ABSTRACT
In this study, we examined the prevalence and molecular characteristics of Cryptosporidium in buffalo, dairy cattle and sheep in different farms at Kafr El Sheikh Province, Egypt. Rectal fecal samples, including 466 samples from buffalo, 1697 from cattle and 120 from sheep, were collected from different ages and screened by modified Ziehl-Neelsen acid-fast microscopy for detection of Cryptosporidium oocysts. All studied farms were positives with an overall prevalence of 1.29% in buffalo (4.17% in claves versus 0.48% in adults), 7.07% in cattle (6.90% in calves versus 10.20% and 6.10% in heifers and adults, respectively) and 2.50% in sheep (4.40% in lambs versus 1.30% in adults). PCR-RFLP analyses of small-subunit rRNA genes from positive specimens revealed the occurrence of C. parvum and C. ryanae in buffalo; C. parvum, C. ryanae, C. bovis and C. andersoni in cattle and only C. xiaoi in sheep. Genotypes distribution showed that C. ryanae was the dominant species (60.0%) followed by C. parvum (40.0%) in buffalo calves. Meanwhile, in cattle calves, C. parvum was the commonest species (74.23%) followed by C. ryanae (16.10%) and C. bovis (9.70%). Subtyping of C. parvum based on sequence analysis of the polymorphic 60 kDa glycoprotein gene locus showed the presence of subtypes IIdA20G1 and IIaA15G1R1 in both buffalo and cattle calves, addressing the potential role of calves in zoonotic cryptosporidiosis in Egypt.
Keywords: Cryptosporidium, Egypt, genotyping, subtyping, zoonosis
Cryptosporidium spp. are obligatory apicomplexans that infect the gastrointestinal epithelium of a wide range of vertebrates [14], resulting in gastroenteritis, manifested by diarrhea of different severities. The infection is acquired orally, usually by routes of direct contact with feces of infected animals or persons and/or waterborne and foodborne transmission [26]. The sequelae of the infection depends on the host immune status, ranging from severe but self-limiting diarrhea in immunocompetents to prolonged, debilitating, life-threatening illness in immunocompromised persons, such as AIDS and cancer patients [11]. Cryptosporidiosis is especially common in developing countries, imposing additional challenges to poorly supported public health infrastructure. Several species are commonly found in humans, and their distribution differs depending on socioeconomic development and the intensity of animal farming. Nevertheless, C. hominis and C. parvum in general are responsible for the majority of human Cryptosporidium infections [51].
Farm animals are usually susceptible to several species of Cryptosporidium, resulting in considerable economic loss either directly due to retarded growth, diminished weight gain or even mortality in neonates/juveniles and decreased milk production in adults [12, 45], or indirectly as a cost of medical intervention [12]. Four Cryptosporidium species are commonly found in cattle; C. parvum, C. bovis, C. ryanae and C. andersoni with age-species related distribution [40]. C. parvum is the major contributor of zoonotic transmission of cryptosporidiosis [51], and dairy calves are the major reservoirs for this species. The high prevalence and great oocysts shedding among calves play an important role as direct source of infection [15] and environmental contamination including rivers [46]. Infected hosts may excrete several millions to billions of oocysts with the feces [32], which may survive and persist in feces and environment for extended periods, ranging from several weeks to many months [36].
The genus Cryptosporidium is composed of multiple genetically distinct forms. In the absence of distinguishable morphologic features of different cryptosporidia, molecular biologic analyses have been used widely in characterizing Cryptosporidium spp. of various animals at both species and subtype levels. Data from these studies have improved our understanding of inter-species transmission and public health significance of Cryptosporidium spp. in animals [51].
Intensive animal farming is a very common practice in Egypt, making zoonosis a major public health concern. Kafr El Sheikh is a coastal province known for intensive farming activities. Monitoring of Cryptosporidium spp. in farm animals in this province is important for both veterinary and public health. Previous studies on the epidemiology of Cryptosporidium in animals in Egypt were mostly performed microscopically [13, 31]. So far, only a few studies have been done on the molecular level to characterize Cryptosporidium in cattle and water buffalo [3,4,5, 21]. This study was done to extend our knowledge on the molecular epidemiology of Cryptosporidium in Kafr El Sheikh Province and to follow up the changes in the infection profile of the animal farms in this area, for establishment of the appropriate measures for control and prevention of the disease.
MATERIALS AND METHODS
Collection of specimens and examination: Samples of this study were collected during June 2010–November 2011 from 5 private farms designated A, B, C, D and E, which were situated at different localities in Kafr El Sheikh (130 km north of Cairo), Egypt (Table 1). In cattle and buffalo farms, the neonates were kept in separate pens with milk feeding of suckling calves performed manually, allowing no direct contact of calves with their dams. Based on animals’ age, the examined cattle and buffalo were classified into 3 groups: less than 3 months of age (calves), 4–24 months (heifers) and older than 2 years (adults). A total of 2,163 fecal samples were collected from 570 calves including 450 cattle and 120 buffalo, 462 heifers comprising 323 cattle and 139 buffalo and 1,131 adults with 924 cattle and 207 buffalo. Additionally, 120 sheep samples (45 neonates and 75 adults) were sampled. Fecal specimens were collected directly from the rectum of each animal with a gloved hand and transferred into plastic cups labeled with the ear tag number and farm name. The fecal samples were screened for Cryptosporidium oocysts by microscopy using the modified Ziehl-Neelsen stain [10]. Positive specimens were stored in 2.50% potassium dichromate at 4.0°C for molecular biologic analyses.
Table 1. Infection rates of Cryptosporidium in different age groups of buffalo, cattle and sheep in Kafr El Sheikh Province, Egypt, based on acid fast staining-microscopy.
| Farm Name | Farm animals | Neonates | Heifers | Adults | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sample size | No. positive | Infection rate (%) | Sample size | No. positive | Infection rate (%) | Sample size | No. positive | Infection rate (%) | ||
| Farm A | Cattle | 154 | 4 | 2.60 | 125 | 2 | 1.60 | 370 | 2 | 0.54 |
| Farm B | Cattle | 159 | 16 | 10.10 | 123 | 13 | 10.60 | 318 | 21 | 6.60 |
| Farm C | Cattle | 137 | 11 | 8.0 | 75 | 18 | 24.0 | 236 | 33 | 14.0 |
| Farm D | Buffalo | 57 | 1 | 1.80 | 89 | 0 | 0.0 | 116 | 0 | 0.0 |
| Farm E | Buffalo | 63 | 4 | 6.40 | 50 | 0 | 0.0 | 91 | 1 | 1.10 |
| Farm C | Sheep | 45 | 2 | 4.40 | - | - | - | 75 | 1 | 1.30 |
DNA extraction and Cryptosporidium genotyping and subtyping: Microscopy-positive specimens were washed off potassium dichromate with distilled water by centrifugation. DNA was extracted using the QIAmp DNA Stool Mini Kit (Qiagen, Hilden, Germany), according to the manufacturer’s protocol. Genotyping of Cryptosporidium was done by PCR-RFLP analysis of SSU rRNA gene as described by Feng et al. [16]. Because 60 kDa glycoprotein (gp60) gene is highly polymorphic, it is favorably used as a good tool for genotyping and subtyping of many Cryptosporidium species including C. parvum, C. hominies,…etc. In the present study, samples from C. parvum were subtyped by nested PCR-sequence analysis of gp60 as described previously [2]. Nomenclature system established by Sulaiman et al. [42] was employed in naming C. parvum subtypes.
DNA sequencing, sequence analysis and phylogenetic analysis: Secondary PCR products of the representative samples of each RFLP pattern were sequenced directly using Big Dye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, U.S.A.) and read on an ABI 3130 Genetic Analyzer (Applied Biosystems). Sequences were assembled using the ChromasPro software (version 1.5) (http://www.technelysium.com.au/ChromasPro.html). The accuracy of data was confirmed by two-directional sequencing. The obtained sequences were aligned with reference sequences in database using ClustalX (ftp://ftp-igbmc.u-trasbg.fr/pub/ClustalX/) to determine the identity of the examined species. Unique nucleotide sequences generated in this study were deposited in GenBank under accession numbers AB922117–AB922125. The neighbor-joining (NJ) method as implemented in the MEGA5 (http://www.megasoftware.net/) was used in a phylogenetic analysis of SSU rRNA sequences, utilizing Eimeria tenella (AF026388) as an out-group.
RESULTS
Cryptosporidium infection: Cryptosporidium oocysts were detected in all examined farms in spite of species or age of the subjected animals (Table 1). In water buffalo, Cryptosporidium oocysts were detected microscopically in 5 out of 120 (4.17%) fecal samples from calves and in one out of 207 of adults (0.48%), while no oocysts were detected in the 139 examined specimens from heifers. Nonetheless, the infection rate in cattle was higher than that detected in buffalo animals, addressing the effect of animal species on the Cryptosporidium infection rate, especially in case of older animals. A total of 31 out of 450 (6.90%) fecal samples from calves, 33 out of 323 (10.20%) from heifers and 56 out of 924 (6.10%) from adult cattle proved to have Cryptosporidium oocysts. Microscopic examination of sheep samples revealed the occurrence of Cryptosporidium oocysts in 2 out of 45 lambs (4.40%) and only one sample out of 75 adult sheep (1.30%).
Cryptosporidium species: RFLP analysis of PCR products of the SSU rRNA gene indicated the presence of C. ryanae in 4 specimens (66.67%, 3 calves and one adult) and C. parvum in 2 specimens (33.33%) of buffalo samples. In addition, results revealed that 3 Cryptosporidium species were detected in cattle calves with varying prevalence; C. parvum (23/31; 74.23%), C. ryanae (5/31; 16.10%) and C. bovis (3/31; 9.70%), indicating the dominance of C. parvum. On the other hand, in heifers and adults, C. andersoni was the overwhelming species accompanied with a minor fraction of C. parvum (2/89; 2.30%). In sheep, C. xiaoi was the only detected species in the 3 positively infected animals. Based on the RFLP patterns, no mixed infections in one animal were detected in the present study.
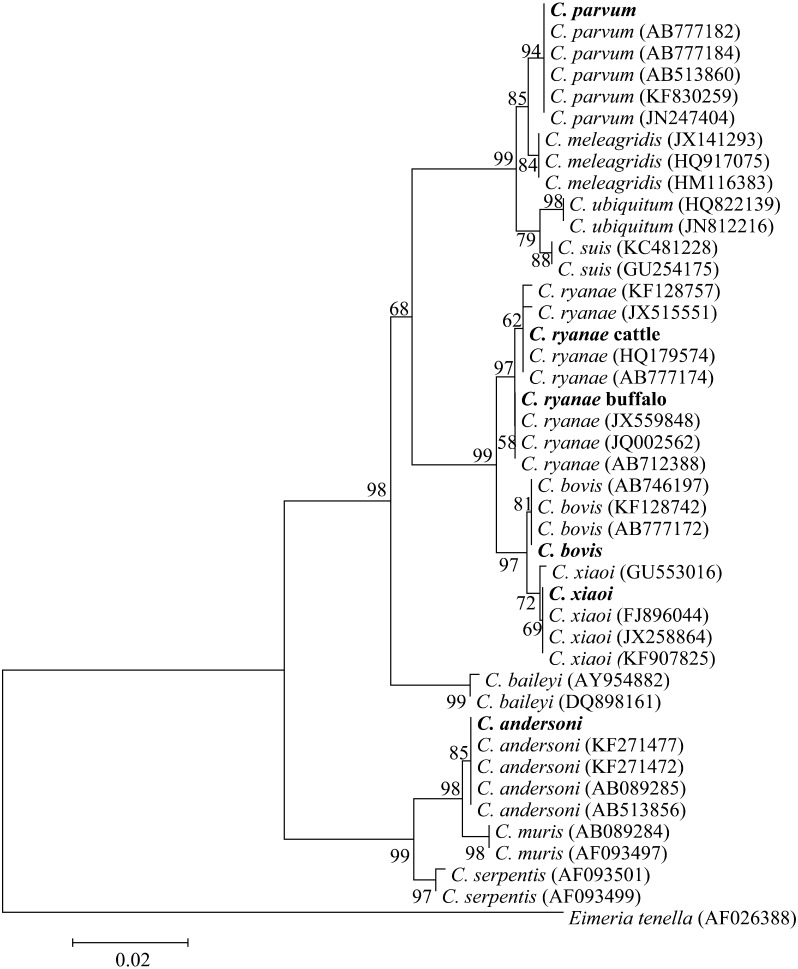
DNA sequencing of representative samples confirmed the identification of the Cryptosporidium species. Blasting the obtained sequences with those in database and the deduced phylogenetic analysis (Fig. 1) demonstrated that out of all the isolated samples, the sequences of C. parvum were 100% identical to each other in spite of the difference in host species and age of the animals. Moreover, these sequences were identical to reference sequences of C. parvum (AB777182) from Egypt and showed one base pair difference from the reference sequences reported from Egypt (AB513881), Iran (KF830257) and Brazil (JN247404). Meanwhile, specimens of C. andersoni generated one sequence that was identical to those from China (KF271477), Japan (AB089285) and Egypt (AB513856).
Fig. 1.
Phylogenetic relationship of Cryptosporidium spp. based on sequences of the partial SSU rRNA gene. Evolutionary relationships of 43 taxa were inferred using the Neighbor-Joining method [39] with the coccidian Eimeria tenella (AF026388) as an out-group. Numbers at the internodes correspond to percent bootstrap values from 2,000 replicates. Egyptian isolates are bolded.
C. ryanae specimens generated two sequence types. The first was derived from two isolates of buffalo and was identical to reference sequences reported from buffalo in Brazil, Egypt and India (JX559848, AB712388 and JQ002562, respectively). The second sequence type was generated from 3 specimens of cattle and was identical to that derived from dairy calves in China (HQ179574) as well as from cattle in Egypt (AB777174), while it has one base pair difference (99.0% homology) to sequences JX515551 isolated from dairy calf and KF128757 isolated from yak in China.
One type of sequence was obtained from C. bovis that was identical to the reference reported from Egypt (AB777172) and 99.0% similarity, with one base pair difference, compared to the sequences described from yak in China (KF128742), from cattle in Japan (AB746197) and from humans in Australia (JQ362488).
C. xiaoi from sheep generated one sequence type that was identical to sequences from sheep in U.S.A. and China (FJ896044 and KF907825). In addition, 99.0% similarity, one base pair difference, was noticed when compared to sequences from goat in Spain and sheep in Romania (GU553016 and JX258864, respectively).
C. parvum subtypes: In the current study, sequence analysis of C. parvum isolates on the gp60 gene locus revealed no difference among subtypes isolated from buffalo or cattle and generated two types of sequences. The first includes 3 sequences and belongs to the subtype IId (IIdA20G1 subtype). This sequence type was identical to sequences obtained from dairy cattle and buffalo in Egypt (AB514086, AB514089 and AB712389) and 99.0% with sequences with those reported from children in Kuwait and Sweden (AY738186 and JQ028866). The second type includes 2 sequences and belongs to IIa (IIaA15G1R1subtype). These sequences were identical to those reported from humans in Kuwait [42], Slovenia [41] and Australia [47] as well as from dairy calves in the Czech Republic [25], dairy and buffalo calves from Egypt [4, 5] and dairy calves and humans from Egypt [21].
DISCUSSION
The coccidian protozoa Cryptosporidium infects a wide range of livestock, resulting in significant economic loss and public health concern. In the present study, the infection rate varies according to the age and species of the host. Cryptosporidium oocysts were identified microscopically in 4.17% fecal samples of water buffalo calves and in 0.48% of adults, while no oocysts were detected in specimens from heifers. The infection rate in calves reported herein is much lower than that reported by Amer et al. [4] (9.50%) and El-Khodery and Osman [13] (14.10%), for water buffalo from Egypt and Bhat et al. [6] (38.30%), Feng et al. [17] (37.50%) and Nasir et al. [30] (24.0%), from India, Nepal and Pakistan, respectively. The differences in the reported infection rates may be attributed to differences in the age and breed of animals, season of specimen collection, environmental settings as well as management and husbandry regimens. Results from older animals indicated the negativity of all heifers’ samples and very low infection rate (0.48%) in adults. These results are comparable to those reported by Abou-Eisha [1] and Amer et al. [4] for adult water buffalo in Egypt and also to Rodriguez et al. [38] for buffalo in Cuba, who reported no oocysts detection in adult water buffalo. Although it is difficult to explain the low incidence of Cryptosporidium in adult buffalo, it might, partially, be attributed to the sensitivity of microscopic investigation or more likely to the resistance of adult buffalo either as species or owing to previous infections.
In cattle, the infection rates (overall 7.07%) were higher than that detected in buffalos, addressing the effect of animal species on the Cryptosporidium infection rate, especially in case of older animals. The prevalence was almost similar in both calves and adults (~6.90%) which relatively increased in case of heifers (10.20%). These results are comparable to those previously reported in dairy cattle in Egypt; 30.20% [3] and 13.60% [5]. In the present study, high differences were detected in the infection rates between different farms; 2.60% versus 10.10% in calves, 1.60% versus 24.0% in heifers and 0.54% versus 14.0% in adults, in low and high rate infected farms, respectively. These differences reflect the effect of farm related factors on the incidence of Cryptosporidium. The management regimen in the farm A imposes very restricting roles on the in and out of the animals, and the suckling calves up to the ~6 weeks of age are reared in ground separated pens with very limited opportunity of calves to contact with the discharged fecal matters. Because farms B and C belong to the same body, the move of animals between the two farms allows introducing new members to the herds. The calves are reared in ground pens, which in turn afford high possibility to contact with the fecal matter. This might be a factor behind the high infection rates in both farms.
Microscopic examination of sheep samples revealed the occurrence of Cryptosporidium oocysts in 4.40% of lambs and 1.30% of adult sheep. Similar results were reported in sheep from Brazil [18] and Egypt [31]. However, this infection rate is remarkably lower than that reported in sheep and lambs in Norway (15–24.0%), Romania (13.70%), Brazil (25.0%) and Australia (16.90%) [23, 34, 37, 53], addressing the habitat on the incidence of the disease.
Cryptosporidium species: RFLP and sequence analyses of SSU rRNA gene fragment (Table 2) indicated the presence of C. ryanae (66.67%) and C. parvum (33.33%) in buffalo samples. Similar results were reported in water buffalo from Egypt [4]. On the other hand, Caccio et al. [9] and Gomez-Couso et al. [20] reported only C. parvum in water buffalo calves in Spain and Italy, respectively. Feng et al. [17] reported only C. ryanae in buffalo calves from Nepal. These discrepancies may be attributed to farm, breed and/or climatic factors.
Table 2. Distribution of Cryptosporidium spp. in different age groups of buffalo, cattle and sheep in Kafr El Sheikh Province, Egypt, based on PCR-RFLP patterns of SSU rRNA gene fragment.
| Farm Name | Neonates | Heifers | Adults | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C. parvum | C. ryanae | C. bovis | C. andersoni | C. parvum | C. ryanae | C. bovis | C. andersoni | C. parvum | C. ryanae | C. bovis | C. andersoni | |
| Farm A | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 2 |
| Farm B | 11 | 3 | 2 | 0 | 0 | 0 | 0 | 13 | 1 | 0 | 0 | 20 |
| Farm C | 9 | 1 | 1 | 0 | 0 | 0 | 0 | 18 | 1 | 0 | 0 | 32 |
| Farm D | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Farm E | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| Farm C | 2 specimens of C. xiaoi | — | — | one specimen of C. xiaoi | ||||||||
C. parvum was the dominant prevailed species in cattle calves (74.23%) versus C. ryanae (16.10%) and C. bovis (9.70%), whereas, in heifers and adults, C. andersoni was the prevailing along with a small fraction of with C. parvum (2.30%). It is evident that the distribution of Cryptosporidium species in pre-weaned calves differs from a country to another and from a study to others, reflecting the profound effect of the environment and husbandry regimen on the infection rate. An exclusive presence of C. parvum, or with a minor fraction of other cryptosporidia, in pre-weaned dairy calves was previously reported in many countries [3, 44], contrasting the predominance of C. bovis and C. ryanae in some other countries [17, 29]. In the present study, C. andersoni appears to be the major species existing in heifers and adult cattle (87/89; 97.80%). A similar dominance was reported in a number of Asian countries, such as China [28, 49], as well as European countries [33]. In the United States and Canada, C. andersoni occurs in adult cattle beside different proportions of C. bovis and C. ryanae[8, 15]. Although C. andersoni represents no public health concerns, it can cause impairment of weight gain in growing animals and milk production in dairy cows and thus reduced feed efficiency [12].
In concordance with results reported herein, C. xiaoi was the dominant Cryptosporidium species in ewes and lambs in China [54] and in sheep in Australia [53]. In contrast, Imre et al. [24] reported the dominance of the zoonotic C. parvum in newborn lambs in Romania, while C. cervine was the major genotype in adult sheep in China [48]. The possible occurrence of periparturient transmission of Cryptosporidium spp., especially C. xiaoi, from ewes to lambs was addressed for cycling of the infection [54]. In spite of the limitation of sample size, the obtained results indicated that Cryptosporidium in sheep has no communal health concerns. Generally, the high identity of the generated sequences of similar type may be due to the semi-conservative nature of the SSU rRNA gene locus [52] common origin of the infection and/or the possibility that the infection was due to epidemic clone (the genetic fitness of this Cryptosporidium strain) [2, 7]. Moreover, geographic differences in the distribution of specific alleles may exist.
C. parvum subtypes: Occurrence of IId and IIa subtypes highlights the public health risk posed by Cryptosporidium parvum recovered in the present study. The incidence of C. parvum subtype families differs widely according to geographic locations and hosts. Subtype IId was reported as the predominant subtype family of C. parvum in cattle in Egypt, Malaysia and China [3, 29, 50], sheep and goats in Spain [35] and human in Kuwait, Jordan, Iran and Egypt [21, 22, 42, 43] in addition to HIV/AIDS patients in Malaysia [27]. However, it was detected at low frequencies in cattle in Europe and is largely absent in cattle and humans in U.S.A. and Canada [51]. Meanwhile, IIa is the most prevalent subtype family of C. parvum in calves worldwide, especially in industrialized countries [19, 25], implicating the cattle as the main source of human IIa infections.
In conclusion, Cryptosporidium oocysts were detected in all of the examined farms in spite of species or age of animals, which signify the widespread of the infection in Egypt. In cattle, the infection rates were higher than that detected in buffalo or sheep, revealing the effect of animal species on Cryptosporidium infection rate, especially in case of older animals. The common occurrence of the C. parvum IIa and IId subtype families in both buffalo and dairy calves in Egypt highlights the potential role of Cryptosporidium zoonotic in these animals.
ETHICS STATEMENT. Fecal samples were obtained with consent from farm owners of subjected animals. The study was approved by the Institutional Committee of the Post Graduate studies and research at Kafr El Sheikh University, Egypt. All efforts were made to minimize animal suffering. No experimentation was done on animals.
Acknowledgments
This study was partially support by Kafr el sheikh University. Thanks to farm owners for approval and facilities provided during collection of samples. Thanks extended to Dr. Abd El Naby Tahoon and Dr. Ehab El Sebaay, National Institute of Animal Health, Kafr El Sheikh Provincial lab, for their immense help.
REFERENCES
- 1.Abou-Eisha A.1994. Cryptosporidial infection in man and farm animals in Ismailia Governorate. Vet. Med. J. Giza 42: 107–111. [Google Scholar]
- 2.Alves M., Xiao L., Sulaiman I., Lal A. A., Matos O., Antunes F.2003. Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. J. Clin. Microbiol. 41: 2744–2747. doi: 10.1128/JCM.41.6.2744-2747.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amer S., Honma H., Ikarashi M., Tada C., Fukuda Y., Suyama Y., Nakai Y.2010. Cryptosporidium genotypes and subtypes in dairy calves in Egypt. Vet. Parasitol. 169: 382–386. doi: 10.1016/j.vetpar.2010.01.017 [DOI] [PubMed] [Google Scholar]
- 4.Amer S., Zidan S., Feng Y., Adamu H., Li N., Xiao L.2013. Identity and public health potential of Cryptosporidium spp. in water buffalo calves in Egypt. Vet. Parasitol. 191: 123–127. doi: 10.1016/j.vetpar.2012.08.015 [DOI] [PubMed] [Google Scholar]
- 5.Amer S., Zidan S., Adamu H., Ye J., Roellig D., Xiao L., Feng Y.2013. Prevalence and characterization of Cryptosporidium spp. in dairy cattle in Nile River delta provinces, Egypt. Exp. Parasitol. 135: 518–523. doi: 10.1016/j.exppara.2013.09.002 [DOI] [PubMed] [Google Scholar]
- 6.Bhat S., Juyal P., Singla L.2012. Prevalence of cryptosporidiosis in neonatal buffalo calves in Ludhiana District of Punjab, India. Asian J. Anim. Vet. Adv. 7: 512–520. doi: 10.3923/ajava.2012.512.520 [DOI] [Google Scholar]
- 7.Brook E. J., Anthony Hart C., French N. P., Christley R. M.2009. Molecular epidemiology of Cryptosporidium subtypes in cattle in England. Vet. J. 179: 378–382. doi: 10.1016/j.tvjl.2007.10.023 [DOI] [PubMed] [Google Scholar]
- 8.Budu-Amoako E., Greenwood S. J., Dixon B. R., Barkema H. W., McClure J. T.2012. Giardia and Cryptosporidium on dairy farms and the role these farms may play in contaminating water sources in Prince Edward Island, Canada. J. Vet. Intern. Med. 26: 668–673. doi: 10.1111/j.1939-1676.2012.00930.x [DOI] [PubMed] [Google Scholar]
- 9.Cacciò S. M., Rinaldi L., Cringoli G., Condoleo R., Pozio E.2007. Molecular identification of Cryptosporidium parvum and Giardia duodenalis in the Italian water buffalo (Bubalus bubalis). Vet. Parasitol. 150: 146–149. doi: 10.1016/j.vetpar.2007.09.013 [DOI] [PubMed] [Google Scholar]
- 10.Casemore D. P., Armstrong M., Sands R. L.1985. Laboratory diagnosis of cryptosporidiosis. J. Clin. Pathol. 38: 1337–1341. doi: 10.1136/jcp.38.12.1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chalmers R. M., Davies A. P.2010. Minireview: clinical cryptosporidiosis. Exp. Parasitol. 124: 138–146. doi: 10.1016/j.exppara.2009.02.003 [DOI] [PubMed] [Google Scholar]
- 12.de Graaf D. C., Vanopdenbosch E., Ortega-Mora L. M., Abbassi H., Peeters J. E.1999. A review of the importance of cryptosporidiosis in farm animals. Int. J. Parasitol. 29: 1269–1287. doi: 10.1016/S0020-7519(99)00076-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El-Khodery S. A., Osman S. A.2008. Cryptosporidiosis in buffalo calves (Bubalus bubalis): prevalence and potential risk factors. Trop. Anim. Health Prod. 40: 419–426. doi: 10.1007/s11250-007-9113-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fayer R.2010. Taxonomy and species delimitation in Cryptosporidium. Exp. Parasitol. 124: 90–97. doi: 10.1016/j.exppara.2009.03.005 [DOI] [PubMed] [Google Scholar]
- 15.Fayer R., Santin M., Trout J. M.2007. Prevalence of Cryptosporidium species and genotypes in mature dairy cattle on farms in eastern United States compared with younger cattle from the same locations. Vet. Parasitol. 145: 260–266. doi: 10.1016/j.vetpar.2006.12.009 [DOI] [PubMed] [Google Scholar]
- 16.Feng Y., Ortega Y., He G., Das P., Xu M., Zhang X., Fayer R., Gatei W., Cama V., Xiao L.2007. Wide geographic distribution of Cryptosporidium bovis and the deer-like genotype in bovines. Vet. Parasitol. 144: 1–9. doi: 10.1016/j.vetpar.2006.10.001 [DOI] [PubMed] [Google Scholar]
- 17.Feng Y., Karna S. R., Dearen T. K., Singh D. K., Adhikari L. N., Shrestha A., Xiao L.2012. Common occurrence of a unique Cryptosporidium ryanae variant in zebu cattle and water buffaloes in the buffer zone of the Chitwan National Park, Nepal. Vet. Parasitol. 185: 309–314. doi: 10.1016/j.vetpar.2011.09.025 [DOI] [PubMed] [Google Scholar]
- 18.Fiuza V. R., Cosendey R. I., Frazão-Teixeira E., Santín M., Fayer R., de Oliveira F. C.2011. Molecular characterization of Cryptosporidium in Brazilian sheep. Vet. Parasitol. 175: 360–362. doi: 10.1016/j.vetpar.2010.10.036 [DOI] [PubMed] [Google Scholar]
- 19.Ghaffari S., Kalantari N., A Hart C.2014. A Multi-Locus Study for Detection of Cryptosporidium Species Isolated from Calves Population, Liverpool; UK. Int. J. Mol. Cell Med. 3: 35–42. [PMC free article] [PubMed] [Google Scholar]
- 20.Gómez-Couso H., Amar C. F., McLauchlin J., Ares-Mazás E.2005. Characterisation of a Cryptosporidium isolate from water buffalo (Bubalus bubalis) by sequencing of a fragment of the Cryptosporidium oocyst wall protein gene (COWP). Vet. Parasitol. 131: 139–144. doi: 10.1016/j.vetpar.2005.04.022 [DOI] [PubMed] [Google Scholar]
- 21.Helmy Y. A., Krücken J., Nöckler K., von Samson-Himmelstjerna G., Zessin K. H.2013. Molecular epidemiology of Cryptosporidium in livestock animals and humans in the Ismailia province of Egypt. Vet. Parasitol. 193: 15–24. doi: 10.1016/j.vetpar.2012.12.015 [DOI] [PubMed] [Google Scholar]
- 22.Hijjawi N., Ng J., Yang R., Atoum M. F., Ryan U.2010. Identification of rare and novel Cryptosporidium GP60 subtypes in human isolates from Jordan. Exp. Parasitol. 125: 161–164. doi: 10.1016/j.exppara.2010.01.011 [DOI] [PubMed] [Google Scholar]
- 23.Imre K., Lobo L. M., Matos O., Popescu C., Genchi C., Dărăbuş G.2011. Molecular characterisation of Cryptosporidium isolates from pre-weaned calves in Romania: is there an actual risk of zoonotic infections? Vet. Parasitol. 181: 321–324. doi: 10.1016/j.vetpar.2011.04.042 [DOI] [PubMed] [Google Scholar]
- 24.Imre K., Luca C., Costache M., Sala C., Morar A., Morariu S., Ilie M. S., Imre M., Dărăbuş G.2013. Zoonotic Cryptosporidium parvum in Romanian newborn lambs (Ovis aries). Vet. Parasitol. 191: 119–122. doi: 10.1016/j.vetpar.2012.08.020 [DOI] [PubMed] [Google Scholar]
- 25.Kváč M., Hromadová N., Květoňová D., Rost M., Sak B.2011. Molecular characterization of Cryptosporidium spp. in pre-weaned dairy calves in the Czech Republic: absence of C. ryanae and management-associated distribution of C. andersoni, C. bovis and C. parvum subtypes. Vet. Parasitol. 177: 378–382. doi: 10.1016/j.vetpar.2010.11.048 [DOI] [PubMed] [Google Scholar]
- 26.Lange H., Johansen O. H., Vold L., Robertson L. J., Anthonisen I. L., Nygard K.2014. Second outbreak of infection with a rare Cryptosporidium parvum genotype in schoolchildren associated with contact with lambs/goat kids at a holiday farm in Norway. Epidemiol. Infect. 142: 2105–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim Y. A., Iqbal A., Surin J., Sim B. L., Jex A. R., Nolan M. J., Smith H. V., Gasser R. B.2011. First genetic classification of Cryptosporidium and Giardia from HIV/AIDS patients in Malaysia. Infect. Genet. Evol. 11: 968–974. doi: 10.1016/j.meegid.2011.03.007 [DOI] [PubMed] [Google Scholar]
- 28.Liu A., Wang R., Li Y., Zhang L., Shu J., Zhang W., Feng Y., Xiao L., Ling H.2009. Prevalence and distribution of Cryptosporidium spp. in dairy cattle in Heilongjiang Province, China. Parasitol. Res. 105: 797–802. doi: 10.1007/s00436-009-1457-2 [DOI] [PubMed] [Google Scholar]
- 29.Muhid A., Robertson I., Ng J., Ryan U.2011. Prevalence of and management factors contributing to Cryptosporidium sp. infection in pre-weaned and post-weaned calves in Johor, Malaysia. Exp. Parasitol. 127: 534–538. doi: 10.1016/j.exppara.2010.10.015 [DOI] [PubMed] [Google Scholar]
- 30.Nasir A., Avais M., Khan M., Ahmad N.2009. Prevalence of Cryptosporidium parvum infection in Lahore (Pakistan) and its association with diarrhea in dairy calves. Int. J. Agri. Biol. 11: 221–224. [Google Scholar]
- 31.Nassif M. N., Amer S., Osman S. A.2002. Some studies on ovine and caprine cryptosporidiosis concerning prevalence and electrophoretic pattern of blood serum proteins. Assiut Vet. Med. J. 47: 249–262. [Google Scholar]
- 32.Nydam D. V., Wade S. E., Schaaf S. L., Mohammed H. O.2001. Number of Cryptosporidium parvum oocysts or Giardia spp cysts shed by dairy calves after natural infection. Am. J. Vet. Res. 62: 1612–1615. doi: 10.2460/ajvr.2001.62.1612 [DOI] [PubMed] [Google Scholar]
- 33.Ondrácková Z., Kvác M., Sak B., Kvetonová D., Rost M.2009. Prevalence and molecular characterization of Cryptosporidium spp. in dairy cattle in South Bohemia, the Czech Republic. Vet. Parasitol. 165: 141–144. doi: 10.1016/j.vetpar.2009.06.035 [DOI] [PubMed] [Google Scholar]
- 34.Paz e Silva F. M., Lopes R. S., Bresciani K. D., Amarante A. F., Araujo J. P., Jr2014. High occurrence of Cryptosporidium ubiquitum and Giardia duodenalis genotype E in sheep from Brazil. Acta Parasitol. 59: 193–196. doi: 10.2478/s11686-014-0223-5 [DOI] [PubMed] [Google Scholar]
- 35.Quílez J., Torres E., Chalmers R. M., Hadfield S. J., Del Cacho E., Sánchez-Acedo C.2008. Cryptosporidium genotypes and subtypes in lambs and goat kids in Spain. Appl. Environ. Microbiol. 74: 6026–6031. doi: 10.1128/AEM.00606-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robertson L. J., Campbell A. T., Smith H. V.1992. Survival of Cryptosporidium parvum oocysts under various environmental pressures. Appl. Environ. Microbiol. 58: 3494–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robertson L. J., Gjerde B. K., Furuseth Hansen E.2010. The zoonotic potential of Giardia and Cryptosporidium in Norwegian sheep: a longitudinal investigation of 6 flocks of lambs. Vet. Parasitol. 171: 140–145. doi: 10.1016/j.vetpar.2010.03.014 [DOI] [PubMed] [Google Scholar]
- 38.Rodriguez Diego J., Abreu J. R., Perez E., Roque E., Cartas O.1991. Presence of Cryptosporidium in buffaloes (Bubalus bubalis) in Cuba. Revista de Salud Animal 13: 78–80. [Google Scholar]
- 39.Saitou N., Nei M.1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425. [DOI] [PubMed] [Google Scholar]
- 40.Santín M., Trout J. M., Xiao L., Zhou L., Greiner E., Fayer R.2004. Prevalence and age-related variation of Cryptosporidium species and genotypes in dairy calves. Vet. Parasitol. 122: 103–117. doi: 10.1016/j.vetpar.2004.03.020 [DOI] [PubMed] [Google Scholar]
- 41.Soba B., Logar J.2008. Genetic classification of Cryptosporidium isolates from humans and calves in Slovenia. Parasitology 135: 1263–1270. doi: 10.1017/S0031182008004800 [DOI] [PubMed] [Google Scholar]
- 42.Sulaiman I. M., Hira P. R., Zhou L., Al-Ali F. M., Al-Shelahi F. A., Shweiki H. M., Iqbal J., Khalid N., Xiao L.2005. Unique endemicity of cryptosporidiosis in children in Kuwait. J. Clin. Microbiol. 43: 2805–2809. doi: 10.1128/JCM.43.6.2805-2809.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taghipour N., Nazemalhosseini-Mojarad E., Haghighi A., Rostami-Nejad M., Romani S., Keshavarz A., Alebouyeh M., Zali M.2011. Molecular epidemiology of cryptosporidiosis in Iranian children, tehran, iran. Iran. J. Parasitol. 6: 41–45. [PMC free article] [PubMed] [Google Scholar]
- 44.Tomazic M. L., Maidana J., Dominguez M., Uriarte E. L., Galarza R., Garro C., Florin-Christensen M., Schnittger L.2013. Molecular characterization of Cryptosporidium isolates from calves in Argentina. Vet. Parasitol. 198: 382–386. doi: 10.1016/j.vetpar.2013.09.022 [DOI] [PubMed] [Google Scholar]
- 45.Torsein M., Lindberg A., Sandgren C. H., Waller K. P., Törnquist M., Svensson C.2011. Risk factors for calf mortality in large Swedish dairy herds. Prev. Vet. Med. 99: 136–147. doi: 10.1016/j.prevetmed.2010.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsushima Y., Karanis P., Kamada T., Makala L., Xuan X., Tohya Y., Akashi H., Nagasawa H.2003. Seasonal change in the number of Cryptosporidium parvum oocysts in water samples from the rivers in Hokkaido, Japan, detected by the ferric sulfate flocculation method. J. Vet. Med. Sci. 65: 121–123. doi: 10.1292/jvms.65.121 [DOI] [PubMed] [Google Scholar]
- 47.Waldron L. S., Dimeski B., Beggs P. J., Ferrari B. C., Power M. L.2011. Molecular epidemiology, spatiotemporal analysis, and ecology of sporadic human cryptosporidiosis in Australia. Appl. Environ. Microbiol. 77: 7757–7765. doi: 10.1128/AEM.00615-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y., Feng Y., Cui B., Jian F., Ning C., Wang R., Zhang L., Xiao L.2010. Cervine genotype is the major Cryptosporidium genotype in sheep in China. Parasitol. Res. 106: 341–347. doi: 10.1007/s00436-009-1664-x [DOI] [PubMed] [Google Scholar]
- 49.Wang R., Ma G., Zhao J., Lu Q., Wang H., Zhang L., Jian F., Ning C., Xiao L.2011a. Cryptosporidium andersoni is the predominant species in post-weaned and adult dairy cattle in China. Parasitol. Int. 60: 1–4. doi: 10.1016/j.parint.2010.09.002 [DOI] [PubMed] [Google Scholar]
- 50.Wang R., Wang H., Sun Y., Zhang L., Jian F., Qi M., Ning C., Xiao L.2011b. Characteristics of Cryptosporidium transmission in preweaned dairy cattle in Henan, China. J. Clin. Microbiol. 49: 1077–1082. doi: 10.1128/JCM.02194-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiao L.2010. Molecular epidemiology of cryptosporidiosis: an update. Exp. Parasitol. 124: 80–89. doi: 10.1016/j.exppara.2009.03.018 [DOI] [PubMed] [Google Scholar]
- 52.Xiao L., Escalante L., Yang C., Sulaiman I., Escalante A. A., Montali R. J., Fayer R., Lal A. A.1999. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl. Environ. Microbiol. 65: 1578–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang R., Jacobson C., Gardner G., Carmichael I., Campbell A. J., Ng-Hublin J., Ryan U.2014. Longitudinal prevalence, oocyst shedding and molecular characterisation of Cryptosporidium species in sheep across four states in Australia. Vet. Parasitol. 200: 50–58. doi: 10.1016/j.vetpar.2013.11.014 [DOI] [PubMed] [Google Scholar]
- 54.Ye J., Xiao L., Wang Y., Wang L., Amer S., Roellig D. M., Guo Y., Feng Y.2013. Periparturient transmission of Cryptosporidium xiaoi from ewes to lambs. Vet. Parasitol. 197: 627–633. doi: 10.1016/j.vetpar.2013.07.021 [DOI] [PubMed] [Google Scholar]