Abstract
Epigenetic control of genome function is an important regulatory mechanism in diverse processes such as lineage commitment and environmental sensing, and in disease etiologies ranging from neuropsychiatric disorders to cancer. Here we report a robust, high-throughput targeted, quantitative mass spectrometry (MS) method to rapidly profile modifications of the core histones of chromatin that compose the epigenetic landscape, enabling comparisons among cells with differing genetic backgrounds, genomic perturbations, and drug treatments.
Keywords: Histones, Chromatin, Epigenetics, Proteomics, Mass spectrometry, SILAC
1. Introduction
The recent explosion of epigenetic research has spawned incredible interest in this layer of control on genomic functionality. Large-scale projects – executed by consortia like ENCODE [1] – have yielded a wealth of data that are beginning to provide insight into the role epigenetic phenomena play in controlling cellular processes such as gene transcription, cell fate, senescence, and pluripotency. Chemical modifications to DNA and histone proteins (“epigenetic marks”) constitute an important effector of epigenetic control. Therapeutics that modulate levels of these modifications have recently been brought to the clinic and further development of drugs in this class is ongoing.
Much of the large-scale data thus far has been derived from chromatin immunoprecipitation experiments followed by DNA sequencing (ChIP-Seq) experiments. This technique typically isolates small segments of chromatin using an antibody directed against a particular chromatin modification, transcription factor, or other DNA binding protein. Sequencing of the associated DNA reveals the genomic loci at which the targeted modification or factor was present. However, this technique is generally limited to interrogating one modification at a time.
The complementary experiment that we describe herein interrogates numerous chromatin modifications – and combinations thereof – in bulk chromatin derived from cells, but without locus determination of the originating sites. Through this technique we can derive a molecular chromatin signature of the cellular state that can be used for comparison against signatures derived from other cellular states. Differing chromatin signatures can arise due to lineage, genotype, mutation, drug treatment, or other laboratory/environmental manipulations. We recently demonstrated how this technique could be used to compare across a collection of >100 cell lines to discover a novel oncogenic mutation in the gene WHSC1 (also known as NSD2) [2].
Building on prior work [3,4], we have developed a completely targeted, quantitative MS assay suitable for profiling large numbers of cellular conditions. The key features of this assay are that it is (1) optimized to minimize spurious derivatization side reactions, (2) amenable to automation, (3) rigorously qualified via synthetic peptide standards, (4) completely deterministic with regard to the chromatin marks that are quantified, enabling a high rate of observation of specific modified histone peptides, (5) highly reproducible, enabling comparison across sample sets, acquired over extended periods of time, (6) compatible with reasonable amounts of biological material, including cultured cells, normal tissue, and tumor samples (1–2 × 106 cells, on the order of a 10 cm culture dish), and (7) high throughput (~1 h/analysis).
2. Methods
2.1. Overview
The overall workflow is depicted in Fig. 1A. Histones are extracted using classical biochemical methods [5]. Reactive amines on histone proteins (free N-termini, unmodified and monomethyl lysines) are gently propionylated using the N-hydroxyl succinimidyl (NHS) ester of propionic acid. Derivatization with this reagent reduces unwanted side reactions (such as propionylation of alcohols and methyl esterification of carboxylic acids) and can be performed using buffers compatible with laboratory automation. Use of 96-well solid phase extraction (SPE) devices to clean up the reaction enables parallelized sample processing. Derivatized histones are digested and subjected to a second round of propionylation, again under conditions conducive to automation. Final clean up is again performed in a 96-well SPE device.
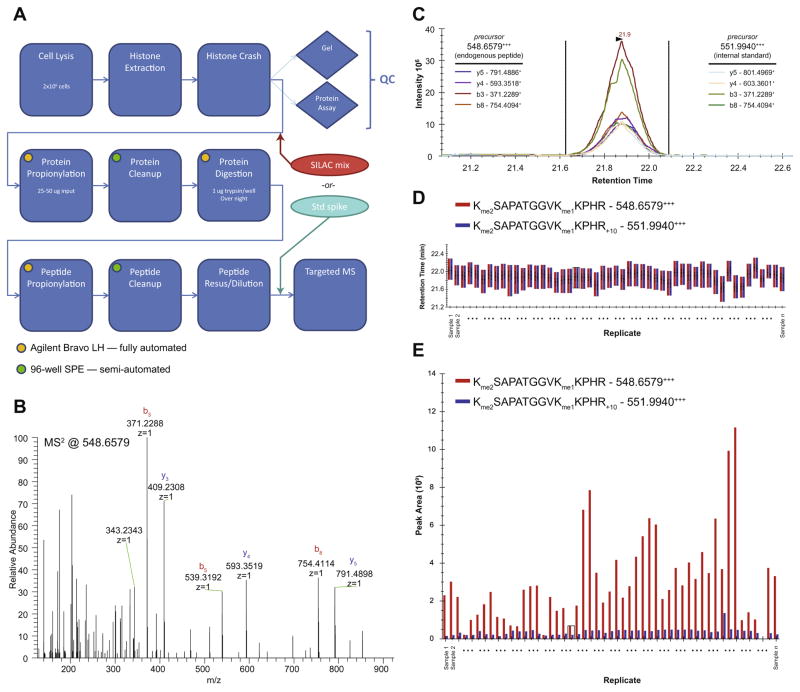
Fig. 1.
Global chromatin profiling targeted proteomics workflow and quality control. (A) Process flow diagram, illustrating points of entry for internal standards and automated steps. In panels (B–E), we follow a peptide corresponding to H3K27me2K36me1 from data acquisition through final ratio determination in a set of samples. Data shown is derived from samples analyzed in Fig. 2A. (B) A single raw MS/MS spectrum targeting a peptide precursor of m/z 548.6579, corresponding to the z = 3 state of the fully derivatized endogenous peptide. (C) Extracted ion chromatograms of MS/MS fragment ions (transitions) for both the endogenous peptide and the internal standard for a single sample in the set (extracted using Skyline). (D) Retention time view of all samples in the set for both the endogenous peptides (red) and internal standards (blue). Consistency illustrates the reproducibility of the chromatographic dimension of the assay. (E) Calculated ion abundances for both the endogenous peptides (red) and internal standards (blue) of all samples in the set, as derived from areas under the curve in (C). Ratios are determined by dividing the ion abundances of the endogenous peptides (red) by the internal standards (blue).
The introduction of internal standards is important to enable comparison across many samples. Internal standards may be introduced at the protein level via SILAC-labeled histones, as described previously [2], or at the peptide level via addition of isotope-labeled synthetic peptides.
For SILAC standardization, a cocktail of histones from 3 cell lines is employed. The principal advantages of the SILAC method are that: (1) the standards undergo nearly the entire biochemical preparation together with the samples of interest, (2) the standard is “pre-normalized” to the levels expected to be observed in the samples, and (3) potentially all chromatin mark combinations are present in the standard itself, allowing re-interrogation of the samples for new analytes (i.e., histone mark combinations) as necessary. The main disadvantages are that: (1) introduction of additional cellular material creates a more complex sample, (2) the absolute quantities of the standards are not known, and (3) it is periodically necessary to produce a new batch of standards (by growing new SILAC labeled cells) that may differ from a prior batch.
As an alternative to SILAC-labeled cells as internal standards, a library of stable isotope-labeled modified peptides for nearly all common combinations of epigenetic marks on H3 (Supplemental Table 1) was synthesized. For example, to quantify the state H3K23ac in the absence of H3K18ac, we synthesized the peptide pr-KprQLATKacAAR10, where R10 is 15N4, 13C6 and “pr” is a propionyl group. Synthesis of these peptides let us construct an unambiguous MS/MS spectral library (Supplemental Item 1) of modified histone peptides to guarantee identification of the proper modified peptide and to establish retention time coordinates for each in the LC–MS/MS assay. We have enumerated almost all common combinations on H3. The principal advantages of the synthetic peptide method are that: (1) every peptide quantified has a guaranteed correct referent, (2) the molar quantities of the peptides standards in the mixture are known, enabling back-calculation of stoichiometry and estimations of the actual amounts of analytes, and (3) the library can be regenerated or amended synthetically at any time. The main disadvantages are that: (1) the formulation of the library must be empirically titrated to match biological levels, (2) not every possible histone mark combination is currently represented, and (3) adding analytes to the assay requires synthesis of novel peptide reagents which can be difficult in the case of long or highly modified peptides.
The assay itself is executed on a high performance LCMS system. In our laboratory it is performed with a Q Exactive mass spectrometer coupled to a Proxeon Easy-nLC UHPLC, although in principle any high resolution/mass accuracy MS instrument capable of collisional fragmentation could be used. High resolution and mass accuracy in the MS/MS spectra are necessary to disambiguate near isobars like the ac and me3 modifications. The assay is implemented as a scheduled, targeted MS data acquisition method, often called an MRM or SRM assay [6]. Targeting is crucial to ensure that we observe each analyte in every sample, rather than relying on stochastic sampling properties of shotgun MS. We use our synthetic peptides or historical samples to assist with the scheduling component of the assay (see Supplemental Item 2). Raw MS data (Fig. 1B) are processed using Skyline [7], an open-source software designed for targeted MS quantification. Example and template Skyline documents for our assay are included (Supplemental Items 3 and 4). Quantification is performed primarily at the level of MS/MS fragment ions (or “transitions”), occasionally supplemented by quantification at the precursor-ion level from the MS scans. The marks H3K4me2 and H3K4me3 require precursor-level supplementation due to the low-abundance nature of transitions for these peptides, and to ensure accurate peak-picking due to their early and wide retention times. We additionally extract the precursor ion current from the H3 normalization peptide, YRPGTVALR, to ensure that similar ratios would be obtained from precursor-base and transition-based quantification. Extracted ion chromatograms (XICs) from selected transitions and precursors are generated for both the sample and the internal standard (Fig. 1C). Skyline uses the ratios of peak areas between the sample and standard of each XIC to determine the overall ratio of each modified peptide that was targeted (sample:standard). Many samples can be aggregated in a single Skyline analysis document to facilitate quality control (Fig. 1D) and comparison (Fig. 1E). Ratio data are exported from Skyline and subjected to minimal additional processing: (1) log2 transformation, (2) sample-to-standard normalization within each sample using the ratio of peptide H341–49, and (3) peptide normalization across all samples using the median ratio for each peptide.
2.2. Reagents, cell culture, and genetic perturbations
2.2.1. Synthetic peptides
All synthetic peptide standards used in the study were synthesized by New England Peptide, Inc. (Gardner, MA, USA). Each peptide bearing a C-terminal arginine residue is synthesized using 15N4, 13C6 arginine as an isotopic label. A complete list of peptides and their formulation into a master mixture can be found in Supplemental Table 1.
2.2.2. Synthesis of NHS propionate
NHS (5 g) and Propionic anhydride (5.8 mL) was dissolved in EtOAc (25 mL) and Et3N (6.1 mL) was added at room temperature slowly over a period of 30 min. Stirring was continued at room temperature for 3 h. Then, saturated NaCl solution (50 mL) was added, and the separated the organic layer was recovered and concentrated to obtain crude NHS-propionate. Hexane (100 mL) was added to this crude product and stirred for 30 min, and the resultant slurry was filtered to obtain 5 g of white solid (67% yield). The synthesis was performed by SAI Life Sciences Ltd. (Hyderabad, India).
2.2.3. SILAC cell culture of HeLa, 293T, and K562 cells
These three cell lines were used in making the R10-labeled histone cocktail or “heavy triple mix” for SILAC standardization. HeLa and K562 cells were cultured in suspension using RPMI 1640 media (Caisson, RPL17) supplemented with 5% (HeLa) or 10% (K562) dialyzed fetal bovine serum (dFBS) (Sigma, F0392), 1% penicillin/streptomycin/glutamine (PSG) (Invitrogen, 10378-016), heavy-labeled 13C6–15N4 L-arginine (R10) at 83.5 mg/L, and lysine (K0) at 40 mg/L. K562 cells were additionally supplemented with proline at 20 mg/L. 293T cells were cultured as an attached cell line using DMEM media (Caisson, DQL16) supplemented with 10% dFBS, 1% PSG, heavy-labeled 13C6–15N4 L-arginine (R10) at 58.5 mg/L, lysine at 146 mg/mL, proline at 20 mg/mL, methionine at 3 mg/mL, and glucose (Sigma, G8769) at 35.1 mg/mL.
After 5 doublings HeLa cells were harvested by centrifuging cells at 1000g for 5 min at 4°C, removing supernatant, and resuspending cells in ice-cold phosphate buffered-saline (Invitrogen, 10010-049) (PBS) in such a way that 1 mL aliquots yielded 2 × 107 cells. Aliquots were centrifuged at 1000g for 5 min at 4 °C, supernatant was removed, and resulting pellets were frozen in liquid nitrogen and stored at −80 °C. K562 cells were harvested in the same manner after 8 doublings.
293T cells were harvested after 7 doublings. Cells were harvested by removing media, washing plates twice with ice-cold PBS, and scraping cells off the plates into a small amount of ice-cold PBS. Cells were combined, centrifuged at 1000g for 5 min at 4 °C, and resuspended in ice-cold PBS in such a way that yielded 2.5 × 107 cells per aliquot. Aliquots were centrifuged at 1000g for 5 min at 4 °C, supernatants were removed, and resulting pellets were flash frozen in liquid nitrogen and stored at −80 °C.
We chose these lines to have representation from solid and hematological tumor types, and for their practicality of large-scale growth. In principle, any cell line or combination of cell lines that could be labeled using SILAC could serve as a standard. It is important to generate enough of this standard so that longitudinal comparisons of samples will be possible.
2.2.4. Knockdowns and knockouts of chromatin modulators in 293T cells and mESCs
Detailed conditions regarding shRNA knockdowns and knockouts are given in Supplemental Methods 1.
2.2.5. Drug treatments in mESCs
Mouse embryonic stem cells were cultured in mESC media (DMEM supplemented with 15% fetal bovine serum, 25 mM Hepes, pH 7.6, 0.1 mM non-essential amino acids, 100 units/mL penicillin/streptomycin, 50 μM β-mercaptoethanol, 1000 units/mL leukemia inhibitory factor) on a layer of irradiated mouse embryonic fibroblasts. Ezh2−/− [8] and G9a−/− [9] mESCs were kindly provided to Dr. Lee by S. Orkin and Y. Shinkai, respectively. For chemical inhibition of Ezh2 or G9a, J1 mESCs were grown in mESC media containing 0.5 μM GSK126 (Xcessbio) or 0.5 μM UNC0646 (Axon Medchem), respectively, for 24 h before harvesting.
2.3. Biochemical preparation of histones
2.3.1. Cell lysis and histone extraction
Pellets of at least two million cells were thawed on ice and lysed in 1 mL of ice-cold “nucleus” buffer (250 mM sucrose, 60 mM KCl, 15 mM NaCl, 15 mM Tris, pH 7.5, 5 mM MgCl2, 1 mM CaCl2, 1mM DTT (Thermo Scientific, 20291), 10 mM sodium butyrate (Sigma, B5887), 0.5 mM AEBSF (Sigma, A8456), 5 nM microcystin LR (Calbiochem, 101500), 0.3% NP40 (USB Corporation, 19628). Nuclei were centrifuged at 4 °C, 10,000g, for 1 min. Supernatants were removed and the nucleus isolation procedure was repeated twice more, removing the supernatant each time. Histones were extracted from the remaining pellet with 800 μL 0.4 N H2SO4 at room temperature for 4 h with shaking.
Samples were centrifuged at 4 °C, 10,000g, for 5 min. Supernatants were saved and pellets discarded. Histones were precipitated from solution with a volumetric concentration of 20% ice-cold trichloroacetic acid (TCA) (BDH, BDH0310) on ice for 30 min. Afterwards samples were centrifuged at 4 °C, maximum speed, for 15 min. Supernatants were discarded and the resulting films (precipitated histone) were washed once with ice-cold acetone. Samples were centrifuged once more at 4 °C, maximum speed, for 5 min. Supernatants were removed and extracted histones were allowed to air dry for 10 min at room temperature. Histones were then resuspended in HPLC-grade water on ice.
Histone purity was assessed via SDS–PAGE gel and total protein concentration was determined using Coomassie Plus Protein Assay (Thermo Scientific, 1856210) with a standard curve created from 2 mg/mL Albumin Standard (Thermo Scientific, 23209) and HPLC-grade H2O as diluent.
If SILAC standardization was used, one or more pellets (of 2e7 cells) from each cell line (HeLa, 293T, K562) were processed along-side the sample set in an identical manner.
2.3.2. Histone derivatization with heavy synthetic peptide standardization
In this workflow 50 μg of histone per sample was used. Samples were adjusted to 100 mM sodium phosphate, pH 8.0 by adding 13 μL 500 mM sodium phosphate, pH 8.0 and bringing the total volume of the sample up to 65 μL with HPLC-grade water. Phosphate-buffered samples were reacted with 200 μL of 400 mM NHS propionate in anhydrous methanol, at room temperature, for 30 min with shaking. Enough 0.1% trifluoroacetic acid (TFA) was added to bring samples to a volumetric concentration of 20% organic solvent. Samples were desalted on an Oasis HLB 30 mg/1 cc 96-well plate (Waters, WAT058951). Wells were activated with 1 × 1 mL 100% acetonitrile (ACN) and conditioned with 2 × 1 mL 20% (ACN)/0.1% TFA. Samples were loaded onto the wells and washed with 2 × 1 mL 20% ACN/0.1% TFA and eluted from the Oasis plate with 500 μL 60% ACN/0.1% TFA. The propionylated, desalted histones were then lyophilized using a centrifugal vacuum concentrator. Samples were resuspended in 100 μL 50mM ammonium bicarbonate, pH 8.0. Trypsin (Promega, V5113) was added in a 1:50 enzyme:substrate ratio and incubated for 16 h at 37 °C with shaking.
After digestion, samples were frozen and then lyophilized using a centrifugal vacuum concentrator. To derivatize newly created N-termini, peptides were resuspended in 100 μL 100 mM NHS propionate/anhydrous methanol and adjusted to 18 mM sodium phosphate buffer, pH 8.0 with 10 μL 200 mM sodium phosphate buffer, pH 8.0. The reaction was incubated for 30 min at room temperature with shaking, and quenched with 5 μL 50% hydroxylamine solution (Sigma, 467804). Samples were then incubated for another 30 min at room temperature with shaking. HPLC-grade water was added to a total volume of 1 mL to aid in freezing of the primarily organic mixture. Samples were subsequently frozen at −80 °C and lyophilized via vacuum concentrator. Samples were brought up in 1 mL 0.1% TFA and desalted on a SepPak (tC18 100 mg/well) 96-well plate (Waters, 186002321). Wells were activated with 1 × 1 mL 100% ACN and conditioned with 2 × 1 mL 0.1% TFA. Samples were loaded onto the wells and washed with 2 × 1 mL 0.1% TFA. Samples were eluted with 500 μL 50% ACN/0.1% TFA and lyophilized using a vacuum concentrator. Desalted, propionylated peptides were resuspended in 50 μL 3% ACN/5% formic acid (FA). Resuspended peptides were diluted twenty-fold in a heavy synthetic peptide “Mastermix” (Supplemental Table 1) and 3% ACN/5% FA solvent before analysis to have a final concentration of 1× Mastermix and 50 fg/μL histone peptides.
Mixing in the peptide standards after peptide cleanup allows for more flexible dilution and repurposing of samples. For instance, if one wants to reanalyze a sample at a higher or lower dilution to the standards it is advantageous. Also, if one ever adds new peptides to the standards mixture we can go back and reanalyze old samples this way without complete re-preparation. We studied the recovery of spiking the standards before and after the final peptide cleanup and found that the recovery is generally quite good (Supplemental Table 1, last column), and thus addition of standards pre- or post-cleanup is feasible.
Details concerning the completeness of propionylation and comparison of the NHS-propionate to Propionic anhydride can be found in Supplemental Tables 2 and 3. We find that our process increases the yield of desired propionylation products by over 60% when compared with the use of propionic anhydride as in Ref. [3].
2.3.3. Histone derivatization with SILAC standardization
For sample sets requiring SILAC standardization, the extracted histones of SILAC-labeled cell lines (HeLa, K562, 293T) were combined in a 1:1:1 protein ratio. This heavy-histone mix was combined with the light sample in a 1:1 protein ratio (25 μg heavy histone and 25 μg histone from the sample). We mix after histone extraction because we find that quantification of histones by protein assay gets a more reliable match than cell counts. Cancer cells can sometimes suffer significant aneuploidy which makes mixing by cell counts unreliable. Workflow for SILAC-standardized samples was identical to the method described above. After derivatized, propionylated peptides were resuspended in 50 μL 3% ACN/5% FA, the samples were further diluted 1:10 in 3% ACN/5% FA before analysis, to have a final concentration of 50 fg/μL histone peptides.
2.3.4. Automating the workflow for high-throughput
To increase efficiency and throughput of the chromatin profiling assay, a Liquid Transfer Bravo robot (LT-BRAVO) (Agilent) and Positive Pressure 96 Apparatus (Waters) were employed to either fully automate or semi-automate large parts of the workflow. Propionylation and digestion reactions were fully automated by placing entire sample sets into 96-well plates (up to 96 samples/plate), with each well holding a maximum volume of 2 mL. Three programs were written for the LT-BRAVO: a primary propionylation program, an overnight trypsin digestion program, and a secondary propionylation program. In each program, the 96-well plate containing samples (1 sample/well) was placed on the deck of the robot, along with required reagents and pipette tips that corresponded to sample layout. The head of the robot dispensed specified volumes of reagents into each well and incubated reactions with tip mixing instead of shaking. For the overnight trypsin digestion program, trypsin was pre-aliquoted in a 96-well v-bottom plate and placed on a section of the robot’s deck kept at 4 °C. After required reagents were dispensed and mixed in the sample plate, the robot head moved the plate to a section of the deck kept at 37 °C with a shaking platform. This 96-well format was also carried through both desalt processes using 2 mL 96-well plates packed with material (Oasis and SepPak plates) instead of individual cartridges. Use of a positive pressure apparatus allows all samples to be desalted simultaneously and keep sample layout the same throughout the workflow. All freezing and lyophilization steps were carried out in 96-well plates in centrifugal vacuum concentrators equipped with appropriate rotors. Automating the work-flow not only increases throughput but also minimizes hands-on sample handling, allowing that time to be allocated elsewhere.
2.4. LC–MS/MS analysis
2.4.1. Assay parameters
All samples were separated on an online Proxeon EASY-nLC 1000 UHPLC system (Thermo Scientific) and analyzed on a Q Exactive mass spectrometer (Thermo Scientific). All samples were injected onto a fused silica capillary column with a 10 μm Picofrit opening and 75 μm diameter (New Objective, info here) packed in-house with 20 cm Reprosil-Pur 120 C18-AQ, 1.9 μm material (Dr. Maisch GmbH, r119.aq). All columns were heated to 50 °C with a heater jacket. In both scheduling and scheduled runs peptides were injected onto the column and separated at a flow rate of 200 nL/min with a 45 min linear gradient from 97% solvent A (3% ACN/0.1% FA) to 40% solvent B (90% ACN/0.1% FA). This gradient was followed by a 5 min linear gradient from 40% solvent B to 90% solvent B, at which point 90% solvent B was held for an additional 5 min. Separated peptides were introduced to the mass spectrometer using ESI with a spray voltage of 2.2 kV. Data were acquired using Xcalibur software in positive ion mode. Including sample loading and column equilibration times each sample took 90 min till completion.
2.4.2. Optimization of assay analytes and creation of spectral libraries for H3 peptides
To ensure that the most sensitive combination of precursor and fragment ions were selected as each peptide’s MRM ion pairs, synthetic peptides were used to generate spectral libraries for generation of the final MRM assays. Each synthetic peptide was prepared as a 1 pmol/μL solution in 0.1% formic acid/50% MeOH and individually introduced into a Q Exactive mass spectrometer (Thermo Scientific) with an Advion TriVersa NanoMate using chip-based nanoelectrospray. The Q Exactive instrument was operated in a targeted manner. In a given duty cycle an MS1 scan was first recorded with a resolution of 35,000, AGC target of 1e6, maximum injection time of 250 ms, and scan range of 300–1800 m/z. Followed by the MS1 scan, consecutive MS2 scans using a collision energy of 10, 21, 23, 25, 27, 29, 31, 33, and 35 were recorded on charge states 1–4 of the peptide if the resulting precursor mass was above 300 m/z. MS2 scans were recorded with a resolution of 17,500, AGC target of 1e5, maximum injection time of 60 ms, and isolation window of 2.0 m/z.
To generate spectral libraries, we exported raw spectra from our infusion experiments in text format. Because the peptides were synthetic, we manually assigned sequences and modifications. Spectra were imported into Skyline using the .ssl/.ms2 files formats as detailed here (https://skyline.gs.washington.edu/labkey/announcements/home/support/thread.view?entityId=86be1b94-d328-102e-a8bb-da20258202b3&_anchor=716#row:716). The resulting spectral library (in .blib format) is available as supplementary material (Supplemental Item 1).
2.4.3. Scheduling for H3 targets
Since the assay is fully targeted, a scheduling method was first employed to determine each peptide’s retention time. An equimolar mix of R10-labeled synthetic peptides targeted in the assay was injected onto the column (15 fmol/peptide). A single MS1 spectra was first obtained at a resolution of 35,000 with an AGC target of 1e6, maximum injection time of 250 ms, and a scan range of 300–950 m/z. MS1 spectra were acquired followed by 17 targeted MS2 scans. Each MS2 scan had a default charge state of 2, resolution of 17,500, AGC target of 1e5, maximum injection time of 60 ms, isolation window of 4.0 m/z, fixed first mass of 100.0 m/z, and NCE of 30. Inclusion list was turned on for each targeted MS2 scan. The list included heavy-labeled versions of each peptide to be observed in the assay, their charge states, and optimized collisional energies. Each peptide was scheduled from start of the method to 59.90 min. Total run time for each scan was 0–60 min. Two technical replicates of the equimolar mix of synthetic peptides were run.
Resulting raw data was imported into Skyline (info here) and extracted ion chromatograms (XIC’s) for each peptide were created. After checking the Skyline file to verify identified peaks, a report was generated including each peptide’s m/z for both heavy and light versions and an averaged retention time (RT). This information was used to create an inclusion list for each peptide, with a specific acquisition window from 3 min to 20 min, based on its chromatographic properties. An example of how to create such an inclusion list is found in Supplemental Item 2.
2.4.4. Scheduled data acquisition
After retention times of all peptides were determined 1 μL of sample was injected onto the same column used for scheduling and peptides were separated using the same gradients as previously described. Peptides were introduced to the mass spectrometer via ESI with a spray voltage of 2.2 kV. A single MS1 scan was acquired with the same specifications as the scheduling method, only differing in run time (55 min). This MS1 scan was followed by 14 targeted MS2 scans with a default charge state of 2, resolution of 17,500, AGC target of 1e5, maximum injection time of 60 ms, isolation window of 2.7 m/z, fixed first mass of 100.0 m/z, and NCE of 30 units. Inclusion list was turned on for each MS2 scan, and included both light and heavy versions of each peptide to be observed, their charge states, new acquisition windows based off the scheduling runs, and optimal collision energies. Each scan had a run time of 0–55 min.
2.5. Data analysis
All raw files were imported into and processed by Skyline Daily (Supplemental Item 3 – note that the “Daily” release or the “Stable” release >= version 2.6 is required to open these files) [7]. Most isobaric precursors have clearly separated elution peaks, but the specific fragment ion transitions selected in the Skyline document allow for disambiguation of even co-eluting isobaric precursors. Peaks identified in the resulting XIC’s of each peptide were verified, and a report was generated including the ratio of light to heavy standard for each peptide, for each sample. Each histone mark/mark combination was normalized to a peptide originating from the same protein that is not typically modified, to account for differences in protein quantity between samples (column normalization for protein load). For example, all histone H3 mark/mark combinations were normalized to peptide YRPGTVALR. These normalized light/heavy standard ratios were transformed into log2 space. To compare the abundance of mark/mark combinations between samples, each peptide was normalized to its median value across the sample set (row normalization). The resulting values were visualized with GENE-E (http://www.broadinstitute.org/cancer/software/GENE-E/) (version 3.0.202 was used). Rows and columns of data sets can undergo hierarchical clustering using a Euclidian distance metric and complete linkage. Heatmaps were colored using absolute values of the cells. An example of a data set that gave rise to Fig. 2A is given as Supplemental Item 4. This supplementary item contains a step-by-step walkthrough of the data reduction procedures in Excel spreadsheet format. GCT files that provide the numerical data before and after processing are provided as part of this Supplementary Item. These files can be visualized and clustered in GENE-E.
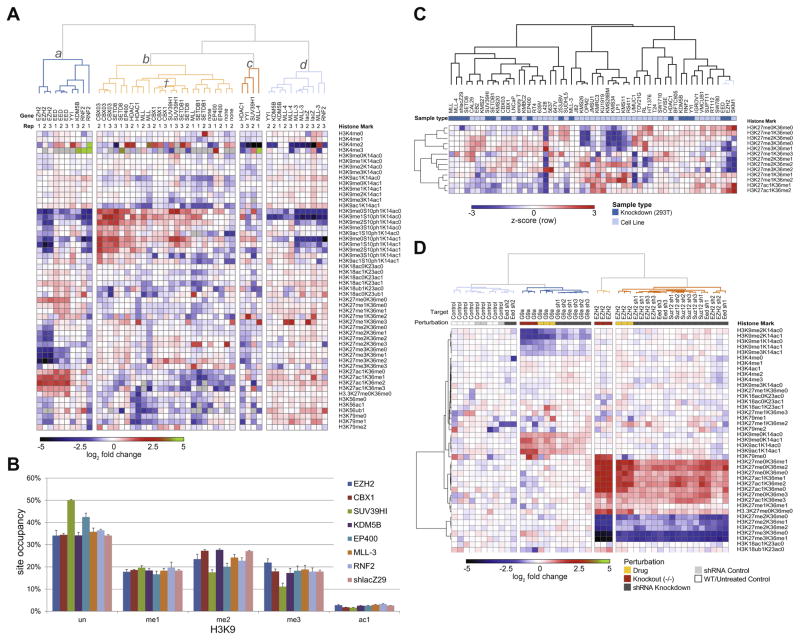
Fig. 2.
Connectivity of cellular perturbations and states through chromatin signatures. Several use cases of molecular chromatin signature data are illustrated. (A) Connectivity of perturbations of gene expression by shRNAs. Genes knocked down and biological replicates are indicated across the top. Perturbations were hierarchically clustered using a Euclidian distance metric. (B) Calculation of site occupancy is possible with synthetic peptide standards. H3K9 site occupancy is calculated for selected shRNA knockdowns depicted in (A). (C) Conflation of shRNA knockdown profiles (from A) with profiles from cells of differing genetic backgrounds (from Ref. [2]) for a subset of H3 marks. This approach allows classification of genetic mutants by comparing with reference gold-standard perturbations. Samples were clustered using a Spearman rank-order metric. (D) Connectivity of knockdowns, knockouts, and drug treatments of cells in murine stem cells. The approach allows facile comparison of multiple modes of perturbations. Samples clustered by Euclidean distance.
Estimates of site occupancy can also be calculated from the observed ratios and known standard peptide concentrations (Workflow 2.3.2). An example of the data reduction procedures that gave rise to Fig. 2B is given in Supplemental Item 5. This supplementary item gives a step-by-step walkthrough of the calculations necessary to estimate the individual site occupancies as an Excel spreadsheet. Briefly, ratios of light-to-heavy standard were exported from Skyline for each histone mark and normalized to peptide YRPGTVALR (peptide normalization). The peptide-normalized sample ratios of each histone mark were scaled to the ratio of its unmodified peptide (scaled peptide-normalization). In this context, “unmodified” means the completely propionylated version of the peptide with no methyls, acetyls, or other post-translational modifications. The concentration of each histone mark in the standard “2× Mastermix” was scaled to the concentration of its unmodified peptide to produce “scaled standard values.” Scaled peptide-normalized sample ratios and scaled standard values of each mark were used to estimate the concentrations of endogenous histone mark combinations in each sample. Estimated site combination occupancy percentages of endogenous histone marks on various histone H3 peptides were calculated and final estimated site occupancies for marks at a given site were collapsed for all samples.
3. Results
Results (Fig. 2) can be depicted as a heatmap and are amenable to techniques typically associated with gene expression analysis, such as clustering and marker selection. As a proof-of-principle, we performed shRNA-based knockdowns in 293T cells of a variety of genes known to be active in epigenetic processes (Fig. 2A). Unsupervised hierarchical clustering yielded three main branches. Cluster a contains profiles from knockdowns of several members of the Polycomb Repressive Complexes (PRC1/PRC2) [10], including EZH2, EED, and RNF2. Thus, chromatin profiling was able to recapitulate known chromatin biology by connecting these genes through their chromatin signatures. As another example, one subcluster (marked with †) in cluster b contains SUV39H1, known to catalyze the formation of H3K9me3, and CBX1, a gene known to bind H3K9me3. Because we profiled these perturbations using our comprehensive collection of synthetic peptides with known abundances, we were able to estimate the percent occupancy of each modification at key lysines. For example, knocking down SUV39H1 transcript levels by ~95% causes a reduction in H3K9me3 from ~20% to 10%; H3K9me2 from ~25% to ~18%; and a concomitant increase in H3K9un from ~33% to 50% (Fig. 2B).
In Fig. 2C, we illustrate how clustering of data from cells of different genotypes together with data from gene knockdowns enables functional annotation of genotypes in specific cellular contexts [2]. Clustering these two data streams by a subset of chromatin modifications revealed an association among knockdowns of EED and EZH2 with the cell line SKM-1 (far right, Fig. 2C). SKM-1 has a known mutation in EZH2 leading to the gene product EZH2 p.Y641C. These data allow us to annotate EZH2 p.Y641C as a loss-of-function mutation in a cellular context. This differs from in vitro data characterizing EZH2 p.Y641C [11] but corroborate other work [12], illustrating the importance of profiling in the proper biological system. Moreover, these results illustrate the flexibility of the assay by allowing comparison of two data sets that were acquired in completely different systems and at different points in time.
Finally, we illustrate the ability to compare across shRNA knockdown, genetic knockout, and drug treatment (Fig. 2D) with chromatin signatures derived from murine stem cells. Here, we show that knockdowns and knockouts of PRC2 members co-cluster with a chemical inhibitor of EZH2 (GSK-126), and likewise knock-downs and knockouts of the H3K9 methyltransferase G9a co-cluster with a chemical inhibitor of G9a (UNC-0646).
4. Discussion
The ability to compare molecular chromatin signatures across many cell types and perturbational conditions potentiates a Connectivity Map [13] of epigenetics. Although this work is focused on H3, the workflow can be extended to all histones. Through these connections, we can functionally annotate genetic mutations, infer mechanisms of actions of small molecules, and discover unexpected associations. The information gleaned by chromatin profiling is complementary to ChIP-Seq and gene expression profiling, and importantly, these direct measurements of chromatin modification states are compatible with sample types and amounts (including tissues) that could have direct clinical impact as novel epigenetically-directed therapeutics emerge.
One current limitation of our method is the requirement for a relatively large number of cells (2e6) compared to typical genomic analyses. We expect that process improvements and reduction in scale could lead to a reduction of the input requirements by as much as 5-fold. This would lead to an equivalent input amount of the number of (average sized) cells that can be grown in a single well of a 6-well plate. Combined with our automated workflow, we could begin to think about using the assay as a serious secondary screen for epigenetically active compound discovery. This technique would have the advantage of being able to ascertain both on-target and off-target epigenetic effects in a single assay.
Another limitation is that standard peptides must be synthesized for novel combinations of marks as they are discovered. This limitation is partially overcome by using the SILAC-based standardization procedure, as long as the novel combinations are present in the cell lines chosen as the SILAC standards. In this way, SILAC could be used as a bridging step until new synthetic peptides can be obtained. In fact, it may be desirable to determine the biological importance and variability of a novel combination of marks before investing in the creation of new synthetic standards.
This assay will be a cornerstone in the NIH Library of Integrated Network-based Cellular Signatures (LINCS) program. We will employ the assay to study the effects of >100 knock-downs and drug-based perturbations of epigenetic regulators across multiple cell types that represent different disease paradigms. This project will formulate the basis for our epigenetic Connectivity Map.
Supplementary Material
Acknowledgments
This work was funded in part by R21 DA025720-02 and U01 CA164186-02 to J.D.J. and by a Thought Leader Award and gift to S.A.C. from the Agilent Technologies Foundation and Agilent Technologies, Inc., respectively.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.ymeth.2014.10.033.
References
- 1.Consortium EP. Science. 2004;306:636–640. [Google Scholar]
- 2.Jaffe JD, Wang Y, Chan HM, Zhang J, Huether R, Kryukov GV, Bhang HE, Taylor JE, Hu M, Englund NP, Yan F, Wang Z, Robert McDonald E, 3rd, Wei L, Ma J, Easton J, Yu Z, deBeaumount R, Gibaja V, Venkatesan K, Schlegel R, Sellers WR, Keen N, Liu J, Caponigro G, Barretina J, Cooke VG, Mullighan C, Carr SA, Downing JR, Garraway LA, Stegmeier F. Nat Genet. 2013;45:1386–1391. doi: 10.1038/ng.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garcia BA, Mollah S, Ueberheide BM, Busby SA, Muratore TL, Shabanowitz J, Hunt DF. Nat Protoc. 2007;2:933–938. doi: 10.1038/nprot.2007.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peach SE, Rudomin EL, Udeshi ND, Carr SA, Jaffe JD. Mol Cell Proteomics. 2012;11:128–137. doi: 10.1074/mcp.M111.015941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomas CE, Kelleher NL, Mizzen CA. J Proteome Res. 2006;5:240–247. doi: 10.1021/pr050266a. [DOI] [PubMed] [Google Scholar]
- 6.Gillette MA, Carr SA. Nat Methods. 2013;10:28–34. doi: 10.1038/nmeth.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH. Mol Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, Shinkai Y. Genes Dev. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Margueron R, Reinberg D. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wigle TJ, Knutson SK, Jin L, Kuntz KW, Pollock RM, Richon VM, Copeland RA, Scott MP. FEBS Lett. 2011;585:3011–3014. doi: 10.1016/j.febslet.2011.08.018. [DOI] [PubMed] [Google Scholar]
- 12.Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, Waghorn K, Zoi K, Ross FM, Reiter A, Hochhaus A, Drexler HG, Duncombe A, Cervantes F, Oscier D, Boultwood J, Grand FH, Cross NC. Nat Genet. 2010;42:722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 13.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, Reich M, Hieronymus H, Wei G, Armstrong SA, Haggarty SJ, Clemons PA, Wei R, Carr SA, Lander ES, Golub TR. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.