Abstract
To test the hypothesis that alternative splicing could be an adaptive mechanism for populations subject to multi-generational estrogenic exposures, we compared estrogen receptor alpha (ERα) splicing variants in two populations of killifish (Fundulus heteroclitus): one resident in an estrogenic polluted environment (New Bedford Harbor, NBH, MA, USA) and one from a relatively uncontaminated reference site (Scorton Creek, SC, MA, USA). In total we identified nineteen ERα variants, each with deletions of one or more coding exons. Four of the variants with potential functional relevance were analyzed by qPCR to test for population differences in expression by tissue type, site, sex, seasonal reproductive status and estrogen treatment. Significantly, a 5′-truncated short form variant (ERαS) was highly expressed in liver and ovary, and was associated with seasonal reproductive activity in SC but not NBH fish. Both ERαS and the full-length long variant (ERαL) were estrogen-inducible (ERαS > ERαL) but the induction response was lower in NBH than in SC fish. In contrast, NBH killifish were hyper-responsive to estrogen as measured by expression of two other estrogen responsive genes: vitellogenin (Vtg) and aromatase B (AroB). Most strikingly, two ERα deletion variants (Δ6 and Δ6–8), lacking ligand binding and activation function domains, were identified in a subset of NBH fish, where they were associated with reduced responsiveness to estrogen treatment. Together, these results support the hypothesis that alternative splicing of the esr1 gene of killifish could be an autoregulatory mechanism by which estrogen modulates the differential expression of ERα, and suggests a novel and adaptive mechanistic response to xenoestrogenic exposure.
Keywords: estrogen receptor alpha, mRNA variants, killifish, alternative splicing, endocrine disruption
1. Introduction1
Estrogen-like endocrine disrupting chemicals (EDCs) or xenoestrogens interact with the ligand-binding site of estrogen receptors (ER) to alter transcription (Diamanti-Kandarakis et al., 2009). However, the extent to which EDCs could alter estrogen signaling though mechanisms other than direct transcriptional regulation remains largely unexplored in both human and wildlife populations. Because such mechanisms could provide adaptive opportunities for populations subject to multi-generational EDC exposures, we chose to study two populations of killifish (Fundulus heteroclitus): one resident in a polluted EPA Superfund site, New Bedford Harbor (NBH), MA, USA, and one resident in a clean reference site, Scorton Creek (SC), MA, USA. The NBH site is highly contaminated with polychlorinated biphenyls (PCBs) whose metabolites are weak ER agonists (DeCastro et al., 2006). Additionally, outflow from wastewater is the presumed source of several natural and synthetic estrogens that have been identified in NBH (Zuo et al., 2006), and other contaminants present in urban estuaries are also estrogenic (Benotti and Brownawell, 2007). The NBH killifish have demonstrated resistance to polycyclic aromatic hydrocarbons (PAHs), dioxins, and dioxin-like PCBs as measured by reduced toxicity and decreased cytochrome P450 1a1 (Cyp1a) expression and activity (Bello et al., 2001; Nacci et al., 1999). This trait is heritable (Nacci et al., 2010), and has been ascribed to a genome-wide disruption of aryl hydrocarbon receptor (AhR) signaling (Oleksiak et al., 2011; Whitehead et al., 2010); however the exact mechanism remains to be determined (Aluru et al., 2011; Hahn et al., 2004; Reitzel et al., 2014).
ERα/β are members of the steroid receptor family of ligand-activated nuclear receptors and interact as homo- or heterodimers on estrogen responsive elements (ERE) of target gene promoters to enhance or repress transcription (Heldring, et al., 2007). In killifish, like other teleost fishes, estrogenic responses are mediated by three ERs (ERα, -βa, -βb) but only ERα is estrogen responsive (Greytak and Callard, 2007). As measured by increased expression of estrogen responsive genes in reproductively inactive adults (vitellogenin, Vtg; and the predominant brain form of cytochrome P450 aromatase, AroB) (Greytak et al., 2005), the NBH environment is highly estrogenic, but ERα mRNA (itself a marker of estrogen exposure) remains unchanged, nor are there changes in the non-estrogen-regulated ERβa and ERβb (Greytak and Callard, 2007). Moreover, seasonal increases in ERα mRNA seen in SC are dampened in NBH fish, for example, in female liver ~3.5-fold increases are reduced to less than 2-fold in NBH (Greytak and Callard, 2007; Greytak et al., 2005). Although this suggests attenuation of the ERα auto-regulatory feedback loop that amplifies estrogenic responses in teleosts, induction of Vtg and AroB mRNAs was the same or greater in NBH males as compared to SC male fish (Greytak et al., 2010). Paradoxically, NBH embryos/larvae overexpress ERα ~five-fold, yet they are hypo-responsive to estrogen treatment as measured by ERα and AroB induction (Greytak and Callard, 2007; Greytak et al., 2010). Together these findings suggest that the mechanism by which NBH killifish may be resistant to estrogenic exposures is gene- and life stage-specific.
Alternative splicing of the esr1 gene encoding ERα, is a plausible candidate mechanism to explain the complexity and diversity of estrogenic responses in NBH killifish. Like other nuclear receptors, ERα is a modular protein with five distinct functional domains (A – F) encoded by 8 exons; the DNA- (DBD, D) and ligand-binding (LBD, E) domains; two activation functions (AF), the constitutively active AF-1 (AB) at the N-terminus and the ligand dependent AF-2 (EF) in the LBD (Heldring, et al., 2007). In humans and mice, differential splicing of the eight coding exons of the ESR1 gene generates an exceptional number of structurally and functionally different ERα splice variants, some of which are associated with disease phenotypes (for a review see, Taylor et al., 2010). Moreover, transcription and splicing are mechanistically coupled and co-regulated (Moore and Proudfoot, 2002; Auboeuf, et al., 2004), suggesting that xenoestrogens that regulate transcription of a gene target also have the potential to regulate its splicing. Initial cloning efforts demonstrated no significant site differences in single nucleotide polymorphisms (SNPs) in the coding region of the killifish esr1 gene (Greytak and Callard, 2007), nor were any identified in a more comprehensive candidate gene scan SNP analysis which included ERα (Proestou et al., 2014). However, one alternatively spliced ERα was cloned from killifish (ERαx), which was present in both SC and NBH populations (Greytak and Callard, 2007). Alternatively spliced ERα variants have been identified in many fish species (Cotter et al., 2013; Pakdel et al., 2000; Patino et al., 2000; Pinto et al., 2012; Seo et al., 2006; Tan et al., 1996; Xia et al., 1999). In particular, the expression of alternative long (ERαL, full-length) and short (ERαS, 5′-truncated) mRNA variants appears to be a conserved feature of teleosts (Cotter et al., 2013). In rainbow trout, where the processing of the esr1 gene and production of alternative ERαS and ERαL isoforms was first described, the two variants have different estrogen binding and transactivation functions (Menuet et al., 2001; Pakdel et al., 2000). Also, in zebrafish, alternatively spliced ERαL and ERαS transcripts are differentially expressed by tissue type, during development and in response to estrogens and xenoestrogens (Cotter et al., 2013).
Here we investigated the hypothesis that the esr1 gene is differentially processed in NBH and SC killifish, and that resultant expression patterns of alternatively spliced ERα mRNAs could account for observed gene-, tissue-, and site-related differences in estrogen responses. We applied a targeted PCR cloning approach to characterize the killifish esr1 gene and to comprehensively survey the spectrum of ERα splice variants in fish from both sites. Four of the most abundant and functionally relevant variants were then analyzed by quantitative (q) and semi-quantitative (sq) PCR to document SC and NBH population differences in expression patterns by tissue type, reproductive status, and estrogen treatment. A total of nineteen alternatively spliced ERα transcripts were identified, some with more than one alternative splicing event. ERαL and ERαS transcripts were differentially expressed by site, tissue type, reproductive status, and estrogen treatment. Strikingly, two deletion variants lacking exons that encode both the ligand binding and second activation domains were specific to a subset of fish from NBH and, in these individuals, were associated with reduced estrogen responsiveness.
2. Materials and Methods
2.1 Collection and treatment of adult killifish
To obtain tissues for PCR cloning and determine the tissue distribution of identified ERα mRNA variants (Section 3.3), adult killifish (six of each sex) were collected monthly from SC and NBH between May (reproductively active) and October (reproductively inactive), as described previously (Greytak and Callard, 2007; Greytak et al., 2005). Size, body weight and gonadosomatic index (GSI) of these fish were also reported in these earlier publications. Previously collected tissues and RNA extracts as an ethanolic precipitate used for cloning and variant analysis, respectively, were stored at −80 °C.
To determine effects of estrogen on expression of ERα, AroB and Vtg mRNAs, (Section 3.4) additional reproductively active male and female killifish were collected from the two sites in June 2013 and kept until August 2013 in separate flow-through tanks with ambient temperature and light cycles at the National Health and Environmental Research Laboratory (Aquatic Ecology Division, US EPA), as previously described (Nacci et al., 1999), at which time they were past the period of seasonal reproductive activity at these sites (Greytak et al., 2005). Fish were then injected intraperitoneally with 5 mg/kg 17β-estradiol (E2; Sigma, St Louis, MO) in sesame oil or vehicle alone 5 days before decapitation. This concentration was chosen based on previous studies (Greytak et al., 2010; Pait and Nelson, 2003; Urushitani et al., 2003), which demonstrated significant induction of Vtg, AroB, and ERα mRNAs with this dosing protocol. Average body weights of these groups were SC males 8.13 g +/− 0.48 g, SC females 10.05 g +/− 0.85 g, NBH males 6.52 g +/− 0.34 g, NBH females 7.39 g +/−0.48 g, and reproductive regression was verified by visual inspection of the gonads. All fish were anesthetized in 0.06% MS-222 (Sigma) before decapitation. Tissues were immediately quick-frozen on dry ice, and stored at −80 °C.
2.2 RNA extraction and reverse transcription
Frozen tissues were homogenized in Tri Reagent (Sigma) and total RNA was extracted according to the manufacturer’s instructions. Extracts were treated with DNase I (Promega, Madison, WI) to minimize gDNA contamination. Concentration and quality of RNA was determined spectrophotometrically and confirmed by gel electrophoresis. Reverse transcription of 3 μg of total RNA (20 μl reaction volume) was performed using oligo(dT) priming and SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA), according to the manufacturer’s instructions.
2.3 Oligonucleotides
Oligonucleotides used for PCR and qPCR are listed in Table 1 and Supplementary Table 2. Primers used in qPCR measurements of total ERα (Q5/Q6, Table 1), AroB (F-ACGAGCACAGTCTGAGCATGAG; R- CTCAGATCCTCGTCATCGTTCA) and Vtg (F- GAGGATCTGTGCTGATGCAGTTGTG; R-GGGTAGAAGGCAGTCTTTCCCAGG) mRNAs were previously published (Greytak and Callard, 2007; Greytak et al., 2005; Greytak et al., 2010). Primers were designed using the Primer Express 2.0 software program (Applied Biosystems, Foster City, CA). Primers for elfa were (F- TGATCCCCCAGAAGCCCATGGT; R-GCCACGGTCTGCCTCATG). All other primers were based on the previously published killifish ERα cDNA (GenBank # AY571785) and a preliminary annotation of the killifish genome (Fundulus.org) or based on homology to the marine medaka ERα cDNA (GenBank # AY917147) and the medaka genome (Ensembl.org, #ENSORLG00000014514).
Table 1.
Oligonucleotide primer sequences. Numbering of nucleotide position corresponds to ERαL (GenBank: HM641843) except for the ERαS-specific primers Q1 and Q2, indicated by asterisk (*), where numbering corresponds to position in ERαS (GenBank: AY571785). With the exception of primer set Q5/Q6 (Greytak and Callard, 2007; Greytak et al., 2010), all other primers were designed in this study. See Supplementary Table 2 for additional primers.
| Primer | Exon Position | Nucleotide Position | Sequence 5′-3′ | |
|---|---|---|---|---|
| PCR cloning | 1 (F) | Exon 2a | 162–186 | GCAAAATKGGAKARRSRAGTCTAAATC |
| 2 (R) | Exon 2b | 294–314 | GGAGAGGGTCTCCAGCTCTGA | |
| 3 (F) | Exon 1 | 1–23 | GCCCCGAGGATGATTCATGTATAA | |
| 4 (F) | Exon 5a | 1138–1157 | GGGACAGATCTTCAGTGGCC | |
| 5 (F) | Exon 5b | 1286–1305 | CAAGGAGCTGGTCCACATGA | |
| 6 (R) | Exon 5b | 1219–1238 | TCGGCTAAGTTTTTGACGGG | |
| 7 (R) | Exon 5b | 1230–1255 | GTGACCTCGGTGTAGGGTCGGCTAAG | |
| 8 (R) | Exon 8 | 1674–1692 | TGAGAGCGTCGGTGATGGT | |
| 9 (R) | Exon 9 | 2074–2092 | TTCAGGCCTCCGACTCACA | |
| qPCR | Q1 (F) | Exons 1/2b | 8–33* | GATGATTCATGTATAAAGGGCAGAAC |
| Q2 (R) | Exon 2b | 94–114* | GGAGAGGGTCTCCAGCTCTGA | |
| Q3 (F) | Exon 2a | 110–132 | GAAAAGGGTGAGACCCAGAGAGA | |
| Q4 (R) | Exon 2a | 191–210 | CACGGCAGCACTGATAAGCA | |
| Q5 (F) | Exon 7 | 1540–1558 | GCATGCTCAAGCTCAAACC | |
| Q6 (R) | Exon 8 | 1611–1627 | GTGCCGGTGCAGAAAGA | |
| Expression vector cloning | SF | Exon 2a | 357–374 | CACCATGTACCCTGAAGAGAGC |
| LF | Exon 2b | 216–233 | CACCATGTTGCTCAGGCAGAAC | |
| R | Exon 9 | 2060–2078 | TCACAGGACCTGGGCACAG |
2.4 Targeted PCR cloning and cDNA analysis
Reverse transcribed cDNA was amplified using exon-specific primers (2 mM each) and GoTaq Flexi polymerase (Promega) under the following conditions: 95 °C for 5 minutes; 40 cycles of 95 °C for 30 seconds, 62 °C for 30 seconds, and 72 °C for 1 min/kb; and 72 °C for 10 minutes. The ERα cDNA products were size separated on a 2% agarose gel, and the bands extracted using the MinElute Gel Extraction Kit (Qiagen, Hilden, Germany; 10 μl final volume). An aliquot (3 μl) of purified product was ligated into pGEM®-T Easy vector (Promega) and cloned using JM109 competent cells (Promega). Colonies were cultured and plasmid DNA was isolated using a Wizard® SV Minipreps DNA Purification System (Promega). Each sample was sequenced (Eurofins MWG Operon, Huntsville, AL or Eton Bioscience, San Diego, CA) in two directions using plasmid specific primers. Sequences were analyzed using BLAST.
2.5 Quantitative (q)PCR analysis
As previously described (Greytak and Callard, 2007; Greytak et al., 2005; Greytak et al., 2010), qPCR analysis was performed using an ABI PRISM 7900 HT (Applied Biosystems, Foster City, CA) instrument with SYBR green fluorescent labeling. Amplifications were performed in triplicate for each cDNA sample in optically clear 384-well plates. Cycling parameters were as follows: 50 °C for 2 minutes, 95 °C for 10 minutes; 40 cycles of 95 °C for 15 seconds, 60 °C for 1 minute. Data were exported to Q-gene (Simon, 2003), adjusted for amplification efficiency, and normalized to elfa as previously described (McCurley and Callard, 2008). Results are expressed as mean normalized expression (MNE +/− SEM) of at least three independent biological replicates per experimental condition (see figure legends).
2.6 In vitro translation
To obtain the full coding region of ERαS, ERαL, and ERαSΔ6–8 end-to-end PCR was applied using specific primers in exons 2a, 2b, and 9 (Table 1) and NBH killifish liver cDNA. Variants were size separated on a 1% agarose gel, and the bands extracted using the MinElute Gel Extraction Kit and cloned into pcDNA3.1 mammalian expression vectors (Invitrogen). [35S]methionine-labeled ERα proteins were generated in vitro with a TNT coupled reticulocyte lysate system using 1 μg of the T7-promoter driven pcDNA template. Proteins were then separated by SDS-PAGE gel and visualized by autoradiography.
2.7 Statistics
Statistical analysis by one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparisons test was performed using JMP 11.0 (SAS, Cary, NC). Normality and equal variance of the data sets were analyzed by Shapiro-Wilk and O’Brien tests respectively, and those that did not pass both tests were transformed using log or arcsine methods as indicated in the figure legends. The dataset for the effects of estrogen on Vtg failed normality testing despite attempted transformations and, therefore, was rank transformed prior to ANOVA. Student’s standardized t-test was performed where specified using the online QuickCalcs calculator (GraphPad). See figure legends for details. Significance was set at P ≤ 0.05.
Statistical analysis by analysis of covariance (ANCOVA) was performed using JMP 11.0 (SAS) to determine the predictors of expression changes in estrogen responsive genes. Utilizing data from individual fish, we investigated the effect of the independent variables, ERα isoform expression (ERαS or ERαL), population (SC, NBH, NBH del), and their interactions on the dependent variable, expression of Vtg, AroB, or ERα. Variables were removed in a backwards, stepwise fashion if their effect was not significant (P < 0.05).
3. Results
3.1 Identification of short and long 5′-end ERα variants and structure of the esr1 gene
Previous research in rainbow trout and zebrafish (see Introduction) described two ERα isoforms, ERαL (long isoform) and an N-truncated ERαS (short isoform), which are generated by alternative use of transcription and translation start sites. In killifish, the previously cloned ERα cDNA (GenBank # AY571785; Greytak and Callard, 2007), (GenBank # AB097197; Urushitani et al., 2003) corresponds to the transcript encoding the short isoform of rainbow trout and zebrafish, and is predicted to be lacking the A functional domain. To determine if killifish have an additional ERαL mRNA, we used a homology cloning approach. In silico analysis of the ERαL cDNA previously cloned from the taxonomically related marine medaka (Japanese killifish; GenBank # AY917147), when aligned with the medaka genome (Ensembl.org, #ENSORLG00000014514), indicated the possibility of two 5′-end variants. We first designed a degenerate forward primer (#1, Table 1) in the presumed exon 2a of the medaka gene and paired it with a reverse primer (#2) targeting sequence downstream of the translation start codon (presumed exon 2b) of the cloned killifish ERαS transcript. A 153 bp cDNA fragment was amplified from killifish liver cDNA, indicating a second 5′-end transcript. Further analysis of killifish liver cDNA using a forward primer (#3) positioned in the 5′-UTR of the cloned killifish ERαS (presumed exon 1) paired with the same reverse primer (#2) in exon 2b generated two products (114 and 314 bp) that were identical in sequence at their 5′- and 3′-ends but the longer clone had a 200 bp insertion (presumed exon 2a). These results were confirmed using genomic DNA, indicating the presence of both ERαS and ERαL in killifish. Unlike rainbow trout and zebrafish, however, the 2092 bp killifish long isoform utilizes the same transcription start site (TSS) as the short isoform, and the two variants differ only in the alternative splicing of exon 1 onto exons 2a (long) or 2b (short) (Figure 1). Translation of the killifish ERαL is predicted to begin at ATGII in exon 2a, resulting in a 620 aa protein containing a complete A domain (Figure 2A). In contrast, the killifish ERαS is predicted to be translated from ATGI in exon 2b, resulting in an N-truncated protein that is lacking the 47 aa that comprise the A domain (Figure 2A). The predicted protein sizes for ERαS and ERαL were confirmed via in vitro translation of the full-length cDNAs resulting in 65 and 70 kDa proteins respectively (Figure 2B). The killifish long ERα cDNA sequence was entered into GenBank as accession number HM641843.3 (Table 2).
Figure 1. Organization and alternative splicing (circled a–n) of the killifish esr1 gene, as determined by targeted PCR cloning and sequence analysis of ERα cDNAs.
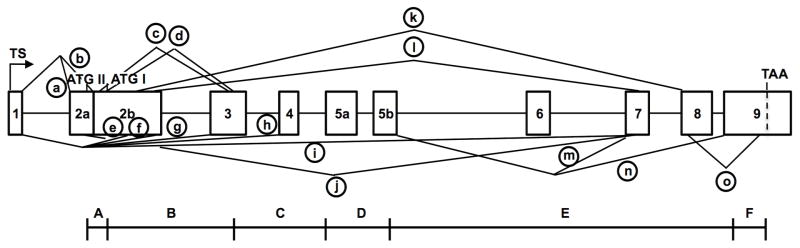
See text for the cloning strategy, Table 1 for primer sequences and Table 2 for mRNA characteristics and GenBank Accession numbers. Cloned cDNAs were assembled and mapped onto the marine medaka genome (Ensembl.org, #ENSORLG00000014514) and subsequently confirmed by mapping onto a preliminary draft of the killifish genome (Fundulus.org) and by PCR cloning and sequencing of selected introns. See Table S1 for comparison of exon and intron sizes in killifish, medaka and zebrafish. A single transcription start site (TS) and two translation start sites (ATGI, ATGII) were identified. A transcript with alternative splicing between exon 1 and an internal acceptor site in exon 2 (circled a) resulted in a short transcript lacking exon 2a and predicting translation from ATGI in exon 2b. A second transcript with alternative splicing between exon 1 and exon 2a (circled b) is predicted to begin translation at ATGII in exon 2a. Additional alternative splicing events (circled c – m) resulted in mRNAs with deletions of one or more internal coding exons. The previously cloned ERαx mRNA splice variant (circled n) is included for reference (Greytak and Callard, 2007). The six functional domains (A–F) of the human ERα protein (Krust et al., 1986) are shown below the corresponding exons. Although the esr1/ESR1 gene has 8 coding exons in all vertebrates, by convention the first coding exon is termed exon 2 in zebrafish and rainbow trout (Menuet et al., 2004; Pakdel et al., 2000) but exon 1 in mouse and human (Ponglikitmongkol et al., 1988; White et al., 1987).
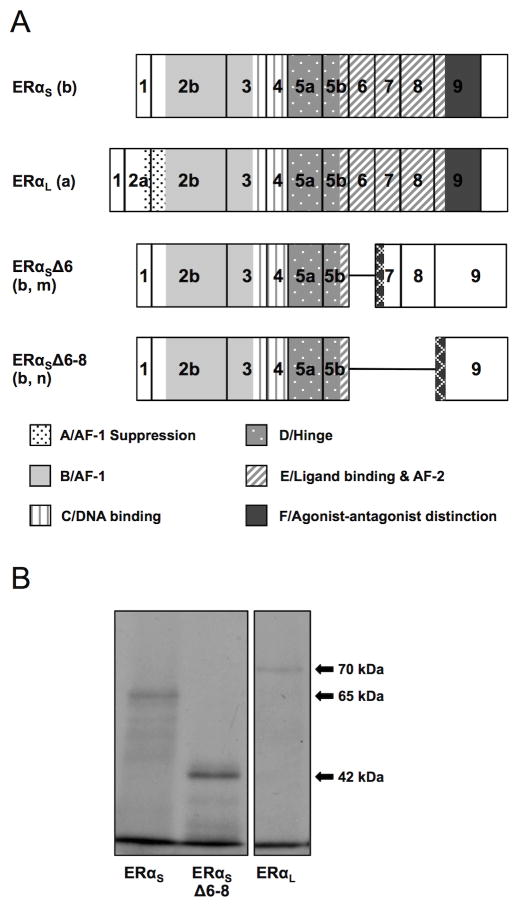
Figure 2. (A) ERαS, ERαL ERαSΔ6 and ERαSΔ6–8 isoform structure as predicted in silico. (B) Short, long and Δ6–8 ERα isoform size as confirmed by in vitro transcription/translation.
(A). Lower case letters in parenthesis are keyed to the splicing events shown in Figure 1. The previously identified ERαS was produced from the splicing of exon 1 to exon 2b (b), and was predicted to use ATGI resulting in a protein of 573 aa lacking the A domain. The newly identified ERαL was produced from the splicing of exon 1 to exon 2a (a), and its translation is predicted from ATGII in exon 2a resulting in a protein of 620 aa containing all six functional domains. A deletion variant lacking exon 6 (Δ6, m) was paired with both the short and long 5′ends predicting a 333 (short) or 380 (long) aa protein, respectively (only the short is shown). A deletion variant lacking exons 6, 7, and 8 (Δ6–8, n) was paired with both the short and long 5′ends, predicting a 334 (short) or 381 (long) aa protein, respectively (only the short is shown, b). Both Δ6 and Δ6–8 are predicted to be missing most of the E and all of the F domains. The key to the functional domains of the ERα protein as defined in human (Krust et al., 1986) and rainbow trout (Pakdel et al., 2000) is shown at the bottom of the panel. Hatched regions indicate novel amino acid sequences generated by frame shifts. Predicted protein structure of additional cloned mRNAs is included in Supplementary materials (Figure S1). Characteristics of the variant mRNAs and predicted proteins are summarized in Table 2.
(B). In vitro translation of ERαS, ERαL, and ERαSΔ6–8 with TnT-coupled reticulocyte lysate in the presence of 35S-labeled methionine resulted in proteins of 65, 70, and 42 kDa, respectively, corresponding with the in silico size predictions.
Table 2.
Summary of cloned killifish ERα mRNA variants.
| Genbank Accession # | mRNA Variant | Splicing Event(s) * | Transcript (nt) | Predicted Protein (aa) | Affected Domain(s) |
|---|---|---|---|---|---|
| AY571785 | ERαS | b | 1941 | 573 | A |
| HM641843.3 | ERαL | a | 2092 | 620 | none |
| HM641845 | ERαΔ2 | g | 582 | 436 | A, B |
| KF931638 | ERαΔ2–3 | h | 1238 | 394 | A, B, C |
| HM641844 | ERαΔ2–6 | i | 636 | 206 | A–E |
| KF931636 | ERαSΔ2–7 | b, k | 740 | 191 | A–E |
| KF931637 | ERαLΔ2–7 | a, k | 837 | 238 | B–E |
| KJ650313 | ERαSΔ3–7.5 | b, l | 828 | 160 | A–F |
| KJ650314 | ERαLΔ3–7.5 | a, l | 926 | 207 | B–F |
| KF931639 | ERαΔ2.5 | e | 1655 | 492 | A,B |
| KJ650315 | ERαΔ2.75 | f | 1525 | 492 | A, B |
| KM040769 | ERαSΔ2–3.5 | b, c | 980 | 436 | A, B |
| KJ650316 | ERαLΔ2b-7.5 | a, j | 741 | 179 | A–E |
| KM040770 | ERαΔ2.5–3.5 | d | 1014 | 394 | A, B, C |
| KJ650320 | ERαSΔ6–8 | b, n | 1428 | 334 | A,E, F |
| KJ650321 | ERαLΔ6–8 | a, n | 1526 | 381 | E, F |
| DQ413179 | ERαx | o | 1611 | 463 | A, E, F |
| KJ650317 | ERαxLΔ2b-7.5 | o, a, j | 411 | 69 | A–F |
| KF931640 | ERαSΔ2–3.5,Δ6–8 | b, c, n | 651 | 197 | A, B, E, F |
| KJ650318 | ERαSΔ6 | m | 1746 | 333 | E |
| KJ650319 | ERαLΔ6 | m | 1836 | 380 | E |
See figure 1
Mapping of our killifish ERαS and ERαL cDNAs onto the medaka genome, and a preliminary annotation of the esr1 gene from the killifish genome (Fundulus.org), revealed a structure similar to the zebrafish esr1 gene (Ensembl.org, #ENSDARG00000004111), with the only major difference being insertion of intronic sequence in exon 5. Cloning of this region using killifish genomic DNA and primers in exon 5 (#4/6) confirmed that there is a 140 bp intron that divides exon 5 into two parts of 179 and 154 bp, respectively termed exons 5a and 5b (Figure 1). The size and sequence of all other introns (with the exception of introns 3, 6, and 8) was confirmed using exon specific primers and killifish genomic DNA (unpublished data) and is shown diagrammatically in Figure 1 and detailed in Supplementary materials (Table S1; Genbank # KM236111).
3.2 Identification of ERα variants with deleted internal exons
Targeted PCR cloning was applied to identify other possible alternative splicing events affecting the killifish esr1 gene, and liver cDNA from killifish from both SC and NBH sites was routinely used. First, deletions exclusive to the N-terminus of the predicted protein were examined using a forward primer in exon 1 and a reverse primer in exon 5b (#3/7, Table 1), resulting in the identification of four ERα variants, which were then extended in the 3′ direction using variant specific forward primers (see Supplementary Table 2) and a reverse primer in the 3′ UTR (#9). The first two (termed ERαΔ2, GenBank: HM641845; and ERαΔ2–3, GenBank: KF931638) resulted from the direct splicing of exon 1 to exons 3 or 4 respectively (Figure 1, circled g/h). The second two (named ERαΔ2.5, GenBank: KF931639; and ERαΔ2.75, GenBank: KJ650315) resulted from the splicing of exon 1 to internal splice acceptor sites approximately one-half or three-quarters into exon 2b (Figure 1, circled e/f). The predicted translation start site for ERαΔ2 is about halfway through exon 3 resulting in a protein missing all of the A and B domains, while translation of ERαΔ2–3 is predicted from an ATG in exon 4 resulting in a protein missing the A, B and the first 38 aa of the C (DNA binding) domains (Supplementary Figure 1, Table 2). Translation of both ERαΔ2.5 and ERαΔ2.75 is predicted to begin at a start codon at the end of exon 2b, resulting in the same 492 aa protein missing the entire A domain (AF-1 Suppression), and the first 81 aa of the B domain (AF-1) (Supplementary Figure 1, Table 2).
Next, possible larger deletions in the ERα mRNA were explored using the same exon 1 forward primer (#3) paired with a reverse primer in exon 8 (#8), and culminated in the discovery of four additional variants, which were then extended in the 3′ direction using variant specific forward primers (see Supplementary Table 2) and a reverse primer in the 3′ UTR (#9). Sequence analysis indicated that the first two of these variants (named ERαSΔ2–3.5, GenBank: KM040769; and ERαSΔ2.5–3.5, GenBank: KM040770) resulted from the splicing of internal regions of exon 2b onto internal regions of exon 3 (Figure 1, circled c/d). The predicted protein translated from ERαΔ2–3.5 is expected to be the same as ERαΔ2, while ERαΔ2.5–3.5 is expected to be the same as ERαΔ2–3 (Supplementary Figure 1, Table 2). The other two variants (termed ERαΔ2–6, GenBank: HM641844; and ERαLΔ2b-7.5, GenBank: KJ650316) resulted from either the direct splicing of exon 1 onto exon 7, or the splicing of the end of exon 2a to an internal splice site halfway through exon 7 (Figure 1, circled i/j). The predicted translation start site for ERαΔ2–6 is at the beginning of exon 7, while ERαLΔ2b-7.5 contains ATGII in exon 2a, resulting in 206 and 179 aa proteins respectively, both missing all of the A–D domains, and one-third of the E domain (Ligand Binding/AF-2) (Supplementary Figure 1, Table 2).
Lastly, we targeted further possible ERα mRNA variants by performing end-to-end PCR using the exon 1 forward primer (#3), and a reverse primer in the 3′ UTR (#9). This technique yielded four remaining variants; two of which (named ERαΔ3–7.5 and ERαΔ2–7) were created by the alternative splicing of a site within exon 2b to a site either halfway through exon 7 or at the beginning of exon 8, and the other two (ERαΔ6 and ERαΔ6–8) consisting of the direct splicing of exon 5b onto exon 9 (Figure 1, circled k/l/m/n). All four have either the short or the long variation at the 5′ end and as such translation is predicted to start from either ATGI or ATGII. Δ3–7.5 would therefore yield protein of two sizes, 160 aa (ERαSΔ3–7.5, GenBank: KJ650313) and 207 aa (ERαLΔ3–7.5, GenBank: KJ650314), both of which are missing half of the B and E domains, and all of the C and D domains (Supplementary Figure 1, Table 2). On the other hand Δ2–7 would yield two proteins that are 191 aa (ERαSΔ2–7, GenBank: KF931636) and 238 aa (ERαLΔ2–7, GenBank: KF931637), both of which are missing most of the B domain, all of the C and D domains, and half of the E domain (Supplementary Figure 1, Table 2). Depending on ATG usage, Δ6–8 would be translated into two proteins of 333 aa (ERαSΔ6–8, GenBank: KJ650320) and 381 aa (ERαLΔ6–8, GenBank: KJ650321) which, due to a frame shift, would be missing almost all of the E domain and the complete F domain (agonist/antagonist distinction) with an additional 10 aa at the C-terminus (Figure 2A, Table 2). The two variants of Δ6 (ERαSΔ6, GenBank: KJ650318 and ERαLΔ6, GenBank: KJ650319) would have a frame shift similar to the Δ6–8 splice variant and would result in similar proteins (Figure 2A). The predicted protein size for ERαSΔ6–8 was confirmed by in vitro translation, which yielded a 42 kDa protein (Figure 2B).
In addition to the combination of internal deletions with either the short or long 5′ ends, we identified several variants produced by more than one splicing event. For instance, Δ2b-7.5 co-occurred with the previously identified ERαx variant (Greytak and Callard, 2007) and predicted a 69 aa protein (GenBank: # KJ650317) containing only small fractions of the E and F domains. Moreover, the Δ2–3.5 and Δ6–8 deletions co-occurred to predict a 197 aa protein (GenBank: KF931640) that would be missing all of the B and F domains and most of the E domain (Supplementary Figure 1, Table 2).
3.3 Differential expression of short, long, and total ERα mRNAs by tissue-type in reproductively active and inactive SC and NBH killifish
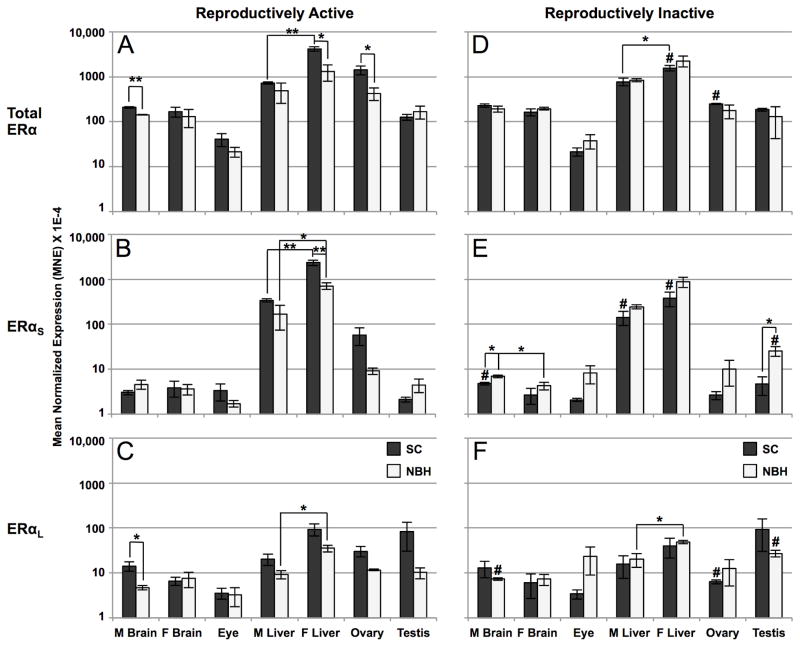
In previous studies, we reported that ERα mRNA expression varies by tissue-type, sex and reproductive status in SC and NBH killifish (Greytak and Callard, 2007; Greytak et al., 2010). To determine whether these variables affect the relative abundance of short and long ERα mRNAs we applied qPCR analysis using variant specific primers. Forward primers (Q1 and Q3) targeted sequences at the 5′ end that are exclusive to either ERαS or ERαL (the exon 1/2b boundary or exon 2a, respectively), while the reverse primers (Q2 and Q4) targeted exon 2b, which is common to both forms (Table 1; Figure 1). As a control, we used the same primer set that was used in our earlier study (Greytak and Callard, 2007). This primer pair (Q5/6) targeted sequence in exons 7 and 8 that is common to both ERαS and ERαL (termed “total” ERα). A similar strategy was used to demonstrate differential regulation of 5′-end ERα variants in zebrafish (Cotter et al., 2013). Confirming our earlier results, total ERα expression in reproductively active SC fish was highest in liver (female > male) and ovary, with ~10-fold lower levels of expression in brain and testis and ~100-fold lower expression in eye (Figure 3A). Overall, the tissue-specific pattern of total ERα mRNA expression was similar for reproductively active SC and NBH killifish but NBH fish had significantly lower mean levels of total ERα mRNA in male brain, female liver, and ovary. In general, the pattern of ERαS expression was similar to that of total ERα: highest in liver (female > male; SC > NBH), with moderate (ovary, ~10-fold lower) or much lower levels (100-fold lower) in other tissues (Figure 3B). In contrast to ERαS, ERαL was more evenly distributed across different tissues, yet highest in female liver and testis (Figure 3C).
Figure 3. Differential expression of total ERα, ERαS, and ERαL mRNAs by tissue type in reproductively active (A, B, C) and inactive (D, E, F) SC and NBH killifish, as determined by qPCR analysis.
Tissues from SC and NBH killifish of both sexes were pooled by site, sex, reproductive condition and tissue type (2 fish per pool) and analyzed using primers specific for ERαS or ERαL. Additional primers targeted sequence common to both variants (“total” ERα). Results are expressed as mean normalized expression (MNE) +/− SEM of 3 (liver, brain, gonads) or 6 (eye) independent pools per tissue type. Sex-related differences in expression were not significant in the eye, so male and female data were combined. Analysis by log-transformed one-way ANOVA separately for each mRNA species showed significant differences by tissue type (total ERα: F = 45.99, P < 0.001; ERαS: F = 81.14, P < 0.001; ERαL: F = 13.52, P < 0.001). Asterisks indicate a significant difference in expression between sites or by sex at a given site as determined by t-test: *, P < 0.05; **, P < 0.01; ***, P < 0.001. Pound signs indicate a significant difference in expression between reproductively active and inactive fish from the same site as determined by t-test: P < 0.05. Note the logarithmic scale of the y-axis.
As shown in Figure 3, the transition to reproductive inactivity in SC killifish significantly decreased expression of total ERα in female liver and ovary and also decreased ERαS in both male and female liver and ERαL in ovary. Interestingly, these fish also demonstrated a small, but significant, 1.5-fold increase in ERαS mRNA in the male brain over their active counterparts (Figure 3E). By contrast, the already low levels of ERα mRNA in reproductively active NBH fish did not further decrease with the transition to reproductive inactivity. Indeed, when reproductively inactive fish are compared at the two sites, NBH killifish had higher levels of ERαS mRNA in male brain and testis (Figure 3E). In addition, the transition from reproductive activity to inactivity in NBH killifish significantly increased ERαS (5.5-fold) and ERαL (2.5-fold) in testis (compare Figure 3 panels B and E, and C and F), which may reflect previously reported differences in testicular size and cellular composition rather than transcriptional activity per se.
3.4 Effects of estrogen on expression of total, short and long ERα mRNAs in the liver of reproductively inactive male and female SC and NBH killifish
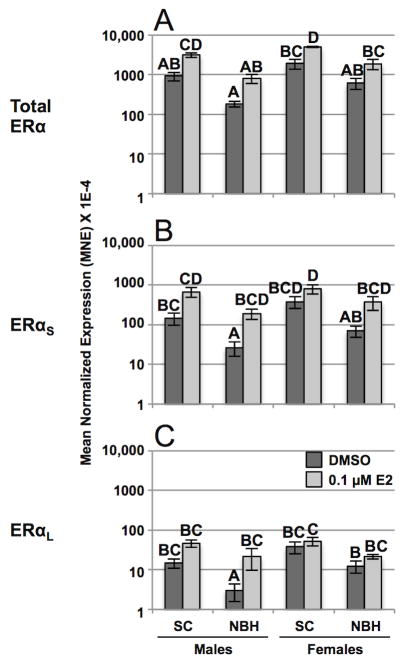
In our earlier study, we reported that a single submaximal estradiol injection upregulates mRNAs of total ERα and Vtg in liver and AroB in brain of reproductively inactive males at both sites after 5 days but there was a high degree of inter-individual variability, especially in NBH fish (Greytak et al., 2010). To confirm these results and determine if estrogen differentially regulates ERαS and ERαL in the liver of killifish, we quantified total, ERαS and ERαL mRNAs in reproductively inactive male and female killifish from both sites using the same treatment paradigm. As measured by one-way ANOVA separately for each mRNA species, total ERα, ERαS and ERαL were estrogen responsive (see legend to Figure 4); however, further analysis using Tukey’s multiple comparisons test revealed a complex pattern of isoform-, sex- and site-related differences in basal and estrogen induced mRNA levels. For example, total ERα was significantly upregulated by estrogen in SC males and females but not in NBH fish of either sex (Figure 4A). Moreover, when basal or estrogen-induced levels of ERαS (Figure 4B) or ERαL (Figure 4C) were compared in males or females at a given site or across sites, mean differences were not significant. Consistent with earlier reports (Greytak et al., 2010), total ERα expression was highly variable but not significantly upregulated by estrogen in the brain and gonads of males or females at either site, nor did estrogen induce expression of ERαS or ERαL in these tissues (Table 3).
Figure 4. Induction effects of estradiol on total ERα, ERαS, and ERαL mRNAs in the liver of reproductively inactive male and female SC and NBH killifish.
Reproductively inactive adult killifish were collected at each site, acclimated in the laboratory for 8 weeks, and then injected with 5 mg/kg of estradiol in sesame oil or vehicle alone. Tissues were collected for analysis 5 days post-injection. Results are expressed as mean normalized expression (MNE) +/− SEM of 5 individuals per sex, site, and treatment type. Analysis by arcsine- (total) or log- (ERαS and ERαL) transformed one-way ANOVA separately for each mRNA species showed that estradiol significantly upregulated total ERα (F = 17.21, P < 0.001), ERαS (F = 18.52, P < 0.001) and ERαL (F = 10.68, P < 0.01) mRNAs in liver. Letters indicate which samples differed significantly (P < 0.05) within an mRNA subtype as determined by Tukey’s multiple comparisons test. Note the logarithmic scale of the y-axis. For effects of estradiol on ERα, ERαS, and ERαL mRNAs in brain and gonads see Table 3. Induction effects of estradiol on hepatic Vtg and brain AroB mRNAs are shown in Supplementary Materials (Fig. S3).
Table 3.
Quantitative PCR analysis of total ERα, ERαS, and ERαL mRNAs in the brain and gonads of reproductively inactive male and female killifish from NBH and SC and the response to estradiol. See legend to Figure 4 for details. Results are normalized to elfa and expressed as mean normalized expression (MNE) +/− SEM of 5 individuals per sex, site, and treatment type. One-way ANOVA separately for each mRNA species demonstrated no significant upregulation with E2 treatment, nor were significant site-related differences detected by paired t-tests.
| Total ERα | ERαS | ERαL | |||
|---|---|---|---|---|---|
| Male Brain | SC | Vehicle | 134.07 ± 23.10 | 3.00 ± 0.67 | 3.97 ± 0.94 |
| E2 | 205.40 ± 77.13 | 3.91 ± 1.25 | 7.79 ± 3.05 | ||
| NBH | Vehicle | 166.69 ± 49.07 | 2.04 ± 0.83 | 3.11 ± 1.14 | |
| E2 | 176.22 ± 53.10 | 2.79 ± 0.50 | 2.59 ± 0.49 | ||
| Female Brain | SC | Vehicle | 1206.03 ± 651.18 | 4.13 ± 0.72 | 3.68 ± 0.88 |
| E2 | 2159.19 ± 1243.15 | 4.69 ± 0.85 | 7.40 ± 3.24 | ||
| NBH | Vehicle | 246.25 ± 85.67 | 2.49 ± 0.80 | 3.29 ± 0.70 | |
| E2 | 754.77 ± 283.57 | 2.97 ± 1.04 | 3.44 ± 0.74 | ||
| Testis | SC | Vehicle | 376.02 ± 41.85 | 14.85 ± 2.23 | 18.15 ± 2.41 |
| E2 | 261.05 ± 59.80 | 12.44 ± 2.59 | 13.43 ± 2.14 | ||
| NBH | Vehicle | 144.84 ± 20.45 | 11.94 ± 2.31 | 7.42 ± 1.16 | |
| E2 | 242.35 ± 45.03 | 16.99 ± 4.53 | 11.15 ± 2.46 | ||
| Ovary | SC | Vehicle | 466.79 ± 241.17 | 22.53 ± 13.81 | 41.94 ± 24.68 |
| E2 | 139.97 ± 29.72 | 2.29 ± 0.52 | 2.94 ± 0.35 | ||
| NBH | Vehicle | 479.17 ± 176.05 | 24.46 ± 14.17 | 36.06 ± 18.47 | |
| E2 | 928.81 ± 666.91 | 40.12 ± 27.68 | 67.45 ± 49.38 |
To ascertain the effectiveness of the estrogen treatment paradigm in this experiment we also measured Vtg and AroB mRNAs, which are known to be more robust than ERα as markers of estrogen responsiveness in many teleosts, including killifish (Greytak et al., 2010). As shown in Figure 5, one-way ANOVA separately for each mRNA species showed a highly significant effect of estradiol on induction of Vtg (50 –2000-fold) and AroB (5 – 10-fold) mRNAs. Interestingly, although estrogen increased mean levels of AroB mRNA in the brain of killifish at both sites, the induction response reached significance only in NBH fish, and was double that seen in SC fish. Similar to qPCR analysis of the three ERα mRNAs (Figure 4 and Table 3), there was a high degree of inter-individual variability in values obtained for Vtg and AroB mRNAs.
Figure 5. Effects of estrogen on expressed levels of (A) Vtg and (B) AroB mRNAs in male and female SC and NBH killifish.
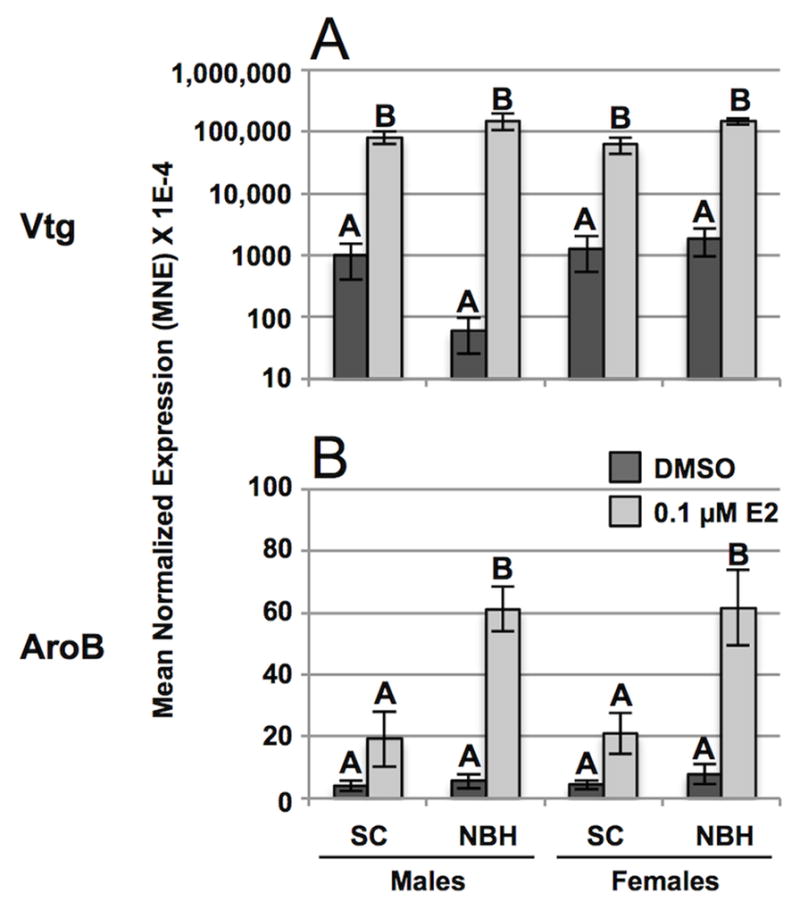
Adult killifish were injected with estradiol or vehicle alone and tissues processed for qPCR analysis (see legend to Figures 4). Results are expressed as mean normalized expression (MNE) +/− SEM of 5 individuals per sex, site, and treatment type. Analysis by rank- (Vtg) or arcsine- (AroB) transformed one-way ANOVA separately for each mRNA species showed a significant effect of estradiol on induction of hepatic vitellogenin (F = 114.29, P < 0.001) and brain aromatase (F = 39.21, P < 0.001). Letters indicate which samples differed significantly (P < 0.05) within an mRNA subtype as determined by Tukey’s multiple comparisons test. Note the logarithmic scale of the y-axis in panel A.
3.5 NBH-specific expression of 3′-end ERα deletion variants
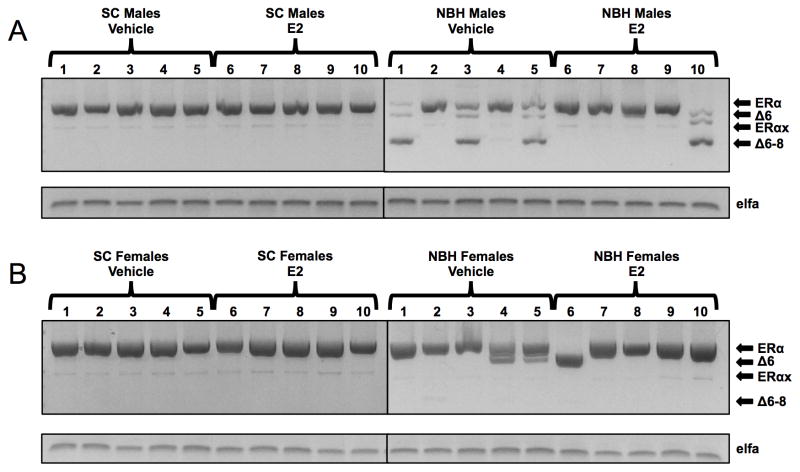
Whereas the majority of the ERα exon deletion variants identified in this report were expressed at very low levels in adult killifish from both sites, ERαΔ6 and ERαΔ6–8 mRNAs were exceptional in that one or both were present in a subset of NBH killifish only and, where present, their PCR products were relatively high in abundance when compared to the normal transcript on agarose gels (Figure 6). Although we were unable to design suitable specific qPCR primers to quantify these variants, PCR analysis with primers that bracketed the region between exon 5b and exon 9 (Figure 6) shows that five out of ten NBH males (#1, 3, 5, 8, 10) and three out of ten NBH females (#4, 5, 6) expressed ERαΔ6 and/or ERαΔ6–8 mRNAs, whereas none of the SC individuals had detectable amounts of these two variants. Interestingly, individuals that expressed one or both of the 3′-end variants in liver also expressed these variants in testis and ovary (Supplementary Figure 2). Smaller amounts of the previously identified ERαx deletion variant (Greytak and Callard, 2007) were detected in both SC and NBH killifish.
Figure 6. Expression of 3′-end ERα variants in the liver of individual male (A) and female (B) killifish after treatment with estradiol or vehicle alone, as determined by PCR analysis and agarose gel electrophoresis.
Adult killifish were injected with estradiol or vehicle alone (see legend to Figure 4) and livers processed to RNA. cDNA from individual male or female killifish livers (lanes 1–10) was amplified with primers in exon 5 (#5) and the untranslated region of exon 9 (#9) in order to generate full-length and possible 3′-end deletion variants of ERα at the same time. Products were size-separated on agarose gels: full-length, 806 bp; ERαΔ6, 668 bp; ERαx, 476 bp; ERαΔ6–8, 350 bp. RNA extraction, reverse transcription, polymerase, primer concentrations, cycling parameters, and agarose gel electrophoresis were exactly as described in Materials & Methods (sections 2.2 and 2.4).
3.6 Correlation between expression of ERα variants and induction of total ERα, Vtg and AroB mRNAs
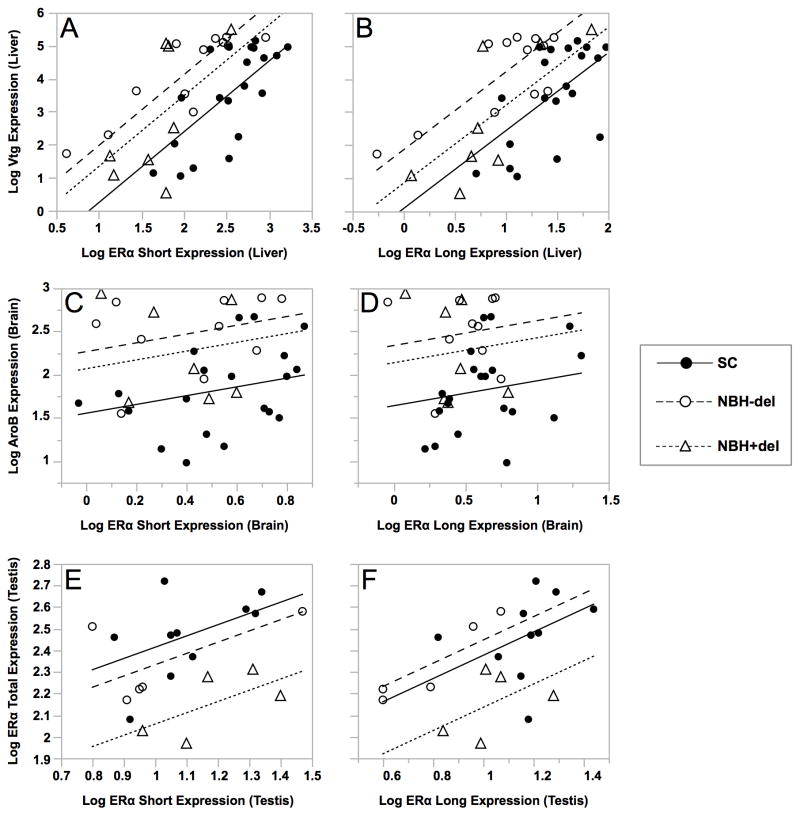
Consistent with our previous study (Greytak et al., 2010) and as shown here (Figures 4 and 5, Table 3), there was a high degree of biological variability in the measured mRNAs, as would be predicted for a wild population. To investigate the possibility that variability within the two populations masks significant differences between populations, we utilized analysis of covariance (ANCOVA) to test correlations between expressed levels of ERαS or ERαL mRNAs (independent variables) and expressed levels of each of the estrogen responsive mRNAs (dependent variables: total ERα, Vtg, AroB) in individual fish, combining males and females, and estrogen treated and untreated individuals, segregated by population. In addition to the SC population, NBH killifish were separated into two sub-populations: NBH+del, expressing one or both of the 3′-end deletion variants (ERαΔ6, ERαΔ6–8), and NBH-del, all remaining NBH killifish. Tables 4 and 5 summarize the statistical comparisons of the regression plots shown in Figure 7 and Supplementary Figure 3.
Table 4.
Effects tests of the relations between mRNAs of estrogen responsive genes (dependent variable) and mRNAs of ERα variants (independent variable) as a function of expression level and population, as determined by analysis of covariance (ANCOVA). For regression plots see Figure 7 and Supplementary Figure 3. NS = not significant (P > 0.05).
| Dependent Variable (mRNA) | Independent Variable (mRNA) | Relation to Expression Level | Relation to Population | |||||
|---|---|---|---|---|---|---|---|---|
| r2 | df | F | P | df | F | P | ||
| Vtg Liver | ERαS Liver | 0.56 | 1 | 38.95 | < 0.0001 | 2 | 8.11 | < 0.01 |
| Vtg Liver | ERαL Liver | 0.52 | 1 | 40.99 | < 0.0001 | 2 | 19.34 | < 0.01 |
| AroB Brain | ERαS Brain | 0.32 | NS | 2 | 7.57 | < 0.01 | ||
| AroB Brain | ERαL Brain | 0.29 | NS | 2 | 6.75 | < 0.01 | ||
| Total ERα Liver | ERαS Liver | 0.86 | 1 | 111.43 | < 0.0001 | NS | ||
| Total ERα Liver | ERαL Liver | 0.73 | 1 | 41.65 | < 0.0001 | NS | ||
| Total ERα Brain | ERαS Brain | 0.17 | 1 | 6.54 | < 0.05 | NS | ||
| Total ERα Brain | ERαL Brain | 0.35 | 1 | 15.92 | < 0.001 | NS | ||
| Total ERα Testis | ERαS Testis | 0.56 | 1 | 7.08 | < 0.05 | 2 | 8.46 | < 0.01 |
| Total ERα Testis | ERαL Testis | 0.54 | 1 | 5.73 | < 0.05 | 2 | 5.32 | < 0.05 |
| Total ERα Ovaries | ERαS Ovaries | 0.89 | 1 | 110.38 | < 0.0001 | NS | ||
| Total ERα Ovaries | ERαL Ovaries | 0.87 | 1 | 93.16 | < 0.0001 | NS | ||
Table 5.
Effects tests of the relations between mRNAs of estrogen responsive genes and mRNAs of ERα variants in different population pairs, as determined by analysis of covariance (ANCOVA). For regression plots see Figure 7 and Supplementary Figure 3. SC = all Scorton Creek killifish; NBH+del = New Bedford Harbor killifish expressing ERαΔ6 and/or ERαΔ6–8 deletion variants; NBH-del = all other NBH killifish excluding NBH+del. NS = not significant (P > 0.05).
| Tissue | Variables | Relation | SC v. NBH-del | SC v. NBH+del | NBH-del v. NBH+del | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| df | F | P | df | F | P | df | F | P | |||
| Liver | Vtg/ERαS | Population | 1 | 19.48 | 0.0001 | 1 | 5.03 | < 0.05 | NS | ||
| Liver | Vtg/ERαL | Population | 1 | 13.74 | < 0.001 | NS | NS | ||||
| Brain | AroB/ERαS | Population | 1 | 16.58 | < 0.001 | 1 | 4.8 | < 0.05 | NS | ||
| Brain | AroB/ERαL | Population | 1 | 14.99 | < 0.001 | 1 | 1.21 | < 0.05 | NS | ||
| Testis | ERα/ERαS | Population | NS | 1 | 18.1 | < 0.01 | 1 | 6.46 | < 0.05 | ||
| Testis | ERα/ERαL | Population | NS | 1 | 6.33 | < 0.05 | 1 | 14.15 | < 0.01 | ||
Figure 7. Relationship between mRNAs of ERαS (A, C, E) or ERαL (B, D, F) and estrogen responsive mRNAs (Vtg: A, B; AroB: C, D; total ERα: E, F) in individual killifish from SC (all) as compared to NBH fish expressing ERαΔ6 and/or ERαΔ6–8 ERα deletion variants (NBH+del) or all other remaining NBH fish (NBH-del).
Adult male and female killifish from NBH or SC were injected with estradiol or vehicle alone and tissues processed for qPCR analysis (see legend to Figure 4). Results were expressed as MNE and log transformed. Analysis of covariance (ANCOVA) was applied separately by site and the presence of deletion variants (SC, NBH+del and NBH-del). For each estrogen responsive mRNA (Y-axis, dependent variable), ERαS or ERαL mRNA was plotted on the X-axis (independent variable). See Table 4 for statistical analysis of the relationships, and Table 5 for significance between regression lines for different population pairs.
As shown in Figure 7 and Supplementary Figure 3, the expression of Vtg mRNA in liver, and total ERα mRNA in liver, brain, testis and ovaries, were all significantly related to the expression of ERαS and ERαL in the same tissue. The relationship between each of the 5′-end variants and Vtg was also significantly related to population (different intercepts), but there was no significant interaction between ERαS/ERαL expression and population (similar slopes) (Figure 7A, B). Specifically, the Vtg/ERαS relationship in SC fish differed significantly from that in both NBH+del and NBH-del fish and, although the Vtg/ERαs relationship did not differ significantly between NBH-del and NBH+del, expression of the deletion variant (NBH+del) tended to shift the intercepts closer to those of the SC population (e.g., attenuated estrogen hyper-responsiveness; Figure 7A). In marked contrast to the Vtg/ERαs relationship, there was no significant relation between the expression of AroB and either ERαS or ERαL in brain of the killifish examined (Figure 7C/D) but AroB expression was related to population. Although expression of total ERα, ERαS or ERαL in liver, brain, and ovary was not significantly related to population (Supplementary Figure 3), testicular expression of these transcripts was significantly related to population, as revealed by different intercepts, but not to an interaction between either of the ERα isoforms and population, as demonstrated by similar slopes for all regression lines (Figure 7E/F). These population effects were specific to the NBH+del population, with no significant differences seen between SC and NBH-del (Table 5).
4. Discussion
To explain the reproductive success of the NBH killifish population, despite intense long-term multi-generational exposure to estrogenic environmental pollutants and evidence of ongoing endocrine disruption, we hypothesized that adaptations in ER-mediated signaling pathways somehow attenuate or neutralize the effects of estrogenic chemicals. In earlier studies, no population-related differences were found in the coding sequences of the three killifish esr genes, or in basal or estrogen induced levels of ERα, –βa or βb mRNAs, that could be interpreted as adaptive (see Introduction). Here we investigated the possibility that dysregulation of normal gene splicing patterns, by altering the spectrum of functionally relevant ER transcripts, could account for maladaptive (endocrine disrupting) as well as adaptive (tolerance, resistance) effects of environmental xenoestrogens. Using a targeted PCR cloning approach, we systematically documented ERα variants in the two killifish populations. A total of nineteen splice variants were identified, most of which were low in abundance and presumably functionally irrelevant. However, two major alternatively spliced 5′-end transcripts, predicting a full length (long, ERαL) and an N-truncated (short, ERαS) isoform, were identified in both NBH and SC fish. In addition, two deletion variants (ERαΔ6–8, ERαΔ6), lacking some or all of the exons encoding functionally critical LBD/AF-2 domains, were unique to a subset of individuals in the NBH population and, where expressed, were relatively abundant as compared to the corresponding exon-retained forms. To understand how these variants might contribute to endocrine disruption or adaptation, we used variant-specific PCR primers to compare expression by tissue type, sex, reproductive condition and estrogen treatment in the two populations, and applied correlational analysis to examine the relationship between expressed levels of the key 5′-and 3′-end ERα variants and estrogen responses as measured by markers of estrogen exposure and effect (ERα, Vtg, AroB) in individual NBH and SC killifish.
4.1 Alternative 5′end ERα transcripts
In addition to the previously cloned “short” ERα transcript (Greytak and Callard, 2007; Urushitani et al., 2003), we identified a second alternatively spliced “long” ERα transcript in killifish. Production of ERαL and ERαS mRNAs in killifish is consistent with findings in rainbow trout (Pakdel et al., 2000) and zebrafish (Cotter et al., 2013) and, together with a survey of publically available ERα mRNA sequences in forty-nine taxonomically diverse fish species (Cotter et al., 2013), indicates that alternative splicing at the 5′ end of the esr1 transcript is a conserved feature of teleosts (see Introduction). In killifish, ERαL and ERαS mRNAs are produced by inclusion or skipping of sequence at the proximal end of exon 2 but originate from a single transcription start site, implying a single promoter and regulatory region. By contrast, alternatively spliced long and short transcripts in trout and zebrafish are produced from two transcription start sites, which is consistent with distinct promoters and regulatory regions. In silico translation of long and short transcripts in all three species predicts use of two different ATG codons and is supported by the sizes of the in vitro translated proteins. Functional analysis of the long and short rainbow trout transcripts after transfection into mammalian cells demonstrates that ERαL is activated only in the presence of ligand, whereas ERαS displays some ligand-independent activity but its maximal transactivation activity is higher (Pakdel et al., 2000). Further studies indicate that the A domain of the rainbow trout long isoform is responsible for suppressing its constitutive ligand-independent activity (Metivier et al., 2000; Métivier et al., 2002), a function that was also demonstrated with human ERα (Nicol-Benoit et al., 2011). Thus, based on ERα structure alone, it is unclear whether the long or the short isoform would be adaptive in an estrogenic environment.
To further understand the potential functional significance of the two N-terminal isoforms in killifish, we designed variant specific primers to measure ERαS and ERαL mRNAs by qPCR in different tissue types from reproductively active and inactive male and female fish at the two sites, and also tested the effects of estradiol treatment. Primers that targeted a region common to both long and short transcripts were used as a control to estimate overall esr1 expression (“total” ERα mRNA). In general, expressed levels of ERαS and ERαL mRNAs reflect differences in total ERα mRNA, which confirm results of our previous studies (Greytak and Callard, 2007; Greytak et al., 2010). Where the two transcripts differ is in the magnitude of tissue-specific differences, and the degree of change when different physiological conditions (male/female, reproductive activity/inactivity, estrogen treatment) and fish populations (SC/NBH) are compared. For example, the tissue distribution of total ERα and ERαS in killifish (female liver > male liver = ovary > other tissues) agrees with our previous analysis of total ERα and ERαS in zebrafish (Cotter et al., 2013). Although the distribution of ERαL to some extent reflects the more robust tissue differences of ERαS in killifish, ERαL mRNA does not differ significantly in different tissue types in zebrafish. These species differences are likely due to the fact that ERαS and ERαL share a single transcription start site in killifish while the two mRNAs utilize separate transcription start sites in zebrafish, and further suggest that tissue factors selectively regulate production of ERαS. Similar to tissue-specific patterns of expression, comparison of expressed levels of ERαS are significantly related to reproductive status, sex or site whereas ERαL mRNA shows little or no change with physiological status.
In zebrafish embryos, ERαS but not ERαL is robustly estrogen-inducible (Cotter et al., 2013), strong evidence that the short isoform is the main component of the autoregulatory feedback loop that advances and amplifies estrogenic responses. We infer from this that promoter switching from the short to the long transcript is a mechanism that could attenuate the autoregulatory feedback loop, which would be adaptive in an estrogenic environment. Contrary to this prediction, both ERαS and ERαL are upregulated by estradiol in killifish at the two sites. Although basal and induced levels of ERαL are lower than ERαS, they generally reflect changes in total ERα, again, consistent with a shared transcription start site.
4.2 3′-end ERα deletion variants
Two of the nineteen internal deletion variants identified, namely, ERαΔ6 and ERαΔ6–8, were cloned exclusively from NBH killifish. In silico translation of these variants indicates a frame-shift after exon 5 and predicts proteins missing the entirety of the E and F domains, including the functionally important LBD and AF-2 domains. These variants were not identified in zebrafish (Cotter et al., 2013), but a similar variant (ERΔE5) with a deletion in exon 5 (corresponding to exon 6 in killifish) has been identified in humans and also results in a frame-shift that translates to a protein similar in size to the killifish ERαΔ6–8 (Bollig and Miksicek, 2000). The human ERΔE5 variant displays a complete inability to bind ligand and, although it lacks transactivating activity of its own, it functions as a dominant negative when co-transfected with full-length ERα. Interestingly, a 3′-end deletion variant (ERαx) previously identified in killifish (Greytak and Callard, 2007) and zebrafish (Cotter et al., 2013) is affected in the E and F domains and corresponds to a splice variant termed ERαΔ7 in human (Herynk and Fuqua, 2004).
Using a PCR approach to co-amplify normal and variant products at the 3′-end, we found that ERαΔ6 and ERαΔ6–8 are expressed in a subset of individuals in the NBH killifish population and are relatively abundant compared to the exon-retained normal product in the same fish. Of twenty vehicle and estrogen-treated male and female NBH fish examined, a total of eight expressed variable amounts of one or both of these deletion variants as indicated by band intensity. Interestingly, individuals expressing ERαΔ6 and/or ERαΔ6–8 mRNA in the liver expressed the same forms in the testis or ovary. This suggests that the 3′-end deletion variants result from a genome-wide change in factors or mechanisms controlling ERα pre-mRNA processing rather than from tissue-, sex- or estrogen-specific regulatory factors. Neither our earlier analysis of exonic sequences (Greytak and Callard, 2007), nor our preliminary analysis of intronic sequences (unpublished data), of esr1 in NBH killifish identified sequence changes that could be implicated in splicing control, but global changes in gene splicing could also be controlled by changes in exonic DNA methylation (Maunakea et al., 2013) or by genetic or epigenetic changes altering the expression of serine/arginine-rich (SR) proteins or other splicing regulatory proteins (Chen and Manley, 2009).
4.3 Relation between ERα variants and estrogen responses in SC and NBH killifish
Based on lower basal levels of ERα mRNA in several tissue types of NBH killifish (Greytak et al., 2010), we initially postulated that tolerance to an estrogenic environment could be explained by acquired hypo-responsiveness to estrogen. Indeed, results here show that the response to administered estrogen is reduced in NBH fish when measured as upregulation of total ERα mRNA (but not ERαS and ERαL). Nonetheless, we confirm here that NBH fish are actually hyper-responsive to estrogen when Vtg is measured in liver and AroB is measured in brain. Although induction of AroB is strictly ERβb mediated, estrogen upregulation of ERα and Vtg mRNAs are ERα-mediated, at least in zebrafish (Griffin et al., 2013). Also, the normal or exaggerated estrogen responses we record in adult male and female killifish are in contrast to the hypo-responsiveness seen when ERα and AroB mRNAs are measured in NBH embryos (Greytak et al., 2010), and when a variety of estrogen markers are measured in killifish from other polluted sites (Bugel et al., 2014; Bugel et al., 2011).
Here and in our previous studies (Greytak et al., 2010) we noted the high degree of variability in the estrogen responses of killifish in general and NBH fish in particular, which may indicate evolving pre-adaptive or adaptive mechanisms. Killifish are known to be highly polymorphic (Burnett et al., 2007), and variability would not be surprising if we consider environmental estrogens as agents of selection for adaptations in estrogen signaling pathways. Consistent with this idea, some but not all NBH killifish express ERα deletion variants with potential to function as dominant negatives.
For all tissues examined using ANCOVA, the expressed levels of ERαS and ERαL were significantly correlated with expressed levels of total ERα mRNA, indicating that there is little regulation of either isoform that is independent of regulation of total ERα mRNA. Expressed levels of Vtg mRNA were also significantly correlated with ERα isoform expression, while the expression of AroB was not. This is consistent with results of our study using morpholino technology to demonstrate that estrogen upregulation of Vtg is mediated, at least in part, by ERα while AroB upregulation is mediated exclusively by ERβ b in zebrafish embryos (Griffin et al., 2013).
Interestingly, significant population effects are revealed by ANCOVA but these are manifested not as differences in slope but as differences in the position of the regression line on the scatter plot (y-intercept). To illustrate, in SC and NBH killifish, every increment in ERαS expression is significantly related to an increase in Vtg expression; however, for each increment in ERαS mRNA there is greater amount of Vtg expressed in NBH than in SC fish. Most importantly, for the relationship between the short and long isoforms and Vtg expression, displacement of the regression line on the scatter plot relative to the SC controls is greater in fish not expressing the deletion variants ERαΔ6 or ERαΔ6–8 (NBH-del) than in killifish in which the variants are present (NBH+del). This suggests that the presence of the variants somehow “normalizes” the ERα/Vtg relationship. Whether ERαΔ6 or ERαΔ6–8 function as dominant negatives in vivo, thereby reducing availability of transcriptionally active ERα, and perhaps the ERβa and ERβb, requires further study.
A possible explanation for the observed population differences in the ERα/Vtg relationship and the hyper-responsiveness of Vtg to estrogen may lie with transcriptional memory as a result of the long-term exposure of the NBH fish to their estrogenic environment. It has been thoroughly documented that prior estrogen exposure substantially increases the rate and magnitude of Vtg upregulation after subsequent exposures, and this phenomenon is true for chickens (Beuving and Gruber, 1971), frogs (Baker and Shapiro, 1978), and fish (Le Guellec et al., 1988). Although the exact mechanism of the Vtg memory effect remains a matter of debate, a recent study in rainbow trout implicates the short ERα isoform in the maintenance of transcriptional memory (Nicol-Benoit et al., 2011). Further, there are numerous studies demonstrating that developmental exposure to EDCs affects estrogen responsiveness later in life (Ceccatelli et al., 2006; Maerkel et al., 2005; Newbold et al., 2004; Roepke et al., 2006) and some of these effects could be mediated by epigenetic changes (Bromer et al., 2010).
An alternative explanation for the observed population differences in the ERα/Vtg relationship and the hyperresponsiveness of Vtg to estrogen is that it compensates for an inhibitory factor in the NBH environment. In mice, AhR transcriptionally represses ERα (Tian et al., 1998), and previous studies have established that there are dioxin-like chemicals and changes in the AhR signaling pathway in NBH killifish (Bello et al., 2001; Nacci et al., 1999; Powell et al., 2000). It remains to be determined whether there are differences in the estrogen response of NBH fish that have been depurated of PCBs and other persistent and bioaccumulative chemicals that may act as EDCs.
It has been previously demonstrated that alternative splicing can act as a mechanism for adaptation to various environmental pressures. In plants, this phenomenon has been very well studied, and changes in splicing have been implicated in adaptation to changes in photoperiod, temperature, salt stress, and disease resistance (Ding et al., 2014; Kwon et al., 2014; Yang et al., 2014). Alternative splicing has also been associated with pesticide resistance in several insect species (Fabrick et al., 2014; He et al., 2012; Xu et al., 2005) and has been widely linked to the adaptive resistance of multiple cancer types to chemotherapy and other treatments (Dehm, 2013; Krett et al., 2013; Sprenger and Plymate, 2014).
In conclusion, these results demonstrate that there is a significant number of alternatively spliced ERα mRNAs in killifish, which are differentially regulated in various tissues and in response to estrogen. Altered expression of these mRNAs, including the induction of two novel NBH-specific variants, could reflect adaptive responses to multi-generational exposures to xenoestrogens and other pollutants. Although formal proof of the dominant negative functions of these variants in vitro and in vivo is required, our results strongly support the idea that deletion variants are negatively regulating (attenuating/neutralizing) the hyper-responsiveness of this population. It is interesting to speculate that the presence of 3′-end deletion variants in some but not all NBH killifish indicates that the population is still evolving. In this context, it will be important to determine if mechanisms that result in the NBH-specific deletion variants are heritable and, if so, whether heritability is genetic or epigenetic in nature. The NBH killifish provide a unique and valuable resource to study molecular mechanisms of adaptation. Their responses to further environmental changes such as remediation or many other factors (e.g., climate change) provides a continually evolving “natural” experiment.
Supplementary Material
Highlights.
Nineteen ERα variants were identified in killifish from NBH and SC
ERαS and ERαL differed by tissue type, site, sex, and reproductive status
ERαS was hypo-responsive to estrogen in NBH fish
Vtg and AroB were hyper-responsive to estrogen in NBH fish
Variants lacking E/AF2 domains were linked to ERαS hypo-responsiveness in NBH fish
Acknowledgments
We thank Ann Tarrant (Woods Hole Oceanographic Institute) for help with the in vitro translation of our variants, and Peter Buston (Boston University) for guidance with the statistical analyses. This work was supported by the National Institutes for Environmental Health Sciences (NIEHS P42ES07381), the US Environmental Protection Agency National Center for Environmental Research (NCER) STAR Program (RD831301), a Boston University, Warren-McLeod Fellowship for Graduate Research in Marine Biology (KC), an EPA STAR Fellowship (FP917445, KC) and a National Science Foundation grant (DEB-1265282; PI, Andrew Whitehead, UC Davis) in support of the killifish genome project.
Footnotes
Abbreviations: AhR, aryl hydrocarbon receptor; AroB, brain aromatase or cytochrome P450 19b; Cyp1a, cytochrome P450 1a; elfa, gene encoding elongation factor alpha; EDC, endocrine disrupting chemical; ER, estrogen receptor; esr1/ESR1, gene encoding ER alpha in teleosts/mammals; NBH, New Bedford Harbor, MA; PAH, polycyclic aromatic hydrocarbons; PCB, polychlorinated biphenyl; qPCR, quantitative PCR; SC, Scorton Creek, MA; SNP, single nucleotide polymorphism; UTR, untranslated region; Vtg, vitellogenin
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aluru N, Karchner SI, Hahn ME. Role of DNA methylation of AHR1 and AHR2 promoters in differential sensitivity to PCBs in Atlantic Killifish, Fundulus heteroclitus. Aquatic Toxicology. 2011;101:288–294. doi: 10.1016/j.aquatox.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auboeuf D, Dowhan DH, Kang YK, Larkin K, Lee JW, Berget SM, O’Malley BW. Differential recruitment of nuclear receptor coactivators may determine alternative RNA splice site choice in target genes. Proceedings of the National Academy of Sciences USA. 2004;101:2270–2274. doi: 10.1073/pnas.0308133100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker HJ, Shapiro DJ. Rapid accumulation of vitellogenin messenger RNA during secondary estrogen stimulation of Xenopus laevis. The Journal of Biological Chemistry. 1978;253:4521–4524. [PubMed] [Google Scholar]
- Bello SM, Franks DG, Stegeman JJ, Hahn ME. Acquired resistance to Ah receptor agonists in a population of Atlantic killifish (Fundulus heteroclitus) inhabiting a marine superfund site: in vivo and in vitro studies on the inducibility of xenobiotic metabolizing enzymes. Toxicological Sciences. 2001;60:77–91. doi: 10.1093/toxsci/60.1.77. [DOI] [PubMed] [Google Scholar]
- Benotti MJ, Brownawell BJ. Distributions of pharmaceuticals in an urban estuary during both dry- and wet-weather conditions. Environmental Science & Technology. 2007;41:5795–5802. doi: 10.1021/es0629965. [DOI] [PubMed] [Google Scholar]
- Beuving G, Gruber M. Induction of phosvitin synthesis in roosters by estradiol injection. Biochimica et Biophysica Acta. 1971;232:529–536. doi: 10.1016/0005-2787(71)90607-1. [DOI] [PubMed] [Google Scholar]
- Bollig A, Miksicek RJ. An estrogen receptor-α splicing variant mediates both positive and negative effects on gene transcription. Molecular Endocrinology. 2000;14:634–649. doi: 10.1210/mend.14.5.0460. [DOI] [PubMed] [Google Scholar]
- Bromer JG, Zhou Y, Taylor MB, Doherty L, Taylor HS. Bisphenol-A exposure in utero leads to epigenetic alterations in the developmental programming of uterine estrogen response. The FASEB Journal. 2010;24:2273–2280. doi: 10.1096/fj.09-140533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugel SM, Bonventre JA, White LA, Tanguay RL, Cooper KR. Chronic exposure of killifish to a highly polluted environment desensitizes estrogen-responsive reproductive and biomarker genes. Aquatic Toxicology. 2014;152:222–231. doi: 10.1016/j.aquatox.2014.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugel SM, White LA, Cooper KR. Decreased vitellogenin inducibility and 17β-estradiol levels correlated with reduced egg production in killifish (Fundulus heteroclitus) from Newark Bay, NJ. Aquatic Toxicology. 2011;105:1–12. doi: 10.1016/j.aquatox.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett KG, Bain LJ, Baldwin WS, Callard GV, Cohen S, Di Giulio RT, Evans DH, Gomez-Chiarri M, Hahn ME, Hoover CA, Karchner SI, Katoh F, Maclatchy DL, Marshall WS, Meyer JN, Nacci DE, Oleksiak MF, Rees BB, Singer TD, Stegeman JJ, Towle DW, Van Veld PA, Vogelbein WK, Whitehead A, Winn RN, Crawford DL. Fundulus as the premier teleost model in environmental biology: opportunities for new insights using genomics. Comparative Biochemistry and Physiology. Part D, Genomics & Proteomics. 2007;2:257–286. doi: 10.1016/j.cbd.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccatelli R, Faass O, Schlumpf M, Lichtensteiger W. Gene expression and estrogen sensitivity in rat uterus after developmental exposure to the polybrominated diphenylether PBDE 99 and PCB. Toxicology. 2006;220:104–116. doi: 10.1016/j.tox.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nature Reviews Molecular Cell Biology. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter KA, Yershov A, Novillo A, Callard GV. Multiple structurally distinct ERα mRNA variants in zebrafish are differentially expressed by tissue type, stage of development and estrogen exposure. General and Comparative Endocrinology. 2013;194:217–229. doi: 10.1016/j.ygcen.2013.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCastro BR, Korrick SA, Spengler JD, Soto AM. Estrogenic activity of polychlorinated biphenyls present in human tissue and the environment. Environmental Science & Technology. 2006;40:2819–2825. doi: 10.1021/es051667u. [DOI] [PubMed] [Google Scholar]
- Dehm SM. mRNA splicing variants: exploiting modularity to outwit cancer therapy. Cancer Research. 2013;73:5309–5314. doi: 10.1158/0008-5472.CAN-13-0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamanti-Kandarakis E, Bourguignon JP, Giudice LC, Hauser R, Prins GS, Soto AM, Zoeller RT, Gore AC. Endocrine-disrupting chemicals: an Endocrine Society scientific statement. Endocrine Reviews. 2009;30:293–342. doi: 10.1210/er.2009-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F, Cui P, Wang Z, Zhang S, Ali S, Xiong L. Genome-wide analysis of alternative splicing of pre-mRNA under salt stress in Arabidopsis. BMC Genomics. 2014;15:431. doi: 10.1186/1471-2164-15-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrick JA, Ponnuraj J, Singh A, Tanwar RK, Unnithan GC, Yelich AJ, Li X, Carrière Y, Tabashnik BE. Alternative splicing and highly variable cadherin transcripts associated with field-evolved resistance of pink bollworm to Bt cotton in India. PLoS ONE. 2014;9:e97900. doi: 10.1371/journal.pone.0097900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greytak SR, Callard GV. Cloning of three estrogen receptors (ER) from killifish (Fundulus heteroclitus): differences in populations from polluted and reference environments. General and Comparative Endocrinology. 2007;150:174–188. doi: 10.1016/j.ygcen.2006.07.017. [DOI] [PubMed] [Google Scholar]
- Greytak SR, Champlin D, Callard GV. Isolation and characterization of two cytochrome P450 aromatase forms in killifish (Fundulus heteroclitus): differential expression in fish from polluted and unpolluted environments. Aquatic Toxicology. 2005;71:371–389. doi: 10.1016/j.aquatox.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Greytak SR, Tarrant AM, Nacci D, Hahn ME, Callard GV. Estrogen responses in killifish (Fundulus heteroclitus) from polluted and unpolluted environments are site- and gene-specific. Aquatic Toxicology. 2010;99:291–299. doi: 10.1016/j.aquatox.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin LB, January KE, Ho KW, Cotter KA, Callard GV. Morpholino-mediated knockdown of ERα, ERβa, and ERβb mRNAs in zebrafish (Danio rerio) embryos reveals differential regulation of estrogen-inducible genes. Endocrinology. 2013;154:4158–4169. doi: 10.1210/en.2013-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn ME, Karchner SI, Franks DG, Merson RR. Aryl hydrocarbon receptor polymorphisms and dioxin resistance in Atlantic killifish (Fundulus heteroclitus) Pharmacogenetics. 2004;14:131–143. doi: 10.1097/00008571-200402000-00007. [DOI] [PubMed] [Google Scholar]
- Hawkins MB, Thornton JW, Crews D, Skipper JK, Dotte A, Thomas P. Identification of a third distinct estrogen receptor and reclassification of estrogen receptors in teleosts. Proceedings of the National Academy of Sciences USA. 2000;97:10751–10756. doi: 10.1073/pnas.97.20.10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Li T, Zhang L, Liu N. Multiple sodium channel variants in the mosquito Culex quinquefasciatus. International Journal of Biological Sciences. 2012;8:1291. doi: 10.7150/ijbs.4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldring N, Pike A, Andersson S, Matthews J, Cheng G, Hartman J, Tujague M, Ström A, Treuter E, Warner M, Gustafsson J-Å. Estrogen receptors: how do they signal and what are their targets. Physiolgical Reviews. 2007;87:905–931. doi: 10.1152/physrev.00026.2006. [DOI] [PubMed] [Google Scholar]
- Herynk MH, Fuqua SA. Estrogen receptor mutations in human disease. Endocrine Reviews. 2004;25:869–898. doi: 10.1210/er.2003-0010. [DOI] [PubMed] [Google Scholar]
- Krett NL, Ma S, Rosen ST. Clinical perspective on chemo-resistance and the role of RNA processing. In: Wu JY, editor. RNA and Cancer. Springer; Berlin Heidelberg: 2013. pp. 235–245. [DOI] [PubMed] [Google Scholar]
- Krust A, Green S, Argos P, Kumar V, Walter P, Bornert JM, Chambon P. The chicken oestrogen receptor sequence: homology with v-erbA and the human oestrogen and glucocorticoid receptors. The EMBO Journal. 1986;5:891–897. doi: 10.1002/j.1460-2075.1986.tb04300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YJ, Park MJ, Kim SG, Baldwin I, Park CM. Alternative splicing and nonsense-mediated decay of circadian clock genes under environmental stress conditions in Arabidopsis. BMC Plant Biology. 2014;14:136. doi: 10.1186/1471-2229-14-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guellec K, Lawless K, Valotaire Y, Kress M, Tenniswood M. Vitellogenin gene expression in male rainbow trout (Salmo gairdneri) General and Comparative Endocrinology. 1988;71:359–371. doi: 10.1016/0016-6480(88)90264-x. [DOI] [PubMed] [Google Scholar]
- Maerkel K, Lichtensteiger W, Durrer S, Conscience M, Schlumpf M. Sex-and region-specific alterations of progesterone receptor mRNA levels and estrogen sensitivity in rat brain following developmental exposure to the estrogenic UV filter 4-methylbenzylidene camphor. Environmental Toxicology and Pharmacology. 2005;19:761–765. doi: 10.1016/j.etap.2004.12.055. [DOI] [PubMed] [Google Scholar]
- Maunakea AK, Chepelev I, Cui K, Zhao K. Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Research. 2013;23:1256–1269. doi: 10.1038/cr.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCurley A, Callard G. Characterization of housekeeping genes in zebrafish: male-female differences and effects of tissue type, developmental stage and chemical treatment. BMC Molecular Biology. 2008;9:102. doi: 10.1186/1471-2199-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menuet A, Anglade I, Flouriot G, Pakdel F, Kah O. Tissue-specific expression of two structurally different estrogen receptor alpha isoforms along the female reproductive axis of an oviparous species, the rainbow trout. Biology of Reproduction. 2001;65:1548–1557. doi: 10.1095/biolreprod65.5.1548. [DOI] [PubMed] [Google Scholar]
- Menuet A, Le Page Y, Torres O, Kern L, Kah O, Pakdel F. Analysis of the estrogen regulation of the zebrafish estrogen receptor (ER) reveals distinct effects of ERα, ERβ1 and ERβ2. Journal of Molecular Endocrinology. 2004;32:975–986. doi: 10.1677/jme.0.0320975. [DOI] [PubMed] [Google Scholar]
- Metivier R, Petit FG, Valotaire Y, Pakdel F. Function of N-terminal transactivation domain of the estrogen receptor requires a potential alpha-helical structure and is negatively regulated by the A domain. Molecular Endocrinology. 2000;14:1849–1871. doi: 10.1210/mend.14.11.0546. [DOI] [PubMed] [Google Scholar]
- Métivier R, Stark A, Flouriot G, Hübner MR, Brand H, Penot G, Manu D, Denger S, Reid G, Koš M, Russell RB, Kah O, Pakdel F, Gannon F. A dynamic structural model for estrogen receptor-α activation by ligands, emphasizing the role of interactions between distant A and E domains. Molecular Cell. 2002;10:1019–1032. doi: 10.1016/s1097-2765(02)00746-3. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Nacci D, Coiro L, Champlin D, Jayaraman S, McKinney R, Gleason TR, Munns WR, Jr, Specker JL, Cooper KR. Adaptations of wild populations of the estuarine fish Fundulus heteroclitus to persistent environmental contaminants. Marine Biology. 1999;134:9–17. [Google Scholar]
- Nacci DE, Champlin D, Jayaraman S. Adaptation of the estuarine fish Fundulus heteroclitus (Atlantic killifish) to polychlorinated biphenyls (PCBs) Estuaries and Coasts. 2010;33:853–864. [Google Scholar]
- Newbold RR, Jefferson WN, Padilla-Banks E, Haseman J. Developmental exposure to diethylstilbestrol (DES) alters uterine response to estrogens in prepubescent mice: low versus high dose effects. Reproductive Toxicology. 2004;18:399–406. doi: 10.1016/j.reprotox.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Nicol-Benoit F, Amon A, Vaillant C, le Goff P, le Dréan Y, Pakdel F, Flouriot G, Valotaire Y, Michel D. A dynamic model of transcriptional imprinting derived from the vitellogenesis memory effect. Biophysical Journal. 2011;101:1557–1568. doi: 10.1016/j.bpj.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleksiak MF, Karchner SI, Jenny MJ, Franks DG, Welch DB, Hahn ME. Transcriptomic assessment of resistance to effects of an aryl hydrocarbon receptor (AHR) agonist in embryos of Atlantic killifish (Fundulus heteroclitus) from a marine Superfund site. BMC Genomics. 2011;12:263. doi: 10.1186/1471-2164-12-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pait AS, Nelson JO. Vitellogenesis in male Fundulus heteroclitus (killifish) induced by selected estrogenic compounds. Aquatic Toxicology. 2003;64:331–342. doi: 10.1016/s0166-445x(03)00060-2. [DOI] [PubMed] [Google Scholar]
- Pakdel F, Metivier R, Flouriot G, Valotaire Y. Two estrogen receptor (ER) isoforms with different estrogen dependencies are generated from the trout ER gene. Endocrinology. 2000;141:571–580. doi: 10.1210/endo.141.2.7296. [DOI] [PubMed] [Google Scholar]
- Patino R, Xia Z, Gale WL, Wu C, Maule AG, Chang X. Novel transcripts of the estrogen receptor alpha gene in channel catfish. General and Comparative Endocrinology. 2000;120:314–325. doi: 10.1006/gcen.2000.7566. [DOI] [PubMed] [Google Scholar]
- Pinto PI, Teodosio R, Socorro S, Power DM, Canario AV. Structure, tissue distribution and estrogen regulation of splice variants of the sea bream estrogen receptor alpha gene. Gene. 2012;503:18–24. doi: 10.1016/j.gene.2012.04.081. [DOI] [PubMed] [Google Scholar]
- Ponglikitmongkol M, Green S, Chambon P. Genomic organization of the human oestrogen receptor gene. The EMBO journal. 1988;7:3385–3388. doi: 10.1002/j.1460-2075.1988.tb03211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell WH, Bright R, Bello SM, Hahn ME. Developmental and tissue-specific expression of AHR1, AHR2, and ARNT2 in dioxin-sensitive and -resistant populations of the marine fish Fundulus heteroclitus. Toxicological Sciences. 2000;57:229–239. doi: 10.1093/toxsci/57.2.229. [DOI] [PubMed] [Google Scholar]
- Proestou DA, Flight P, Champlin D, Nacci D. Targeted approach to identify genetic loci associated with evolved dioxin tolerance in Atlantic killifish (Fundulus heteroclitus) BMC Evolutionary Biology. 2014;14:7. doi: 10.1186/1471-2148-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzel AM, Karchner SI, Franks DG, Evans BR, Nacci D, Champlin D, Vieira VM, Hahn ME. Genetic variation at aryl hydrocarbon receptor (AHR) loci in populations of Atlantic killifish (Fundulus heteroclitus) inhabiting polluted and reference habitats. BMC Evolutionary Biology. 2014;14:6. doi: 10.1186/1471-2148-14-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roepke TA, Chang ES, Cherr GN. Maternal exposure to estradiol and endocrine disrupting compounds alters the sensitivity of sea urchin embryos and the expression of an orphan steroid receptor. Journal of Experimental Zoology. 2006;305A:830–841. doi: 10.1002/jez.a.320. [DOI] [PubMed] [Google Scholar]
- Seo JS, Lee YM, Jung SO, Kim IC, Yoon YD, Lee JS. Nonylphenol modulates expression of androgen receptor and estrogen receptor genes differently in gender types of the hermaphroditic fish Rivulus marmoratus. Biochemical and Biophysical Research Communications. 2006;346:213–223. doi: 10.1016/j.bbrc.2006.05.123. [DOI] [PubMed] [Google Scholar]
- Simon P. Q-Gene: processing quantitative real-time RT–PCR data. Bioinformatics. 2003;19:1439–1440. doi: 10.1093/bioinformatics/btg157. [DOI] [PubMed] [Google Scholar]
- Sprenger CT, Plymate S. The link between androgen receptor splice variants and castration-resistant prostate cancer. Hormones and Cancer. 2014;5:207–217. doi: 10.1007/s12672-014-0177-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan NS, Lam TJ, Ding JL. The first contiguous estrogen receptor gene from a fish, Oreochromis aureus: evidence for multiple transcripts. Molecular and Cellular Endocrinology. 1996;120:177–192. doi: 10.1016/0303-7207(96)03836-1. [DOI] [PubMed] [Google Scholar]
- Taylor SE, Martin-Hirsch PL, Martin FL. Oestrogen receptor splice variants in the pathogenesis of disease. Cancer Letters. 2010;288:133–148. doi: 10.1016/j.canlet.2009.06.017. [DOI] [PubMed] [Google Scholar]
- Tian Y, Ke S, Thomas T, Meeker RJ, Gallo MA. Transcriptional suppression of estrogen receptor gene expression by 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) The Journal of Steroid Biochemistry and Molecular Biology. 1998;67:17–24. doi: 10.1016/s0960-0760(98)00067-3. [DOI] [PubMed] [Google Scholar]
- Urushitani H, Nakai M, Inanaga H, Shimohigashi Y, Shimizu A, Katsu Y, Iguchi T. Cloning and characterization of estrogen receptor α in mummichog, Fundulus heteroclitus. Molecular and Cellular Endocrinology. 2003;203:41–50. doi: 10.1016/s0303-7207(03)00118-7. [DOI] [PubMed] [Google Scholar]
- White R, Lees JA, Needham M, Ham J, Parker M. Structural organization and expression of the mouse estrogen receptor. Molecular Endocrinology. 1987;1:735–744. doi: 10.1210/mend-1-10-735. [DOI] [PubMed] [Google Scholar]
- Whitehead A, Triant DA, Champlin D, Nacci D. Comparative transcriptomics implicates mechanisms of evolved pollution tolerance in a killifish population. Molecular Ecology. 2010;19:5186–5203. doi: 10.1111/j.1365-294X.2010.04829.x. [DOI] [PubMed] [Google Scholar]
- Xia Z, Patino R, Gale WL, Maule AG, Densmore LD. Cloning, in vitro expression, and novel phylogenetic classification of a channel catfish estrogen receptor. General and Comparative Endocrinology. 1999;113:360–368. doi: 10.1006/gcen.1999.7196. [DOI] [PubMed] [Google Scholar]
- Xu Q, Liu H, Zhang L, Liu N. Resistance in the mosquito, Culex quinquefasciatus, and possible mechanisms for resistance. Pest Management Science. 2005;61:1096–1102. doi: 10.1002/ps.1090. [DOI] [PubMed] [Google Scholar]
- Yang S, Tang F, Zhu H. Alternative splicing in plant immunity. International Journal of Molecular Sciences. 2014;15:10424–10445. doi: 10.3390/ijms150610424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y, Zhang K, Deng Y. Occurrence and photochemical degradation of 17α-ethinylestradiol in Acushnet River Estuary. Chemosphere. 2006;63:1583–1590. doi: 10.1016/j.chemosphere.2005.08.063. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.