Abstract
Multiple sclerosis is an autoimmune disease of the CNS resulting in degeneration of myelin sheaths and loss of oligodendrocytes, which means that protection and electrical insulation of axons and rapid signal propagation are impaired, leading to axonal damage and permanent disabilities. Partial replacement of lost oligodendrocytes and remyelination can occur as a result of activation and recruitment of resident oligodendroglial precursor cells. However, the overall remyelination capacity remains inefficient because precursor cells often fail to generate new oligodendrocytes. Increasing evidence points to the existence of several molecular inhibitors that act on these cells and interfere with their cellular maturation. The p57kip2 gene encodes one such potent inhibitor of oligodendroglial differentiation and this study sheds light on the underlying mode of action. We found that subcellular distribution of the p57kip2 protein changed during differentiation of rat, mouse, and human oligodendroglial cells both in vivo and in vitro. Nuclear export of p57kip2 was correlated with promoted myelin expression, higher morphological phenotypes, and enhanced myelination in vitro. In contrast, nuclear accumulation of p57kip2 resulted in blocked oligodendroglial differentiation. Experimental evidence suggests that the inhibitory role of p57kip2 depends on specific interactions with binding proteins such as LIMK-1, CDK2, Mash1, and Hes5 either by controlling their site of action or their activity. Because functional restoration in demyelinating diseases critically depends on the successful generation of oligodendroglial cells, a therapeutic need that is currently unmet, the regulatory mechanism described here might be of particular interest for identifying suitable drug targets and devising novel therapeutic approaches.
Keywords: inhibitor, mode of action, myelin repair, nuclear export, p57kip2, regeneration
Introduction
In inflammatory demyelinating diseases such as multiple sclerosis (MS), myelin sheaths and oligodendrocytes are attacked and damaged as a result of a misguided immune response. This autoimmune process creates demyelinated lesions, which have impaired electrical signal transduction and are prone to axonal degeneration throughout the CNS (Lassmann et al., 2007). Due to the restricted regeneration capacity of the CNS, recurring demyelination inevitably results in permanent damage and functional loss. However, in light of the increasing number of immunomodulatory treatments available for the treatment of MS disease activity, one of the main future treatment objectives is the functional restoration of such demyelinated areas (Mullard, 2011). Endogenous myelin repair activities exist and, particularly in early MS stages, demyelination can be counteracted by the recruitment, activation, and differentiation of resident oligodendroglial precursor cells (OPCs) (Chang et al., 2000). Nevertheless, remyelination efficiency is generally low and repair gradually fails as the disease progresses over time, ultimately leading to significant clinical disability. This failure of remyelination was shown to be mainly due to a blockade of oligodendroglial differentiation and maturation processes (Wolswijk, 1998; Chang et al., 2002; Kuhlmann et al., 2008; Kremer et al., 2011). A promising tactic in the search for regenerative approaches is to exploit features of the naturally occurring repair process. Therefore, identifying endogenous inhibitory pathways that can be targeted for future remyelination therapies is a step toward this goal.
We previously identified a negative regulator of oligodendrogenesis encoded by the p57kip2 gene (Kremer et al., 2009). As a member of the cip/kip family, p57kip2 was originally described as a cyclin-dependent kinase inhibitor, but several additional cellular processes were shown to depend on cip/kip proteins (Besson et al., 2008). Importantly, our observations revealed that p57kip2 is a potent inhibitor of both peripheral nervous system and CNS glial cell differentiation and it appears to regulate adult neural stem cell fate decisions (Heinen et al., 2008; Kremer et al., 2009; Jadasz et al., 2012). In these studies, short hairpin RNA-mediated gene suppression of p57kip2 was found to accelerate morphological maturation and to promote myelin expression. However, the mode of action of p57kip2 in interfering with cellular maturation remains unknown.
In this study, we investigated the underlying molecular mechanism and found that the specific relocation of p57kip2 from the nucleus to cytoplasm appears to be a key functional process. Moreover, we identified four proteins that interact directly with p57kip2 in oligodendroglial cells. We found that LIM kinase-1 (LIMK-1) and cyclin-dependent kinase 2 (CDK2) bind to p57kip2 and their activity and role in the differentiation process appear to depend on their subcellular localization. Conversely, we detected interactions between p57kip2 and the transcription factors Mash1 and Hes5. For Mash1, we demonstrated p57kip2-dependent transactivation properties, whereas Hes5 was found to be shuttled out of the nucleus along with p57kip2, thus neutralizing its predominantly negative impact on myelin gene transcription.
Materials and Methods
Oligodendroglial cell culture.
Generation of primary OPCs from postnatal day zero (P0) cerebral rat cortices (Wistar rats of either sex) was performed as described previously (Göttle et al., 2010). Anti-A2B5 staining (catalog #MAB312R RRID:AB_11213098; Merck) revealed that the cultures consisted of 98% oligodendroglial cells (data not shown). OPCs were either kept in proliferation-supporting high-glucose DMEM-based Sato medium (Life Technologies), supplemented with 10 ng/ml recombinant human bFGF (PeproTech) and 10 ng/ml recombinant human PDGF-AA (R&D Systems), or differentiation was initiated by Sato medium depleted of growth factors and supplemented with 0.5% fetal calf serum (PAA Laboratories). Recombinant human CXCL12 (R&D Systems) stimulation was performed at concentrations of 100 ng/ml in differentiation medium. Ratjadone (Enzo Life Sciences) blocking experiments were performed at a concentration of 0.5 ng/ml. Primary A2B5-positive OPCs of human origin were purchased from 3H Biomedical and cultured according to the supplier's protocols.
Myelinating coculture.
Dissociated neuron–oligodendrocyte cocultures were obtained from embryonic day 16 (E16) rat cerebral cortex (Wistar rats of either sex) (Pang et al., 2012). Cortical cells were plated on 15 mm poly-d-lysine (0.1 mg/ml)-coated coverslips (65,000 cells per coverslip) and kept in myelination medium consisting of N2 and neurobasal medium (ratio 1:1; Life Technologies) including NGF (50 ng/ml) and NT-3 (10 ng/ml) (both R&D Systems). After 10 d in vitro (DIV10), insulin was excluded and the ratio of the insulin-free N2 to neurobasal medium including B27 supplement was adjusted to 4:1. This myelination medium was further supplemented with 60 ng/ml tri-iodo-thyronine (T3; Sigma). Final concentrations of individual N2 medium components (DMEM-F12 based, high glucose; Life Technologies) were insulin (10 μg/ml), transferrin (50 μg/ml), sodium selenite (5.2 ng/ml), hydrocortisone (18 ng/ml), putrescine (16 μg/ml), progesterone (6.3 ng/ml), biotin (10 ng/ml), N-acetyl-l-cysteine (5 μg/ml) (all Sigma), bovine serum albumin (0.1%; Roth) and penicillin–streptomycin (50 units/ml; Life Technologies). At DIV26, coverslips were washed with PBS, fixed with 4% paraformaldehyde, and processed for immunofluorescent staining.
OPC transfection and assessment of morphology.
OPC transfections were performed using NanoJuice transfection reagent (Merck) in proliferation medium as described previously (Göttle et al., 2010). For transplantation of primary OPCs onto neuron–oligodendrocyte cocultures, cells were transfected using the Nucleofector II device and the Basic Neuron Small Cell Number Nucleofector Kit (all Lonza). At DIV16, transfected primary oligodendrocyte precursors (100,000 cells per coverslip) were added to the coculture. Transfection with overexpression constructs pIRES (empty control vector), pIRES_p57 (wild-type p57kip2 overexpression vector), and pIRES_NLS [overexpression of p57kip2 with mutated nuclear localization sequence (NLS)] were performed in combination with the pcDNA3.1-HygB-citrine expression vector (yellow fluorescent protein; Heikal et al., 2000) in a 5:1 ratio (Heinen et al., 2008) to visualize cells. In general, when gene modulation was applied, only transfected cells were scored and these cells were identified by means of the cotransfected citrine (yellow fluorescent protein) expression vector. To assess the degree of cellular maturation, transfected (yellow fluorescent) cells only were scored and analyzed by fluorescence microscopy (Zeiss Axioplan) while applying an evaluation tool based on morphological cell parameters (processes, branches) and distinguishing simple, medium, and high morphologies, as described previously (Kremer et al., 2009; Göttle et al., 2010). Overexpression of Mash1 (pIRES_Mash1) and p57kip2 (pIRES_p57) was performed in the presence of a 10-fold smaller amount of the reporter plasmid tdTomato (Castro et al., 2006). This combination allowed us to assess the transactivation properties of this transcription factor directly because only transfected cells were analyzed. Experiments with transfected cells were performed at least three times (exact numbers are specified in the figure legends) and, on average, 763 cells were counted for each experiment (exact numbers are indicated in the figure legends).
Immunostaining.
Fixed cells were permeabilized with PBS containing 0.01% Triton X-100 (Sigma) and unspecific staining was blocked with 10% normal goat serum or donkey serum (Sigma) for 40 min (Göttle et al., 2010). Cells were then incubated with primary antibodies in PBS overnight at 4°C using the following dilutions: mouse anti-adenomatosis polyposis coli [clone CC1, 1/1000 (rat), 1/300 (human); GeneTex, catalog #GTX16794 RRID:AB_422404], mouse anti-galactocerebroside C (GalC, 1/500; catalog #MAB342, RRID:AB_94857; Millipore), mouse anti-myelin oligodendrocyte glycoprotein (MOG, 1/1000, catalog #MAB5680 RRID:AB_1587278; Millipore), mouse anti-myelin basic protein (MBP) and anti-2′,3′-cyclic-nucleotide 3′-phosphodiesterase (CNPase) antibodies (1/500, catalog #SMI-94R-500 RRID:AB_10124143, Covance, and 1/1000, catalog #SMI-91R-500 RRID:AB_510038, Covance, respectively), rabbit anti-p57kip2 [1/500 (rat, human); catalog #P0357 RRID:AB_260850; Sigma-Aldrich], rabbit anti-LIMK-1 (1/500; catalog #611748 RRID:AB_399224; BD Biosciences), mouse anti-CDK2 (1/75, catalog #GTX22363 RRID:AB_369268; GeneTex), mouse anti-Sox10 (1/100; catalog #MAB2864 RRID:AB_2195180; R&D Systems), rabbit anti-Hes5 (1/500; catalog #ab25374 RRID:AB_448776; Abcam), mouse anti-Hes5 (1/200, catalog #CABT-18331MH RRID:AB_11397757; Creative Biomart), rat anti-platelet-derived growth factor receptor α (PDGFRα, 1/500, catalog #16-1401-82 RRID:AB_529483; eBioscience), goat anti-PDGFRα (1/400, catalog #GT15150-100 RRID:AB_1610460; Acris Antibodies), and mouse anti-Mash1 (1/200; Nakatani et al., 2013). Fixed cocultures were blocked with PBS containing 0.5% Triton X-100 and 2% normal goat serum and then incubated overnight in 0.1% Triton X-100 and 2% normal goat serum containing primary antibodies anti-MBP (1/250), anti-p57kip2 (1/500), and chicken anti-βIII-tubulin (1/1000; catalog #TUJ RRID:AB_2313564; Aves Labs). After 24 h, coverslips were washed with PBS and then incubated with secondary antibodies in PBS for 2 h: Alexa Fluor 488 (catalog #A11001 RRID:AB_10566289 and catalog #A11034 RRID:AB_10562715; Life Technologies), Alexa Fluor 405 (catalog #A31553 RRID:AB_221604; Invitrogen), Alexa Fluor 594 (catalog #A11005 RRID:AB_10561507 and catalog #A11037 RRID:AB_10561549; Life Technologies), and DyLight 405-conjugated antibodies (catalog #603-146-126 RRID:AB_1961602; Rockland Immunochemicals), all dilutions 1/500. Nuclei were stained with 4′,6-diamidin-2-phenylindol (DAPI; Roche). Images (20×; Zeiss Axionplan2 microscope) were captured using the same light intensity and filters for all images to be compared and processed with Axiovision 4.2 software (RRID:SciRes_000111; Zeiss). The analysis was done using Java software (ImageJ, RRID:nif-0000-30467/Wright Cell Imaging Facility, RRID:nif-0000-30471). Immunopositive cells were counted in nine randomly chosen fields per coverslip. The total number of cells per field was determined by DAPI staining. Experiments were performed at least three times (exact numbers are specified in the figure legends) and, on average, 7242 cells were counted for each experiment (exact numbers are indicated in the figure legends). For quantification, the number of immunopositive cells was compared with the total cell number and expressed as percentage (mean ± SEM).
Proximity ligation assay.
Direct protein–protein interactions were investigated by means of a proximity ligation assay (PLA) (Söderberg et al., 2006) using Duolink In Situ Red Starter Kit Mouse/Rabbit (Sigma). Cells were rapidly washed with ice-cold PBS and then fixed at room temperature for 10 min in 4% paraformaldehyde. Cells were further rinsed 3 × 3 min with PBS containing 0.1% Triton X-100 (Sigma) and then washed 3 × 2 min with 0.05% Tween 20 in TBS to allow permeabilization. Next, one droplet (40 μl) of Duolink II blocking solution was added to each coverslip, followed by incubation in a preheated humidity chamber for 1 h at 37°C. The blocking solution was then tapped off and cells were incubated for 1 h at 37°C with primary antibodies (dilutions as described above). Duolink anti-rabbit PLUS and anti-mouse MINUS secondary antibodies and red detection reagents were used and antibody incubation, ligation, amplification, and washing steps were performed according to the supplier's manual. Coverslips were then incubated with 50 μg/ml phalloidin (Sigma) for visualization of cells, dried, mounted using Duolink Mounting Media with DAPI, and analyzed using Axiovision 4.2 software (Zeiss) and image processing and analysis in Java software (ImageJ, RRID:nif-0000-30467/Wright Cell Imaging Facility, RRID:nif-0000-30471).
RNA preparation, cDNA synthesis, and quantitative RT-PCR.
RNA purification, cDNA synthesis, and determination of gene expression levels by means of quantitative real-time RT-PCR were all performed as described previously (Göttle et al., 2010). Primer sequences were determined using PrimerExpress 2.0 software (Life Technologies) and tested for the generation of specific amplicons (sequences are available upon request). GAPDH and ODC were used as reference genes and relative gene expression levels were determined according to the ΔΔCt method (Life Technologies). Each sample was measured in quadruplicate.
Cuprizone-mediated demyelination in the corpus callosum.
Cuprizone-mediated demyelination of adult female C57BL/6 mice was induced as described previously (Cate et al., 2010). After 4 weeks of cuprizone challenge, recombinant human BMP4, mouse noggin (both R&D Systems) dissolved in artificial CSF at a dose of 400 ng/d, or artificial CSF only was delivered into the lateral ventricle by mini-osmotic pumps (ALZET model 1007;; Durect) at a 0.50 μl/h flow rate using Brain Infusion Kit III for 5 d of infusion with continued cuprizone challenge, as described previously (Cate et al., 2010). Brain tissue was collected, cryosectioned, and stained as described previously (Cate et al., 2010). All cell counts and area analyses were performed blind to experimental treatment. Immunopositive cells and area measures were quantified as described previously (Cate et al., 2010) and data are expressed as mean per square millimeter ± SEM. Images (20×) were taken from 3–6 sections 50 μm apart for each animal at locations between −0.46 and −1.22 mm bregma with the midline corpus callosum (CC) at the epicenter and lateral borders determined by the image width.
Plasmid construction.
Overexpression of wild-type rat p57kip2/Cdkn1c (GenBank accession number gi: 76257399) was achieved using the pIRES_p57 expression vector containing an EcoRI-EcoRI fragment of the rat p57kip2 cDNA cloned into pIRES2-EGFP vector (BD Biosciences) as described previously (Heinen et al., 2008). Point mutations (887-CAAGCG-892 to 887-GCGGCG-892) were induced by means of artificial gene generation (custom design manufactured by GeneArt). This mutated fragment was cloned into pIRES2-EGFP vector to generate the pIRES_NLS expression vector harboring substituted amino acids K291 and R292 to A291 and A292, respectively, within the predicted NLS sequence 291-KRKR-294. Sequence verification was done by GeneArt.
Yeast two-hybrid analysis.
Yeast two-hybrid screening was performed by Hybrigenics Services (http://www.hybrigenics-services.com). The coding sequence for rat p57kip2/Cdkn1c (GenBank accession number gi: 76257399) was PCR amplified and cloned into pB29 as an N-terminal fusion to LexA (N-Cdkn1c-LexA-C). The construct was verified by sequencing the entire insert and used as a bait to screen a random-primed rat hippocampus cDNA library constructed into pP6. pB29 and pP6 derive from the original pBTM116 (Vojtek and Hollenberg, 1995; Béranger et al., 1997) and pGADGH (Bartel, 1993) plasmids, respectively. We screened 116 × 106 clones (12-fold the complexity of the library) using a mating approach with YHGX13 (Y187 ade2-101::loxP-kanMX-loxP, matα) and L40ΔGal4 (mata) yeast strains as described previously (Fromont-Racine et al., 1997). We selected 63 His+ colonies on a medium lacking tryptophan, leucine, and histidine. The prey fragments of the positive clones were amplified by PCR and sequenced at their 5′ and 3′ junctions. The resulting sequences were used to identify the corresponding interacting proteins in the GenBank database using a fully automated procedure. A confidence score was attributed to each interaction as described previously (Formstecher et al., 2005).
Statistical analysis.
Data are presented as mean ± SEM and significance was assessed by two-sided Student's t test, unpaired comparison for means (GraphPad Prism, RRID:rid_000081). The experimental groups were considered significantly different at *p < 0.05, **p < 0.01, and ***p < 0.001; n represents the number of independent experiments.
Results
In addition to a transient downregulation of p57kip2 expression within the spinal cord during the course of MOG-induced experimental autoimmune encephalomyelitis, nuclear export and cytoplasmic accumulation of p57kip2 protein was observed in oligodendroglial cells (Kremer et al., 2009). We examined the CC of mice using confocal laser-scanning microscopy to evaluate p57kip2 protein expression in resident PDGFRα-positive OPCs (Fig. 1). This demonstrated that resident OPCs either exhibited nuclear (Fig. 1A–A′′′′) or, more rarely, cytoplasmic p57kip2 signals (Fig. 1B–B′′′′) in healthy adult mice. Collectively, these findings suggest that, in addition to gene regulation, nucleocytoplasmic protein shuttling might also be involved as a regulatory mechanism of p57kip2's inhibitory function.
Figure 1.
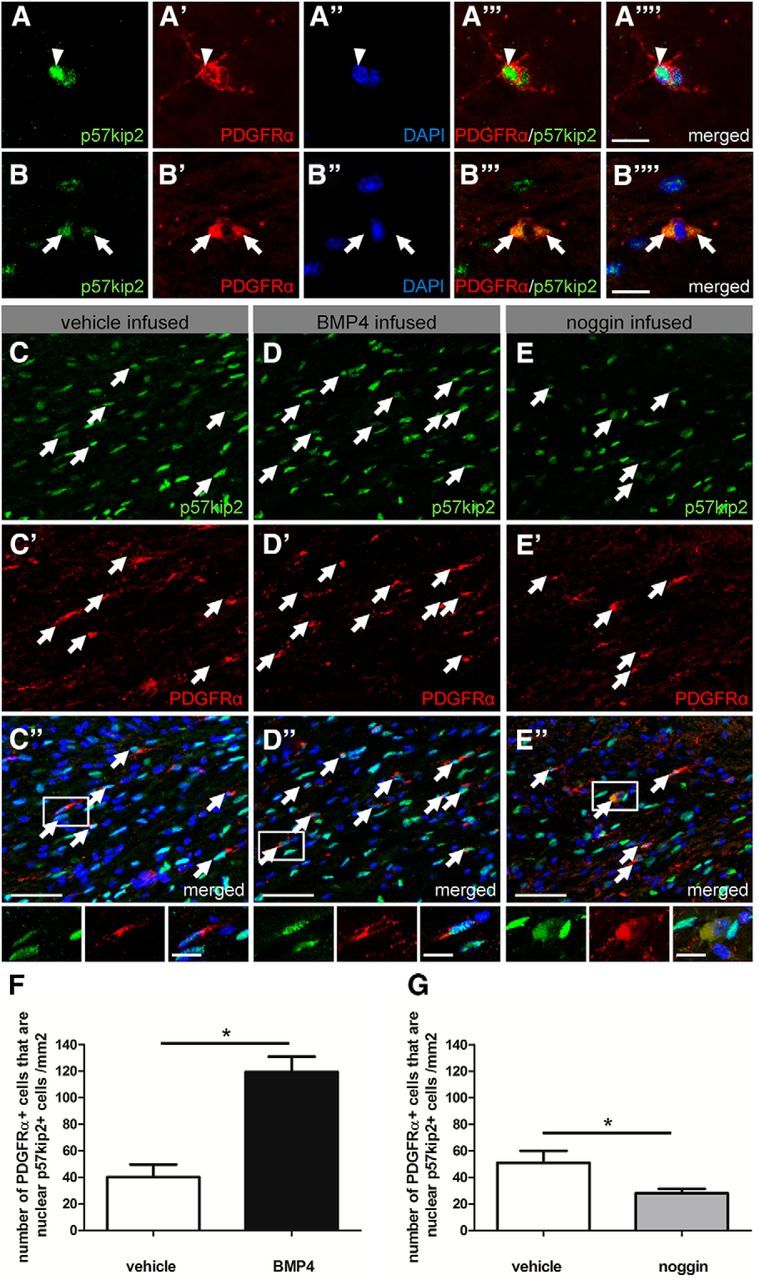
Subcellular localization of p57kip2 in the CC of healthy and diseased adult mouse brain. Immunostaining of p57kip2+/PDGFRα+ OPCs in healthy adult wild-type C57BL/6 mouse CC. OPCs either displayed nuclear (A–A′′′′; arrowheads) or cytoplasmic p57kip2 localization (B–B′′′′; arrows). Immunostaining of p57kip2+/PDGFRα+ OPCs in the CC of cuprizone-challenged mice infused with vehicle (C–C′′), BMP4 (D–D′′), or noggin (E–E′′). Arrows indicate PDGFRα+ OPCs with nuclear p57kip2. F, G, Numbers of OPCs displaying a nuclear p57kip2 signal in the CC of mice treated with cuprizone for 4 weeks after infusion of vehicle, BMP4 (400 ng/d) or noggin (400 ng/d) for 5 d. F, Upon BMP4 infusion, an increase in the number of OPCs with nuclear p57kip2 signals was observed, whereas noggin infusion led to a decreased number of OPCs with strong nuclear p57kip2 signals (G). Scale bars: A–B′′′′ 10 μm; C–E′′ 50 μm. Data are means ± SEM (n = 3, 4 in F, n = 5, 4 in G; total number of counted cells: 8750; t test: *p < 0.05).
To investigate whether such a functional correlation exists, we examined protein localization in the context of the cuprizone-induced demyelination model, an established in vivo mouse model of demyelination and remyelination (Matsushima and Morell, 2001). Previous studies revealed that, after cuprizone-induced demyelination of wild-type C57BL/6 mice CC, bone morphogenetic proteins (BMPs) interfere with remyelination, whereas BMP antagonists such as noggin can promote oligodendrocyte generation and remyelination (Sabo et al., 2011). We therefore determined the consequences of modulated BMP signaling on p57kip2 expression in the demyelinated CC. To generate a correlation with oligodendroglial differentiation, we determined whether the properties of p57kip2 expression changed during cuprizone-mediated demyelination and remyelination with BMP and noggin infusion. After 4 weeks of cuprizone-induced demyelination, mini-osmotic pumps were implanted and BMP4, noggin, or vehicle was infused into lateral ventricles for another 5 d. This analysis revealed an increase in the number of OPCs with strong nuclear p57kip2 signals in BMP4-infused versus vehicle-treated mice (Fig. 1C–C′′,D–D′′,F). In contrast, noggin-infused mice displayed a significantly lower number of cells with nuclear p57kip2 signals (Fig. 1E–E′′,G), indicating that, in this paradigm as well, nuclear export of p57kip2 correlates with progression along the oligodendroglial lineage.
Subcellular redistribution of p57kip2 during spontaneous differentiation of cultured oligodendroglial cells
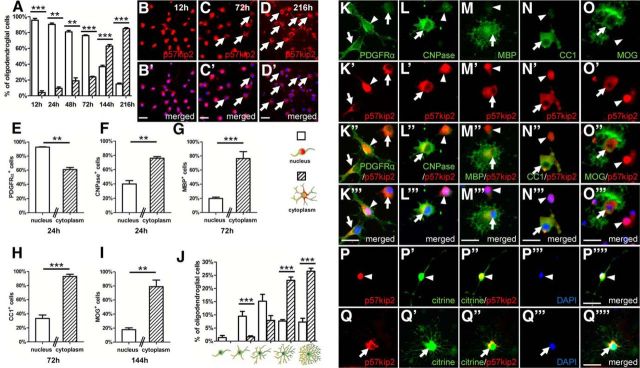
We then studied the dynamics of p57kip2 protein localization during spontaneous differentiation of cultured primary oligodendroglial cells. Differentiation was induced by means of growth factor withdrawal, which demonstrated that, over the course of 9 d, the number of cells with p57kip2 expelled from the nucleus steadily increased (Fig. 2A–D′). Because oligodendroglial differentiation is reflected by the regulation of specific markers, we performed double immunofluorescent staining for p57kip2 in combination with the oligodendroglial precursor marker PDGFRα (Fig. 2E,K–K′′′), the early myelin marker CNPase (Fig. 2F, L–L′′′), the late myelin marker MBP (Fig. 2G, M–M′′′), and the oligodendrocyte marker adenomatosis polyposis coli gene product (clone CC1; Fig. 2H,N–N′′′) as well as MOG (Fig. 2I,O–O′′′). In culture, OPCs with nuclear and with cytoplasmic p57kip2 signals were observed (Fig. 2K–K′′′). However, our experiment clearly demonstrated that the number of PDGFRα-positive cells that displayed cytoplasmic p57kip2 signals decreased during the differentiation process because of the previously described downregulation of PDGFRα during differentiation (Boulanger and Messier, 2014). This process was accompanied by the accumulation of maturation markers such as CNPase, MBP, CC1, and MOG (Fig. 2L–O′′′). In particular, we observed that, among those oligodendroglial cells that expressed maturation markers, the percentage of cells with cytoplasmic p57kip2 protein was significantly increased (Fig. 2F–I). We then looked at the distribution of cellular morphologies after 6 d in culture because primary oligodendroglial cells are heterogeneous in terms of their maturation degree. Cellular morphology determination (as previously introduced by Kremer et al., 2009) revealed that, among morphologically advanced cells, the number of cells with translocated p57kip2 was substantially increased (Fig. 2J,P–Q′′′′). Therefore, we concluded that spontaneous oligodendroglial differentiation in vitro is accompanied by nucleocytoplasmic transport of p57kip2.
Figure 2.
Distribution of p57kip2 during spontaneous differentiation of cultured OPCs. A, Percentage of oligodendroglial cells displaying nuclear (white bars) or cytoplasmic (dashed bars) p57kip2 localization over the course of 9 d. The number of oligodendroglial cells with p57kip2 expelled from the nucleus steadily increased (B–D′). Double staining for p57kip2/PDGFRα after 1 d (K–K′′′), p57kip2/CNPase after 1 d (L–L′′′′), p57kip2/MBP (M–M′′′), and p57kip2/CC1 (N–N′′′) after 3 d and p57kip2/MOG (O–O′′′) after 6 d in culture. p57kip2 translocation from the nucleus (arrowheads) to the cytoplasm (arrows) could be observed in some precursor cells (K–K′′′) and during differentiation in cells having acquired the expression of maturation markers (L–O′′′). E, Significantly fewer PDGFRα-positive OPCs display cytoplasmic p57kip2 signals as opposed to nuclear signals (white bars: cells with nuclear p57kip2; dashed bars: cells with cytoplasmic p57kip2), whereas the percentage of oligodendroglial cells with CNPase, MBP, MOG, or CC1 expression was significantly increased in cells with translocated p57kip2 protein (F–I). J, Morphological classification of oligodendroglial cells cultured for 6 d and determination of the percentage of cells with nuclear (white bars) and translocated/cytoplasmic p57kip2 signals (dashed bars). P–Q′′′′, Representative citrine (yellow fluorescent protein) expressing oligodendroglial cells with “simple” and “high” morphological complexity, respectively, and corresponding p57kip2 localizations (red). Data are means derived from n = 4 experiments ± SEM (total number of counted cells: 27,937 in A; 12,997 in E; 2450 in F; 2291 in G; 3653 in H; 2828 in I; 425 in J; t test: **p < 0.01, ***p < 0.001). Scale bars, 20 μm.
p57kip2 localization in myelinating neuron–oligodendrocyte cocultures
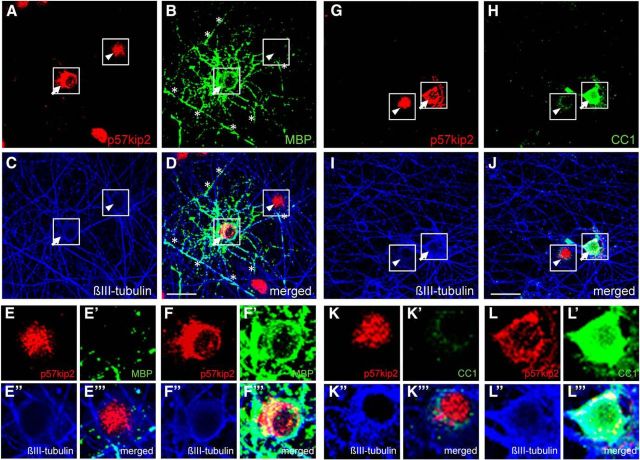
We further examined whether p57kip2 nuclear export correlates with myelin sheath generation. To this end, in vitro myelination-competent neuron–oligodendrocyte cocultures were established (Pang et al., 2012). According to the published protocol, cocultures were fixed after DIV26 and then processed for staining with antibodies directed against the neuronal marker βIII-tubulin, as well as the markers MBP, CC1, and p57kip2 (Fig. 3). Immunofluorescence revealed that mature cells either expressing CC1 (Fig. 3G–L′′′) or having established MBP-positive myelinated segments (Fig. 3A–F′′′) exhibited cytoplasmic p57kip2 signals, whereas CC1- and MBP-negative cells had strong nuclear p57kip2 signals.
Figure 3.
In vitro myelinating cocultures. A–J, Representative pictures of neuron–oligodendrocyte cocultures fixed and stained at DIV26, displaying active myelination of βIII-tubulin-positive axons (C,I). B, MBP-positive oligodendrocyte (arrow) and MBP-positive myelinated segments (asterisks). Myelin was formed by oligodendrocytes with cytoplasmic p57kip2 signals (arrow in D,F–F′′′), but not by cells with nuclear accumulation of p57kip2 (arrowhead in D,E–E′′′). G–J, Moreover, in this coculture p57kip2 expression could be confined to oligodendroglial cells and CC1-positive oligodendrocytes also exhibited cytoplasmic p57kip2 localization (J, arrow, L–L′′′). Scale bars, 30 μm.
Nucleocytoplasmic p57kip2 translocation correlates with promoted OPC differentiation in culture
In a subsequent series of experiments, we determined whether cytoplasmic p57kip2 is also a feature of chemokine-promoted OPC differentiation and to what extent modulation of nuclear shuttling directly affects oligodendroglial maturation processes. We identified a putative nuclear export sequence (NES; 43-LGRELRMRLAEL-56) within the CDK inhibitory domain of p57kip2 (NESbase analysis; la Cour et al., 2003). NESs are recognized by the chromosome region maintenance 1 (CRM1, exportin 1; Ossareh-Nazari et al., 1997). OPCs were stimulated with the chemokine CXCL12 to examine the subcellular distribution of p57kip2 under differentiation-promoting conditions (Göttle et al., 2010) and, in parallel, ratjadone, a specific CRM1 inhibitor (Köster et al., 2003), was applied to the cells. The bars shown in Figure 4, E and J, illustrate the percentage of cells expressing either CNPase, marking very late stage precursors and immature oligodendrocytes, or CC1, a marker for mature oligodendrocytes, under four treatment conditions. Upon CXCL12 stimulation, the expression of CNPase and CC1 was increased. Within each bar, the dashed area indicates the subfraction of cells with cytoplasmic p57kip2 localization. Under control conditions, almost similar percentages displayed either p57kip2 phenotype, whereas more CXCL12-stimulated cells expelled p57kip2 from the nucleus (the dashed area is 2.6-fold the size of the nondashed area for CNPase-positive cells and 4.3-fold larger for CC1-positive cells). Importantly, pharmacological blockade of CRM1-mediated nuclear export inhibited spontaneous (i.e., nonstimulated) and CXCL12-promoted OPC differentiation and was accompanied by a nuclear accumulation of p57kip2.
Figure 4.
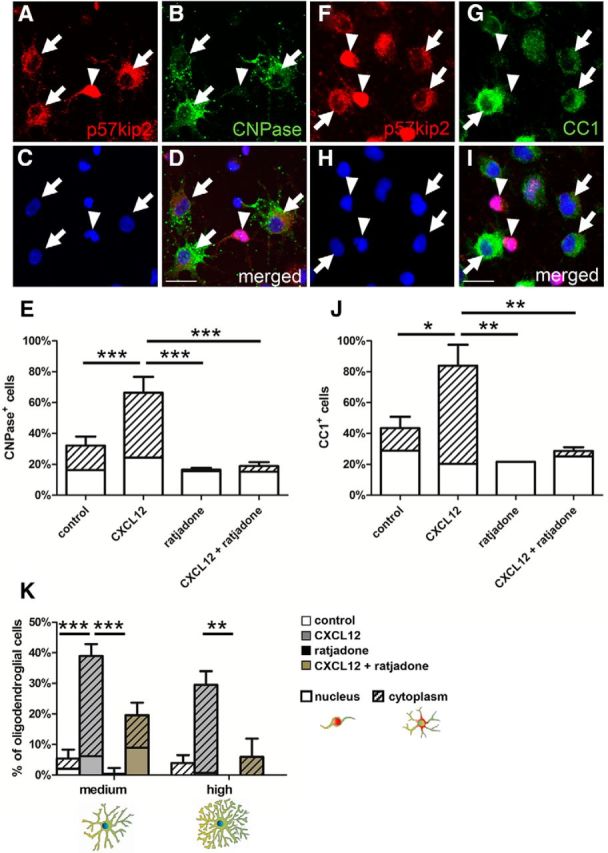
Pharmacological blockade of CRM1-mediated nuclear export. Immunofluorescent staining of cultured oligodendroglial cells for p57kip2 and CNPase (A–D) and p57kip2 and CC1 (F–I) after 1 d in culture, revealing a correlation between p57kip2 translocation from the nucleus (arrowheads) to the cytoplasm (arrows) and the abundance of CNPase and CC1 expression. E, J, Determination of the percentage of CNPase- or CC1-positive cells upon CXCL12 exposure in the absence or presence of ratjadone. This demonstrated a CXCL12-dependent promotion of cellular maturation and of cytoplasmic localization of p57kip2 [compare white areas (nucleus) with dashed areas (cytoplasm) within the bar], which was abolished in the presence of ratjadone. In the presence of ratjadone, most oligodendroglial cells expressing marker proteins exhibited a nuclear accumulation of p57kip2. K, Determination of the percentage of oligodendroglial cells with “medium” and “high” morphological complexity after 3 d in culture in response to CXCL12 stimulation (gray bars), or (co)application of ratjadone (brown bars), revealing that nuclear export blockade efficiently interfered with morphological maturation. Data are means derived from n = 3 experiments ± SEM (total number of counted cells: 15,649 in E; 12656 in J; 625 in K; t test: *p < 0.05, **p < 0.01, ***p < 0.001). Scale bars, 20 μm.
To exclude that the observed effects were driven by apoptosis, we also investigated cell death rates. No differences occurred among control and treated (CXCL12, ratjadone) cells (data not shown). With respect to our previous results demonstrating that CXCL12 also accelerated morphological maturation, we then investigated the association between p57kip2 translocation and morphometric changes. We focused our attention on oligodendroglial cells featuring “medium” and “high” morphology stages (see panel below Fig. 2), given that these were mainly displayed in the presence of CXCL12 (Fig. 4K). Again, CXCL12 exposure increased the percentage of cells displaying a higher morphological phenotype compared with control cells, and this increase was accompanied by a greater percentage of p57kip2-translocated cells. Both morphological maturation and p57kip2 translocation were efficiently blocked by ratjadone. These observations therefore suggested that oligodendroglial differentiation relies on functional export activities including translocation of p57kip2.
Colocalization of p57kip2 with binding proteins during OPC differentiation in culture
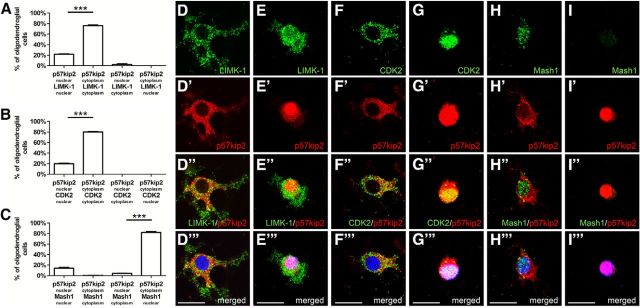
Because cytoplasmic localization of p57kip2 appears to exert a beneficial effect on oligodendroglial differentiation, we investigated the underlying mode of action. We studied two known protein-binding partners of p57kip2, LIMK-1 (Yokoo et al., 2003) and CDK2 (Caillava et al., 2011). Additional glial binding partners were identified by performing a yeast two-hybrid screen of a rat hippocampus cDNA library according to Fromont-Racine et al. (1997). Using the full-length construct of p57kip2 (aa 14-343; gi: 76257399) as bait, library screening of 116 × 106 clones (12-fold the complexity of the library) revealed 63 interactions representing 6 additional preys (binding proteins). Among these proteins, the most promising candidate was Mash1 (gi: 11693147) due to its previous description as a transcription factor involved in oligodendrogenesis (Nakatani et al., 2013). Description of the remaining five identified binding proteins awaits experimental confirmation and will be presented elsewhere. To determine whether the impact of p57kip2 on OPC differentiation is attributed to the interaction with these specific binding partners either within the cytoplasm or the nucleus, double immunofluorescent staining was performed and analyzed by confocal laser-scanning microscopy (Fig. 5). This analysis revealed almost completely overlapping subcellular distributions of p57kip2 together with LIMK-1 (Fig. 5A,D–D′′′,E–E′′′) and p57kip2 with CDK2 (Fig. 5B,F–F′′,G–G′′′), indicating that these two proteins cosegregate with p57kip2. Subcellular localizations of LIMK-1 and CDK2 also overlapped with p57kip2 in CXCL12-stimulated and ratjadone-treated cells (data not shown). However, p57kip2 and Mash1 were only overlapping in immature oligodendroglial cells with nuclear p57kip2 signals, whereas upon p57kip2 translocation, Mash1 remained in the nucleus (Fig. 5C,H–H′′′,I–I′′′). This could be interpreted as meaning that the separation of Mash1 and p57kip2 could influence the transactivation properties of Mash1, whereas p57kip2-mediated translocation of LIMK-1 and CDK2 could regulate kinase substrate accessibility.
Figure 5.
Colocalization of p57kip2 with oligodendroglial binding proteins. A–C, Evaluation of the percentage of oligodendroglial cells displaying different subcellular distribution pattern of p57kip2 in combination with nuclear and/or cytoplasmic accumulation of LIMK-1 (A), CDK2 (B), and Mash1 (C), respectively, after 6 d in culture. The following distribution patterns of p57kip2 and binding partner (bp) were examined: p57kip2nuclear/bpnuclear; p57kip2cytoplasm/bpcytoplasm; p57kip2nuclear/bpcytoplasm; p57kip2cytoplasm/bpnuclear. D–I′′′, Representative images illustrating nuclear (arrowheads) and cytoplasmic p57kip2 localization (arrows) within oligodendroglial cells after 6 d under differentiation conditions. Colocalization with both LIMK-1 (D–D′′′,E–E′′′) and CDK2 (F–F′′′,G–G′′′) was observed, whereas only nuclear coexpression of Mash1 and p57kip2 in immature oligodendroglial cells (H–H′′′, I–I′′′) was found. Scale bars, 20 μm. Data are means derived from n = 6 experiments ± SEM (total number of counted cells: 3353 in A; 3858 in B; 2378 in C; t test: ***p < 0.001).
Proximity ligation assay for p57kip2 and binding partners
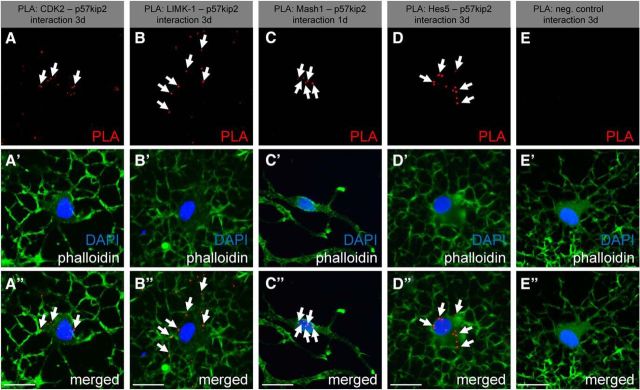
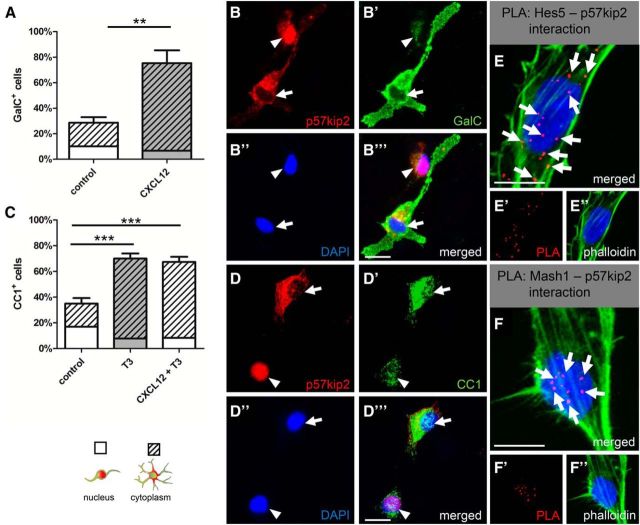
To confirm the physical interactions between p57kip2 and the identified binding partners in situ, a PLA (Söderberg et al., 2006) was performed. PLA is an extension of coimmunofluorescence in which secondary antibodies are conjugated to short DNA sequences. These can then be ligated by the addition of two oligonucleotides given that they are within 40 nm distance of each other, amplified, and detected by labeled complementary DNA probes. For these experiments, cells were either kept in differentiation medium for 1 d (Mash1) or stimulated with CXCL12 for 3 d (CDK2, LIMK-1, and Hes5). Positive PLA events resulted in distinct red fluorescent spots and confirmed that p57kip2 and CDK2 (Fig. 6A–A′′) and p57kip2 and LIMK-1 (Fig. 6B–B′′) interact with each other in both the cytoplasm and the nucleus. Nuclear PLA signals for p57kip2 and Mash1 could also be detected in immature oligodendroglial cells compared with negative single-antibody background controls (Fig. 6C–C′′). Control experiments using an antibody directed against the oligodendroglial O4 surface antigen did not generate PLA signals (data not shown).
Figure 6.
PLA-confirmed interactions among p57kip2 and CDK2, LIMK-1, Mash1, and Hes5. PLA of cultured oligodendroglial cells after 1 d (Mash1) or 3 d (CDK2, LIMK-1, Hes5) under differentiating culture conditions. Positive PLA events (red dots) are indicated by arrows and could be detected for CDK2/p57kip2 (A–A′′), LIMK-1/p57kip2 (B–B′′), Mash1/p57kip2 (C–C′′), and Hes5/p57kip2 (D–D′′). In negative antibody controls (E–E′′), no PLA events could be detected. Phalloidin was used to visualize cell bodies and processes. Scale bars, 20 μm.
p57kip2-mediated reduction of Mash1 transactivation properties
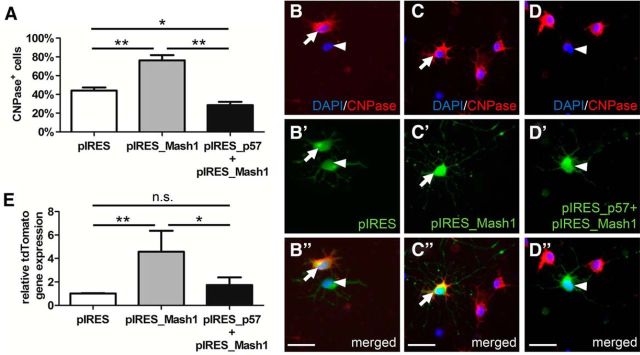
Because Mash1 was previously shown to be an important transcriptional regulator of oligodendroglial specification and differentiation (Parras et al., 2007; Sugimori et al., 2008; Nakatani et al., 2013), we investigated whether p57kip2 could control Mash1 activity. Upon forced expression of this transcription factor (pIRES_Mash1), primary oligodendroglial cells displayed elevated levels of CNPase positivity (Fig. 7A–D′′) and MBP positivity (data not shown), confirming that this transcriptional regulator positively influences cellular maturation. Mash1-induced differentiation was prevented by the forced expression of p57kip2 (pIRES_p57). To assess whether p57kip2 can inhibit Mash1-mediated transcriptional activation, we used a reporter plasmid in which tdTomato expression was under the control of DeltaM enhancer directly responding to Mash1 (Castro et al., 2006). We quantitated tdTomato transcript levels by means of real-time RT-PCR (Fig. 7E) and demonstrated that Mash1-induced reporter activity was significantly decreased in the presence of p57kip2.
Figure 7.
Functional evaluation of Mash1 transactivation properties upon overexpression of p57kip2. A, Determination of the percentage of CNPase-positive oligodendroglial cells upon combinatorial overexpression of an empty vector as control (pIRES), overexpression of Mash1 (pIRES_Mash1), and overexpression of p57kip2 (pIRES_p57) after 3 d in culture. B–D′′, Representative immunofluorescent CNPase staining of cultured cells upon combinatorial gene overexpression. E, Quantitative real-time RT-PCR illustrating the relative expression levels of the Mash1 reporter plasmid tdTomato upon combinatorial transfection of expression vectors pIRES (empty control), pIRES_Mash1, and pIRES_p57kip2. Data are means derived from n = 3 experiments ± SEM (total number of counted cells: 1177 in A; t test: *p < 0.05, **p < 0.01). Scale bars, 30 μm.
Overexpression of a mutated p57kip2 version promotes OPC differentiation and in vitro myelination
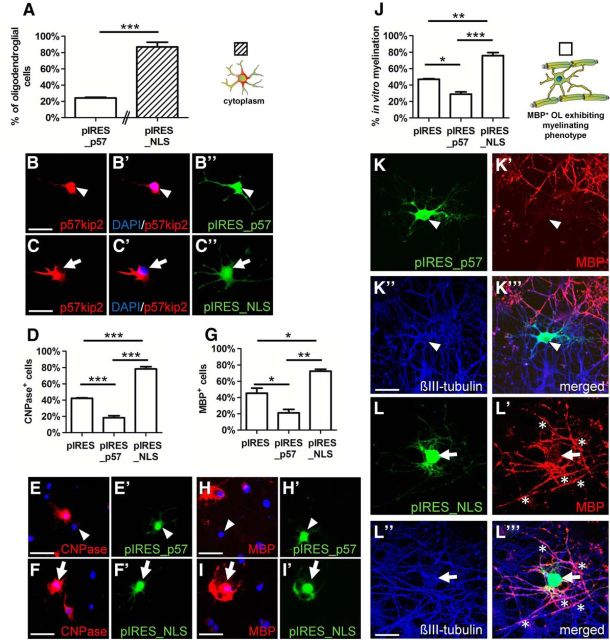
Because ratjadone treatment targets CRM1 and thereby several proteins carrying a NES are presumably missorted, we measured to what degree the localization of p57kip2 protein only affects OPC differentiation. For this purpose, we mutated the putative NLS of p57kip2, which is limited to a classical monopartite KRKR sequence inside the QT domain (Boulikas, 1993; Potikha et al., 2005), to specifically prevent the entry of p57kip2 into the nucleus. Within the predicted NLS 290-KRKR-295, amino acids K291 and R292 were substituted with A291 and A292, respectively, and the resulting mutant coding frame was used to generate the mammalian expression vector pIRES_NLS. OPCs were transfected with an empty control vector (pIRES), the full-length wild-type p57kip2 overexpression construct (pIRES_p57), or pIRES_NLS together with a citrine expression vector (yellow fluorescent protein; Heikal et al., 2000) to label transfected cells. Mitogen withdrawal was used to induce OPC differentiation; cells were then fixed and subjected to immunofluorescent staining. Cells transfected with the NLS mutant mainly displayed a cytoplasmic expression of p57kip2 (Fig. 8A,C–C′′), confirming that the mutation abolished NLS functionality. Conversely, overexpression of the wild-type p57kip2 construct (pIRES_p57) resulted in nuclear accumulation of p57kip2 proteins (Fig. 8A,B–B′′). Transfected cells were then scored for CNPase (Fig. 8D,E–F′) and MBP (Fig. 8G,H–I′) protein expression. Whereas overexpression of the wild-type p57kip2 protein decreased the percentages of cells with myelin protein expression, overexpression of the NLS mutant (pIRES_NLS) resulted in even higher degrees of myelin positivity compared with control transfected cells.
Figure 8.
p57kip2 overexpression during OPC differentiation and in vitro myelination. A, Cytoplasmic p57kip2 localization in oligodendroglial cells either transfected with the wild-type overexpression construct for p57kip2 (pIRES_p57; B–B′′) or with the NLS mutant p57kip2 construct (pIRES_NLS; C–C′′) after 24 h. D, G, Determination of CNPase- and MBP-positive cells after transfection of control, wild-type, and NLS mutant p57kip2 constructs after 24 h (CNPase) and 72 h (MBP), respectively. Representative cells double stained for p57kip2/CNPase (E–F′) and p57kip2/MBP (H–I′); negative (arrowheads) and positive (arrows) for myelin expression. J, Immunostaining revealed that overexpression of the p57kip2 NLS mutant protein increased the number of oligodendrocytes generating MBP-positive internodes (asterisks) when seeded on neuron–oligodendrocyte cocultures compared with control or wild-type p57kip2 transfected cells. (K–L′′′) Representative oligodendroglial cells without (pIRES_p57) and with myelin formation (pIRES_NLS) capacity. Data are means derived from n = 3 experiments ± SEM (total number of counted cells: 475 in A; 1471 in D; 630 in G; 2024 in J; t test: *p < 0.05, **p < 0.01, ***p < 0.001). Scale bars, 20 μm. OL, Oligodendrocyte.
To study the effect of the NLS mutant protein on the cellular capacity to generate myelin sheaths, transfected cells were seeded onto neuron–oligodendrocyte cocultures at a time point before endogenous in vitro myelination could be observed (DIV17). Nine days later, cocultures were fixed and stained for βIII-tubulin and MBP. Transfected (and transplanted) cells were visualized by means of citrine vector cotransfection and the generation of MBP-positive internodes by these cells was determined (Fig. 8J–L′′′). Cells overexpressing NLS mutant formed significantly more MBP-positive myelinated segments compared with those overexpressing wild-type p57kip2 and even compared with control transfected cells.
Colocalization of binding proteins upon overexpression of wild-type or mutant p57kip2 proteins
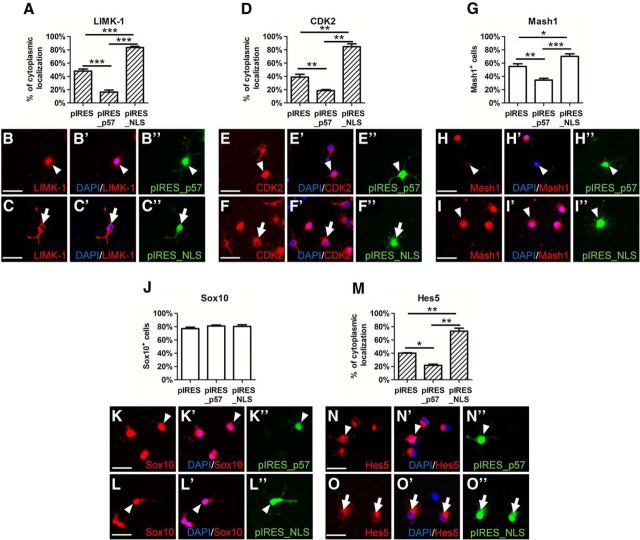
We next investigated whether the subcellular localization of binding proteins was altered upon overexpression of either wild-type or NLS mutant p57kip2 proteins. Immunofluorescent staining revealed that all cells expressed LIMK-1 (Fig. 9A–C′′) and CDK2 (Fig. 9D–F′′) and that they strictly followed the localization pattern of p57kip2 proteins such as a cytoplasmic accumulation after pIRES_NLS overexpression or a nuclear localization upon overexpression of pIRES_p57. We also looked at the distribution of Mash1 along with two other transcription factors that were previously described to be functionally implicated in oligodendroglial cells. Importantly, under all conditions, no cytoplasmic localization of Mash1 could be observed. However, upon overexpression of wild-type p57kip2, the number of Mash1-expressing cells was significantly decreased, whereas overexpression of NLS mutant significantly increased the number of Mash1-expressing cells (Fig. 9G–I′′), suggesting an additional effect on expression levels. The percentage of Sox10-transcription-factor-expressing cells was not affected by the different constructs and served as a negative control (Fig. 9J–L′′). Finally, expression of the oligodendroglial-differentiation-associated inhibitory transcription factor Hes5 was studied to provide another control. Unexpectedly, the staining revealed that it was also dependent on the localization of transfected p57kip2 proteins (Fig. 9M–O′′). Similar to CDK2 and LIMK-1, this regulator was also found in both the cytoplasm and nucleus and shuttled along with the NLS mutant p57kip2 protein. The forced accumulation of wild-type p57kip2 protein, formerly shown to be inhibitory to oligodendroglial differentiation (Kremer et al., 2009), strongly increased the nuclear presence of this transcriptional inhibitor. A direct interaction between p57kip2 and Hes5 could then be confirmed using PLA (Fig. 6D–D′′).
Figure 9.
Colocalization of binding proteins upon overexpression of wild-type or NLS mutant p57kip2 proteins. Examination of cytoplasmic localization of LIMK-1 (A–C′′), CDK2 (D–F′′), and transcription factor Hes5 (M–O′′), and percentage of cells positive for Sox10 (J–L′′) and Mash1 (G–I′′). LIMK-1, CDK2, and Hes5 follow subcellular translocation of p57kip2, whereas Sox10 and Mash1 remained in the nucleus, the latter of which was downregulated and upregulated upon wild-type and NLS mutant p57kip2 overexpression, respectively. Data are means derived from n = 4 experiments ± SEM (total number of counted cells: 677 in A; 569 in D; 541 in G; 546 in J; 415 in M; t test: *p < 0.05, **p < 0.01, ***p < 0.001). Scale bars, 20 μm.
p57kip2 localization in primary human oligodendroglial cells
Given the observed p57kip2 localization dynamics in primary rodent oligodendroglial cells, we wondered whether a similar regulation could also be observed for primary human oligodendroglial cells (i.e., whether the assumed function is conserved in the human p57kip2 protein). To determine this, we performed stimulatory experiments with primary OPCs of human origin. In general, human OPCs tend to mature slowly, which is why they were stimulated for 7 d with differentiation-promoting factors such as the chemokine CXCL12 (Göttle et al., 2010) and tri-iodo-thyronine (Barres et al., 1994). We clearly demonstrated that these stimuli significantly increased the percentage of cells positive for GalC (Fig. 10A–B′′′) and/or CC1 (Fig. 10C–D′′′), thus labeling maturing human oligodendroglial cells as described previously (Cui et al., 2012). Expression of GalC and CC1 was found to be concomitant with an increased fraction of cells exhibiting cytoplasmic p57kip2 localization (Fig. 10A,C). In addition, positive PLA events (red dots) were detected, confirming that p57kip2 and Hes5 (Fig. 10E–E′′) interact with each other in both the cytoplasm and nucleus. Nuclear PLA signals for p57kip2 and Mash1 were also detected in immature oligodendroglial cells (Fig. 10F–F′′) compared with negative single-antibody background controls (data not shown). This observation strongly suggests that this posttranscriptional regulatory process is also active in human oligodendroglial cells.
Figure 10.
p57kip2 subcellular localization in primary human oligodendroglial cells. A, Quantification of the percentage of GalC- (C) and CC1-positive cells in absence or presence of stimuli such as CXCL12 and/or tri-iodo-thyronine. The fraction of oligodendroglial cells with cytoplasmic p57kip2 localization is represented by the dashed areas. Data are means derived from n = 3 experiments ± SEM (total number of counted cells: 3553 in A; 6612 in C; t test: **p < 0.01). B–B′′′, D–D′′′, Representative cells displaying the association between maturation marker expression and nuclear export of p57kip2. Arrowheads mark immature and arrows mark mature oligodendroglial cells. Scale bars, 20 μm. E–F′′, PLA of cultured oligodendroglial cells after 3 d in culture. Positive PLA events (red dots) are indicated by arrows and could be detected for Hes5/p57kip2 (E–E′′) and Mash1/p57kip2 (F–F′′). Phalloidin was used to visualize cell bodies and processes. Scale bars, 10 μm.
Discussion
Several inhibitors associated with oligodendroglial differentiation have been identified and are supposed to constrain myelin repair in the adult CNS (Chang et al., 2002; Kuhlmann et al., 2008; Kremer et al., 2011). Despite this increasing knowledge, little is currently known about the functional properties of these regulators. Data presented in this study reveal the mode of action underlying the previously described inhibitory function of p57kip2 (Kremer et al., 2009) and suggest that the subcellular localization of p57kip2 is key to this inhibitory role. Moreover, the observed interactions with at least four different proteins indicate that multiple cellular processes are controlled by p57kip2.
Our experiments revealed interactions with CDK2, LIMK-1, and Hes5 and showed that these partners tightly follow p57kip2 between nucleus and cytoplasm. In differentiating oligodendroglial cells, CDK2 protein levels and kinase activity were shown to be decreased compared with those of proliferating OPCs and CDK2 loss was found to accelerate OPC differentiation in the adult CNS (Caillava et al., 2011). This was interpreted as a functional correlation between cell cycle exit and initiation of differentiation. Nuclear exclusion, as we observed it, is likely to also result in a differentiation boost and it remains to be shown to what degree a directed CDK2 protein relocation precedes CDK2 downregulation during the differentiation process (Caillava and Baron-Van Evercooren, 2012).
p57kip2 was described previously to interact directly with LIMK-1 and thus affect filament turnover and dynamics (Yokoo et al., 2003; Vlachos and Joseph, 2009) and we gathered preliminary evidence that, in myelinating glial cells of both peripheral nervous system and CNS, this particular protein–protein interaction occurs (Heinen et al., 2008; Kremer et al., 2009). Here, we confirmed this direct interaction and demonstrated that p57kip2 firmly controls LIMK-1 compartmental localization. Because nuclear LIMK-1 is unable to inhibit cytoplasmic cofilin, a negative regulator of actin filament turnover, actin depolymerization is enhanced. Conversely, actin polymerization was demonstrated to be fundamental for Schwann and oligodendroglial cell maturation in that pharmacological inhibition of actin polymerization resulted in impaired process extension, axonal ensheathment, differentiation, and myelination (Fernandez-Valle et al., 1997; Bacon et al., 2007; Wang et al., 2008). Unexpectedly, evidence of an additional interaction with the transcriptional regulator Hes5 emerged during our investigations. Hes5 was shown to prevent terminal oligodendroglial differentiation and to repress the activity of CNS myelin promoters (Liu et al., 2006). Because it was found to translocate from the nucleus to cytoplasm along with p57kip2, this indicates that regulatory gene regions will be deprived of this transcriptional inhibitor, resulting in a transcriptional activation of myelin and other genes.
A different behavior was observed for Mash1, which appears to be bound and inhibited by p57kip2, but was not found to translocate out of the nucleus. Nevertheless, p57kip2 was clearly shown to block Mash1 transcriptional activity, thus providing a functional explanation of how maturation-related gene expression can be controlled by its nuclear presence. Moreover, the observed counter-regulation of Mash1 expression upon wild-type versus NLS mutant p57kip2 overexpression (Fig. 9) suggests that a positive feedback loop exists and that accessory factors such as p57kip2 (or others) influence Mash1 protein levels.
Our observations thus suggest a dual mode of action. Kinase-binding partners such as LIMK-1 and CDK2 appear to travel with p57kip2 and, as a result, their site of action and access to substrates will change and affect their role. Conversely, interactions with the transcription factors Mash1 and Hes5 appear to control target gene transcription. Because they both belong to the basic helix-loop-helix family of transcriptional regulators, we speculate that they compete for the same DNA-binding sites and that activation or inhibition of target genes is controlled by p57kip2.
To what degree these processes reflect the whole or main mode of action is currently unresolved. Further functional validation of additional detected binding partners will clarify whether other proteins centered around the p57kip2 protein are functional constituents of this negative regulatory process.
Our experiments also revealed that overexpression of a cytoplasm-restricted variant of p57kip2 (NLS) is as effective in promoting the differentiation process as the previously shown short hairpin RNA-dependent gene suppression (Kremer et al., 2009). This suggests that the prevention of localization of cytoplasmic proteins in the nucleus is as important as the control of gene transcription. Which of the two mechanisms is rate limiting in regard to the inhibitory role of p57kip2 remains to be clarified in future experiments.
We clearly demonstrated that this mechanism is also valid for primary human oligodendroglial cells in that their maturation is also accompanied by nuclear export of p57kip2. This observation is of particular interest because, in a recent study of established MS lesions, no evidence of a p57kip2 translocation in white matter OPCs could be found (Pfeifenbring et al., 2013). Apart from the possibility that MS tissue preservation might interfere with the antibody-mediated detection of the lower density p57kip2 signals, this could also have a functional implication and indicate that prevented or slowed myelin repair could suffer from averted p57kip2 export. It remains to be clarified whether in the adult human brain only certain conditions, time windows (disease-course or age related), or areas (white vs gray matter) allow for subcellular relocation of p57kip2 in oligodendroglial cells to occur or if this capacity is restricted to cultured human cells only (Fig. 10). If p57kip2 translocation is restricted or blocked, the ongoing presence of p57kip2 is likely to interfere with myelin repair activities in the adult brain, even more so because protein–protein interaction data showed that in human cells as well, p57kip2 binds to Mash1 and to Hes5 (Fig. 10E–F′′). Therefore, if translocation cannot take place anymore, such cells would definitely be stalled in the differentiation process.
Regulated protein localization appears to be a more general mechanism involved in glial cell differentiation. Of particular interest is a recent report on inadequate nucleocytoplasmic transport activities that affect MS progression. TAT-interacting protein 30 kDa (TIP30) was found to be induced within OPCs in MS lesions and to block nucleocytoplasmic transport of Notch1-intracellular domain, leading to inhibited OPC differentiation and subsequent remyelination failure (Nakahara et al., 2009). Other proteins in addition to TIP30 and p57kip2 also display multiple intracellular distributions and data on changes in subcellular localization of p27kip1, Olig2, or Olig1 indicate a functional involvement in the glial repair process (Arnett et al., 2004; Magnus et al., 2007; Shen et al., 2008). It therefore remains to be shown how the specific nuclear export of p57kip2 is regulated on the molecular level and to what degree posttranslational modifications account for this process. Identification of rate-limiting signals is also of biomedical interest because such knowledge could aid the use of our findings in the development of therapies promoting myelin repair. Currently, it can only be speculated whether protein import (NLS) or export (NES) motifs or specific protein–protein interfaces (between p57kip2 and binding partners) could serve as suitable targets. Nevertheless, the disclosed regulatory mechanism underlying the role of p57kip2 is noteworthy. Apart from the functional implication, subcellular localization of oligodendroglial p57kip2 could also serve as a biomarker for differentiation-competent cells. This must certainly be evaluated in greater detail and using more sophisticated human tissue analyses featuring, for example, cortical remyelination (Chang et al., 2012). Finally, regulated protein shuttling and subsequent redistribution or regulation of binding partners must also be considered in the ongoing analysis of the in vivo function of p57kip2.
In conclusion, our data provide strong evidence that nuclear exclusion of p57kip2 is a crucial process in oligodendroglial differentiation either during CNS development or for the regeneration of injured or diseased fiber tracts. Pharmacological means to specifically address p57kip2 localization or its physical interaction with partner proteins will therefore constitute an important step toward the development of a remyelination therapy.
Footnotes
This work was supported primarily by a grant from the Hertie Foundation (to P.K.). The MS Center at the Department of Neurology is supported in part by the Walter and Ilse Rose Foundation and the James and Elisabeth Cloppenburg, Peek and Cloppenburg Düsseldorf Stiftung. J.K.S. and H.S.C. are supported by MS Research Australia and the Australian National Health and Medical Research Council. Confocal imaging of in vivo experiments was performed at the Biological Optical Microscopy Platform, the University of Melbourne. We thank Brigida Ziegler and Aysel Jafarova for help with OPC culture and the generation of myelinating cocultures and Marcia Gasis for assisting with confocal microscopy.
The authors declare no competing financial interests.
This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/3.0), which permits unrestricted use, distribution and reproduction in any medium provided that the original work is properly attributed.
References
- Arnett HA, Fancy SP, Alberta JA, Zhao C, Plant SR, Kaing S, Raine CS, Rowitch DH, Franklin RJ, Stiles CD. bHLH transcription factor Olig1 is required to repair demyelinated lesions in the CNS. Science. 2004;306:2111–2115. doi: 10.1126/science.1103709. [DOI] [PubMed] [Google Scholar]
- Bacon C, Lakics V, Machesky L, Rumsby M. N-WASP regulates extension of filopodia and processes by oligodendrocyte progenitors, oligodendrocytes, and Schwann cells: implications for axon ensheathment at myelination. Glia. 2007;55:844–858. doi: 10.1002/glia.20505. [DOI] [PubMed] [Google Scholar]
- Barres BA, Lazar MA, Raff MC. A novel role for thyroid hormone, glucocorticoids and retinoic acid in timing oligodendrocyte development. Development. 1994;120:1097–1108. doi: 10.1242/dev.120.5.1097. [DOI] [PubMed] [Google Scholar]
- Bartel PL. Using the two-hybrid system to detect protein-protein interactions. In: Hartley DA, editor. Cellular interactions in development: a practical approach. Oxford: Oxford UP; 1993. pp. 153–179. [Google Scholar]
- Béranger F, Aresta S, de Gunzburg J, Camonis J. Getting more from the two-hybrid system: N-terminal fusions to LexA are efficient and sensitive baits for two-hybrid studies. Nucleic Acids Res. 1997;25:2035–2036. doi: 10.1093/nar/25.10.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besson A, Dowdy SF, Roberts JM. CDK inhibitors: cell cycle regulators and beyond. Dev Cell. 2008;14:159–169. doi: 10.1016/j.devcel.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Boulanger JJ, Messier C. From precursors to myelinating oligodendrocytes: contribution of intrinsic and extrinsic factors to white matter plasticity in the adult brain. Neuroscience. 2014;269:343–366. doi: 10.1016/j.neuroscience.2014.03.063. [DOI] [PubMed] [Google Scholar]
- Boulikas T. Nuclear localization signals (NLS) Crit Rev Eukaryot Gene Expr. 1993;3:193–227. [PubMed] [Google Scholar]
- Caillava C, Baron-Van Evercooren A. Differential requirement of cyclin-dependent kinase 2 for oligodendrocyte progenitor cell proliferation and differentiation. Cell Div. 2012;7:14. doi: 10.1186/1747-1028-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillava C, Vandenbosch R, Jablonska B, Deboux C, Spigoni G, Gallo V, Malgrange B, Baron-Van Evercooren A. Cdk2 loss accelerates precursor differentiation and remyelination in the adult central nervous system. J Cell Biol. 2011;193:397–407. doi: 10.1083/jcb.201004146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro DS, Skowronska-Krawczyk D, Armant O, Donaldson IJ, Parras C, Hunt C, Critchley JA, Nguyen L, Gossler A, Göttgens B, Matter JM, Guillemot F. Proneural bHLH and Brn proteins coregulate a neurogenic program through cooperative binding to a conserved DNA motif. Dev Cell. 2006;11:831–844. doi: 10.1016/j.devcel.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Cate HS, Sabo JK, Merlo D, Kemper D, Aumann TD, Robinson J, Merson TD, Emery B, Perreau VM, Kilpatrick TJ. Modulation of bone morphogenic protein signalling alters numbers of astrocytes and oligodendroglia in the subventricular zone during cuprizone-induced demyelination. J Neurochem. 2010;115:11–22. doi: 10.1111/j.1471-4159.2010.06660.x. [DOI] [PubMed] [Google Scholar]
- Chang A, Nishiyama A, Peterson J, Prineas J, Trapp BD. NG2-positive oligodendrocyte progenitor cells in adult human brain and multiple sclerosis lesions. J Neurosci. 2000;20:6404–6412. doi: 10.1523/JNEUROSCI.20-17-06404.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A, Tourtellotte WW, Rudick R, Trapp BD. Premyelinating oligodendrocytes in chronic lesions of multiple sclerosis. N Engl J Med. 2002;346:165–173. doi: 10.1056/NEJMoa010994. [DOI] [PubMed] [Google Scholar]
- Chang A, Staugaitis SM, Dutta R, Batt CE, Easley KE, Chomyk AM, Yong VW, Fox RJ, Kidd GJ, Trapp BD. Cortical remyelination: a new target for repair therapies in multiple sclerosis. Ann Neurol. 2012;72:918–926. doi: 10.1002/ana.23693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui QL, D'Abate L, Fang J, Leong SY, Ludwin S, Kennedy TE, Antel J, Almazan G. Human fetal oligodendrocyte progenitor cells from different gestational stages exhibit substantially different potential to myelinate. Stem Cells Dev. 2012;21:1831–1837. doi: 10.1089/scd.2011.0494. [DOI] [PubMed] [Google Scholar]
- Fernandez-Valle C, Gorman D, Gomez AM, Bunge MB. Actin plays a role in both changes in cell shape and gene-expression associated with Schwann cell myelination. J Neurosci. 1997;17:241–250. doi: 10.1523/JNEUROSCI.17-01-00241.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formstecher E, Aresta S, Collura V, Hamburger A, Meil A, Trehin A, Reverdy C, Betin V, Maire S, Brun C, Jacq B, Arpin M, Bellaiche Y, Bellusci S, Benaroch P, Bornens M, Chanet R, Chavrier P, Delattre O, Doye V, et al. Protein interaction mapping: a Drosophila case study. Genome Res. 2005;15:376–384. doi: 10.1101/gr.2659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine M, Rain JC, Legrain P. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat Genet. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- Göttle P, Kremer D, Jander S, Odemis V, Engele J, Hartung HP, Küry P. Activation of CXCR7 receptor promotes oligodendroglial cell maturation. Ann Neurol. 2010;68:915–924. doi: 10.1002/ana.22214. [DOI] [PubMed] [Google Scholar]
- Heikal AA, Hess ST, Baird GS, Tsien RY, Webb WW. Molecular spectroscopy and dynamics of intrinsically fluorescent proteins: coral red (dsRed) and yellow (Citrine) Proc Natl Acad Sci U S A. 2000;97:11996–12001. doi: 10.1073/pnas.97.22.11996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinen A, Kremer D, Göttle P, Kruse F, Hasse B, Lehmann H, Hartung HP, Küry P. The cyclin-dependent kinase inhibitor p57kip2 is a negative regulator of Schwann cell differentiation and in vitro myelination. Proc Natl Acad Sci U S A. 2008;105:8748–8753. doi: 10.1073/pnas.0802659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadasz JJ, Rivera FJ, Taubert A, Kandasamy M, Sandner B, Weidner N, Aktas O, Hartung HP, Aigner L, Küry P. p57kip2 regulates glial fate decision in adult neural stem cells. Development. 2012;139:3306–3315. doi: 10.1242/dev.074518. [DOI] [PubMed] [Google Scholar]
- Köster M, Lykke-Andersen S, Elnakady YA, Gerth K, Washausen P, Höfle G, Sasse F, Kjems J, Hauser H. Ratjadones inhibit nuclear export by blocking CRM1/exportin 1. Exp Cell Res. 2003;286:321–331. doi: 10.1016/S0014-4827(03)00100-9. [DOI] [PubMed] [Google Scholar]
- Kremer D, Heinen A, Jadasz J, Göttle P, Zimmermann K, Zickler P, Jander S, Hartung HP, Küry P. p57kip2 is dynamically regulated in experimental autoimmune encephalomyelitis and interferes with oligodendroglial maturation. Proc Natl Acad Sci U S A. 2009;106:9087–9092. doi: 10.1073/pnas.0900204106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer D, Aktas O, Hartung HP, Küry P. The complex world of oligodendroglial differentiation inhibitors. Ann Neurol. 2011;69:602–618. doi: 10.1002/ana.22415. [DOI] [PubMed] [Google Scholar]
- Kuhlmann T, Miron V, Cui Q, Wegner C, Antel J, Brück W. Differentiation block of oligodendroglial progenitor cells as a cause for remyelination failure in chronic multiple sclerosis. Brain. 2008;131:1749–1758. doi: 10.1093/brain/awn096. [DOI] [PubMed] [Google Scholar]
- la Cour T, Gupta R, Rapacki K, Skriver K, Poulsen FM, Brunak S. NESbase version 1.0: a database of nuclear export signals. Nucleic Acids Res. 2003;31:393–396. doi: 10.1093/nar/gkg101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassmann H, Brück W, Lucchinetti CF. The immunopathology of multiple sclerosis: an overview. Brain Pathol. 2007;17:210–218. doi: 10.1111/j.1750-3639.2007.00064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Li J, Marin-Husstege M, Kageyama R, Fan Y, Gelinas C, Casaccia-Bonnefil P. A molecular insight of Hes5-dependent inhibition of myelin gene expression: old partners and new players. EMBO J. 2006;25:4833–4842. doi: 10.1038/sj.emboj.7601352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnus T, Coksaygan T, Korn T, Xue H, Arumugam TV, Mughal MR, Eckley DM, Tang SC, Detolla L, Rao MS, Cassiani-Ingoni R, Mattson MP. Evidence that nucleocytoplasmic Olig2 translocation mediates brain-injury-induced differentiation of glial precursors to astrocytes. J Neurosci Res. 2007;85:2126–2137. doi: 10.1002/jnr.21368. [DOI] [PubMed] [Google Scholar]
- Matsushima GK, Morell P. The neurotoxicant, cuprizone, as a model to study demyelination and remyelination in the central nervous system. Brain Pathol. 2001;11:107–116. doi: 10.1111/j.1750-3639.2001.tb00385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullard A. Success of immunomodulators in MS shifts discovery focus to neuroprotection. Nat Rev Drug Discov. 2011;10:885–887. doi: 10.1038/nrd3610. [DOI] [PubMed] [Google Scholar]
- Nakahara J, Kanekura K, Nawa M, Aiso S, Suzuki N. Abnormal expression of TIP30 and arrested nucleocytoplasmic transport within oligodendrocyte precursor cells in multiple sclerosis. J Clin Invest. 2009;119:169–181. doi: 10.1172/JCI35440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani H, Martin E, Hassani H, Clavairoly A, Maire CL, Viadieu A, Kerninon C, Delmasure A, Frah M, Weber M, Nakafuku M, Zalc B, Thomas JL, Guillemot F, Nait-Oumesmar B, Parras C. Ascl1/Mash1 promotes brain oligodendrogenesis during myelination and remyelination. J Neurosci. 2013;33:9752–9768. doi: 10.1523/JNEUROSCI.0805-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- Pang Y, Zheng B, Kimberly SL, Cai Z, Rhodes PG, Lin RC. Neuron-oligodendrocyte myelination co-culture derived from embryonic rat spinal cord and cerebral cortex. Brain Behav. 2012;2:53–67. doi: 10.1002/brb3.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parras CM, Hunt C, Sugimori M, Nakafuku M, Rowitch D, Guillemot F. The proneural gene Mash1 specifies an early population of telencephalic oligodendrocytes. J Neurosci. 2007;27:4233–4242. doi: 10.1523/JNEUROSCI.0126-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifenbring S, Metz I, Kremer D, Küry P, Hartung HP, Brück W. Oligodendroglial lineage cells express nuclear p57kip2 in multiple sclerosis lesions. Glia. 2013;61:1250–1260. doi: 10.1002/glia.22512. [DOI] [PubMed] [Google Scholar]
- Potikha T, Kassem S, Haber EP, Ariel I, Glaser B. p57Kip2 (cdkn1c): sequence, splice variants and unique temporal and spatial expression pattern in the rat pancreas. Lab Invest. 2005;85:364–375. doi: 10.1038/labinvest.3700229. [DOI] [PubMed] [Google Scholar]
- Sabo JK, Aumann TD, Merlo D, Kilpatrick TJ, Cate HS. Remyelination is altered by bone morphogenic protein signaling in demyelinated lesions. J Neurosci. 2011;31:4504–4510. doi: 10.1523/JNEUROSCI.5859-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen A, Liu Y, Zhao J, Qin J, Shi S, Chen M, Gao S, Xiao F, Lu Q, Cheng C. Temporal-spatial expressions of p27kip1 and its phosphorylation on Serine-10 after acute spinal cord injury in adult rat: implications for post-traumatic glial proliferation. Neurochem Int. 2008;52:1266–1275. doi: 10.1016/j.neuint.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Söderberg O, Gullberg M, Jarvius M, Ridderstråle K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson LG, Landegren U. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- Sugimori M, Nagao M, Parras CM, Nakatani H, Lebel M, Guillemot F, Nakafuku M. Ascl1 is required for oligodendrocyte development in the spinal cord. Development. 2008;135:1271–1281. doi: 10.1242/dev.015370. [DOI] [PubMed] [Google Scholar]
- Vlachos P, Joseph B. The Cdk inhibitor p57(Kip2) controls LIM-kinase 1 activity and regulates actin cytoskeleton dynamics. Oncogene. 2009;28:4175–4188. doi: 10.1038/onc.2009.269. [DOI] [PubMed] [Google Scholar]
- Vojtek AB, Hollenberg SM. Ras-Raf interaction: two-hybrid analysis. Methods Enzymol. 1995;255:331–342. doi: 10.1016/S0076-6879(95)55036-4. [DOI] [PubMed] [Google Scholar]
- Wang H, Tewari A, Einheber S, Salzer JL, Melendez-Vasquez CV. Myosin II has distinct functions in PNS and CNS myelin sheath formation. J Cell Biol. 2008;182:1171–1184. doi: 10.1083/jcb.200802091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolswijk G. Chronic stage multiple sclerosis lesions contain a relatively quiescent population of oligodendrocyte precursor cells. J Neurosci. 1998;18:601–609. doi: 10.1523/JNEUROSCI.18-02-00601.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoo T, Toyoshima H, Miura M, Wang Y, Iida KT, Suzuki H, Sone H, Shimano H, Gotoda T, Nishimori S, Tanaka K, Yamada N. p57Kip2 regulates actin dynamics by binding and translocating LIM-kinase 1 to the nucleus. J Biol Chem. 2003;278:52919–52923. doi: 10.1074/jbc.M309334200. [DOI] [PubMed] [Google Scholar]