound and determined: A computer program for making buffers of defined ion concentrations. Analytical Biochemistry. 1992;201:119–126. doi: 10.1016/0003-2697(92)90183-8. [DOI] [PubMed] [Google Scholar]
- Burnett KG, Bain LJ, Baldwin WS, Callard GV, Cohen S, Di Giulio RT, Evans DH, Gómez-Chiarri M, Hahn ME, Hoover CA, Karchner SI, Katoh F, MacLatchy DL, Marshall WS, Meyer JN, Nacci DE, Oleksiak MF, Rees BB, Singer TD, Stegeman JJ, Towle DW, Van Veld PA, Vogelbein WK, Whitehead A, Winn RN, Crawford DL. Fundulus as the premier teleost model in environmental biology: Opportunities for new insights using genomics. Comparative Biochemistry and Physiology Part D: Genomics and Proteomics. 2007;2:257–286. doi: 10.1016/j.cbd.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connon RE, D’Abronzo LS, Hostetter NJ, Javidmehr A, Roby DD, Evans AF, Loge FJ, Werner I. Transcription Profiling in Environmental Diagnostics: Health Assessments in Columbia River Basin Steelhead (Oncorhynchus mykiss) Environmental Science & Technology. 2012;46:6081–6087. doi: 10.1021/es3005128. [DOI] [PubMed] [Google Scholar]
- Darbandi S, Franck JPC. A comparative study of ryanodine receptor (RyR) gene expression levels in a basal ray-finned fish, bichir (Polypterus ornatipinnis) and the derived euteleost zebrafish (Danio rerio) Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology. 2009;154:443–448. doi: 10.1016/j.cbpb.2009.09.003. [DOI] [PubMed] [Google Scholar]
- Feng W, Tu J, Yang T, Vernon PS, Allen PD, Worley PF, Pessah IN. Homer Regulates Gain of Ryanodine Receptor Type 1 Channel Complex. Journal of Biological Chemistry. 2002;277:44722–44730. doi: 10.1074/jbc.M207675200. [DOI] [PubMed] [Google Scholar]
- Franck JPC, Morrissette J, Keen JE, Londraville RL, Beamsley M, Block BA. Cloning and characterization of fiber type-specific ryanodine receptor isoforms in skeletal muscles of fish. American Journal of Physiology - Cell Physiology. 1998;275:C401–C415. doi: 10.1152/ajpcell.1998.275.2.C401. [DOI] [PubMed] [Google Scholar]
- Fritsch EB, Connon RE, Werner I, Davies RE, Beggel S, Feng W, Pessah IN. Triclosan Impairs Swimming Behavior and Alters Expression of Excitation-Contraction Coupling Proteins in Fathead Minnow (Pimephales promelas) Environmental Science & Technology. 2013;47:2008–2017. doi: 10.1021/es303790b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch EB, Pessah IN. Structure-activity relationship of non-coplanar polychlorinated biphenyls toward skeletal muscle ryanodine receptors in rainbow trout (Oncorhynchus mykiss) Aquatic Toxicology. 2013;140–141:204–212. doi: 10.1016/j.aquatox.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giesy JP, Kannan K. Dioxin-Like and Non-Dioxin-Like Toxic Effects of Polychlorinated Biphenyls (PCBs): Implications For Risk Assessment. Critical Reviews in Toxicology. 1998;28:511–569. doi: 10.1080/10408449891344263. [DOI] [PubMed] [Google Scholar]
- Gutjahr-Gobell RE, Black DE, Mills LJ, Pruell RJ, Taplin BK, Jayaraman S. Feeding the mummichog (Fundulus heteroclitus) a diet spiked with non-ortho-and mono-ortho-substituted Polychlorinated biphenyls: Accumulation and effects. Environmental Toxicology and Chemistry. 1999;18:699–707. [Google Scholar]
- Harvey J, Harwell L, Summers JK. Contaminant concentrations in whole-body fish and shellfish from US estuaries. Environmental Monitoring and Assessment. 2008;137:403–412. doi: 10.1007/s10661-007-9776-1. [DOI] [PubMed] [Google Scholar]
- Hoeffer CA, Klann E. mTOR signaling: at the crossroads of plasticity, memory and disease. Trends in Neurosciences. 2010;33:67–75. doi: 10.1016/j.tins.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Kim JY, Dehoff M, Mizuno Y, Kamm KE, Worley PF, Muallem S, Zeng W. Ca2+ Signaling in Microdomains: Homer1 MEDIATES THE INTERACTION BETWEEN RyR2 AND Cav1.2 TO REGULATE EXCITATION-CONTRACTION COUPLING. Journal of Biological Chemistry. 2007;282:14283–14290. doi: 10.1074/jbc.M611529200. [DOI] [PubMed] [Google Scholar]
- Hwang SA, Yang BZ, Fitzgerald EF, Bush B, Cook K. Fingerprinting PCB patterns among Mohawk women. Journal of Exposure Analysis and Environmental Epidemiology. 2001;11:184–192. doi: 10.1038/sj.jea.7500159. [DOI] [PubMed] [Google Scholar]
- James RS, Walter I, Seebacher F. Variation in expression of calcium-handling proteins is associated with inter-individual differences in mechanical performance of rat (Rattus norvegicus) skeletal muscle. The Journal of Experimental Biology. 2011;214:3542–3548. doi: 10.1242/jeb.058305. [DOI] [PubMed] [Google Scholar]
- Jayaraman S, Pruell RJ, McKinney R. Extraction of organic contaminants from marine sediments and tissues using microwave energy. Chemosphere. 2001;44:181–191. doi: 10.1016/s0045-6535(00)00201-0. [DOI] [PubMed] [Google Scholar]
- Jeyakumar LH, Ballester L, Cheng DS, McIntyre JO, Chang P, Olivey HE, Rollins-Smith L, Barnett JV, Murray K, Xin H-B, Fleischer S. FKBP Binding Characteristics of Cardiac Microsomes from Diverse Vertebrates. Biochemical and Biophysical Research Communications. 2001;281:979–986. doi: 10.1006/bbrc.2001.4444. [DOI] [PubMed] [Google Scholar]
- Kenet T, Froemke RC, Schreiner CE, Pessah IN, Merzenich MM. Perinatal exposure to a noncoplanar polychlorinated biphenyl alters tonotopy, receptive fields, and plasticity in rat primary auditory cortex. Proceedings of the National Academy of Sciences. 2007;104:7646–7651. doi: 10.1073/pnas.0701944104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Pelloux V, Guyot E, Tordjman J, Bui L-C, Chevallier A, Forest C, Benelli C, Clément K, Barouki R. Inflammatory pathway genes belong to major targets of persistent organic pollutants in adipose cells. Environmental Health Perspectives. 2012;120:508–514. doi: 10.1289/ehp.1104282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake JL, McKinney R, Lake CA, Osterman FA, Heltshe J. Comparisons of patterns of polychlorinated biphenyl congeners in water, sediment, and indigenous organisms from New Bedford Harbor, Massachusetts. Archives of Environmental Contamination and Toxicology. 1995;29:207–220. [Google Scholar]
- Linnert M, Lin Y-J, Manns A, Haupt K, Paschke A-K, Fischer G, Weiwad M, Lücke C. The FKBP-type domain of the human aryl hydrocarbon receptor-interacting protein reveals an unusual Hsp90 interaction. Biochemistry. 2013;52:2097–2107. doi: 10.1021/bi301649m. [DOI] [PubMed] [Google Scholar]
- Martín-Cano FE, Camello-Almaraz C, Hernandez D, Pozo MJ, Camello PJ. mTOR pathway and Ca2+ stores mobilization in aged smooth muscle cells. Aging (Albany NY) 2013;5:339. doi: 10.18632/aging.100555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacci D, Coiro L, Champlin D, Jayaraman S, McKinney R, Gleason TR, Munns WR, Jr, Specker JL, Cooper KR. Adaptations of wild populations of the estuarine fish Fundulus heteroclitus to persistent environmental contaminants. Marine Biology. 1999;134:9–17. [Google Scholar]
- Nacci DE, Champlin D, Coiro L, McKinney R, Jayaraman S. Predicting the occurrence of genetic adaptation to dioxinlike compounds in populations of the estuarine fish Fundulus heteroclitus. Environmental Toxicology and Chemistry. 2002;21:1525–1532. [PubMed] [Google Scholar]
- Nacci DE, Champlin D, Jayaraman S. Adaptation of the estuarine fish Fundulus heteroclitus (Atlantic killifish) to polychlorinated biphenyls (PCBs) Estuaries and Coasts. 2010;33:853–864. [Google Scholar]
- Niknam Y, Feng W, Cherednichenko G, Dong Y, Joshi SN, Vyas SM, Lehmler H-J, Pessah IN. Structure-Activity Relationship of Selected Meta- and Para-Hydroxylated Non-Dioxin Like Polychlorinated Biphenyls: From Single RyR1 Channels to Muscle Dysfunction. Toxicological Sciences. 2013;136:500–513. doi: 10.1093/toxsci/kft202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien J, Valdivia HH, Block BA. Physiological differences between the alpha and beta ryanodine receptors of fish skeletal muscle. Biophysical Journal. 1995;68:471–482. doi: 10.1016/S0006-3495(95)80208-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessah IN, Cherednichenko G, Lein PJ. Minding the calcium store: Ryanodine receptor activation as a convergent mechanism of PCB toxicity. Pharmacology & Therapeutics. 2010;125:260–285. doi: 10.1016/j.pharmthera.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessah IN, Hansen LG, Albertson TE, Garner CE, Ta TA, Do Z, Kim KH, Wong PW. Structure-Activity Relationship for Noncoplanar Polychlorinated Biphenyl Congeners toward the Ryanodine Receptor-Ca2+ Channel Complex Type 1 (RyR1) Chemical Research in Toxicology. 2006;19:92–101. doi: 10.1021/tx050196m. [DOI] [PubMed] [Google Scholar]
- Pessah IN, Lehmler H-J, Robertson LW, Perez CF, Cabrales E, Bose DD, Feng W. Enantiomeric Specificity of (−)-2,2′,3,3′,6,6′-Hexachlorobiphenyl toward Ryanodine Receptor Types 1 and 2. Chemical Research in Toxicology. 2008;22:201–207. doi: 10.1021/tx800328u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessah IN, Stambuk RA, Casida JE. Ca2+-activated ryanodine binding: mechanisms of sensitivity and intensity modulation by Mg2+, caffeine, and adenine nucleotides. Molecular Pharmacology. 1987;31:232–238. [PubMed] [Google Scholar]
- Pessah IN, Waterhouse AL, Casida JE. The calcium-Ryanodine receptor complex of skeletal and cardiac muscle. Biochemical and Biophysical Research Communications. 1985;128:449–456. doi: 10.1016/0006-291x(85)91699-7. [DOI] [PubMed] [Google Scholar]
- Pessah IN, Zimanyi I. Characterization of multiple [3H]ryanodine binding sites on the Ca2+ release channel of sarcoplasmic reticulum from skeletal and cardiac muscle: evidence for a sequential mechanism in ryanodine action. Molecular Pharmacology. 1991;39:679–689. [PubMed] [Google Scholar]
- Pouliquin P, Dulhunty A. Homer and the ryanodine receptor. European Biophysics Journal. 2009;39:91–102. doi: 10.1007/s00249-009-0494-1. [DOI] [PubMed] [Google Scholar]
- Qi Y, Ogunbunmi EM, Freund EA, Timerman AP, Fleischer S. FK-binding Protein Is Associated with the Ryanodine Receptor of Skeletal Muscle in Vertebrate Animals. Journal of Biological Chemistry. 1998;273:34813–34819. doi: 10.1074/jbc.273.52.34813. [DOI] [PubMed] [Google Scholar]
- Samsó M, Feng W, Pessah IN, Allen P. Coordinated movement of cytoplasmic and transmembrane domains of RyR1 upon gating. PLoS biology. 2009;7:e1000085. doi: 10.1371/journal.pbio.1000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini E, Klann E. Dysregulated mTORC1-dependent translational control: from brain disorders to psychoactive drugs. Frontiers in behavioral neuroscience. 2011:5. doi: 10.3389/fnbeh.2011.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schredelseker J, Di Biase V, Obermair GJ, Felder ET, Flucher BE, Franzini-Armstrong C, Grabner M. The β1a subunit is essential for the assembly of dihydropyridine-receptor arrays in skeletal muscle. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:17219–17224. doi: 10.1073/pnas.0508710102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seebacher F, Pollard SR, James RS. How well do muscle biomechanics predict whole-animal locomotor performance? The role of Ca2+ handling. The Journal of Experimental Biology. 2012;215:1847–1853. doi: 10.1242/jeb.067918. [DOI] [PubMed] [Google Scholar]
- Simon T, Britt JK, James RC. Development of a neurotoxic equivalence scheme of relative potency for assessing the risk of PCB mixtures. Regulatory Toxicology and Pharmacology. 2007;48:148–170. doi: 10.1016/j.yrtph.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Soloviev MM, Ciruela F, Chan W-Y, McIlhinney RAJ. Mouse brain and muscle tissues constitutively express high levels of Homer proteins. European Journal of Biochemistry. 2000;267:634–639. doi: 10.1046/j.1432-1327.2000.01078.x. [DOI] [PubMed] [Google Scholar]
- Stahl L, Snyder B, Olsen A, Pitt J. Contaminants in fish tissue from US lakes and reservoirs: a national probabilistic study. Environmental Monitoring and Assessment. 2009;150:3–19. doi: 10.1007/s10661-008-0669-8. [DOI] [PubMed] [Google Scholar]
- Van Dongen AM, Hoeffer CA, Klann E. NMDA Receptors and Translational Control. 2009 [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology. 2002:3. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayman GA, Bose DD, Yang D, Lesiak A, Bruun D, Impey S, Ledoux V, Pessah IN, Lein PJ. PCB-95 Modulates the Calcium-Dependent Signaling Pathway Responsible for Activity-Dependent Dendritic Growth. Environ Health Perspect. 2012a:120. doi: 10.1289/ehp.1104833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayman GA, Yang D, Bose DD, Lesiak A, Ledoux V, Bruun D, Pessah IN, Lein PJ. PCB-95 Promotes Dendritic Growth via Ryanodine Receptor–Dependent Mechanisms. Environ Health Perspect. 2012b:120. doi: 10.1289/ehp.1104832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams L, Oleksiak M. Signatures of selection in natural populations adapted to chronic pollution. BMC Evolutionary Biology. 2008;8:282. doi: 10.1186/1471-2148-8-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirgin I, Roy NK, Loftus M, Chambers RC, Franks DG, Hahn ME. Mechanistic Basis of Resistance to PCBs in Atlantic Tomcod from the Hudson River. Science. 2011;331:1322–1325. doi: 10.1126/science.1197296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirgin I, Waldman JR. Resistance to contaminants in North American fish populations. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 2004;552:73–100. doi: 10.1016/j.mrfmmm.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Wong PW, Pessah IN. Noncoplanar PCB 95 Alters Microsomal Calcium Transport by an Immunophilin FKBP12-Dependent Mechanism. Molecular Pharmacology. 1997;51:693–702. doi: 10.1124/mol.51.5.693. [DOI] [PubMed] [Google Scholar]
- Yang D, Kim KH, Phimister A, Bachstetter AD, Ward TR, Stackman RW, Mervis RF, Wisniewski AB, Klein SL, Kodavanti PRS. Developmental exposure to polychlorinated biphenyls interferes with experience-dependent dendritic plasticity and ryanodine receptor expression in weanling rats. Environmental Health Perspectives. 2009;117:426. doi: 10.1289/ehp.11771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalk R, Lehnart SE, Marks AR. Modulation of the ryanodine receptor and intracellular calcium. Annual review of biochemistry. 2007;76:367–385. doi: 10.1146/annurev.biochem.76.053105.094237. [DOI] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nature reviews Molecular cell biology. 2010;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]