Abstract
Ultrasound at 5.0 MHz was noted to be chondro-inductive, with improved SOX-9 gene and COL2A1 protein expression in constructs that allowed for cell-to-cell contact. To achieve tissue-engineered cartilage using macroporous scaffolds, it is hypothesized that a combination of ultrasound at 5.0 MHz and transforming growth factor-β3 induces human mesenchymal stem cell differentiation to chondrocytes. Expression of miR-145 was used as a metric to qualitatively assess the efficacy of human mesenchymal stem cell conversion. Our results suggest that in group 1 (no transforming growth factor-β3, no ultrasound), as anticipated, human mesenchymal stem cells were not efficiently differentiated into chondrocytes, judging by the lack of decrease in the level of miR-145 expression. Human mesenchymal stem cells differentiated into chondrocytes in group 2 (transforming growth factor-β3, no ultrasound) and group 3 (transforming growth factor-β3, ultrasound) with group 3 having a 2-fold lower miR-145 when compared to group 2 at day 7, indicating a higher conversion to chondrocytes. Transforming growth factor-β3–induced chondrogenesis with and without ultrasound stimulation for 14 days in the ultrasound-assisted bioreactor was compared and followed by additional culture in the absence of growth factors. The combination of growth factor and ultrasound stimulation (group 3) resulted in enhanced COL2A1, SOX-9, and ACAN protein expression when compared to growth factor alone (group 2). No COL10A1 protein expression was noted. Enhanced cell proliferation and glycosaminoglycan deposition was noted with the combination of growth factor and ultrasound stimulation. These results suggest that ultrasound at 5.0 MHz could be used to induce chondrogenic differentiation of mesenchymal stem cells for cartilage tissue engineering.
Keywords: Low-intensity continuous ultrasound, Transforming growth factor-β3, miR-145, chondrogenesis, mesenchymal stem cells
Introduction
Functional biological tissue-substitutes can potentially serve as a replacement to help regenerate damaged or defective tissues.1,2 Initial attempts to generate tissue-engineered cartilage (TEC) involved the usage of adult autologous chondrocyte as these cells can be isolated from the less weight-bearing areas of the joint, amplified in vitro, seeded, and cultured on a scaffold in a bioreactor to form cartilage that can be implanted without compromising the patient’s immune system.1 In the last 10 years, the use of stem cells, which can potentially differentiate into chondrocytes under appropriate conditions, has been explored as a promising alternative.3 It is widely accepted that human mesenchymal stem cells (hMSCs) provide a better starting cell source than adult human chondrocytes for a variety of reasons.3–5 The ability to acquire hMSCs autologously and their potential for multilineage differentiation and proliferative potential in vitro make hMSCs an attractive source of cells for tissue engineering.4,5 Various studies have reported on the feasibility of using hMSCs to form cartilage-like tissue.6–11
Chondro-induction of mesenchymal stem cell (MSC) is influenced by many factors, including transforming growth factor beta (TGFβ), ascorbic acid, as well as external mechanical stimuli such as three-dimensional (3D) culture in scaffolds, hydrostatic pressure, dynamic compression, and ultrasound (US).12–14 Typically, hMSCs on scaffolds or hydrogels are directed toward a chondrocytic lineage in vitro using chondrogenic medium along with a combination of physical stimuli and then are released, re-passaged, and re-seeded onto scaffolds and further cultured to obtain TEC.15,16 In an attempt to merge the two steps, recently, hMSCs seeded onto focal-defect-sized PLGA scaffold were chondro-induced for 8 weeks in a chondrogenic medium and then either implanted directly (one-step) or released, passaged, and seeded onto PLGA scaffolds and implanted (two-step).17 Our long-term goal is to first promote the chondrogenic differentiation of hMSCs in vitro using US, 3D scaffolds and growth factors and then seamlessly transition to expand and culture hMSC-derived chondrocytes on 3D scaffolds using US in a US-assisted bioreactor. We hypothesize that the US bioreactor creates a microenvironment in the seeded scaffold that assists the differentiation of hMSCs into chondrocytes and aids the maintenance of the hMSC-derived chondrocytes along with a uniform cellular distribution throughout the scaffold volume.
A bioreactor configuration that uses US to stimulate in vitro cultures over a range of US stimulations has been designed and developed at the University of Nebraska–Lincoln.18,19 Aspects of US that would negatively affect cells, including temperature and cavitation, were shown to be insignificant for the US protocols used covering a wide range of frequencies and pressure amplitudes, including the ones used in this study.18 We conclude that any US effects in the bioreactor, aside from cellular responses, are negligible. Furthermore, we have shown that the response of cells to US is frequency dependent, with a primary resonant frequency at 5.0 MHz where the cells mostly undergo dilatation.20
The success of any bioreactor that seeks to attain lineage-dependent conversion of hMSCs to chondrocytes depends upon the ability of the bioreactor to mediate the conversion to chondrocytic lineage by enhancing specific cellular pathways that control the cell-specific differentiation markers. For example, in the generation of hMSC-derived chondrocytes, bioreactors have to provide the mechanical conditioning to cells and enable the coupling of the external stimuli to nuclear process that controls chondrocytes’ fate via SOX-9-dependent processes.21 We have recently established that exposure of adult chondrocytes to US at 5.0 MHz significantly modulates the level of gene expression of a variety of chondrocyte-specific genes (i.e. SOX-9) and that this likely occurs through signals transmitted to chondrocytes.22,23
Current protocols use ≤10 ng/mL of TGFβ (1 or 3) in the chondrogenic differentiation medium.6,24 Further inductive molecules (i.e. TGFβ3 alone) when acting alone may have limited capacity to direct specific differentiation pathways, and we further hypothesize that the synergism between US-mediated cell processes and signaling provided by TGFβ may augment the cell fate processes.
We hypothesize that by combining continuous, low-intensity US with 3D culture techniques and growth factors, we can engineer an optimized microenvironment to induce MSC differentiation to chondrocytes, and the microenvironment can promote the MSC-derived chondrocytes to thrive and produce cartilage-specific markers. Thus, in this study, we seeded hMSCs on macroporous scaffolds, cultured hMSC-seeded constructs in the presence of TGFβ3 for 14 days in the US-assisted bioreactor, and continued the culture in the US-assisted bioreactor for an additional 7 days in the absence of any exogenously added growth factors. During culture, cell-seeded scaffolds were retrieved from respective groups and chondrogenic differentiation of hMSCs was assessed as follows: proliferation by Picogreen™ assay, total collagen content by hydroxyproline assay, total glycosaminoglycan (GAG) by dimethylmethylene blue (DMMB) assay, microRNA (miRNA) and messenger RNA (mRNA) expression by quantitative real-time polymerase chain reaction (qRT-PCR), and protein expression by western blotting, histology, and immunohistochemistry.
Materials and methods
Polyurethane-urea-based macroporous 3D scaffolds (denoted as BM) were received as generous gift from Biomerix Corporation, Somerset, NJ, USA. Unless otherwise stated, all chemicals were ACS grade and purchased from Sigma (St. Louis, MO, USA).
hMSC culture and generation of cell-seeded constructs
Bone marrow–derived hMSCs were obtained from Lonza (Walkersville, MD, USA) and expanded in medium-1 (M1) consisting of alpha-Minimum Essential Medium (MEM) supplemented with 10% mesenchymal stem cells qualified fetal bovine serum (FBS), 1X Glutamax™, and 1X antibiotic–antimyotic™ solution. Passage 5 hMSCs were used in all experiments. hMSC pellet cultures (~3 × 105 cells/pellet) were prepared using established protocols25 and were transferred to ultra-low attachment 6-well plates. hMSCs were also encapsulated in Hystem-C™ matrices (ESI BIO, Alameda, CA, USA) using instructions provided by the manufacturer.26–28 Typically, ~105 cells/mL were encapsulated in Hystem-C matrices and were transferred to ultra-low attachment 6-well plates.
To obtain prewetted scaffolds, BM scaffolds (5 mm × 2.5 mm) were sterilized with sequential treatments of 70% ethanol for 1 h followed by sterile 1X phosphate buffered saline (PBS) rinse and then incubated for 12 h in medium (high-glucose Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% FBS). One side of the prewetted scaffold disks was seeded with hMSCs (passage 5) at a seeding density of 2 × 105 cells/scaffold using procedures outlined elsewhere29 and were transferred to a new six-well tissue culture plate (TCP) housing a cell crown™ insert/well with 15–18 scaffolds/insert.
Medium 2 (denoted as M2) containing high-glucose DMEM, 10% FBS, 100 nM dexamethasone, 50 µg/mL of L-ascorbic Acid, and 1X antibiotic–antimyotic solution was further supplemented with TGFβ3 according to Figure 1 for chondrogenic differentiation and denoted as chondrogenic differentiation media (CDM; M2 + TGFβ3). After 14 days of culture, M2 or CDM was removed and cell-constructs were cultured for an additional 1 week in Medium 3 (M3) comprising M2 without dexamethasone (Table 1). All supplements and media for cell culture were purchased from Life Technologies (Grand Island, NY, USA).
Figure 1.

TGFβ3 dose in CDM over 1–21 days. hMSCs were seeded on Biomerix™ scaffolds (2 × 105 cells/scaffold, 5 mm × 2.5 mm) and cultured in the US-assisted bioreactor. The following independent study groups (Table 1) were adopted: Group 1—No TGFβ3, No US (Control); Group 2—TGFβ3, No US; and Group 3—TGFβ3, US. TGFβ3 concentrations used are indicated above. After 14 days of culture in M2 or CDM, cell-constructs were cultured for an additional week in M3.
Table 1.
Construct culture condition.
| Study groups | Media usage over 21 days |
US regimen over 21 days | |
|---|---|---|---|
| 1–14 days | 15–21 days | ||
| Group 1 | M2a | M3b | – |
| Group 2 | CDMc | M3b | – |
| Group 3 | CDMc | M3b | 14 kPa 5 min/application, 6X/day |
US: ultrasound; CDM: chondrogenic differentiation media; FBS: fetal bovine serum; DMEM: Dulbecco’s Modified Eagle Medium; TGFβ3: transforming growth factor-β3.
M2: high-glucose DMEM, 10% FBS, 100 nM dexamethasone, 50 µg/mL of L-ascorbic acid, and 1X antibiotic–antimyotic™.
M3: M2 without dexamethasone.
CDM: M2 with TGFβ3 (Figure 1).
Experimental methods followed in US-assisted bioreactor
hMSC-seeded scaffolds were either cultured in CDM (group 2) or in combination of CDM and US stimulation (group 3) for 21 days according to Table 1. hMSC-seeded scaffolds cultured in M2 and no US stimulation (group 1) served as control (Table 1). Six-well TCPs containing MSC pellets, MSCs in Glycosan™ matrices, and MSC-seeded scaffolds (group 3 only) were placed in plate holder of the US-assisted bioreactor developed at the Department of Chemical and Biomolecular Engineering, University of Nebraska–Lincoln, Lincoln, NE, USA.18 The experimental setting was described elsewhere.29 Automated US stimulation was provided in the near field at the following regimen: 14 kPa (5.0 MHz, 2.5 Vpp), 5 min/application and 6 applications/day at a regular interval. At indicated time points, cell-seeded scaffolds were retrieved randomly from respective groups and assayed as indicated.
Cell viability
Cell-seeded scaffolds were treated with LIVE/DEAD® Viability/Cytotoxicity Kit (Life Technologies) according to previously published protocol19 and visualized with inverted Confocal Microscope (Olympus IX 81) at the Center of Biotechnology, University of Nebraska–Lincoln. All the images were collected at 20× magnification (z step size = 10 µm).
Biochemical analysis
Randomly selected scaffolds (n = 5–8) per study group were first washed with sterile 1X PBS and incubated with papain digestion buffer (5 mM L-cysteine, 100 mM Na2HPO4, 5 mM EDTA, 125 µg/mL papain, pH = 7.5) for 16–18 h at 70°C. Upon completion of the incubation, scaffolds were spun down, and supernatants were collected and subjected to DNA, GAG, and Hydroxyproline content measurements.30 Double-stranded DNA (dsDNA) was measured using Quant-iT™ PicoGreen® dsDNA Assay Kit (Life Technologies) according to manufacturer’s instructions. All quantifications were based on a λ DNA standard. Total GAG content was determined with DMMB assay on papain-digested supernatant according to the protocol described elsewhere31 and quantified based on shark chondroitin sulfate standard. Hydroxyproline content was measured on the papain-digested supernatant accordingly using Hydroxyproline Assay Kit (Sigma) according to manufacturer’s instructions. Deionized (DI) water was used as a blank in the assay measurements. Total collagen was extrapolated from hydroxyproline content as hydroxyproline amino acid comprises 13.5% of total collagen.32
Cell release from BM scaffolds and isolation of small and large RNA fractions
After desired treatment, cell-seeded scaffolds were retrieved immediately, rinsed with ice-cold HBSS, and finally, cells were released by incubating with 1X trypsin. Followed by scaffold removal, trypsin was neutralized with 10% FBS supplemented media, and the cell suspension was centrifuged at 1000×g for 10 min to obtain the cell pellet. Cell pellet obtained after releasing cells from scaffolds was treated to isolate small and large RNA fraction using the miRNA isolation kit (Life Technologies) as per manufacturer’s instructions. The small RNA fraction contained RNAs with size less than 200 nucleotides and used for miRNA gene expression analysis, whereas large RNA fraction was used for mRNA gene expression analysis.
miRNA reverse transcription and qRT-PCR analysis
The miRNA level was quantified using TaqMan-based qRT-PCR. All reagents and primers (detailed in Table 2) were purchased from Applied Biosystems (Foster City, CA, USA). The qRT-PCR analysis was carried out using two-step method. In step 1, reverse transcription reaction was performed; 20 ng of each small RNA fraction sample was mixed with MultiScribe reverse transcriptase, RNase inhibitor, and nuclease-free water as per manufacturer’s instruction, and mixtures were incubated for 30 min at 16°C, 30 min at 42°C, and then 5 min at 85°C. In step 2, qRT-PCR was carried out using Eppendorf’s thermocycler RealPlex real-time PCR system (Eppendorf North America, Hauppauge, NY, USA). The PCR master mix containing TaqMan 2X Universal PCR Master Mix (No AmpErase UNG), 10X TaqMan assay, and RT products in 20 µL volume was processed as follows: 95°C for 10 min and then 40 cycles of 95°C for 15 s and 60°C for 60 s (n = 3). The amplified expression of miR-145 transcript was normalized to SnU6 expression; 2−ΔΔCt method was used to calculate relative expression levels.
Table 2.
List of primers.
| S. no. | Gene | Species | TaqMan probe dye | Catalog no./assay ID |
|---|---|---|---|---|
| 1 | U6 snRNA | Human | FAM-6 | 4427975/001973 |
| 2 | miR-145 | Human | FAM-6 | 4427975/002278 |
| 3 | GAPDH | Human | FAM-6 | Hs03929097_g1 |
| 4 | ACAN | Human | FAM-6 | Hs00153936_m1 |
| 5 | COL10A1 | Human | FAM-6 | Hs00164004_m1 |
| 6 | TGFβ1 | Human | FAM-6 | Hs00998133_m1 |
| 7 | TGFβ3 | Human | FAM-6 | Hs01086000_m1 |
TGF: transforming growth factor.
mRNA gene expression analysis
Large RNA fraction was collected as described above and quantified by qRT-PCR using QuantiFast Probe RT-PCR Kit (Qiagen, Valencia, CA, USA); 50 ng of total RNA was added per 10 µL reaction vial with RT mix, RT-PCR master mix, sequence-specific primers, and TaqMan probes. Sequences of primers (Table 2) are proprietary to Applied Biosystems and not disclosed. qRT-PCR assays were carried out in triplicate on Eppendorf’s Mastercycler® RealPlex real-time PCR system (Eppendorf North America). Cycling was initiated by 10 min for complementary DNA (cDNA) formation by reverse transcriptase enzyme at 50°C and 5 min polymerase activation at 95°C, followed by 40 cycles at 95°C for 30 s, at 55°C for 30 s, and at 72°C for 1 min. The threshold was set above the non-template control background and within the linear phase of target gene amplification to calculate the cycle number at which the transcript was detected. The amplified expression of mRNA transcripts was normalized to GAPDH expression; 2−ΔΔCt method was used to calculate relative expression levels.
Protein isolation and western blotting analysis
At the end of 14 days of culture, cells were released from scaffolds with 1X Trypsin followed by pelleting cells at 1000×g and finally lysing the pellet with Pierce IP lysis buffer supplemented with 1X Halt protease and phosphatase inhibitor cocktail (Thermo Scientific, Rockford, IL, USA). After centrifugation of the lysate at 15,000×g for 10 min at 4°C, supernatants were collected and the protein concentration was measured by the bicinchoninic acid (BCA) method. A volume equivalent to a total protein of 8 µg (for COL1A1, COL2A1, COL9A1, ACAN protein expression) and 16 µg (for COL10A1, SOX-9, p-SOX-9 protein expression) of all samples were subjected to SDS-PAGE analysis on a 10% NuPAGE Bis-tris gel (Life Technologies), followed by western blotting to PVDF using the NuPAGE system. After the transfer, the membranes were blocked with 0.5% casein in 1X TBST and probed separately with COL1A1 (Santa Cruz Biotechnology CA, USA; 80565), COL2A1 (ABCAM; ab34712), COL9A1 (Santa Cruz Biotechnology; 376969), COL10A1 (ABCAM; ab182563), SOX-9 (Millipore-ab5535), phospho S181-SOX-9 or p-SOX-9 (ABCAM; ab59252), and Aggrecan (ABCAM; ab3773). β-Actin was used as the respective loading control. After washing the membranes with 1X TBST and incubating with respective horseradish peroxidase (HRP)–linked secondary antibodies incubation procedures, protein bands were visualized using an Immun-star HRP substrate kit (Bio-Rad Laboratories, Hercules, CA, USA) and captured with GE Healthcare Amersham Hyperfilm ECL (GE Healthcare, Piscataway, NJ, USA).
Histology and immunohistochemistry
Cell-seeded constructs were rinsed with 1X PBS, fixed in 4% formalin for 24 h, and embedded in paraffin. Histological sections of 15 µm thickness were prepared from selected construct using standard histological procedures with Leica Bond III at the Tissue Science Facility, University of Nebraska Medical Center (Omaha, NE, USA). Sections were separately stained with hematoxylin and eosin (H&E), Alcian Blue (AB), and Masson Trichrome (MTC) and also subjected to immunohistochemical staining using antibodies specific to COL1A1 (ab138492; ABCAM, MA) and COL2A1 (ab34712; ABCAM, MA). Bond DAB enhancer reaction was performed using the Bond polymer refine detection kit (Leica Biosystems, Richmond, IL, USA) to visualize protein expression on the respective scaffolds.
Statistical analysis
All results were expressed as a mean with standard deviations (SDs). Sample sizes (n) specific to individual analysis were indicated in the associated methods and figure legends. One-way analysis of variance (ANOVA) with replication was used to compare all study groups/scaffold type. A pairwise Student’s t-test with unequal variance was used to observe significant changes among either stimulated or non-stimulated scaffold type at each sampling day. Difference was considered significant when p < 0.05 (denoted as *), p < 0.01 (denoted as **), and p < 0.001 (denoted as ***).
Results and discussion
Lineage-dependent conversion of MSCs into chondrocytes, in vitro, is impacted by a multitude of factors and stands as a special culture system that seeks to mimic the critical steps of limb bud chondrogenesis.33 Culture conditions, growth factors, scaffold microstructure, and stiffness and mechanical microenvironment have all shown to impact the chondrogenesis of hMSCs.13,14,24 Previous reports have shown that certain distinct cell-specific transcription factors (i.e. master genes) control cell lineage commitment of MSCs into chondrocytes, and SOX-9 has been identified as the only transcriptional factor that controls COL2A1 expression.34 SOX-9 has been shown to play a pivotal role in the commitment of MSCs to a chondrogenic cell lineage, and in cooperation with SOX-5 and SOX-6, it regulates chondrocyte proliferation, maturation, and matrix production.21,34
US has been previously employed to promote chondrogenesis of hMSCs.35–37 However, previous studies involving US have used low-intensity pulsed US (1.5 MHz, 1.0 kHz repeat, 6–40 min) to stimulate in vitro MSC cultures.35,36,38–40 As a significant departure from such strategies, we have employed low-intensity continuous US of 5.0 MHz to stimulate hMSCs seeded in 3D matrices. What also sets us apart is the fact that we are stimulating the chondrocytes at 5.0 MHz, the primary resonant frequency at which the cytoplasmic and nuclear stress is maximized and resulting in enhanced mechanotransduction.19,23 Previously, we have modeled mammalian cell dynamics in response to US, and a primary resonant frequency of 5.2 ± 0.8 MHz was predicted.22 The bioeffects of US are a function of geometry of the culture chamber, transducer placement, scaffold properties, and the set-up, to name a few variables.20 We have undertaken a theoretical evaluation of the US-assisted bioreactor used in this study and systematically evaluated the power delivery to the bioreactor and calculated the US field in the different media that constitute the bioreactor, including the porous scaffold that is used in this article.29,41 We have also recently established via modeling and experimentation that the open pore architecture and pore size of BM scaffolds permit an even distribution of US field within the scaffold volume and minimize attenuation, thus allowing for the maximum coupling of mechanical energy to the cells.29
In an attempt to isolate the impact of US-induced MSC chondrogenesis from growth factors and to ascertain the chondro-inductive ability of US at 5.0 MHz, MSC media (M2, no exogenous growth factors) was used to culture hMSC pellets, MSCs encapsulated in Glycosan, and MSCs seeded on macroporous BM scaffolds in these initial sets of experiments. The SOX-9 gene expression was assessed and shown in Figure 2. In comparison to controls (day-0), MSC pellets had a 7-fold higher expression of SOX-9 and MSC pellets exposed to US stimulation had an 11-fold higher expression of SOX-9. MSC-Glycosan exposed to US stimulation had 2.4-fold higher gene expression of SOX-9 when compared to unstimulated MSCs encapsulated in Glycosan, where MSC-Glycosan had a 13-fold higher expression compared to day-0 control. SOX-9 gene expression levels were similar between day-0 controls and unstimulated cell-seeded BM constructs; however, an 11-fold higher gene expression of SOX-9 was noted on US-stimulated cell-seeded BM constructs. Inserts show the protein expression of COL2A1 by western blotting in MSCs encapsulated in Glycosan and MSCs seeded in BM constructs. The successful differentiation of MSCs to chondrocytes in pellet cultures in the presence of TGFβ3 (10 ng/mL) is also demonstrated. Irrespective of the cellular microenvironment, Figure 2 shows that SOX-9 (a type-II collagen transcription factor) expression was enhanced by US, in the absence of exogenously added growth factors. The ability of US to impact hMSC proliferation in the absence of TGFβ3 was also evaluated and depicted in Figure 3(a). Our results indicate that US impacts hMSC proliferation in the absence of exogenously added growth factors. Although the standard method of pellet culture for in vitro MSC chondrogenesis is effective and mimics the MSC condensation phase in the limb bud (i.e. high density and cell-to-cell contact), the requirement for pellet culture makes scale-up problematic in addition to the limitations in pellet size, diffusional limitations, and hypertrophy in the pellet core. As an alternative to pellet cultures, hydrogels have been developed as they can provide higher cell density upon encapsulation and mimic the initial phase of mesenchymal condensation. Indeed, our results with MSC-Glycosan show that the gene expression of SOX-9 was comparable to pellet cultures, with additional increase in SOX-9 gene expression upon US stimulation.
Figure 2.
SOX-9 expression is increased with culture environment and US application. MSC pellets (~5 × 105 cells/pellet, 1 mm diameter), MSCs encapsulated in Glycosan (Hystem-C™) hydrogel matrices (105 cell/mL), and MSCs seeded on macroporous Biomerix™ scaffolds (2 × 104 cells/scaffold, 5 mm × 2.5 mm) were cultured in M2 for 21 days and treated with and without US (5.0 MHz, 2.5 Vpp, 3 min, 4 times/day). M2: high-glucose DMEM, 10% FBS, 100 nM dexamethasone, 50 µg/mL of L-ascorbic acid, and 1X antibiotic–antimyotic™; CDM: M2 with TGFβ3; M3: M2 without dexamethasone. Control is mRNA collected from a MSC pellet at the start of the experiment, 0 days. qRT-PCR analysis for SOX-9 gene expression was performed. Inserts show the protein expression of COL2A1. (a) Cell pellet was stained for COL2A1 (red; chondrocytes) and PPARγ (green; adipocytes); (b) cell pellet was stained with Alcian Blue 8GX to demonstrate glycosaminoglycans—COL2A1 expression at 130 kDa detected by Western blotting, cultured on Glycosan matrix in M2; (c) without US; (d) with US—on BM matrix in M2; (e) without US; and (f) with US.
Figure 3.

(a) Enhancement of hMSC proliferation using US. MSCs were grown on glass coverslips for 4 days in M2 medium with and without US (14 kPa; 3 min; 1, 2, or 4 times/day). Cells were fixed and stained for Ki67 (mitosis marker) and Hoechst (nuclear marker). Five pictures were randomly taken on three coverslips per condition (n = 15), and Ki67 and Hoechst positive cells were counted using ImageJ™. Data were expressed as percent Ki67 positive. (b) hMSC proliferation is increased with US application. hMSC-seeded scaffolds were cultured in CDM for 14 days and in M3 media for another 7 days in the US-assisted bioreactor according to the culture conditions outlined in Table 2. dsDNA was assayed by Picogreen assay. Data were presented as average ± standard deviation (n = 5–8 constructs). Statistically significant data were accounted with respect to respective control and shown as *p < 0.05, **p < 0.01, and ***p < 0.001.
While encouraging findings with respect to chondrogenic differentiation of hMSCs were noted in pellet cultures and in hydrogel-based matrices, in this article, we focused on the chondrogenic differentiation of hMSCs on scaffolds where the cellular milieu is different from pellet cultures, as lineage-dependent conversion of hMSCs on scaffolds is more relevant to tissue engineering applications that aim to develop macroscopic amounts or larger constructs to treat full thickness defects. Previous studies have presented that various types of biomaterial substrates can promote chondrogenesis without the need for pellet culture or supplemented growth factors.42 It is not surprising that SOX-9 expression levels similar to control were noted in MSC-seeded BM constructs without US stimulation, perhaps owing to poor cell-to-cell contact as it is difficult to attain higher cell densities in scaffolds (Figure 2). We have observed the induction of mesenchymal stem cell chondrogenesis under US, notably in the absence of TGFβ supplementation. A key question arising from our study is how US can induce chondrogenesis in the absence of TGFβ. MSCs cultured in vitro are known to express their own TGFβ;43,44 thus, it is possible that TGFβ secreted reaches fairly high concentrations in the supernatant of MSCs cultured in vitro and promotes signaling. However, higher levels of COL2A1 protein expression were noted in MSC-Glycosan cultures when compared to MSC-seeded BM scaffolds, both under US stimulation and no US stimulation; expression levels between non-stimulated MSC-Glycosan and US-stimulated MSC-seeded BM constructs were similar qualitatively. It may then be instructive to attain a higher seeding density in porous scaffolds; hence, in subsequent studies, we have used higher starting seeding densities. These results are consistent with previous studies showing that when MSCs are cultured on scaffolds that do not promote aggregation, chondrogenesis can still proceed, suggesting that with the appropriate artificial environment, aggregate formation or inclusion of TGFβ may not be absolutely necessary to induce chondrogenesis.42 However, the levels of COL2A1 expression are lower on BM scaffolds, leaving room for further enhancement. Chondrogenic differentiation of MSCs, in vivo, is accurately regulated by essential transcription factors and signaling cascades.21,34 For example, during chondrogenesis in vivo, in limb bud mesenchyme, TGFβ signaling is important in the initial stages, where it regulates the expression of SOX-9; hence, many in vitro cultures include TGFβ to induce chondrogenesis.34 Thus, moving forward, we have supplemented the media with TGFβ3 to aid MSC chondrogenesis, in synergy with US stimulation.
Previous studies have documented the use of TGFβ1 or TGFβ3 to aid chondrogenesis in in vitro cultures, and mostly TGFβ concentrations in the ranges of 1–10 ng/mL were employed.24 Prolonged exposure to TGFβ in the culture medium is known to induce hypertrophy or increased COL10A1 expression in MSC cultures.45 We surmise that the presence of TGFβ is essential during the initial phase of hMSC chondrogenesis as compared to later stages when co-cultures of hMSCs and hMSC-derived chondrocytes might exist; hence, culture conditions as depicted in Figure 1 were adopted.
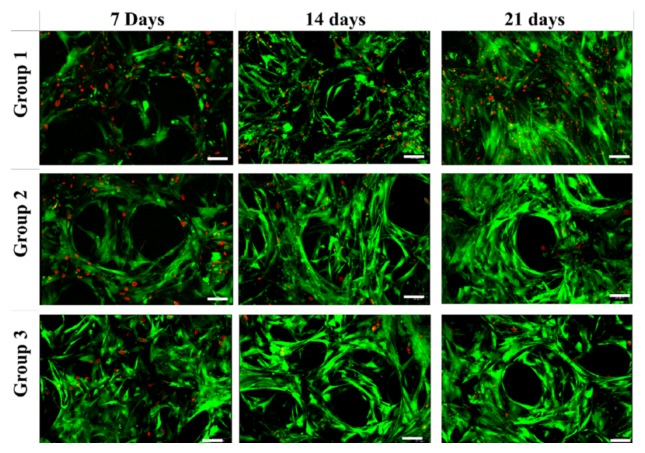
The proliferation and viability of hMSCs and hMSC-derived chondrocytes were assessed and shown in Figure 3(b) and Figure 4, respectively. At day 21, Group 3 had significantly higher cell proliferation (p < 0.01) when compared to groups 1 and 2. Over the culture period, enhanced cell viability was noted in groups 2 and 3 with respect to group 1.
Figure 4.
Cellular viability. hMSC-seeded constructs were cultured in CDM media for 14 days and in M3 for another 7 days in the US-assisted bioreactor according to culture conditions listed in Table 2. Live cells were stained with calceinAM (green color) and dead cells with ethidium (red color) and imaged at 20× magnification (scale bar: 50 µm).
Chondrogenic differentiation of hMSCs seeded on BM scaffolds, cultured for 21 days, was assessed by means of total GAG and collagen content (Figure 5(a) and (b)). Significantly high GAG/DNA (p < 0.01) was observed in group 3 constructs on both day 14 and 21 samples with respect to groups 2 and 3. Collagen/DNA was significantly high for group 3 on day 21 compared to groups 1 and 2.
Figure 5.
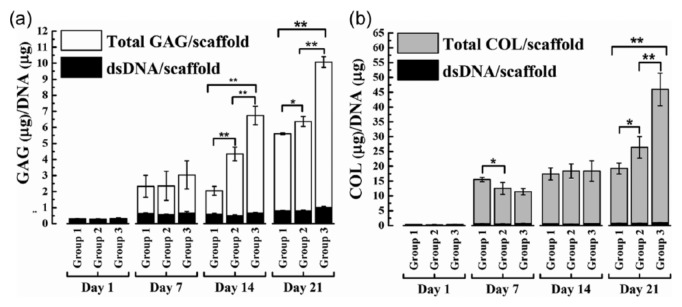
Total glycosaminoglycan (GAG) and collagen (COL) assay: hMSC-seeded scaffolds were cultured for 21 days under mentioned conditions. Total GAG and COL (extrapolated from hydroxyproline content) content per scaffold were estimated at 1, 7, 14, and 21 days. Relative content of (a) GAG to DNA and (b) COL to DNA of group 1, 2, and 3 constructs were plotted. Average with standard deviations are represented (n = 5–8 constructs) (*p < 0.05; **p < 0.01).
miRNA gene regulation is often not a decisive on and off switch but a subtle function that fine-tunes cellular phenotypes.46 As miR-145 is significantly downregulated during MSC chondrogenesis and is silenced in differentiated MSCs,46 we have used miR-145 expression to track the differentiation process under US and TGFβ3. Figure 6 shows the differential expression of miRNA 145 during chondrogenic differentiation of hMSCs under US. Our results suggest that in group 1 (No TGFβ3, No US), as anticipated, hMSCs were not efficiently differentiated into chondrocytes, judging by the lack of decrease in the level of miR-145 expression. hMSCs differentiated into chondrocytes in group 2 (TGFβ3, No US) and group 3 (TGFβ3, US), with group 3 having a 2-fold lower miR-145 when compared to group 2 at day 7, indicating a higher conversion to chondrocytes. Overall, a 20% higher rate of conversion may be inferred based on a linear fit to the data.
Figure 6.

miR-145 gene expression was quantified after 7, 14, and 21 days of culture according to culture conditions listed in Table 2. Gene expression was analyzed by qRT-PCR and normalized to the expression of the house keeping gene snU6. Each bar represents the mean ± standard deviation (n = 3; *p < 0.05, **p < 0.01).
qRT-PCR: quantitative real-time polymerase chain reaction.
The impact of US stimulation on the expression of COL10A1 gene as a function of culture duration was examined by qRT-PCR and is shown in Figure 7. On day 7, the gene expression of hypertrophic marker, COL10A1, was higher in group 2 (TGFβ3, No US) when compared to group 3 (TGFβ3, US) and considerably greater when compared to group 1 (No TGFβ3, No US). The gene expression of COL10A1 in group 1 stayed relatively unchanged during the course of this experiment. On day 21, groups 2 and 3 had similar levels of COL10A1 mRNA levels and were significantly different from mRNA levels in group 1. The relative gene expression of ACAN gene, a member of aggrecan/versican proteoglycan (ACAN), was also carried out. At day 21, compared to group 1, higher levels of ACAN expression were observed on groups 2 and 3; with group 3 presenting the highest gene expression of ACAN. No significant difference was noted between the groups with respect to the gene expression of TGFβ1.
Figure 7.
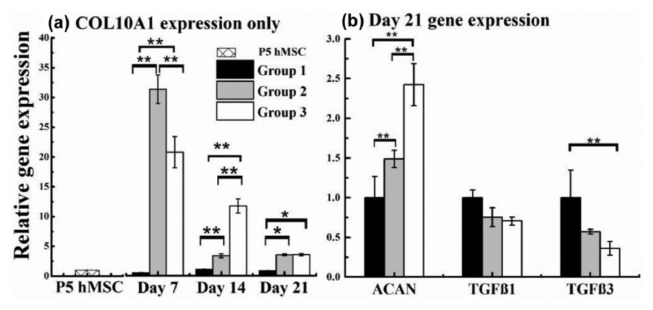
hMSC-seeded Biomerix™ scaffolds were cultured according to culture conditions listed in Table 2. (a) COL10A1 gene expression was presented after 7, 14, and 21 days of culture. (b) Expression for ACAN, TGFβ1, and TGFβ3 genes was shown at day 21. Gene expression was analyzed by qRT-PCR and normalized to the expression of the house keeping gene GAPDH. Each bar represents the mean ± standard deviation (n = 3; *p < 0.05; **p < 0.01).
The survival, proliferation, and ECM production of MSC-derived chondrocyte in response to US was examined by allowing the experiment to progress for an additional week after the removal of TGFβ3 (i.e. CDM) and culturing in M3. In a separate study, we have shown that adult chondrocytes can be successfully cultured over scaffolds in the US-assisted bioreactor for 3 weeks.29 Compared to non-stimulated controls, US-stimulated constructs had a higher cell density throughout the scaffold volume, higher total cell proliferation, a ~13/1 ratio of COL2A1/COL1A1, no detectable COL10A1 gene expression, and uniform deposition of COL2A1 protein. Thus, we are confident that in a future study, we will be able to culture hMSC-derived chondrocytes for an extended period of time and evaluate tissue properties.
The protein expression of ACAN, COL1A1, COL2A2, COL9A1, SOX-9, p-SOX-9 (i.e. p-S181-SOX-9), and COL10A1 were assayed by western blotting and shown in Figure 8. COL10A1 was undetectable. When compared to groups 1 and 2, group 3 showed higher levels of COL2A1 and ACAN. COL1A1 expression was similar between all groups. Protein expression of both SOX-9 and p-SOX-9 was slightly higher in group 3 compared to groups 2 and 1. Our combined gene and protein expression analyses at the end of day 14 indicate that US is able to mediate chondrogenesis of hMSCs seeded on BM scaffolds and this likely happens via SOX-9 mediated pathways.
Figure 8.

Analysis of protein expression. Cell-seeded scaffolds were randomly collected, trypsinized, lysed with IP lysis buffer—1X Halt protease and phosphatase inhibitor cocktail and cell lysates were subjected to SDS-PAGE followed by western blotting. (a) In separate experiments, the membranes were incubated with primary antibodies against ACAN, COL1A1, COL2A2, COL9A1, SOX-9, p-SOX-9 (i.e. p-S181-SOX-9). β-Actin was used as the loading control. (b) All protein bands were quantified by ImageJ™ software and relative protein expression levels were computed with respect to β-actin.
Histological findings on scaffolds harvested from day 21 show cellularity (H&E) and deposition of GAG (AB) and collagen (MTC), shown in Figure 9. Relative expressions of COL1A1 and 2A1 detected by immunohistochemistry (Figure 9) mirrored the changes detected by western blot. COL2A1 expression markedly increased in group 3 with the combination of TGFβ3 and US treatment compared to groups 1 and 2.
Figure 9.
Histological and immunohistochemical analyses. Scaffolds were retrieved on day 21 and subject to histological and immunochemical analyses. H&E, Alcian Blue, and Masson trichrome stains were imaged at 10×, and COL1A1 and COL2A1 were imaged at 4× magnification.
While in this article we have evaluated the gene or protein expression profiles from a mixed population of hMSCs and hMSCs that were converted to chondrocytes, we are able to partly comment of the efficacy of conversion based on miRNA expression. In our ongoing work, we plan to quantify the efficacy and uniformity of chondrogenesis by histological and immunohistochemical staining to ascertain the total number of cells that stain positively for proteoglycan/chondroitin sulfate relative to the total cell number (DAPI).
In summary, our results show that US stimulation at 5.0 MHz can direct chondrogenesis of hMSCs in the absence of exogenous TGFβ. Our study also highlights the importance of the scaffold microenvironment in regulating the lineage-specific conversion of hMSCs. In the absence of growth factors, chondrogenic differentiation (as inferred from COL2A1 expression) on the macroporous scaffolds appeared to be lower to that which occurs in standard pellet cultures, suggesting the use of higher starting seeding densities. We have also demonstrated that US acts synergistically with TGFβ to promote efficient chondrogenesis of MSCs seeded on polymeric scaffolds that limit cell-to-cell contact. Our future efforts will be focused on the further culture of hMSC-derived chondrocytes for longer duration, and a comprehensive evaluation will be carried out. Also, we will study the underlying molecular mechanisms that impact MSC chondrogenesis in hydrogel-based and macroporous polymeric scaffolds under US stimulation and non-stimulation. The ultimate goal is to connect the cellular microscopic inputs to macroscopic inputs like tissue properties.
Acknowledgments
We thank Teresa Fangman from the Morrison Microscopy Core Research Facility at University of Nebraska–Lincoln and Aubre Phillips from the Tissue Science Facility at University of Nebraska Medical Center. We also acknowledge Dr Nicholas Whitney for preliminary discussions and experiments. We also thank the Nebraska Research Initiative and the UNL Center for Biotechnology for support.
Footnotes
Declaration of conflicting interests: The authors declare that there is no conflict of interest.
Funding: This work was supported in part by the Stem Cell-2012-08 from the Nebraska Department of Health and Human Services.
References
- 1. Fulco I, Largo RD, Miot S, et al. Toward clinical application of tissue-engineered cartilage. Facial Plast Surg 2013; 29: 99–105. [DOI] [PubMed] [Google Scholar]
- 2. Mhashilkar AM, Atala A. Advent and maturation of regenerative medicine. Curr Stem Cell Res Ther 2012; 7: 430–445. [DOI] [PubMed] [Google Scholar]
- 3. Oldershaw RA. Cell sources for the regeneration of articular cartilage: the past, the horizon and the future. Int J Exp Pathol 2012; 93: 389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jung S, Panchalingam KM, Wuerth RD, et al. Large-scale production of human mesenchymal stem cells for clinical applications. Biotechnol Appl Bioc 2012; 59: 106–120. [DOI] [PubMed] [Google Scholar]
- 5. Kshitiz, Park J, Kim P, et al. Control of stem cell fate and function by engineering physical microenvironments. Integr Biol 2012; 4: 1008–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dahlin RL, Ni M, Meretoja VV, et al. TGF-beta3-induced chondrogenesis in co-cultures of chondrocytes and mesenchymal stem cells on biodegradable scaffolds. Biomaterials 2014; 35: 123–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Solorio LD, Vieregge EL, Dhami CD, et al. Engineered cartilage via self-assembled hMSC sheets with incorporated biodegradable gelatin microspheres releasing transforming growth factor-beta1. J Control Release 2012; 158: 224–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hui TY, Cheung KM, Cheung WL, et al. In vitro chondrogenic differentiation of human mesenchymal stem cells in collagen microspheres: influence of cell seeding density and collagen concentration. Biomaterials 2008; 29: 3201–3212. [DOI] [PubMed] [Google Scholar]
- 9. Mahmood TA, Shastri VP, van Blitterswijk CA, et al. Tissue engineering of bovine articular cartilage within porous poly(ether ester) copolymer scaffolds with different structures. Tissue Eng 2005; 11: 1244–1253. [DOI] [PubMed] [Google Scholar]
- 10. Xin X, Hussain M, Mao JJ. Continuing differentiation of human mesenchymal stem cells and induced chondrogenic and osteogenic lineages in electrospun PLGA nanofiber scaffold. Biomaterials 2007; 28: 316–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Meinel L, Hofmann S, Karageorgiou V, et al. Engineering cartilage-like tissue using human mesenchymal stem cells and silk protein scaffolds. Biotechnol Bioeng 2004; 88: 379–391. [DOI] [PubMed] [Google Scholar]
- 12. Zhang X, Zhang Y, Wang Z, et al. The effect of non-growth factors on chondrogenic differentiation of mesenchymal stem cells. Cell Tissue Bank 2013; 15; 319–327. [DOI] [PubMed] [Google Scholar]
- 13. Danisovic L, Varga I, Polak S. Growth factors and chondrogenic differentiation of mesenchymal stem cells. Tissue Cell 2012; 44: 69–73. [DOI] [PubMed] [Google Scholar]
- 14. Indrawattana N, Chen G, Tadokoro M, et al. Growth factor combination for chondrogenic induction from human mesenchymal stem cell. Biochem Biophys Res Commun 2004; 320: 914–919. [DOI] [PubMed] [Google Scholar]
- 15. Betre H, Chilkoti A, Setton LA. A two-step chondrocyte recovery system based on thermally sensitive elastin-like polypeptide scaffolds for cartilage tissue engineering. In: Proceedings of the second joint EMBS-BMES conference, Houston, TX, 23–26 October 2002. [Google Scholar]
- 16. Masuda K, Sah RL, Hejna MJ, et al. A novel two-step method for the formation of tissue-engineered cartilage by mature bovine chondrocytes: the alginate-recovered-chondrocyte (ARC) method. J Orthop Res 2003; 21: 139–148. [DOI] [PubMed] [Google Scholar]
- 17. Xue K, Qi L, Zhou G, et al. A two-step method of constructing mature cartilage using bone marrow-derived mesenchymal stem cells. Cells Tissues Organs 2013; 197: 484–495. [DOI] [PubMed] [Google Scholar]
- 18. Subramanian A, Turner JA, Budhiraja G, et al. Ultrasonic bioreactor as a platform for studying cellular response. Tissue Eng Part C Methods 2013; 19: 244–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Noriega S, Mamedov T, Turner JA, et al. Intermittent applications of continuous ultrasound on the viability, proliferation, morphology, and matrix production of chondrocytes in 3D matrices. Tissue Eng 2007; 13: 611–618. [DOI] [PubMed] [Google Scholar]
- 20. Louw TM. Mathematical modeling of ultrasound in tissue engineering: from bioreactors to the cellular scale. Lincoln, NE: Chemical & Biomolecular Engineering, University of Nebraska–Lincoln, 2013, 315 pp. [Google Scholar]
- 21. Akiyama H. Control of chondrogenesis by the transcription factor Sox9. Mod Rheumatol 2008; 18: 213–219. [DOI] [PubMed] [Google Scholar]
- 22. Louw TM, Budhiraja G, Viljoen HJ, et al. Mechanotransduction of ultrasound is frequency dependent below the cavitation threshold. Ultrasound Med Biol 2013; 39: 1303–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Whitney NP, Lamb AC, Louw TM, et al. Integrin-mediated mechanotransduction pathway of low-intensity continuous ultrasound in human chondrocytes. Ultrasound Med Biol 2012; 38: 1734–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li Z, Kupcsik L, Yao S-J, et al. Mechanical load modulates chondrogenesis of human mesenchymal stem cells through the TGF-beta pathway. J Cell Mol Med 2010; 14: 1338–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang L, Su P, Xu C, et al. Chondrogenic differentiation of human mesenchymal stem cells: a comparison between micromass and pellet culture systems. Biotechnol Lett 2010; 32: 1339–1346. [DOI] [PubMed] [Google Scholar]
- 26. Burdick JA, Prestwich GD. Hyaluronic acid hydrogels for biomedical applications. Adv Mater 2011; 23: H41–H56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Serban MA, Scott A, Prestwich GD. Use of hyaluronan-derived hydrogels for three-dimensional cell culture and tumor xenografts. Curr Protoc Cell Biol 2008; Chapter 10: Unit 10.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zheng Shu X, Liu Y, Palumbo FS, et al. In situ crosslinkable hyaluronan hydrogels for tissue engineering. Biomaterials 2004; 25: 1339–1348. [DOI] [PubMed] [Google Scholar]
- 29. Guha Thakurta S, Kraft M, Viljoen HJ, et al. Enhanced depth-independent chondrocyte proliferation and phenotype maintenance in an ultrasound bioreactor and an assessment of ultrasound dampening in the scaffold. Acta Biomater 2014; 10: 4798–4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hoemann CD, Sun J, Chrzanowski V, et al. A multivalent assay to detect glycosaminoglycan, protein, collagen, RNA, and DNA content in milligram samples of cartilage or hydrogel-based repair cartilage. Anal Biochem 2002; 300: 1–10. [DOI] [PubMed] [Google Scholar]
- 31. Barbosa I, Garcia S, Barbier-Chassefiere V, et al. Improved and simple microassay for sulfated GAGs quantification in biological extracts and its use in skin and muscle tissue studies. Glycobiology 2003; 13: 647–653. [DOI] [PubMed] [Google Scholar]
- 32. Kliment CR, Englert JM, Crum LP, et al. A novel method for accurate collagen and biochemical assessment of pulmonary tissue utilizing one animal. Int J Clin Exp Pathol 2011; 4: 349–355. [PMC free article] [PubMed] [Google Scholar]
- 33. Egawa S, Miura S, Yokoyama H, et al. Growth and differentiation of a long bone in limb development, repair and regeneration. Dev Growth Differ 2014; 56: 410–424. [DOI] [PubMed] [Google Scholar]
- 34. De Crombrugghe B, Lefebvre V, Behringer RR, et al. Transcriptional mechanisms of chondrocyte differentiation. Matrix Biol 2000; 19: 389–394. [DOI] [PubMed] [Google Scholar]
- 35. Ebisawa K, Hata K, Okada K, et al. Ultrasound enhances transforming growth factor beta-mediated chondrocyte differentiation of human mesenchymal stem cells. Tissue Eng 2004; 10: 921–929. [DOI] [PubMed] [Google Scholar]
- 36. Schumann D, Kujat R, Zellner J, et al. Treatment of human mesenchymal stem cells with pulsed low intensity ultrasound enhances the chondrogenic phenotype in vitro. Biorheology 2006; 43: 431–443. [PubMed] [Google Scholar]
- 37. Kang KS, Hong JM, Kang JA, et al. Osteogenic differentiation of human adipose-derived stem cells can be accelerated by controlling the frequency of continuous ultrasound. J Ultrasound Med 2013; 32: 1461–1470. [DOI] [PubMed] [Google Scholar]
- 38. Lai CH, Chen SC, Chiu LH, et al. Effects of low-intensity pulsed ultrasound, dexamethasone/TGF-beta1 and/or BMP-2 on the transcriptional expression of genes in human mesenchymal stem cells: chondrogenic vs. osteogenic differentiation. Ultrasound Med Biol 2010; 36: 1022–1033. [DOI] [PubMed] [Google Scholar]
- 39. Cui JH, Park SR, Park K, et al. Preconditioning of mesenchymal stem cells with low-intensity ultrasound for cartilage formation in vivo. Tissue Eng 2007; 13: 351–360. [DOI] [PubMed] [Google Scholar]
- 40. Lee HJ, Choi BH, Min BH, et al. Low-intensity ultrasound stimulation enhances chondrogenic differentiation in alginate culture of mesenchymal stem cells. Artif Organs 2006; 30: 707–715. [DOI] [PubMed] [Google Scholar]
- 41. Louw T, Subramanian A, Viljoen HJ. Theoretical evaluation of ultrasonic bioreactors. Ultrasound Med Biol, in press. DOI: 10.1016/j.ultrasmedbio.2014.12.015. [DOI] [PubMed] [Google Scholar]
- 42. Glennon-Alty L, Williams R, Dixon S, et al. Induction of mesenchymal stem cell chondrogenesis by polyacrylate substrates. Acta Biomater 2013; 9: 6041–6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sawada R, Ito T, Tsuchiya T. Changes in expression of genes related to cell proliferation in human mesenchymal stem cells during in vitro culture in comparison with cancer cells. J Artif Organs 2006; 9: 179–184. [DOI] [PubMed] [Google Scholar]
- 44. Goessler UR, Bugert P, Bieback K, et al. In-vitro analysis of the expression of TGFbeta-superfamily-members during chondrogenic differentiation of mesenchymal stem cells and chondrocytes during dedifferentiation in cell culture. Cell Mol Biol Lett 2005; 10: 345–362. [PubMed] [Google Scholar]
- 45. James CG, Appleton CT, Ulici V, et al. Microarray analyses of gene expression during chondrocyte differentiation identifies novel regulators of hypertrophy. Mol Biol Cell 2005; 16: 5316–5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yang B, Guo H, Zhang Y, et al. MicroRNA-145 regulates chondrogenic differentiation of mesenchymal stem cells by targeting Sox9. PLoS ONE 2011; 6: e21679. [DOI] [PMC free article] [PubMed] [Google Scholar]