Abstract
Although the MAP kinase-interacting kinases (MNKs) have been known for >15 years, their roles in the regulation of protein synthesis have remained obscure. Here, we explore the involvement of the MNKs in brain-derived neurotrophic factor (BDNF)-stimulated protein synthesis in cortical neurons from mice. Using a combination of pharmacological and genetic approaches, we show that BDNF-induced upregulation of protein synthesis requires MEK/ERK signaling and the downstream kinase, MNK1, which phosphorylates eukaryotic initiation factor (eIF) 4E. Translation initiation is mediated by the interaction of eIF4E with the m7GTP cap of mRNA and with eIF4G. The latter interaction is inhibited by the interactions of eIF4E with partner proteins, such as CYFIP1, which acts as a translational repressor. We find that BDNF induces the release of CYFIP1 from eIF4E, and that this depends on MNK1. Finally, using a novel combination of BONCAT and SILAC, we identify a subset of proteins whose synthesis is upregulated by BDNF signaling via MNK1 in neurons. Interestingly, this subset of MNK1-sensitive proteins is enriched for functions involved in neurotransmission and synaptic plasticity. Additionally, we find significant overlap between our subset of proteins whose synthesis is regulated by MNK1 and those encoded by known FMRP-binding mRNAs. Together, our data implicate MNK1 as a key component of BDNF-mediated translational regulation in neurons.
Keywords: BDNF, BONCAT, CYFIP1, eIF4E, MNK, SILAC
Introduction
Regulation of mRNA translation is crucial in determining the proteome (Schwanhausser et al., 2011). In neurons, regulating gene expression at translation, rather than transcription, allows localized control of synthesis of specific proteins at postsynaptic sites, contributing to synaptic plasticity (Liu-Yesucevitz et al., 2011).
Brain-derived neurotrophic factor (BDNF) is implicated in diverse neurological processes (Karpova, 2014; Panja and Bramham, 2014). Stimulation of neurons with BDNF increases overall protein synthesis (Takei et al., 2001, 2004, 2009) and alters the neuronal proteome (Liao et al., 2007; Spellman et al., 2008; Manadas et al., 2009; Leal et al., 2014). Furthermore, BDNF activates components of the translational machinery via the MEK/ERK and mammalian target-of-rapamycin, complex 1 (mTORC1) pathways (Takei et al., 2001, 2004, 2009; Kanhema et al., 2006).
Eukaryotic initiation factor eIF4E binds the 5′-cap structure (containing 7-methylGTP; m7GTP) of cytoplasmic mRNAs and is crucial for their translation. eIF4E is regulated by the mTORC1 and MEK/ERK pathways and brings additional proteins to the mRNA to facilitate translation initiation. eIF4E is phosphorylated exclusively on Ser209 (Flynn and Proud, 1995) by the MAP kinase-interacting kinases, MNKs (Fukunaga and Hunter, 1997; Waskiewicz et al., 1997, 1999; Ueda et al., 2004). MNKs are phosphorylated and thus activated by MAP kinases, e.g., ERK (Fig. 1A). Mice express two MNKs, MNK1 and MNK2. MNK1 shows low basal activity which increases following stimulation of ERK signaling (Waskiewicz et al., 1999; Scheper et al., 2001), whereas MNK2 shows high basal activity, independent of ERK signaling. BDNF induces MNK1 phosphorylation (Takei et al., 2001) and a MEK/ERK-dependent increase in phospho-eIF4E (Takei et al., 2001; Schratt et al., 2004; Kanhema et al., 2006). Although they are implicated in cell transformation/tumorigenesis, the MNKs' physiological roles remain unclear (Joshi and Platanias, 2014).
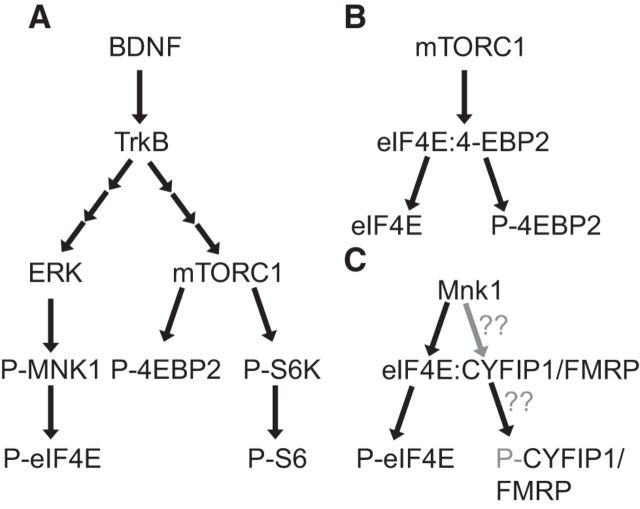
Figure 1.
Signaling connections. A, BDNF stimulates ERK signaling, and thus MNK1 and eIF4E phosphorylation, as well as mTORC1 and the phosphorylation of 4E-BP2. B, Phosphorylation of 4E-BP2 by mTORC1 leads to its dissociation from eIF4E, which can then bind other partners. C, MNK1 phosphorylates eIF4E, and causes the dissociation of eIF4E from CYFIP1. It is not clear whether phosphorylation of eIF4E itself regulates this interaction or whether MNK-mediated phosphorylation of, e.g., CYFIP1, is also involved. Gray color indicates potential, rather than established, links.
The availability of eIF4E to bind to eIF4G and initiate translation is inhibited by its interaction with eIF4E-binding proteins, 4E-BPs, of which the most abundant in brain is 4E-BP2 (Bidinosti et al., 2010). 4E-BP2 is phosphorylated by mTORC1 (Fig. 1B). eIF4E also interacts in a translationally repressive manner with cytoplasmic fragile X mental retardation protein (FMRP)-interacting protein, CYFIP1 (Napoli et al., 2008). CYFIP1 binds FMRP (Fig. 1C), a translational repressor protein which interacts with numerous mRNAs involved in regulating synaptic function (Darnell and Klann, 2013). In neuronal cells, BDNF causes release of CYFIP1 from eIF4E, thereby regulating mRNA translation (Napoli et al., 2008).
Although the MNKs have been implicated in protein synthesis-dependent synaptic plasticity (Panja et al., 2009, 2014), their function in translational regulation remains obscure. Here, we show that BDNF-induced stimulation of protein synthesis in primary cortical neurons requires MEK/ERK signaling and MNK1. Furthermore, MNK1 is necessary for BDNF-induced release of CYFIP1 from an m7GTP resin, which captures eIF4E and associated proteins. Finally, using a novel combination of bio-orthogonal noncanonical amino acid tagging (BONCAT; Dieterich et al., 2006) and stable-isotope labeling in cell culture (SILAC; Huo et al., 2012), we identify a subset of proteins whose synthesis is regulated by BDNF in an MNK1-dependent manner. Strikingly, many are encoded by mRNAs that bind FMRP.
Materials and Methods
Animals
Animals used were C57Bl/6J mice or MNK-WT, MNK1-KO (MNK1−/− MNK2+/+), MNK2-KO (MNK1+/+ MNK2−/−), and MNK-DKO (MNK1−/− MNK2−/−) mice of either gender that were kindly provided by Dr Rikiro Fukunaga (Osaka University, Japan; Ueda et al., 2004).
Reagents
Triton X and Tween were obtained from Acros Organics. Ethylene-diamine tetra-acetic acid (EDTA) and PBS were from Fisher Scientific. Hank's Balanced Salt Solution (HBSS), Neurobasal, B27 Supplement, l-glutamine, penicillin/streptomycin were from Life Technologies. Papain was from Worthington Biochemical. l-cysteine, bovine serum albumin (BSA), soybean trypsin inhibitor (STI), glycerol 2-phosphate disodium salt hydrate (β-GP), sodium fluoride (NaF), dithiothreitol (DTT), sodium metavanadate (NaVO3), and poly-d-lysine were obtained from Sigma-Aldrich. Mycozap was from Lorne and ethylene glycol tetra-acetic acid (EGTA) was from Melford.
Primary cultures
Primary cultures of cortical neurons were isolated as previously described (Wong et al., 2005). Cortices were isolated in ice-cold HBSS from P0 or P1 mice. Tissue was gently broken up and digested in neurobasal medium containing papain (20 U/ml) and l-cysteine (0.32 mg/ml) at 37°C for 20 min followed by 31°C for an additional 20 min. Tissue was then washed once in a light inhibiting solution containing 1 mg/ml each of BSA and STI then a heavy inhibiting solution containing 10 mg/ml each of BSA and STI. The dissociated tissue was washed once and triturated with fire-polished glass pipettes. After another wash, cells were then passed through a 40 μm cell strainer and 1.5–1.7 × 106 cells were plated onto poly-d-lysine-coated 35 mm culture dishes. After 1 h, overlying medium was removed and replaced with neurobasal medium containing 2% B27 supplement, 0.5 mm l-glutamine, and either 100 U/ml penicillin/streptomycin or 1× Mycozap. Half of the medium was then changed and replaced by fresh medium every 3–4 d before use at 10–12 DIV.
Treatment of neuronal cells
Primary cultures were treated with following drugs and concentrations: BDNF (25 ng/ml, Alomone Labs), CGP57380 (50 μm, Sigma-Aldrich), AZD6244 (10 μm, Selleck Chemicals), rapamycin (100 nm, Millipore), or MNK-I1 (0.1 μm, MRC-Technology). Inhibitors were added 30 min before BDNF treatment to allow sufficient time for them to enter the cells and exert their effects.
Western blot
Cells were lysed on ice using lysis buffer (50 mm Tris-HCl, 50 mm β-GP,1 mm EGTA, 1 mm EDTA, 1% Triton X, 10 mm NaF, 1 mm dithiothreitol, 0.5 mm NaVO3, 1× protease inhibitor cocktail; Roche). Cell lysates were then centrifuged for 15 min at 13,000 × g and supernatants transferred to new tubes and sample buffer was added to a final concentration of 1× and samples boiled for 5 min. Equal amounts of samples were separated on a 12.5% polyacrylamide gel and then transferred to a nitrocellulose membrane (0.45 μm pore size) via electro-blotting. Membranes were blocked in PBS containing 0.1% Tween (PBST) and 5% (w/v) BSA for 45 min, washed in PBST and then incubated with primary antibody in PBST containing 2% (w/v) BSA for 1 h at room temperature or 4°C overnight. After washing, membranes were incubated with appropriate secondary antibody for 1 h at room temperature and imaged using the LI-COR Odyssey infrared imaging system.
The following antibodies from Cell Signaling Technology were used at the indicated dilutions: eIF4E (9742), 1:1000; phospho-eIF4E (Ser209; 9741), 1:500; 4E-BP2 (2845) 1:1000; phospho-ERK1/2 (T202/204; 4370), 1:1000; p-PKB (T408; 4056), 1:1000; p-S6 (S240/244; 2215), 1:1000. Antibody for CYFIP, 1:500 (07–531) was from Millipore, total S6, 1:2000 (sc-74459) was from Santa Cruz Biotechnology, and GAPDH 1:500 (G8795) was from Sigma-Aldrich. Fluorescently tagged secondary antibodies were from Fisher Scientific.
Immunoblotting data were quantified using LI-COR Odyssey software (v3.0) and experimental treatments are expressed relative to untreated neurons. Blots for p-eIF4E, CYFIP1, and 4E-BP2 were normalized to eIF4E following enrichment using m7GTP beads. Blots for p-S6 were normalized to S6, and blots for p-PKB and p-ERK were normalized to GAPDH.
m7GTP pull-down assay
m7GTP sepharose 4B beads (7.5 μl, GE Healthcare; diluted with 7.5 μl of sepharose CL-4B beads, Sigma Aldrich) or 20 μl of m7GTP agarose beads (Jena Laboratories) were used. Before usage, the beads were washed twice in ice-cold lysis buffer. Primary cortical neurons were lysed on ice using lysis buffer. Cell lysates were then centrifuged for 15 min at 13,000 × g and supernatant transferred to new tubes. Equal amounts of samples were incubated with beads at 4°C for 120 min with rotation. The beads were then washed twice with ice-cold lysis buffer. Bound proteins were eluted with 20 μl 2× sample buffer followed by Western blotting.
[35S]methionine incorporation
10 μCi [35S]methionine and 25 ng/ml BDNF were added to cells for 60 min before lysing in 1× sample buffer. Lysates were heated to 95°C for 5 min, vortexed and spun down at 13,000 × g for 15 min before removing the supernatant to a new tube. Five microliters of supernatant (in triplicate) were added to filter paper, and boiled twice in 5% TCA for 1 min. Filter papers were then washed once in 5% TCA and thereafter in 100% ethanol before drying for 1–2 h. Quantification of the [35S]methionine incorporation was performed using a Micro Plate counter 2450 MicroBeta2 scintillation counter (PerkinElmer) with counting for 15 min. Protein quantification was performed using the BCA assay (Fisher Scientific) and samples were normalized to protein content. Cells treated with 10 μm cycloheximide (Fisher Scientific) were used as background.
Combined SILAC and AHA labeling
Primary cortical neurons were isolated and cultured as described above except that the neurobasal was replaced with a Neurobasal SILAC medium (Dundee Cell Products) containing either 89 mg/L heavy-arginine (13C6 + 15N4) and 154 mg/L heavy-lysine (13C6 + 15N2), or 87 mg/L medium-arginine (13C6) and 150 mg/L medium-lysine (2H4). At 10–12 DIV cells were starved of methionine for 30 min, then stimulated with 25 ng/ml of BDNF in the presence of 2 mm azidohomoalanine (AHA) for 2 h. Cells were washed twice in cold PBS and lysed in urea lysis buffer (8 m urea, 300 mm Tris pH 8, 4% CHAPS, 1 m NaCI) containing a 2× concentration of protease inhibitors (Roche). Lysates were then sonicated on ice and protein concentrations were determined using a 660 assay (Pierce). Equal amounts of protein from medium and heavy labeled neurons were then mixed before performing click-chemistry to an alkyne agarose resin overnight with agitation at room temperature following the manufacturer's instructions (Life Technologies; C-10416). After incubation, the resin was washed once in H2O and resuspended in 1 ml of SDS wash buffer (100 mm Tris, pH 8, 1% SDS, 250 mm NaCI, 5 mm EDTA) with 10 mm DTT and incubated at 70°C for 15 min. The resin was then centrifuged at 1000 × g for 5 min, supernatant discarded, and then resuspended in SDS wash buffer with 40 mm iodoacetamide for 30 min in the dark at room temperature.
The resin was then transferred to a spin column (Thermo Scientific, 89868) and washed eight times each with SDS wash buffer, urea wash buffer (8 m urea/100 mm Tris, pH 8) and 20% acetonitrile followed by three washes with 50 mm ammonium bicarbonate. After the last wash, the resin was resuspended in 50 mm ammonium bicarbonate, transferred to a fresh tube, and urea was added (∼1.5 m final concentration). The resin was centrifuged, supernatant removed, and 0.1 μg Lys-C (Roche) protease was added and the resin incubated at 37°C for 4 h with agitation. Then calcium chloride (final concentration 0.1 mm) and 1 μg sequencing grade trypsin (Promega) were added and the resin incubated at 37°C overnight with agitation. The resin digest was then added to a spin column, centrifuged at 1000 × g for 2 min, and the flow through was kept. Formic acid was added (∼1% final concentration) and samples were stored at −80°C until used for mass spectrometry analysis.
Mass spectrometry analysis
Peptide mixtures were cleaned with 100 μl capacity C18 tips (Thermo Scientific) with three incremental iterations from 2 to 98% HPLC grade acetonitrile (Fisher Scientific), 0.1% analytical grade formic acid (Fisher Scientific). Eluates were lyophilized in a vacuum concentrator for 4 h at room temperature (Eppendorf).
Lyophilized peptides were reconstituted in 30 μl of loading solution (2% acetonitrile, 0.1% formic acid) and 7 μl were loaded by a Dionex Ultimate 3000 (Thermo Scientific) at 20 μl/min for 6 min onto a C18 PepMap100 trapping cartridge (100 μm × 300 μm internal diameter, 5 μm particle; Thermo Scientific). Peptides were eluted at 300 nl/min over a gradient of 2–18% (355 min), 18–35% (45 min), and 35–85% (25 min) organic phase (95% acetonitrile, 5% DMSO (Sigma-Aldrich), 0.1% formic acid) in aqueous phase (2% acetonitrile, 5% DMSO, 0.1% FA) and resolved on an Acclaim PepMap 100 column (C18, 75 μm × 50 cm, 2 μm particle) retrofitted to a PicoTip nESI emitter (New Objective). Electrospray ionization was conducted at 2.4 kV and ions were characterized with an Orbitrap Elite (Thermo Scientific) at 240,000 mass resolution. The top 12 + 2 and +3 precursor ions per mass spectrometry (MS) scan (minimum intensity 1000) were characterized by high-energy collisional dissociation (HCD; 30,000 mass resolution, 1.2 Da isolation window, 40 keV collision energy) and collision-induced dissociation (CID; ion trap MS, 2 Da isolation window, 35 keV) with a dynamic exclusion (±5 ppm) of 200 s.
MS data processing.
Peptide spectrum matching and quantifications were performed with Proteome Discoverer 1.4 (Thermo Scientific) with SequestHT against the UniprotKB SwissProt mouse proteome (downloaded 03/2014). For matching and quantitation, precursor tolerance was set at 10 and 3 ppm, respectively. Fragment matching was set at 0.02 and 0.5 Da for HCD and CID, respectively. Target–decoy searching allowed for 1 missed cleavage, a minimum length of 6 aa and a maximum of three variable (1 equal) modifications of; Met->Aha (M), deamidation (Asn, Gln), or phosphorylation (Ser,Thr or Tyr). Carbamidomethyl (Cys), was set as a fixed modifications with Lys (+4 Da), Arg (+6 Da), and Lys (+8 Da), Arg (+10 Da) searched to determine medium and heavy labeled peptides, respectively. False discovery rate (FDR) was estimated with the percolator. All three replicates were analyzed separately and opened in unison (results were filtered at <5% peptide FDR) to enable parallel protein grouping and quantitation. The data have been deposited to the ProteomeXchange with identifier PXD001074 (Vizcaíno et al., 2013, 2014).
Statistical analysis
Western blot data from independent experimental replicates were analyzed using independent samples t tests, one-way ANOVAs, or two-way ANOVAs as appropriate. Significant ANOVAs were followed up with Dunnet's t post hoc tests using untreated neurons as the control. Instances of significant interactions or significance of both main effects from 3 × 2 ANOVAs from experiments using neurons derived from MNK transgenic mice were also followed up with orthogonal linear contrasts (i.e., t tests) within genotype. Significance was set to α = 0.05. All statistical analysis was done using SPSS, V.21 (IBM).
Results
BDNF stimulation of primary neurons elicited an increase in protein synthesis, as assessed by measuring [35S]methionine incorporation (Fig. 2A). BDNF also caused a rapid increase in the phosphorylation of eIF4E (Fig. 2B), a readout of MNK activity. eIF4E phosphorylation was maximal after 30–60 min of BDNF treatment (Fig. 2B), after which it declined almost to basal levels. BDNF also caused rapid increases in the phosphorylation (activity) of ERK (Fig. 2C), which is upstream of MNK1, and in the phosphorylation of protein kinase B (PKB) (Fig. 2D), and ribosomal protein S6, a substrate for the S6 kinases which are activated by mTORC1 (Fig. 2E).
Figure 2.
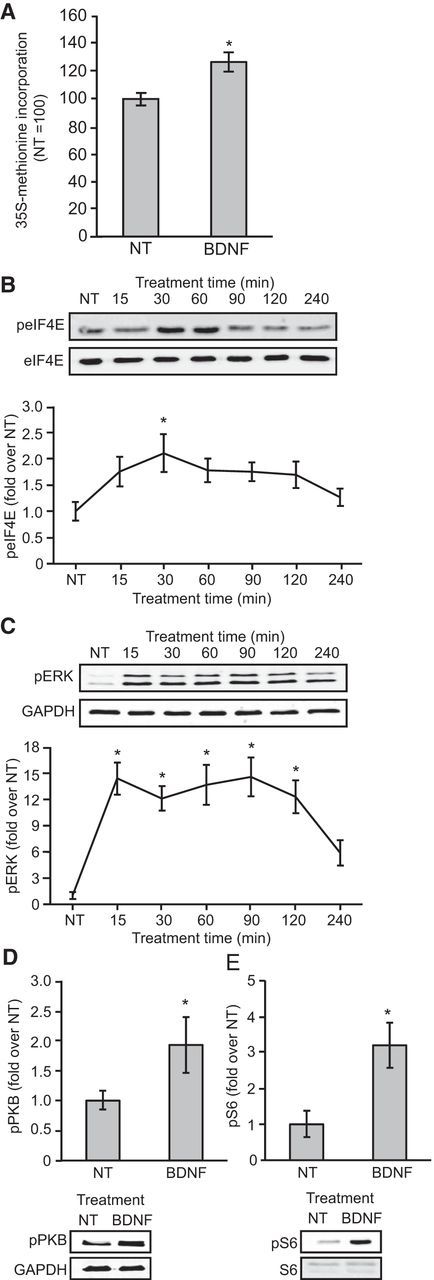
BDNF stimulates global protein synthesis and increases the phosphorylation of ERK, eIF4E, PKB, and S6. A, Cortical neurons were stimulated with BDNF for 60 min in the presence of [35S]methionine; samples of lysate were then processed to measure the incorporation of radiolabel. An unpaired t test indicated an effect of BDNF on protein synthesis (t(16) = 3.3, p = 0.04); n = 9. B, C, Time courses for the effects of BDNF on eIF4E and ERK phosphorylation. Neurons were stimulated with BDNF for 15, 30, 60, 90, 120, or 240 min. One-way ANOVAs revealed effects of stimulation-time on both phospho-eIF4E (F(6,71) = 2.764, p = 0.02) and phospho-ERK (F(6,61) = 11.8, p < 0.001); n = 9–11. D, E, The effect of BDNF stimulation for 30 min on phosphorylation of PKB and S6. There was an effect of BDNF on both phospho-PKB (t(11) = 2.8, p = 0.02) and phospho-S6 (t(12) = 5.0 p < 0.001); n = 6; *p < 0.05 compared with NT. Error bars represent SEM.
AZD6244 is a recently developed MEK inhibitor with greater potency and specificity than UO126 and PD98059 (Yeh et al., 2007), which have previously been used as MEK inhibitors. AZD6244 completely inhibited the BDNF-induced phosphorylation of ERK and the increase in eIF4E phosphorylation (Fig. 3A–C), showing that they are dependent on MEK/ERK signaling. In contrast, BDNF still stimulated the phosphorylation of PKB in the presence of AZD6244, showing that this effect is not dependent on MEK (Fig. 3D). AZD6244 also reduced basal and BDNF-stimulated S6 phosphorylation (Fig. 3A,E), consistent with previous reports that MEK/ERK signaling can contribute to the control of mTORC1 (Ma et al., 2005; Carriere et al., 2011; Fonseca et al., 2011). Finally, AZD6244 prevented the BDNF-induced increase in overall rates of protein synthesis (Fig. 3F).
Figure 3.
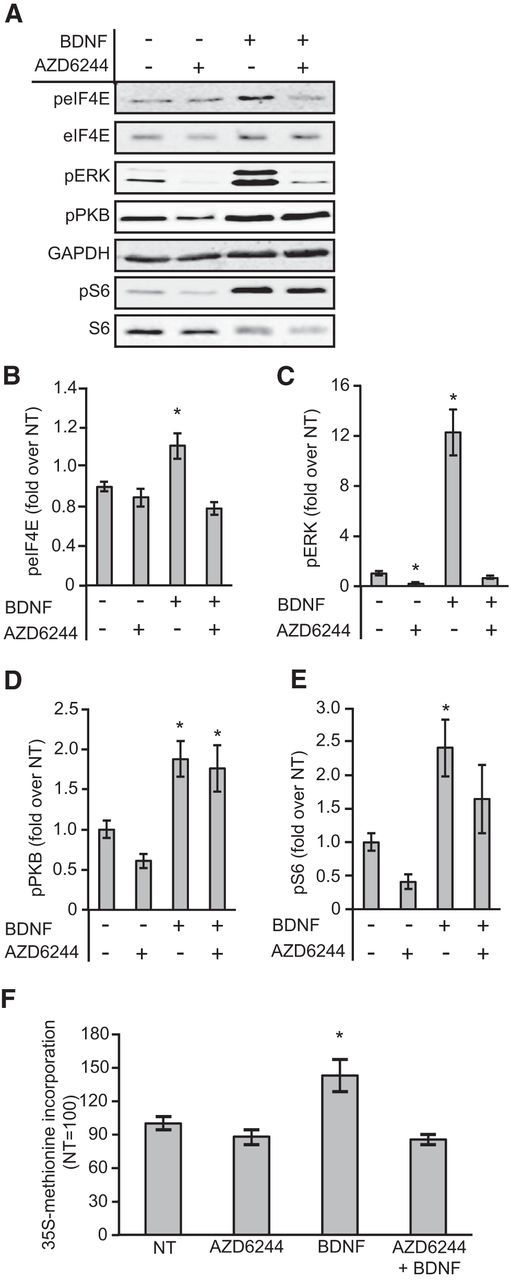
Inhibition of MEK by AZD6244 blocks the BDNF-induced increases in eIF4E phosphorylation and global protein synthesis. A, Representative Western blots for the indicated proteins or phosphoproteins. B–E, The effect of treating neurons with AZD6244 before BDNF stimulation on the phosphorylation of eIF4E, ERK, PKB, and S6. Two-way (BDNF × AZD6244) ANOVAs revealed main effects of BDNF on phospho-eIF4E (F(1,24) = 5.6, p = 0.026),phospho-ERK (F(1,20) = 101.9, p < 0.001), phospho-PKB (F(1,16) = 29.6, p < 0.001), and phospho-S6 (F(1,16) = 15.5, p = 0.001), main effects of AZD6244 on phospho-eIF4E (F(1,24) = 28.8, p < 0.001), phospho-ERK (F(1,20) = 115.1, p < 0.001), and phospho-S6 (F(1,16) = 4.07, p = 0.05), but not phospho-PKB (p = 0.2) and interactions between BDNF and AZD6244 on phospho-eIF4E (F(1,24) = 13.1, p = 0.001) and phospho-ERK (F(1,20) = 86.8, p < 0.001) but not phospho-PKB (p = 0.47) or phospho-S6 (p = 0.80); n = 5–7. F, Neurons were treated with AZD6244 before BDNF stimulation in the presence of [35S]methionine, and samples were then processed to measure incorporation of radiolabel. A two-way (BDNF × AZD6244) ANOVA revealed a main effect of BDNF (F(1,43) = 4.7, p = 0.036), a main effect of AZD6244 (F(1,43) = 10.1, p = 0.003), and an interaction between BDNF and AZD6244 administration (F(1,43) = 5.6, p = 0.023); n = 11–12; *p < 0.05 compared with NT. Error bars represent SEM.
To assess whether the BDNF-induced increase in protein synthesis required MNK activity, CGP57380, a widely used MNK inhibitor (Tschopp et al., 2000), was applied to neurons before stimulation with BDNF. CGP57380 decreased basal eIF4E phosphorylation and prevented the BDNF-induced increase, without affecting ERK phosphorylation (Fig. 4A–C). Furthermore, CGP57380 also prevented the BDNF-induced increase in protein synthesis (Fig. 4D). This finding suggests that MNK activity plays a key role in the activation of protein synthesis by BDNF.
Figure 4.
Inhibition of MNKs by CGP57380 or MNK-I1 blocks the BDNF-induced increases in phospho-eIF4E and global protein synthesis. A, Representative Western blots for the indicated proteins or phosphoproteins. B, C, The effect of treating neurons with CGP57380 before BDNF stimulation on eIF4E and ERK phoshporylation. two-way (BDNF × CGP57380) ANOVAs revealed main effects of BDNF on phospho-eIF4E (F(1,28) = 4.9, p = 0.03) and phospho-ERK (F(1,20) = 205, p < 0.001), a main effect of CGP57380 on phospho-eIF4E (F(1,28) = 142, p < 0.001) but not phospho-ERK (p = 0.78), and an interaction between BDNF and CGP57380 administration on phospho-eIF4E (F(1,28) = 9.6, p = 0.004) but not phospho-ERK (p = 0.94); n = 6–8. D, Neurons were treated with CGP57380 before BDNF stimulation in the presence of [35S]methionine. A two-way (BDNF × CGP57380) ANOVA revealed a main effect of BDNF (F(1,45) = 4.1, p = 0.05), a main effect of CGP57380 (F(1,45) = 18.9, p < 0.001), and an interaction between BDNF and CGP57380 administration (F(1,45) = 4.7, p = 0.04); n = 11–12. E, Representative Western blots for parts (F) and (G). F, G, The effect of treating neurons with MNK-I1 before BDNF stimulation on eIF4E and ERK phosphorylation. Two-way (BDNF × MNK-I1) ANOVAs revealed main effects of BDNF on phospho-eIF4E (F(1,16) = 12.7, p = 0.003) and phospho-ERK (F(1,24) = 194, p < 0.001), a main effect of MNK-I1 on phospho-eIF4E (F(1,16) = 55.8, p < 0.001) but not phospho-ERK (p = 0.36), and an interaction between BDNF and MNK-I1 administration on phospho-eIF4E (F(1,16) = 9.8, p = 0.006) but not phospho-ERK (p = 0.37); n = 5. H, Neurons were treated with MNK-I1 before BDNF stimulation in the presence of [35S]methionine. A two-way (BDNF × MNK-I1) ANOVA revealed a main effect of BDNF (F(1,56) = 8.8, p = 0.005), a main effect of MNK-I1 (F(1,56) = 42.0, p < 0.001), and an interaction between BDNF and MNK-I1 administration (F(1,56) = 5.0, p = 0.028); n = 12; *p < 0.05 compared with NT. Error bars represent SEM.
Although CGP57380 is often used as a MNK inhibitor, it also inhibits several other kinases at similar concentrations in vitro (Bain et al., 2007). Thus, we used the more recently characterized compound, MNK-I1, that is both more potent and has greater specificity than CGP57380 (J. Beggs, G.G. Jones, S. Tian, J. Xie, V. Iadevaia, V. Jenei, G. Thomas, C.G. Proud, unpublished observations). As observed for CGP57380, MNK-I1 decreased basal eIF4E phosphorylation and prevented the BDNF-induced increase in eIF4E phosphorylation (Fig. 4E) without affecting ERK, PKB, or mTORC1 signaling (Fig. 4E,G). MNK-I1 also prevented the BDNF-induced increase in protein synthesis (Fig. 4H).). Interestingly, we noticed a potential small decrease in overall levels of eIF4E by the combined application of BDNF and MNK-I1 (Fig. 4F; see also 6D,G), although we did not consistently see this throughout our experiments (data not shown). Although it is not altogether clear what might be mediating this subtle decrease, it does not affect the interpretation of the data as the m7-GTP pull-downs are normalized to total eIF4E levels.
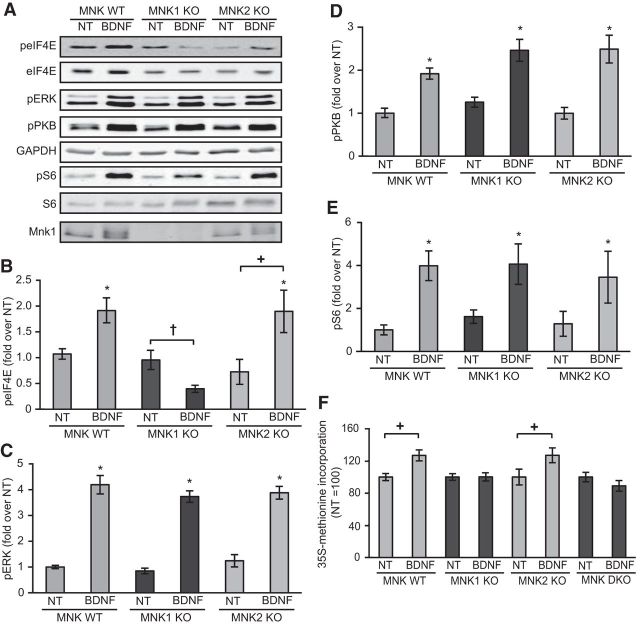
The data obtained using pharmacological inhibitors indicated that the MNKs are important in the BDNF-induced increase in protein synthesis in neuronal cells. To study this further, without the need to use such compounds and to examine potentially unique contributions of MNK1 and MNK2 to the observed effects, we made use of cortical neurons derived from WT, MNK1 KO, MNK2 KO, or MNK1/2 DKO mice (Ueda et al., 2004). BDNF caused an increase in eIF4E phosphorylation in neurons from WT or MNK2 KO mice, but not in cells from MNK1 KO mice (Fig. 5A,B). As expected, no phosphorylation of eIF4E was seen in cells from DKO animals (data not shown). The BDNF-induced increases in ERK, PKB, and S6 phosphorylation were not affected in either MNK1 or MNK2 KO neurons (Fig. 5A,C–E). Interestingly, BDNF treatment resulted in a trend toward a decrease in eIF4E phosphorylation in neurons from MNK1 KO mice (Fig. 5A,B) suggesting that BDNF may also stimulate a negative regulator of eIF4E phosphorylation. Finally, the BDNF-induced increase in overall rates of protein synthesis was observed in neurons derived from WT and MNK2 KO mice, but no increase was seen in cells from MNK1 KO or MNK1/2 DKO mice (Fig. 5F).
Figure 5.
Knocking out MNK1 blocks the BDNF-induced increase of phospho-eIF4E and global protein synthesis. A, Representative Western blots for the indicated proteins or phosphoproteins. B–E, Effect of 30 min of BDNF stimulation on eIF4E, ERK, PKB, and S6 phosphorylation in cortical neurons derived from MNK1 or MNK2 KO mice. ANOVAs (3 × 2; genotype × BDNF) revealed main effects of BDNF on phospho-eIF4E (F(1,33) = 6.99, p = 0.012), phospho-ERK (F(1,33) = 240, p < 0.001), phospho-PKB (F(1,33) = 58, p < 0.001), and phospho-S6 (F(1,33) = 16.9, p < 0.001); an effect of genotype on phospho-eIF4E (F(2,33) = 7.86, p = 0.002) but not phospho-ERK (p = 0.33), phospho-PKB (p = 0.25), or phospho-S6 (p = 0.81); and an interaction between BDNF and genotype administration on phospho-eIF4E (F(2,33) = 8.55, p = 0.001), but not phospho-ERK (p = 0.50), phospho-PKB (p = 0.36) or phospho-S6 (p = 0.86); n = 6–7. F, Neurons derived from WT, MNK1 KO, MNK2 KO, or MNK1/2 DKO mice were stimulated with BDNF for 60 min in the presence of [35S]methionine. Unpaired t tests revealed differences due to BDNF treatment in neurons derived from MNK WT (t(16) = 3.3, p = 0.04) and MNK2 KO mice (t(22) = 2.7, p = 0.012) but no difference due to BDNF treatment in MNK1 KO (t(20) = 0.03, p = 0.98) or MNK1/2 DKO mice (t(10) = 1.24, p = 0.24); n = 6–12;*p < 0.05 compared with NT; +p < 0.05, †p < 0.10 compared with NT within genotype. Error bars represent SEM.
Translation initiation can be regulated via alterations in the binding of eIF4E with small 4E-binding phosphoproteins (4E-BPs) that block its ability to bind to eIF4G (Gingras et al., 1999). Furthermore, BDNF stimulation has been found to result in both an increase in 4E-BP2 phosphorylation (Takei et al., 2001), a mechanism that is important for the release of 4E-BP2 from eIF4E, and in a decrease in the binding of eIF4E with CYFIP1 (Napoli et al., 2008; De Rubeis et al., 2013). Using affinity chromatography on immobilized m7GTP to isolate eIF4E and associated proteins, we observed that BDNF caused a decrease in the binding of 4E-BP2 and CYFIP1 to the m7GTP resin (Fig. 6A,B). BDNF-induced 4E-BP2 release was prevented by prior incubation of neurons with rapamycin (Fig. 6C), an mTORC1 inhibitor, but this effect was insensitive to inhibition of the MNKs by MNK-I1 (Fig. 6D), and was intact in neurons derived from WT, MNK1 KO or MNK2 KO mice (Fig. 6E).
Figure 6.
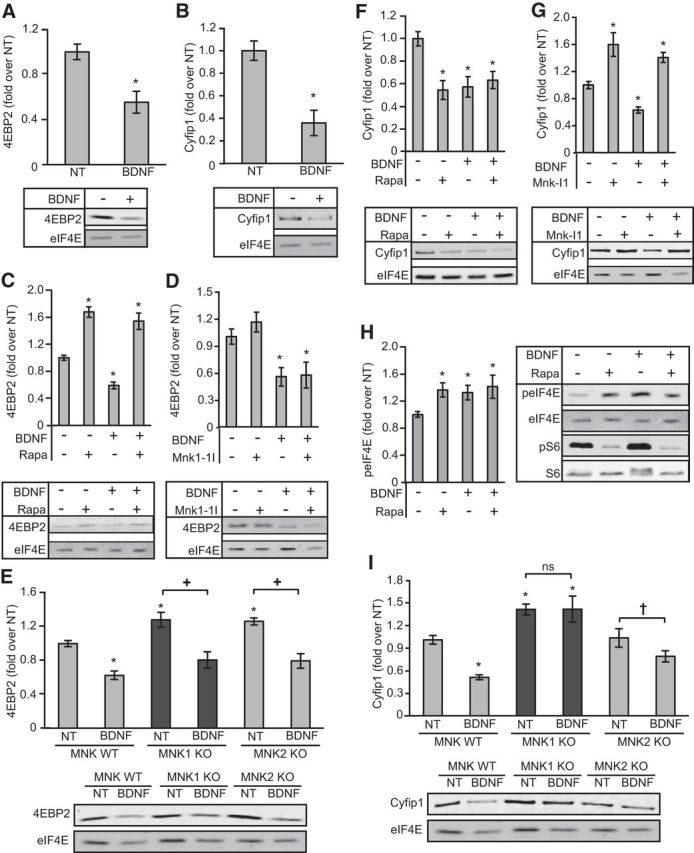
BDNF-induced release of CYFIP1, but not 4E-BP2, from m7GTP cap binding requires MNK1. A, The effect of 30 min BDNF treatment on the binding of 4E-BP2 to an m7GTP cap analog. An unpaired t test revealed an effect of BDNF (t(6) = 3.8, p = 0.009); n = 4. B, The effect of BDNF treatment on CYFIP1 binding to a m7GTP cap analog. An unpaired t test revealed a significant effect of BDNF (t(10) = 7.9, p = 0.01); n = 6. C, Effect of rapamycin on BDNF-induced release of 4E-BP2 from m7GTP binding. A two-way (BDNF × rapamycin) ANOVA revealed a main effect of BDNF (F(1,20) = 13.5, p = 0.002), a main effect of rapamycin (F(1,20) = 119, p < 0.001), and a trend toward an interaction between BDNF and rapamycin administration (F(1,20) = 3.34, p = 0.083); n = 6. D, Effect of MNK-I1 on BDNF-induced release of 4E-BP2 from m7GTP binding. A two-way (BDNF × MNK-I1) ANOVA revealed a main effect of BDNF (F(1,16) = 22, p < 0.001) but no main effect of MNK-I1 (p = 0.43) and no interaction between BDNF and MNK-I1 administration (p = 0.52); n = 5. E, Effect on BDNF-induced release of 4E-BP2 from m7GTP binding in neurons derived from WT, MNK1 or MNK2 KO mice. A 3 × 2 (genotype × BDNF) ANOVA revealed a main effect of BDNF (F(1,30) = 62.8, p < 0.001) an effect of genotype (F(2,30) = 7.32, p = 0.003) but no interaction between BDNF and genotype administration (p = 0.72); n = 6. F, Effect of rapamycin on BDNF-induced release of CYFIP1 from m7GTP binding. A two-way (BDNF × rapamycin) ANOVA revealed a main effect of BDNF (F(1,20) = 5.8, p = 0.025), a main effect of rapamycin (F(1,20) = 7.9, p = 0.011), and an interaction between BDNF and rapamycin administration (F(1,20) = 13.4, p = 0.002); n = 6. G, Effect of MNK-I1 on BDNF-induced release of CYFIP1 from m7GTP binding. A two-way (BDNF × MNK-I1) ANOVA revealed a main effect of BDNF (F(1,16) = 8.8, p = 0.009), a main effect of MNK-I1 (F(1,16) = 82.3, p < 0.001) but no interaction between BDNF and MNK-I1 administration (p = 0.36); n = 5. H, Effect on eIF4E phosphorylation of treating neurons with rapamycin and/or BDNF. A two-way (BDNF × rapamycin) ANOVA revealed a main effect of BDNF (F(1,28) = 9.5, p = 0.005), a main effect of rapamycin (F(1,28) = 11.1, p = 0.002) but no interaction between BDNF and rapamycin administration (p = 0.25); n = 8. I, Effect on BDNF-induced release of CYFIP1 from m7GTP binding in neurons derived from WT, MNK1, or MNK2 KO mice. A 3 × 2 (genotype × BDNF) ANOVA revealed a main effect of BDNF (F(1,30) = 9.63, p = 0.004) a main effect of genotype (F(2,30) = 25.62, p < 0.001), and an interaction between genotype and BDNF (F(2,30) = 3.24, p = 0.05); n = 6; *p < 0.05 compared with NT; +p < 0.05, †p < 0.10 compared with NT within genotype. Error bars represent SEM.
In marked contrast, the BDNF-induced release of CYFIP1 from eIF4E was not prevented by prior incubation of neurons with rapamycin (Fig. 6F), but was prevented by MNK-I1 (Fig. 6G). There was also an increase in CYFIP1 association with the m7GTP-resin following administration of MNK-I1 alone (Fig. 6G). Interestingly, rapamycin alone, which results in an increase in eIF4E phosphorylation (Fig. 6H), also resulted in the release of CYFIP1 from the m7GTP-resin (Fig. 6F). BDNF also failed to induce the release of CYFIP1 in neurons from MNK1 KO mice (Fig. 6I), and higher basal levels of CYFIP1 binding were also seen in MNK1-KO neurons (Fig. 6I). Together, the data suggest that MNK1 is important for regulating the levels of association of CYFIP1 with the m7GTP resin in response to BDNF. The release of CYFIP1 from eIF4E might reflect phosphorylation of eIF4E on the MNK site, Ser209 (J. Beggs, G.G. Jones, S. Tian, J. Xie, V. Iadevaia, V. Jenei, G. Thomas, C.G. Proud, unpublished observations), although, because both CYFIP1 and FRMP are phosphoproteins, it is possible that they are substrates for the MNKs and this contributes to their release from eIF4E. Extensive further work is needed to address this.
Identification of proteins whose synthesis is regulated by MNK1
The data showing that MNK1 mediates the effects of BDNF on overall protein synthesis and on CYFIP1/eIF4E binding raised the question: does MNK1 regulate general protein synthesis or does it selectively alter the synthesis of specific proteins? To examine the rapid changes in protein synthesis induced by BDNF, we combined two complementary labeling techniques: BONCAT (Dieterich et al., 2006) and SILAC (Ong et al., 2002), a combination that has previously been applied and validated in a cancer cell line and T cells (Somasekharan et al., 2012; Howden et al., 2013), but not previously used in primary neurons. Primary neurons derived from MNK1 WT or KO mice were plated in SILAC neurobasal medium in which arginine and lysine were replaced with medium or heavy stable isotope versions of the amino acids, respectively. Neurons were then treated with BDNF and pulse labeled for 2 h with AHA, a methionine analog that contains an azide functional group. AHA incorporation allowed us to selectively isolate and analyze the proteins synthesized in the presence of BDNF only during this brief time window (Howden et al., 2013). We only analyzed BDNF-treated WT and MNK1 KO neurons to specifically determine the subset of proteins regulated by BDNF in an MNK1-dependent manner.
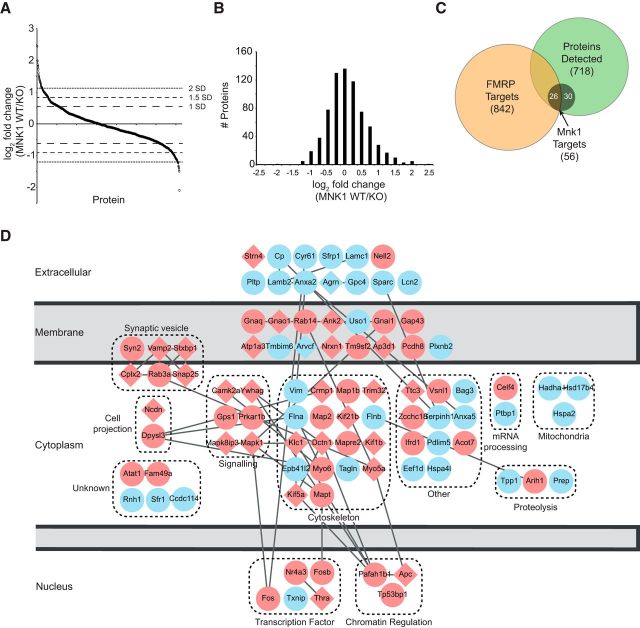
The experiment was performed in triplicate, and 718 proteins were identified in at least two of the three replicates (Fig. 7; a full list of proteins and raw data are available at the ProteomeXchange; ID: PXD001074). To determine whether specific biological processes were regulated via changes in protein synthesis mediated through BDNF stimulation of MNK1, we performed gene ontology (GO) analysis using the DAVID (database for annotation, visualization, and integrated discovery) bioinformatics tool (Huang et al., 2009), and using the 718 identified proteins as background. Given that a variety of cutoffs for analyzing SILAC data have been reported, we looked for consistency in GO analysis using three cutoffs with increasing stringency: 1, 1.5, or 2 SD above the median M/H ratio from the SILAC data which correspond to 1.46, 1.78, and 2.18 M/H ratios, and results in lists of 26, 56, and 113 proteins, respectively (Fig. 7A,B). The GO analysis examining biological processes, molecular function, and cellular components is consistent across the different thresholds in finding that proteins involved in synaptic plasticity, neurotransmitter release, and vesicular transport/exocytosis are selectively upregulated by BDNF in an MNK1 dependent manner (Table 1).
Figure 7.
Summary of proteins identified as sensitive to MNK1 inhibition. A, All proteins identified in two of three replicates and their corresponding heavy medium (H/M) (MNK1 WT/KO) ratio. Dashed lines indicate thresholds for 1.46, 1.78, and 2.18 enrichment of samples in MNK1 WTs over KOs, which correspond to cutoffs of 1, 1.5, and 2 SD over the mean of all H/M ratios. B, Histogram summary of SILAC data. C, Venn diagram illustrating the overlap of the 718 proteins identified in the present study, the 842 high confidence FMRP targets from (Darnell et al., 2011), and the MNK1-sensitive proteins above the 1.5 SD threshold. D, Proteins increased (red) or decreased (blue) by at least 1.5 SD in response to BDNF in WT versus MNK1 KO neurons. Proteins that have also been identified as high-confidence FMRP targets are represented by diamonds. Interactions were determined using the STRING database (v9.1) with a high confidence (0.7) threshold (Franceschini et al., 2013).
Table 1.
Gene ontology analysis
| GO description | 1 SD |
1.5 SD |
2 SD |
|||
|---|---|---|---|---|---|---|
| Fold enrichment | p | Fold enrichment | p | Fold enrichment | p | |
| Biological process | ||||||
| Regulation of synaptic plasticity | 6.7 | 1.9E-03 | 13.0 | 1.3E-04 | — | — |
| Neurotransmitter transport | 5.1 | 2.4E-03 | 9.8 | 9.1E-05 | 13.9 | 1.7E-03 |
| Regulation of neurotransmitter levels | 4.8 | 1.1E-02 | 9.3 | 8.3E-04 | 15.9 | 1.1E-03 |
| Secretion by cell | 4.3 | 2.3E-04 | 8.4 | 1.2E-06 | 10.0 | 8.2E-04 |
| Exocytosis | 4.2 | 1.9E-02 | 8.2 | 1.6E-03 | 10.4 | 2.8E-02 |
| Molecular function | ||||||
| Syntaxin-1 binding | 6.5 | 1.2E-02 | 12.5 | 1.7E-03 | — | — |
| SNARE binding | 5.2 | 2.7E-02 | 10.0 | 4.1E-03 | — | — |
| Syntaxin binding | 5.2 | 2.7E-02 | 10.0 | 4.1E-03 | — | — |
| Microtubule binding | 4.1 | 2.6E-03 | 6.8 | 8.0E-04 | — | — |
| Sequence-specific DNA binding | 3.5 | 1.6E-02 | 5.7 | 7.4E-03 | — | — |
| Cellular component | ||||||
| Synaptic vesicle | — | — | 12.8 | 1.7E-02 | 28.8 | 3.1E-03 |
| Cilium | 4.0 | 2.3E-02 | 8.0 | 1.7E-03 | — | — |
| Synapse | 2.9 | 3.2E-03 | 5.2 | 9.9E-05 | 6.6 | 4.5E-03 |
| Coated vesicle | 5.1 | 2.9E-02 | 5.1 | 3.5E-02 | 8.6 | 4.0E-02 |
| Neuron projection | 3.9 | 2.6E-08 | 4.9 | 5.0E-06 | — | — |
Identification of top five gene ontologies enriched in BDNF-stimulated MNK1 WT over MNK1 KO neurons classified under biological processes, molecular function, and cellular component. Analysis was performed using DAVID (Huang et al., 2009). Fold enrichment indicates the enrichment of the specific GO classifications over the background of the 718 proteins identified in at least two of three replicates using cut-offs from Figure 6A. Reported p values are EASE/modified Fisher's exact p values calculated by DAVID.
Previous work has found that CYFIP1 interacts with FMRP, which binds to and suppresses, the translation of FMRP target mRNA (Napoli et al., 2008). Thus, given that MNK1 mediates the BDNF-induced release of CYFIP1 from eIF4E (Fig. 6), we hypothesized that there would be an enrichment in proteins encoded by FMRP target mRNAs among those proteins whose synthesis is sensitive to genetic removal of MNK1. Using a list of 842 high-confidence FMRP targets (Darnell et al., 2011) we found that 107 of these proteins overlapped with the 718 proteins identified in at least two of three replicates from our study (Fig. 7C). Of the 56 proteins above the 1.5 SD threshold (Fig. 7D, diamonds), 26 were FMRP targets, which is 3.1-fold greater than the 8.3 proteins that would be expected by chance (χ(1)2 = 47.6, p < 0.001). This enrichment was also present using a threshold of either 2 SD (χ(1)2 = 11.8, p = 0.001) or 1 SD (χ(1)2 = 33.6, p < 0.001).
Discussion
The present study provides the first evidence that the MNKs, and in particular MNK1, are required for the activation of overall protein synthesis in response to any physiological stimulus, in this case BDNF. Importantly, our data also reveal that, both basally and in response to BDNF, MNK1 regulates the association between eIF4E and CYFIP1. BDNF normally brings about the release of CYFIP1 from eIF4E, but this effect is abrogated either by treatment of neurons with a novel and specific MNK inhibitor (MNK-I1) and in MNK1-KO neurons. This combined pharmacological and genetic evidence provides strong support for the conclusion that MNKs control eIF4E-CYFIP1 binding. Finally, using a novel combination of BONCAT and SILAC, we identify a subset of proteins whose synthesis is upregulated by BDNF-induced MNK1 signaling and that are involved in synaptic plasticity and neurotransmitter release. This labeling approach has the advantage that, unlike another recently developed method, ribosome profiling (Ingolia et al., 2009), it provides information on the rates of accumulation (synthesis) of specific proteins. The format we used earlier (Huo et al., 2012) suffered limitations, relative to that technique, in terms of proteome coverage; however, by selectively isolating newly synthesized proteins, their improved enrichment and in-depth bottom-up LC-MS analysis using ultra-high performance 50 cm nanocapillary chromatography over extended gradients to enhance tryptic peptide separation, thus enabled optimized ultra-high resolution MS analysis, we have now substantially enhanced the depth of coverage and also recover more peptides from each parent protein.
Given the established role for BDNF in a variety of neurophysiological phenomena, such as synaptic plasticity, memory, development, and psychiatric disorders (Bekinschtein et al., 2014; Karpova, 2014; Panja and Bramham, 2014), our data suggest that MNK1 signaling may also be an important player in these processes. In particular, our proteomic data and GO analysis suggests that MNK1 is an important mediator of the well known effect of BDNF on neurotransmitter release (Kang and Schuman, 1995; Takei et al., 1997; Li et al., 1998) and is consistent with the involvement of the MNKs in synaptic plasticity (Panja et al., 2009, 2014). Furthermore, the involvement of BDNF in fragile X syndrome (FXS), which involves cognitive deficits, has been suggested (Castrén and Castrén, 2014). Our data indicating that proteins whose synthesis is regulated by MNK1 are enriched for FMRP targets imply that MNK1 may be an important mediator of the involvement of BDNF in FXS. Together, these findings suggest that exploring the involvement of the MNKs in the many functions of BDNF is warranted.
The present study suggests that the BDNF/MEK/ERK signaling axis provides a distinct mode of translation regulation in addition to that provided by the BDNF/mTORC1 axis. Although previous work has found the MEK/ERK pathway to be integral for many of the effects of BDNF on translation (Takei et al., 2001; Kelleher et al., 2004; Kanhema et al., 2006), distinguishing the specific contribution of MEK/ERK from mTORC1 is challenging given that mTORC1 is positively regulated via MEK/ERK in neurons (Kelleher et al., 2004; Fig. 2E). By interfering with the MNKs, which are downstream from MEK/ERK and do not affect mTORC1 signaling, we have uncovered distinct contributions of these two BDNF signaling axes to translational control. BDNF-stimulated MNK1 activation is necessary for the release of CYFIP1, but not 4E-BP2, from a m7GTP cap analog, whereas BDNF-to-mTORC1 signaling selectively alters 4E-BP2 binding with no effect on CYFIP1. However, there does appear to be some crosstalk at the level of the MNKs, as inhibition of mTORC1 by rapamycin results in an increase in eIF4E phosphorylation, although this effect has been largely suggested to depend on MNK2, not MNK1 (Wang et al., 2007; Stead and Proud, 2013). Furthermore, BDNF results in a decrease in eIF4E phosphorylation in MNK1-KO neurons, suggesting that in the absence of MNK1 the negative regulation of eIF4E phosphorylation via the mTORC1 pathway may predominate, whereas in WT neurons the MNK1 pathway is dominant.
A variety of neurological disorders, such as autism and fragile X syndrome, have been linked to the dysregulation of mRNA translation (Kelleher and Bear, 2008; Darnell and Klann, 2013). Deregulation of the eIF4E/CYFIP1/FMRP complex—by artificial overexpression of eIF4E (Santini et al., 2013), mutations in the promoter of the EIF4E gene (Neves-Pereira et al., 2009), deletion of 4E-BP2 (Gkogkas et al., 2013), or alterations in FMRP expression (Darnell and Klann, 2013)—leads to autism-related disorders. Furthermore, levels of BDNF and of ERK signaling, which activates MNK1, is elevated in human patients and mouse models with autism-related disorders, and may contribute to their pathogenesis (Nelson et al., 2001; Kalkman, 2012; Yang et al., 2013; Almeida et al., 2014; Taurines et al., 2014; Yin et al., 2014). Our data implicate MNK1 in the effects of BDNF on both general translation and the release of the translational repressor, CYFIP1, binding to an m7GTP cap analog, suggests that MNK1 may also be an important contributor to these diseases.
Several additional lines of evidence suggest that identifying MNK1 as a key regulator of BDNF/MEK/ERK signaling and CYFIP1/eIF4E interactions may have important clinical implications. Stimulation of metabotropic glutamate receptors (mGluRs) results in elevated MNK1 and eIF4E phosphorylation via the MEK/ERK pathway (Banko et al., 2006) and mGluR antagonists appear to have considerable potential in treating autism and FXS (Silverman et al., 2012; Pop et al., 2014) including the reversal of elevated ERK phosphorylation (Seese et al., 2014). Furthermore, CYFIP1± mice have numerous behavioral abnormalities and deficits in synaptic plasticity that can be reversed using an mGluR antagonist (Bozdagi et al., 2012) and MNK1-KO mice have been recently reported to have deficits in LTP (Panja et al., 2014).The MNK1-dependent release of CYFIP1 from eIF4E is expected to derepress the translation of FMRP-bound mRNAs (Napoli et al., 2008); consistent with this, our data reveal that BDNF-induced synthesis of a set of these proteins is impaired in MNK1-KO cells. These include proteins such as SNAP25, a number of kinesin-interacting proteins, neurexin, and MAPK1 (i.e., ERK1). CYFIP1 has also been implicated in maintenance of dendritic complexity (Pathania et al., 2014) and coordinating BDNF-induced cytoskeletal remodeling (De Rubeis et al., 2013), suggesting that MNK1 may also play an important role in regulating CYFIP1 involvement in these processes as well.
It remains to be established whether the observed effects of inhibiting BDNF-to-MNK1 signaling is solely a consequence of the phosphorylation of eIF4E or also involves MNK-catalyzed phosphorylation of other substrates; e.g., CYFIP1 or FMRP, both of which are phosphoproteins (Narayanan et al., 2007, 2008). Known MNK substrates do include some RNA-binding proteins, whose ability to bind the mRNA for tumor necrosis factor α was respectively decreased and increased by MNK-catalyzed phosphorylation (HnRNP A1; Buxadé et al., 2005) and PSF (Buxadé et al., 2008). Thus, it remains to be determined whether the effects of the MNKs on these proteins contribute to any of the effects observed here. Nonetheless, we find a correlation between manipulations that affect eIF4E phosphorylation and consequent binding of CYFIP1 to a m7GTP cap analog. For example, inhibition of mTORC1 by rapamycin resulted in an increase in eIF4E phosphorylation, as has been previously reported (Stead and Proud, 2013), and also a release of CYFIP1 from the m7GTP resin, further supporting the hypothesis that eIF4E phosphorylation is important for mediating the interaction of CYFIP1 with the m7GTP mRNA cap.
Although the MNKs were discovered >15 years ago, little has been discovered regarding their physiological roles. Data from MNK knock-out mice or animals where Ser209 in eIF4E has been mutated to alanine (which cannot be phosphorylated) indicate roles for the MNKs/eIF4E phosphorylation in tumorigenesis (Wendel et al., 2007; Furic et al., 2010; Ueda et al., 2010). However, eIF4E phosphorylation did not affect overall protein synthesis rates. Ours, and other recently published data (Panja et al., 2014), now show that the MNKs play a key role in regulating neuronal cell protein synthesis, including synthesis of proteins encoded by mRNAs that bind FMRP. Given the importance of new protein synthesis in synaptic plasticity and of BDNF and FMRP in neurologic disorders, it is likely the MNKs play a key role in normal and abnormal neuronal cell function. It will be important to learn more of the roles of the MNKs in neurologic processes including learning, memory and other aspects of behavior.
Notes
Supplemental material for this article is available at http://www.ebi.ac.uk/pride/archive/projects/PXD001074. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (Vizcaíno et al., 2014) via the PRIDE partner repository with the dataset identifier PXD001074. This material has not been peer reviewed.
Footnotes
This work was supported by a Project Grant (BB/I004483) from the BBSRC and a Wellcome Trust Programme Grant (086688), both to C.G.P. We thank Roger Allsopp and Derek Coates for their enthusiasm and fund raising and vision in promoting the FT-MS proteomics platform at the University of Southampton, the Wessex Cancer Trust and Medical Research, UK, Hope for Guernsey, and the University of Southampton Annual Adventures in Research Grant; the use of the IRIDIS High Performance Computing Facility, and associated support services at the University of Southampton; and the kind assistance of Tobias Ternent and the PRIDE team in the submission of our data to the PRIDE database.
The authors declare no competing financial interests.
This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/3.0), which permits unrestricted use, distribution and reproduction in any medium provided that the original work is properly attributed.
References
- Almeida LE, Roby CD, Krueger BK. Increased BDNF expression in fetal brain in the valproic acid model of autism. Mol Cell Neurosci. 2014;59:57–62. doi: 10.1016/j.mcn.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, McLauchlan H, Klevernic I, Arthur JS, Alessi DR, Cohen P. The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banko JL, Hou L, Poulin F, Sonenberg N, Klann E. Regulation of eukaryotic initiation factor 4E by converging signaling pathways during metabotropic glutamate receptor-dependent long-term depression. J Neurosci. 2006;26:2167–2173. doi: 10.1523/JNEUROSCI.5196-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekinschtein P, Cammarota M, Medina JH. BDNF and memory processing. Neuropharmacology. 2014;76:677–683. doi: 10.1016/j.neuropharm.2013.04.024. [DOI] [PubMed] [Google Scholar]
- Bidinosti M, Ran I, Sanchez-Carbente MR, Martineau Y, Gingras AC, Gkogkas C, Raught B, Bramham CR, Sossin WS, Costa-Mattioli M, DesGroseillers L, Lacaille JC, Sonenberg N. Postnatal deamidation of 4E-BP2 in brain enhances its association with raptor and alters kinetics of excitatory synaptic transmission. Mol Cell. 2010;37:797–808. doi: 10.1016/j.molcel.2010.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdagi O, Sakurai T, Dorr N, Pilorge M, Takahashi N, Buxbaum JD. Haploinsufficiency of Cyfip1 produces fragile X-like phenotypes in mice. PLoS One. 2012;7:e42422. doi: 10.1371/journal.pone.0042422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxadé M, Parra JL, Rousseau S, Shpiro N, Marquez R, Morrice N, Bain J, Espel E, Proud CG. The Mnks are novel components in the control of TNFalpha biosynthesis and phosphorylate and regulate hnRNP A1. Immunity. 2005;23:177–189. doi: 10.1016/j.immuni.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Buxadé M, Morrice N, Krebs DL, Proud CG. The PSF.p54nrb complex is a novel Mnk substrate that binds the mRNA for tumor necrosis factor alpha. J Biol Chem. 2008;283:57–65. doi: 10.1074/jbc.M705286200. [DOI] [PubMed] [Google Scholar]
- Carriere A, Romeo Y, Acosta-Jaquez HA, Moreau J, Bonneil E, Thibault P, Fingar DC, Roux PP. ERK1/2 phosphorylate Raptor to promote Ras-dependent activation of mTOR complex 1 (mTORC1) J Biol Chem. 2011;286:567–577. doi: 10.1074/jbc.M110.159046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrén ML, Castrén E. BDNF in fragile X syndrome. Neuropharmacology. 2014;76:729–736. doi: 10.1016/j.neuropharm.2013.05.018. [DOI] [PubMed] [Google Scholar]
- Darnell JC, Klann E. The translation of translational control by FMRP: therapeutic targets for FXS. Nat Neurosci. 2013;16:1530–1536. doi: 10.1038/nn.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, Licatalosi DD, Richter JD, Darnell RB. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis S, Pasciuto E, Li KW, Fernández E, Di Marino D, Buzzi A, Ostroff LE, Klann E, Zwartkruis FJ, Komiyama NH, Grant SG, Poujol C, Choquet D, Achsel T, Posthuma D, Smit AB, Bagni C. CYFIP1 coordinates mRNA translation and cytoskeleton remodeling to ensure proper dendritic spine formation. Neuron. 2013;79:1169–1182. doi: 10.1016/j.neuron.2013.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT) Proc Natl Acad Sci U S A. 2006;103:9482–9487. doi: 10.1073/pnas.0601637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn A, Proud CG. Serine 209, not serine 53, is the major site of phosphorylation in initiation factor eIF-4E in serum-treated Chinese hamster ovary cells. J Biol Chem. 1995;270:21684–21688. doi: 10.1074/jbc.270.37.21684. [DOI] [PubMed] [Google Scholar]
- Fonseca BD, Alain T, Finestone LK, Huang BP, Rolfe M, Jiang T, Yao Z, Hernandez G, Bennett CF, Proud CG. Pharmacological and genetic evaluation of proposed roles of mitogen-activated protein kinase/extracellular signal-regulated kinase kinase (MEK), extracellular signal-regulated kinase (ERK), and p90RSK in the control of mTORC1 protein signaling by phorbol esters. J Biol Chem. 2011;286:27111–27122. doi: 10.1074/jbc.M111.260794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9.1: protein–protein interaction networks, with increased coverage and integration. Nucleic Acid Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga R, Hunter T. Mnk1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J. 1997;16:1921–1933. doi: 10.1093/emboj/16.8.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furic L, Rong L, Larsson O, Koumakpayi IH, Yoshida K, Brueschke A, Petroulakis E, Robichaud N, Pollak M, Gaboury LA, Pandolfi PP, Saad F, Sonenberg N. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc Natl Acad Sci U S A. 2010;107:14134–14139. doi: 10.1073/pnas.1005320107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N. eIF4 translation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- Gkogkas CG, Khoutorsky A, Ran I, Rampakakis E, Nevarko T, Weatherill DB, Vasuta C, Yee S, Truitt M, Dallaire P, Major F, Lasko P, Ruggero D, Nader K, Lacaille JC, Sonenberg N. Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature. 2013;493:371–377. doi: 10.1038/nature11628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden AJ, Geoghegan V, Katsch K, Efstathiou G, Bhushan B, Boutureira O, Thomas B, Trudgian DC, Kessler BM, Dieterich DC, Davis BG, Acuto O. QuaNCAT: quantitating proteome dynamics in primary cells. Nat Methods. 2013;10:343–346. doi: 10.1038/nmeth.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huo Y, Iadevaia V, Yao Z, Kelly I, Cosulich S, Guichard S, Foster LJ, Proud CG. Stable isotope-labelling analysis of the impact of inhibition of the mammalian target of rapamycin on protein synthesis. Biochem J. 2012;444:141–151. doi: 10.1042/BJ20112107. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi S, Platanias LC. Mnk kinase pathway: cellular functions and biological outcomes. World J Biol Chem. 2014;5:321–333. doi: 10.4331/wjbc.v5.i3.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkman HO. Potential opposite roles of the extracellular signal-regulated kinase (ERK) pathway in autism spectrum and bipolar disorders. Neurosci Biobehav Rev. 2012;36:2206–2213. doi: 10.1016/j.neubiorev.2012.07.008. [DOI] [PubMed] [Google Scholar]
- Kang H, Schuman EM. Long-lasting neurotrophin-induced enhancement of synaptic transmission in the adult hippocampus. Science. 1995;267:1658–1662. doi: 10.1126/science.7886457. [DOI] [PubMed] [Google Scholar]
- Kanhema T, Dagestad G, Panja D, Tiron A, Messaoudi E, Håvik B, Ying SW, Nairn AC, Sonenberg N, Bramham CR. Dual regulation of translation initiation and peptide chain elongation during BDNF-induced LTP in vivo: evidence for compartment-specific translation control. J Neurochem. 2006;99:1328–1337. doi: 10.1111/j.1471-4159.2006.04158.x. [DOI] [PubMed] [Google Scholar]
- Karpova NN. Role of BDNF epigenetics in activity-dependent neuronal plasticity. Neuropharmacology. 2014;76:709–718. doi: 10.1016/j.neuropharm.2013.04.002. [DOI] [PubMed] [Google Scholar]
- Kelleher RJ, 3rd, Govindarajan A, Jung HY, Kang H, Tonegawa S. Translational control by MAPK signaling in long-term synaptic plasticity and memory. Cell. 2004;116:467–479. doi: 10.1016/S0092-8674(04)00115-1. [DOI] [PubMed] [Google Scholar]
- Kelleher RJ, 3rd, Bear MF. The autistic neuron: troubled translation? Cell. 2008;135:401–406. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Leal G, Comprido D, Duarte CB. BDNF-induced local protein synthesis and synaptic plasticity. Neuropharmacology. 2014;76:639–656. doi: 10.1016/j.neuropharm.2013.04.005. [DOI] [PubMed] [Google Scholar]
- Liao L, Pilotte J, Xu T, Wong CC, Edelman GM, Vanderklish P, Yates JR., 3rd BDNF induces widespread changes in synaptic protein content and up-regulates components of the translation machinery: an analysis using high-throughput proteomics. J Proteome Res. 2007;6:1059–1071. doi: 10.1021/pr060358f. [DOI] [PubMed] [Google Scholar]
- Li YX, Zhang Y, Lester HA, Schuman EM, Davidson N. Enhancement of neurotransmitter release induced by brain-derived neurotrophic factor in cultured hippocampal neurons. J Neurosci. 1998;18:10231–10240. doi: 10.1523/JNEUROSCI.18-24-10231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Yesucevitz L, Bassell GJ, Gitler AD, Hart AC, Klann E, Richter JD, Warren ST, Wolozin B. Local RNA translation at the synapse and in disease. J Neurosci. 2011;31:16086–16093. doi: 10.1523/JNEUROSCI.4105-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Chen Z, Erdjument-Bromage H, Tempst P, Pandolfi PP. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell. 2005;121:179–193. doi: 10.1016/j.cell.2005.02.031. [DOI] [PubMed] [Google Scholar]
- Manadas B, Santos AR, Szabadfi K, Gomes JR, Garbis SD, Fountoulakis M, Duarte CB. BDNF-induced changes in the expression of the translation machinery in hippocampal neurons: protein levels and dendritic mRNA. J Proteome Res. 2009;8:4536–4552. doi: 10.1021/pr900366x. [DOI] [PubMed] [Google Scholar]
- Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, De Rubeis S, Di Marino D, Mohr E, Massimi M, Falconi M, Witke W, Costa-Mattioli M, Sonenberg N, Achsel T, Bagni C. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134:1042–1054. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- Narayanan U, Nalavadi V, Nakamoto M, Pallas DC, Ceman S, Bassell GJ, Warren ST. FMRP phosphorylation reveals an immediate-early signaling pathway triggered by group I mGluR and mediated by PP2A. J Neurosci. 2007;27:14349–14357. doi: 10.1523/JNEUROSCI.2969-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan U, Nalavadi V, Nakamoto M, Thomas G, Ceman S, Bassell GJ, Warren ST. S6K1 phosphorylates and regulates fragile X mental retardation protein (FMRP) with the neuronal protein synthesis-dependent mammalian target of rapamycin (mTOR) signaling cascade. J Biol Chem. 2008;283:18478–18482. doi: 10.1074/jbc.C800055200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson KB, Grether JK, Croen LA, Dambrosia JM, Dickens BF, Jelliffe LL, Hansen RL, Phillips TM. Neuropeptides and neurotrophins in neonatal blood of children with autism or mental retardation. Ann Neurol. 2001;49:597–606. doi: 10.1002/ana.1024.abs. [DOI] [PubMed] [Google Scholar]
- Neves-Pereira M, Müller B, Massie D, Williams JH, O'Brien PC, Hughes A, Shen SB, St CD, Miedzybrodzka Z. Deregulation of EIF4E: a novel mechanism for autism. J Med Genet. 2009;46:759–765. doi: 10.1136/jmg.2009.066852. [DOI] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.M200025-MCP200. [DOI] [PubMed] [Google Scholar]
- Panja D, Bramham CR. BDNF mechanisms in late LTP formation: a synthesis and breakdown. Neuropharmacology. 2014;76:664–676. doi: 10.1016/j.neuropharm.2013.06.024. [DOI] [PubMed] [Google Scholar]
- Panja D, Dagyte G, Bidinosti M, Wibrand K, Kristiansen AM, Sonenberg N, Bramham CR. Novel translational control in Arc-dependent long term potentiation consolidation in vivo. J Biol Chem. 2009;284:31498–31511. doi: 10.1074/jbc.M109.056077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panja D, Kenney JW, D'Andrea L, Zalfa F, Vedeler A, Wibrand K, Fukunaga R, Bagni C, Proud CG, Bramham CR. Two-stage translational control of dentate gyrus LTP consolidation mediated by sustained BDNF-TrkB signaling to MNK. Cell Rep. 2014;9:1430–1445. doi: 10.1016/j.celrep.2014.10.016. [DOI] [PubMed] [Google Scholar]
- Pathania M, Davenport EC, Muir J, Sheehan DF, López-Doménech G, Kittler JT. The autism and schizophrenia associated gene CYFIP1 is critical for the maintenance of dendritic complexity and the stabilization of mature spines. Transl Psychiatry. 2014;4:e374. doi: 10.1038/tp.2014.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pop AS, Gomez-Mancilla B, Neri G, Willemsen R, Gasparini F. Fragile X syndrome: a preclinical review on metabotropic glutamate receptor 5 (mGluR5) antagonists and drug development. Psychopharmacology (Berl) 2014;231:1217–1226. doi: 10.1007/s00213-013-3330-3. [DOI] [PubMed] [Google Scholar]
- Santini E, Huynh TN, MacAskill AF, Carter AG, Pierre P, Ruggero D, Kaphzan H, Klann E. Exaggerated translation causes synaptic and behavioural aberrations associated with autism. Nature. 2013;493:411–415. doi: 10.1038/nature11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheper GC, Morrice NA, Kleijn M, Proud CG. The MAP kinase signal-integrating kinase Mnk2 is an eIF4E kinase with high basal activity in mammalian cells. Mol Cell Biol. 2001;21:743–754. doi: 10.1128/MCB.21.3.743-754.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt GM, Nigh EA, Chen WG, Hu L, Greenberg ME. BDNF regulates the translation of a select group of mRNAs by a mammalian target of rapamycin-phosphatidylinositol 3-kinase-dependent pathway during neuronal development. J Neurosci. 2004;24:7366–7377. doi: 10.1523/JNEUROSCI.1739-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Seese RR, Maske AR, Lynch G, Gall CM. Long-term memory deficits are associated with elevated synaptic ERK1/2 activation and reversed by mGluR5 antagonism in an animal model of autism. Neuropsychopharmacology. 2014;39:1664–1673. doi: 10.1038/npp.2014.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JL, Smith DG, Rizzo SJ, Karras MN, Turner SM, Tolu SS, Bryce DK, Smith DL, Fonseca K, Ring RH, Crawley JN. Negative allosteric modulation of the mGluR5 receptor reduces repetitive behaviors and rescues social deficits in mouse models of autism. Sci Transl Med. 2012;4:131ra51. doi: 10.1126/scitranslmed.3003501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somasekharan SP, Stoynov N, Rotblat B, Leprivier G, Galpin JD, Ahern CA, Foster LJ, Sorensen PH. Identification and quantification of newly synthesized proteins translationally regulated by YB-1 using a novel click-SILAC approach. J Proteomics. 2012;77:e1–10. doi: 10.1016/j.jprot.2012.08.019. [DOI] [PubMed] [Google Scholar]
- Spellman DS, Deinhardt K, Darie CC, Chao MV, Neubert TA. Stable isotopic labeling by amino acids in cultured primary neurons: application to brain-derived neurotrophic factor-dependent phosphotyrosine-associated signaling. Mol Cell Proteomics. 2008;7:1067–1076. doi: 10.1074/mcp.M700387-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead RL, Proud CG. Rapamycin enhances eIF4E phosphorylation by activating MAP kinase-interacting kinase 2a (Mnk2a) FEBS Lett. 2013;587:2623–2628. doi: 10.1016/j.febslet.2013.06.045. [DOI] [PubMed] [Google Scholar]
- Takei N, Sasaoka K, Inoue K, Takahashi M, Endo Y, Hatanaka H. Brain-derived neurotrophic factor increases the stimulation-evoked release of glutamate and the levels of exocytosis-associated proteins in cultured cortical neurons from embryonic rats. J Neurochem. 1997;68:370–375. doi: 10.1046/j.1471-4159.1997.68010370.x. [DOI] [PubMed] [Google Scholar]
- Takei N, Kawamura M, Hara K, Yonezawa K, Nawa H. Brain-derived neurotrophic factor enhances neuronal translation by activating multiple initiation processes: comparison with the effects of insulin. J Biol Chem. 2001;276:42818–42825. doi: 10.1074/jbc.M103237200. [DOI] [PubMed] [Google Scholar]
- Takei N, Inamura N, Kawamura M, Namba H, Hara K, Yonezawa K, Nawa H. Brain-derived neurotrophic factor induces mammalian target of rapamycin-dependent local activation of translation machinery and protein synthesis in neuronal dendrites. J Neurosci. 2004;24:9760–9769. doi: 10.1523/JNEUROSCI.1427-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei N, Kawamura M, Ishizuka Y, Kakiya N, Inamura N, Namba H, Nawa H. Brain-derived neurotrophic factor enhances the basal rate of protein synthesis by increasing active eukaryotic elongation factor 2 levels and promoting translation elongation in cortical neurons. J Biol Chem. 2009;284:26340–26348. doi: 10.1074/jbc.M109.023010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taurines R, Segura M, Schecklmann M, Albantakis L, Grünblatt E, Walitza S, Jans T, Lyttwin B, Haberhausen M, Theisen FM, Martin B, Briegel W, Thome J, Schwenck C, Romanos M, Gerlach M. Altered peripheral BDNF mRNA expression and BDNF protein concentrations in blood of children and adolescents with autism spectrum disorder. J Neural Transm. 2014;121:1117–1128. doi: 10.1007/s00702-014-1162-x. [DOI] [PubMed] [Google Scholar]
- Tschopp C, Knauf U, Brauchle M, Zurini M, Ramage P, Glueck D, New L, Han J, Gram H. Phosphorylation of eIF-4E on Ser 209 in response to mitogenic and inflammatory stimuli is faithfully detected by specific antibodies. Mol Cell Biol Res Commun. 2000;3:205–211. doi: 10.1006/mcbr.2000.0217. [DOI] [PubMed] [Google Scholar]
- Ueda T, Watanabe-Fukunaga R, Fukuyama H, Nagata S, Fukunaga R. Mnk2 and Mnk1 are essential for constitutive and inducible phosphorylation of eukaryotic initiation factor 4E but not for cell growth or development. Mol Cell Biol. 2004;24:6539–6549. doi: 10.1128/MCB.24.15.6539-6549.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Sasaki M, Elia AJ, Chio II, Hamada K, Fukunaga R, Mak TW. Combined deficiency for MAP kinase-interacting kinase 1 and 2 (Mnk1 and Mnk2) delays tumor development. Proc Natl Acad Sci U S A. 2010;107:13984–13990. doi: 10.1073/pnas.1008136107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno JA, Côté RG, Csordas A, Dianes JA, Fabregat A, Foster JM, Griss J, Alpi E, Birim M, Contell J, O'Kelly G, Schoenegger A, Ovelleiro D, Pérez-Riverol Y, Reisinger F, Ríos D, Wang R, Hermjakob H. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;41:D1063–D1069. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Ríos D, Dianes JA, Sun Z, Farrah T, Bandeira N, Binz PA, Xenarios I, Eisenacher M, Mayer G, Gatto L, Campos A, Chalkley RJ, Kraus HJ, Albar JP, Martinez-Bartolomé S, et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol. 2014;32:223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Yue P, Chan CB, Ye K, Ueda T, Watanabe-Fukunaga R, Fukunaga R, Fu H, Khuri FR, Sun SY. Inhibition of mammalian target of rapamycin induces phosphatidylinositol 3-kinase-dependent and Mnk-mediated eukaryotic translation initiation factor 4E phosphorylation. Mol Cell Biol. 2007;27:7405–7413. doi: 10.1128/MCB.00760-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskiewicz AJ, Flynn A, Proud CG, Cooper JA. Mitogen-activated kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J. 1997;16:1909–1920. doi: 10.1093/emboj/16.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskiewicz AJ, Johnson JC, Penn B, Mahalingam M, Kimball SR, Cooper JA. Phosphorylation of the cap-binding protein eukaryotic translation factor 4E by protein kinase Mnk1 in vivo. Mol Cell Biol. 1999;19:1871–1880. doi: 10.1128/mcb.19.3.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel HG, Silva RL, Malina A, Mills JR, Zhu H, Ueda T, Watanabe-Fukunaga R, Fukunaga R, Teruya-Feldstein J, Pelletier J, Lowe SW. Dissecting eIF4E action in tumorigenesis. Genes Dev. 2007;21:3232–3237. doi: 10.1101/gad.1604407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong HK, Fricker M, Wyttenbach A, Villunger A, Michalak EM, Strasser A, Tolkovsky AM. Mutually exclusive subsets of BH3-only proteins are activated by the p53 and c-Jun N-terminal kinase/c-Jun signaling pathways during cortical neuron apoptosis induced by arsenite. Mol Cell Biol. 2005;25:8732–8747. doi: 10.1128/MCB.25.19.8732-8747.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Cao F, Sheikh AM, Malik M, Wen G, Wei H, Ted Brown W, Li X. Up-regulation of Ras/Raf/ERK1/2 signaling impairs cultured neuronal cell migration, neurogenesis, synapse formation, and dendritic spine development. Brain Struct Funct. 2013;218:669–682. doi: 10.1007/s00429-012-0420-7. [DOI] [PubMed] [Google Scholar]
- Yeh TC, Marsh V, Bernat BA, Ballard J, Colwell H, Evans RJ, Parry J, Smith D, Brandhuber BJ, Gross S, Marlow A, Hurley B, Lyssikatos J, Lee PA, Winkler JD, Koch K, Wallace E. Biological characterization of ARRY-142886 (AZD6244), a potent, highly selective mitogen-activated protein kinase kinase 1/2 inhibitor. Clin Cancer Res. 2007;13:1576–1583. doi: 10.1158/1078-0432.CCR-06-1150. [DOI] [PubMed] [Google Scholar]
- Yin A, Qiu Y, Jia B, Song T, Yu Y, Alberts I, Zhong M. The developmental pattern of the RAS/RAF/Erk1/2 pathway in the BTBR autism mouse model. Int J Dev Neurosci. 2014;39:2–8. doi: 10.1016/j.ijdevneu.2014.01.006. [DOI] [PubMed] [Google Scholar]