Abstract
During interphase, chromatin hosts fundamental cellular processes, such as gene expression, DNA replication and DNA damage repair. To analyze chromatin on a proteomic scale, we have developed chromatin enrichment for proteomics (ChEP), which is a simple biochemical procedure that enriches interphase chromatin in all its complexity. It enables researchers to take a ‘snapshot’ of chromatin and to isolate and identify even transiently bound factors. In ChEP, cells are fixed with formaldehyde; subsequently, DNA together with all cross-linked proteins is isolated by centrifugation under denaturing conditions. This approach enables the analysis of global chromatin composition and its changes, which is in contrast with existing chromatin enrichment procedures, which either focus on specific chromatin loci (e.g., affinity purification) or are limited in specificity, such as the analysis of the chromatin pellet (i.e., analysis of all insoluble nuclear material). ChEP takes half a day to complete and requires no specialized laboratory skills or equipment. ChEP enables the characterization of chromatin response to drug treatment or physiological processes. Beyond proteomics, ChEP may preclear chromatin for chromatin immunoprecipitation (ChIP) analyses.
INTRODUCTION
Development of the protocol
Chromatin contains, by mass, about twice as much protein as DNA, and these chromatin-associated proteins have crucial roles in all biological processes that take place in the eukaryotic genome. Unbiased proteomic interrogation of this ‘chromatome’ could provide important information when studying processes that take place in chromatin. Proteomic studies on the chromatome could include, for example, the identification of transcription factors binding to chromatin in response to a physiological stimulus or the discovery of novel DNA repair factors after induction of DNA damage. To make such studies possible, we sought to develop a procedure for chromatin enrichment that satisfied the following three requirements: first, the procedure should enrich whole chromatin in an unbiased manner, i.e., neither lose out on transiently bound factors nor preferentially target any specific chromosome region; second, it should ideally provide a chromatin snapshot enabling one to avoid obscuring results through secondary effects during the purification procedure; and, finally, it should be quick and simple to implement, so that it can be carried out in every molecular biology laboratory. We called the resulting biochemical procedure ChEP1.
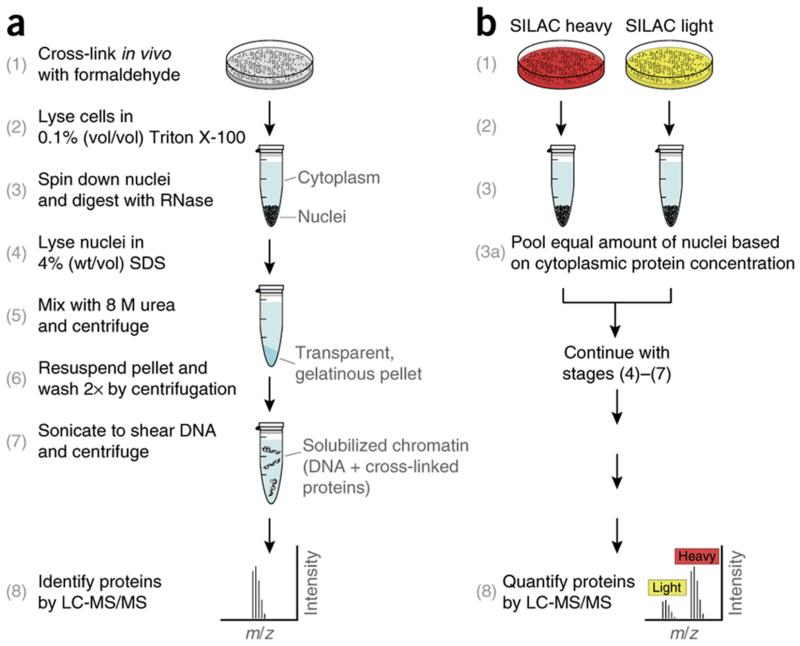
ChEP starts by in vivo cross-linking of chromatin proteins to DNA with formaldehyde, under the same conditions used for ChIP2. After cell lysis, nuclei are digested with RNase and non-cross-linked proteins are washed away using highly denaturing extraction buffers containing 4% (wt/vol) SDS and 8 M urea. We found that these distinct hypotonic buffer conditions force chromatin to precipitate as a transparent, gelatinous pellet. A particularly valuable property of this pellet is that it can be resuspended completely and washed repeatedly (Fig. 1a).
Figure 1.
Outline of the ChEP procedure. (a) The key stages (numbers in parentheses) for chromatin enrichment of a single sample. (b) Scheme for a quantitative version of ChEP, comparing chromatin from different samples based on SILAC. Adapted from Kustatscher et al.1, © 2014 The Authors.
The purpose of cross-linking is to induce the covalent binding of all chromatin-associated proteins either directly or indirectly to DNA. In principle, this covalent linking allows one to use very stringent conditions to wash away proteins that are neither permanently nor transiently associated with chromatin. However, a number of optimization steps have had to be implemented to remove non-cross-linked proteins efficiently. Chromatin is a highly charged biopolymer owing to the phosphate backbone of DNA and, upon cell lysis, it may bind proteins through nonspecific electrostatic interactions. Although this tendency can be counteracted by using high salt concentrations in the extraction buffers, we found that elevated ionic strength in these buffers resulted in the formation of hard, opaque chromatin pellets that cannot be resuspended, making any washing steps and the subsequent handling inefficient (data not shown). We have therefore opted to use denaturing but salt-free extraction conditions that yield a gelatinous, transparent pellet, which can be fully and repeatedly resuspended without any apparent loss of material when using hydrophobic pipette tips. These conditions rely on the detergent SDS as well as urea, a nonionic chaotropic agent, which enable the researcher to solubilize and therefore remove proteins that are not covalently bound to DNA.
Experimental design
Under the highly denaturing conditions used in ChEP, and in the absence of salt and alcohol, precipitation of chromatin depends on distinct cross-linking conditions. Chromatin that has undergone cross-linking with 1% (wt/vol) formaldehyde for 10 min at 37 °C can be pelleted readily, but it cannot be precipitated when lower formaldehyde concentrations or temperatures are used. Formaldehyde creates both protein-to-DNA and protein-to-protein cross-links. We experimented with dual cross-linking conditions, combining formaldehyde with glutaraldehyde or disuccinimidyl glutarate, which preferentially cross-link proteins, but the resulting pellets were resistant to sonication and could not be analyzed efficiently. Thus, the cross-linking conditions typically used for ChIP also appear to be optimal for the purification of chromatin-associated proteins.
We included an RNase digestion step in ChEP to avoid the enrichment of proteins associated with nascent RNA rather than directly with chromatin, and to disintegrate and reduce the cofractionation of ribosomes, a common contaminant of chromatin purifications. Indeed, we found the RNase step to be beneficial for both of these goals (Supplementary Fig. 1).
We routinely use ChEP to quantitatively compare different chromatin states, e.g., chromatin from drug-treated cells with an untreated (mock) control based on stable isotope labeling by amino acids in cell culture (SILAC)3. A key advantage of this metabolic labeling approach is that tissue culture cells grown in different conditions can be pooled together immediately after collection, thus avoiding artifacts introduced by differential downstream handling of samples. However, when confluent monolayers of cells are cross-linked with formaldehyde, they can form clumps, making it difficult to accurately determine cell numbers and pool cells accordingly. In our experience, the best way to pool SILAC samples for ChEP is to perform cell lysis and RNase digestion of nuclei from cells grown in light and heavy SILAC medium separately. While nuclei are being digested, the protein concentration in their respective cytoplasmic supernatants can be determined, and nuclei can then be pooled accurately on the basis of this measurement (Fig. 1b). Alternatively, for quantitative analysis of chromatin from cells that do not grow in SILAC medium (e.g., primary cell lines), we compare unlabeled chromatin samples using SILAC-labeled chromatin from a reference cell line or pool of cell lines as an internal standard. The use of an internal SILAC standard has been described previously, including culture-derived isotope tags4, super-SILAC5 and spike-in SILAC6.
We found that resuspending nuclei or chromatin in SDS buffer first, and subsequently mixing in the urea buffer, was more practical than adding SDS buffer and urea buffer concomitantly, as the high SDS concentration prevents chromatin from sticking to the pipette tips (data not shown). In addition, urea disintegrates SDS micelles and disrupts SDS-protein interactions7, so the application of urea after SDS buffer is likely to be a more effective solubilization strategy. Note that careful but thorough mixing of the SDS-resuspended nuclei or chromatin with the urea buffer is absolutely essential in the present protocol. Overlaying SDS-resuspended nuclei on a urea gradient would require overnight ultracentrifugation in order to pellet chromatin8.
One problem with urea is that when its mixture with proteins is heated above 37 °C, it promotes protein carbamylation, a modification that interferes with protein identification by mass spectrometry. For this reason, we include in the PROCEDURE a wash step to remove urea from the mixture before boiling the samples to reverse formaldehyde cross-links. Cross-link reversal enables proteins to efficiently enter the SDS-PAGE gel. We have not observed any influence of formaldehyde cross-linking on the mass spectrometric analysis of proteins.
To shear genomic DNA and to solubilize chromatin, this protocol makes use of the same equipment and conditions that are often used during ChIP. Although precise sonication conditions may need to be established for ChIP, in order to obtain DNA fragments with optimal length for immunoprecipitation, sonication in ChEP has the sole purpose of solubilizing chromatin, and we have found DNA fragment size to have no effect on subsequent steps of the protocol. Therefore, any sonication conditions may be used that solubilize chromatin completely, as made evident by the disappearance of the gelatinous pellet.
We originally planned to include an additional purification step in the PROCEDURE, whereby chromatin would be sheared, adsorbed to hydroxyapatite resin and then washed with high molar concentrations of NaCl to remove any proteins that might still be bound to DNA in an electrostatic, noncovalent manner. However, after implementation of the additional purification step just described, we did not observe any additional reduction in the amount of co-purifying nonrelevant proteins, i.e., proteins without reported chromatin function (G.K. and J.R., unpublished data). A second step that we dismissed after testing was the removal of histones through ion exchange chromatography before proteomic analysis. Histones are highly enriched as a consequence of chromatin purification and can obscure detection of less abundant proteins by mass spectrometry. However, we found ion exchange chromatography not to be specific and robust enough in histone retention as would be required for a simple, straightforward chromatin enrichment procedure (G.K. and J.R., unpublished data). Instead, trypsin digestion of bulk chromatin and subsequent in-depth proteomic analysis of all peptides appears to be a more efficient way to ensure the detection of chromatin proteins that are less abundant than histones.
Applications of ChEP
We have already used ChEP to analyze chromatin from various cell types in different biological conditions1. In principle, ChEP could be used to conduct chromatin proteomics studies of any cell or tissue sample that can be cross-linked with formaldehyde. So far, we have successfully used ChEP in a range of commonly used human tumor cell lines, such as HEK293, HepG2, HeLa, U2OS and MCF-7 cells1. We have also applied ChEP to MCF-10A, BT-474, BT-549, HCC1954, HBL-100, T-47D, ZR-75-1, SUM159PT, LY2, Hs578T, MDA-MB-468, SUM1315MO2, MDA-MB-453 and SK-BR-3 cells (K.L.H.W. and J.R., unpublished data); to human primary cell lines such as MRC-5 (C.F. and J.R., unpublished data); and to nonhuman cell lines such as chicken DT40 cells1. Note that ChEP can be used also for cells growing in suspension rather than as monolayers.
On the basis of ChEP experiments, we recently presented the first comprehensive inventory of human interphase chromatin1. As part of the same study, we have also applied ChEP and quantitative mass spectrometry to the identification of proteins that associate with chicken chromatin, depending on cyclin-dependent kinase (Cdk)-driven cell cycle progression. We have also used ChEP to purify mitotic chromosomes or to clean up mitotic chromosome fractions prepared using alternative methods (K.L.H.W., C.F. and J.R., unpublished data; note that mitotic chromosomes are highly compacted and can be purified using gradient centrifugation methods9, 10). In principle, ChEP could be used as a front-end process for any procedure requiring the isolation of cross-linked chromatin. ChEP-isolated chromatin could arguably be described as the ‘ChIP-able’ part of the proteome, as it contains all proteins that are cross-linked, directly or indirectly, to DNA by 1% (wt/vol) formaldehyde (i.e., the starting conditions of ChIP experiments). Therefore, the results of ChEP may provide a list of any ChIP-able protein. Combining ChEP with ChIP could be useful to reduce background, and thus could increase the specificity and sensitivity of ChIPs. An additional potential nonproteomic application of ChEP could be the study of chromatin-associated RNAs, in which case the RNase treatment would have to be omitted.
Comparison with other methods
A variety of proteomic approaches have been developed to study chromatin functions, but, in contrast to ChEP, they generally focus on specific subsets of chromatin. Most methods could broadly be characterized as either locus-specific or protein-specific affinity purifications. The classic strategy of isolating sequence-specific DNA-binding proteins using their cognate sites as bait for in vitro pull-downs11 has been complemented with quantitative proteomics readouts12, 13. Proteomics methods to analyze proteins binding to specific loci in vivo rely on genomic insertion of recognition sequences such as LacO, TetO or LexA sites, which enable the researcher to perform affinity purification via their binding proteins (LacI, TetR or LexA DB, respectively)14-19. More recently, purification of specific chromatin domains without genomic insertion has been made possible by targeting endogenous loci using transcription activator–like (TAL) proteins or the clustered regularly interspaced short palindromic repeats (CRISPR) system20, 21. A strategy to enrich repetitive wild-type loci by hybridization with tagged oligonucleotides proved to be successful in conducting telomere proteomics studies22. We and others have reported purification procedures for newly replicated chromatin, based on the incorporation of affinity-tagged DNA base analogs23-25. Additional proteomic approaches have been developed to reveal chromatin-bound interaction partners of tagged proteins of interest26, 27 and to identify heterochromatin or euchromatin-specific factors by immunoprecipitation of regions methylated at H3K9 or H3K4, respectively28. Histone modifications themselves have long been studied by mass spectrometry29, and proteins binding these marks have been identified using histone peptides30, 31 or nucleosomes32, 33 as baits in pull-down assays. A similar ‘pull-down and quantitative proteomics’ technique using modified DNA oligonucleotides identified proteins recognizing methylated and hydroxymethylated cytosine residues34.
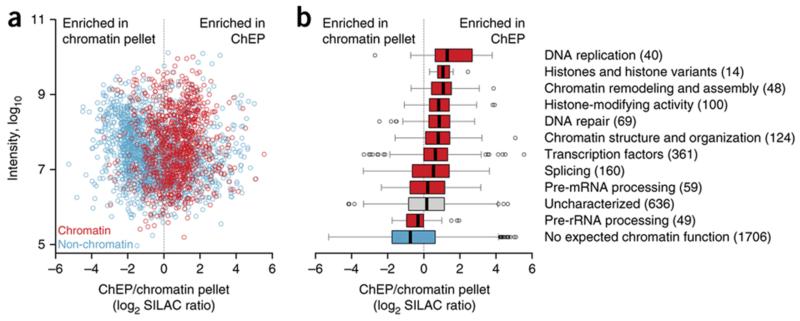
Several proteomics studies have aimed, such as ChEP, to analyze whole chromatin. Shiio et al.35 reported the proteomic analysis of the chromatin pellet, which essentially corresponds to the insoluble part of the nucleus. Although the majority of chromatin-bound proteins can be expected to be in this fraction, this approach suffers from somewhat low specificity, as insolubility is not a feature that is exclusive to chromatin. Indeed, compared with a ChEP-obtained gelatinous chromatin pellet, the classical chromatin pellet is highly enriched in non-chromatin-associated proteins (Fig. 2a,b; ref. 1). Modified procedures to obtain chromatin pellets for proteomics have been reported, including the use of detergents to enhance purification stringency36-38. Chromatin pellets have also been subjected to acid, salt or DNase extraction to enrich them further with chromatin factors39-41. Note that the mentioned additional biochemical enrichment procedures may skew the analysis toward preferentially identifying subsets of chromatin proteins, such as proteins that are soluble at a particular salt concentration or those that remain associated with chromatin even in the presence of detergents. ChEP enriches chromatin on the basis of ‘cross-linkability’ rather than solubility, thus minimizing losses of transiently chromatin-bound factors and reducing the risk of purification artifacts introduced after cell lysis. For example, among the most highly enriched proteins in ChEP compared with the chromatin pellet defined above are the four subunits of DNA polymerase α, which is notorious for leaking into the cytoplasm upon cell disruption42 but is stabilized by cross-linking.
Figure 2.
Comparison of protein enrichment after isolation of ChEP-based gelatinous chromatin and the ‘classical’ chromatin pellet isolated according to Shiio et al.35. A ChEP sample from cells grown in heavy SILAC medium is mixed with a chromatin pellet from unlabeled cells. (a) SILAC ratios are plotted against protein intensities to show the enrichment of known chromatin-associated proteins (red) and depletion of proteins without reported chromatin function (blue) from ChEP-isolated fractions compared with those isolated from a classical chromatin pellet. Proteins were manually annotated on the basis of UniProt and literature evidence1. Only proteins that could clearly be assigned to either category are plotted. (b) Box plot breaking down the same experiment into functional categories, including uncharacterized proteins not shown in a. The number of proteins per category is shown in parentheses. Proteins depleted in the ChEP fractions include those without expected chromatin function (i.e., contaminants) and proteins related to RNA processing, probably depleted after RNase treatment. Some genuine chromatin proteins are also depleted after ChEP implementation. These are generally multifunctional proteins, such as the mitochondrial prohibitins 1 and 2, which have additional roles as modulators of nuclear transcription. Note that such proteins may be depleted in ChEP fractions simply because their main pool is not chromatin-associated under the biological condition in question. The box plot was generated using BoxPlotR53; whiskers extend to data points that are <1.5 interquartile ranges away from the first and third quartile (‘Tukey’ whiskers). Adapted from Kustatscher et al.1, © 2014 The Authors.
Chromatin enriched via ChEP contains fewer contaminants than the classical chromatin pellet. Nevertheless, the presence of a protein in ChEP fractions is not sufficient to conclude that it has a function in chromatin, as numerous proteins without expected chromatin function remain in these samples. However, we would argue that completeness and preservation of in vivo chromatin features through cross-linking are essential for studying chromatin-resident processes by proteomics. We have recently provided a comprehensive inventory of interphase chromatin-associated proteins, but for this we did not rely on a single ChEP purification1. Instead, we used a machine-learning approach to integrate the composition of ChEP fractions from various cell types and biological conditions into the overall probability of any of 7,635 human proteins to have a chromatin-based function1. These probabilities can be used as a filter to focus data sets on likely chromatin factors. As a typical example, we have compared ChEP fractions from cells with or without Cdk activity and then selected uncharacterized proteins that were predicted to have a chromatin-based function for follow-up studies1.
Limitations
ChEP is designed for the global analysis of the protein composition of chromatin. Changes that are confined to small chromatin regions, such as a single locus, may not be apparent in a chromatomics study, if the bulk of a protein remains unaffected. Similarly, proteins may respond to a particular stimulus by re-localizing within chromatin, leaving their overall chromatin association unaffected. However, the high accuracy of SILAC-based quantitative proteomics means that even marginal overall changes can be reliably quantified.
A second potential limitation of ChEP is that formaldehyde cross-linking may not be effective for all chromatin factors. For example, it has been shown that formaldehyde cross-linking is insufficient to preserve chromatin association of several known chromatin factors for robust ChIP43-46. However, out of the eight proteins mentioned in these studies, we find six in a typical ChEP analysis covering ~3,500 proteins, which is still far from being an exhaustive chromatin analysis. In contrast to ChIP, we therefore believe that the vast majority of chromatin factors can be observed using the ChEP protocol.
Finally, as discussed above, a complete separation of chromatin-associated proteins from non-chromatin-associated proteins cannot be achieved by ChEP or by any other fractionation procedure that we are aware of. The presence of a protein in ChEP fractions is therefore not sufficient to conclude that it truly associates with chromatin. However, ChEP enriches all chromatin-associated proteins while removing contaminants more efficiently than an analogous procedure (Fig. 2), thus ensuring optimal detection of chromatin factors by mass spectrometry. To further enhance the value of any detected candidate proteins, they can be sorted according to their reported interphase chromatin probabilities1, which integrate the collective data of many chromatin analyses.
MATERIALS
REAGENTS
-
Cells as starting material, for example, transformed human cell lines such as HEK293, HepG2, HeLa, U2OS, MCF-7, MCF-10A, BT-474, BT-549, HCC1954, HBL-100, T-47D, ZR-75-1, SUM159PT, LY2, Hs578T, MDA-MB-468, SUM1315MO2, MDA-MB-453 or SK-BR-3, and primary human cell lines such as MRC-5 or chicken DT40 cells. Cell lines can be obtained from the American Type Culture Collection (ATCC)
▲ CRITICAL: Given the difficulty of accurately counting cells after the cross-linking procedure has been implemented, we typically refer to cell culture plate sizes and numbers as starting material. For a standard experiment, we prepare one confluent 150-cm2 Petri dish of monolayer cells (or the equivalent if using different-sized plates or flasks). Depending on the cell type, one such dish contains 0.5–2 × 107 cells. For quantitative experiments, we combine two 150-cm2 plates grown in SILAC light and heavy medium, respectively. More starting material may be required for primary cell lines, e.g., 1.5–2 plates per condition (see TROUBLESHOOTING section). For chicken DT40 cells, which grow in suspension and can therefore be counted with a hemocytometer before cross-linking, we use 108 cells per condition. These are general guidelines only. The use of considerably more cells will decrease the purification quality, possibly because background proteins will not be completely solubilized. Please note that many aliquots of ChEP chromatin can be prepared in parallel.
Standard cell culture medium, depending on the cell type (for optimal growth conditions refer to instructions of cell line manufacturer or repository, e.g., ATCC)
For quantitative analyses: SILAC cell culture medium, light and heavy (for instructions on how to prepare SILAC medium and how to adapt cells to it, see Ong and Mann47)
PBS (Life Technologies, cat. no. 10010-056)
Formaldehyde, 37% (wt/wt) (Sigma, cat. no. F1268)
Glycine (Sigma, cat. no. G8898)
RNase A (Thermo Scientific, cat. no. EN0531)
Tris buffer (Sigma, cat. no. 93352)
Triton X-100 (Sigma, cat. no. T8787)
KCl (Sigma, cat. no. 60132)
NaCl (Sigma, cat. no. S7653)
EDTA (Fisher Scientific, cat. no. BPE118-500)
SDS (Sigma, cat. no. 71725)
Urea (Sigma, cat. no. U5378)
Glycerol (Fisher Scientific, cat. no. BPE229-1)
Roche protease inhibitor tablets (Roche, cat. no. 11836170001)
Any Bradford or bicinchoninic acid (BCA) protein assay kit
SDS-PAGE loading buffer, 4x (Life Technologies, cat. no. NP0008)
EQUIPMENT
Hydrophobic pipette tips (200 μl) ▲ CRITICAL: The use of these tips where indicated is crucial to avoid complete sample loss. Use uncoated tips such as epT.I.P.S LoRetention (Eppendorf, cat. no. 0030 072.022).
CO2 incubator for cell culture (e.g., Thermo Scientific, Heracell 150 or equivalent)
Cell culture plates (e.g., Fisher Scientific, cat. no. TKT-310-450U)
Water bath (e.g., Fisherbrand, cat. no. FB51691)
Cell culture centrifuge
Hemocytometer (e.g., Sigma, cat. no. Z359629)
Benchtop microcentrifuge with refrigeration capacity
Bioruptor sonicator (Diagenode)
Liquid chromatography–tandem mass spectrometry (LC-MS/MS) equipment, including nanoflow HPLC system and mass spectrometer, e.g., Q-Exactive from Thermo Scientific
MaxQuant software48, freely downloadable through http://www.maxquant.org/
Perseus software, freely downloadable through http://www.perseus-framework.org/
REAGENT SETUP
1% (wt/vol) formaldehyde in PBS (cross-linking solution)
Dilute the 37% (wt/wt) formaldehyde stock solution 1:37 with PBS. ▲CRITICAL: Freshly prepare the solution before use. ▲ CRITICAL: Make sure that PBS is warmed up in a water bath at 37 °C before it is mixed with formaldehyde.
Cell lysis buffer
This buffer is composed of 25 mM Tris (pH 7.4 at room temperature (20–25 °C)), 0.1% (vol/vol) Triton X-100, 85 mM KCl and Roche protease inhibitor tablets (one tablet per 10 ml of buffer solution). ▲CRITICAL: Store the buffer at room temperature for up to 1 month, but add protease inhibitors only before use. Check the pH after prolonged storage.
SDS buffer
This buffer is composed of 50 mM Tris (pH 7.4 at room temperature), 10 mM EDTA, 4% (wt/vol) SDS and Roche protease inhibitor tablets (one tablet per 10 ml of buffer solution). ▲ CRITICAL: Store the buffer at room temperature for up to 1 month, but only add protease inhibitors before use. Check the pH after prolonged storage.
Urea buffer
This buffer is composed of 10 mM Tris (pH 7.4 at room temperature), 1 mM EDTA and 8 M urea. ▲ CRITICAL: Freshly prepare the buffer before use.
Storage buffer
This buffer is composed of 10 mM Tris (pH 7.4 at room temperature), 1 mM EDTA, 25 mM NaCl, 10% (vol/vol) glycerol and Roche protease inhibitor tablets (one tablet per 10 ml of buffer solution). ▲ CRITICAL: Store the buffer at room temperature for up to 1 month, but only add protease inhibitors before use. Check the pH after prolonged storage.
PROCEDURE
Formaldehyde cross-linking and cell harvesting ● TIMING: 45 min
-
▲
CRITICAL: Please note that ChEP can be applied to either a single sample of cells grown in an ‘unlabeled’ medium (Fig. 1a) or for quantitative chromatin proteomics comparing the composition of two samples with each other (Fig. 1b), the latter approach being described in this PROCEDURE. To analyze chromatin from a single growth condition, simply skip Steps 15 and 16.
-
▲
CRITICAL: Directions in Steps 1–8 apply to adherent cell lines. When growing cells in suspension, adapt Steps 2 and 5 to rinse cells with PBS by centrifugation, using centrifugation conditions that are appropriate for the cell line in question (e.g., for DT40 cells, use 375g for 3 min at room temperature). In addition, perform the incubations at Steps 3 and 4 in tubes on a rotating wheel rather than on plates, and skip Steps 6 and 7, which are specific for adherent cells.
1. For each condition and/or treatment to be analyzed, prepare a confluent 150-cm2 plate of cells using light and heavy SILAC medium, respectively (see Ong and Mann47 for how to perform a SILAC experiment, including growing cells in SILAC medium).
▲ CRITICAL STEP: We recommend performing this step in triplicate to obtain three independent biological replicate samples and ensure reproducibility of the results. Ideally, one of the replicates is a ‘label-swap’ experiment. In other words, if condition A was such that the culture medium contained unlabeled amino acids, whereas condition B involved the presence of isotopically labeled amino acids, then in one replica labeling is inverted such that condition A becomes the labeled condition and condition B becomes the unlabeled one.
2. Rinse the cells with PBS.
3. Add 1% (wt/vol) formaldehyde in PBS to the plates, using 10 ml per 150-cm2 plate, and incubate the plates for 10 min at 37 °C in the CO2 incubator. After 5 min, briefly remove the plates from the incubator and rock them gently by hand at room temperature for a few seconds to ensure even exposure of cells to formaldehyde. Return the plates into the incubator to complete the 10-min incubation at 37 °C.
▲ CRITICAL STEP: The cross-linking conditions are crucial for successful chromatin enrichment. In particular, make sure that PBS is warmed up in a water bath before it is mixed with formaldehyde. The use of a cold solution will decrease the cross-linking efficiency and interfere with subsequent steps.
4. Halt the cross-linking reaction by adding glycine to a final concentration of 0.25 M (as estimated based on the volume of cross-linking solution used), and incubate the plates at room temperature for 5 min. After 2.5 min, gently rock the plates by hand for a few seconds, and then resume the 5-min incubation until completion.
5. Rinse the plates with PBS.
6. Scrape the cells off into 10–15 ml of PBS.
7. Transfer the cell suspensions obtained in Step 6 from SILAC light and heavy cultures to separate conical tubes, and top up each tube to a total volume of 50 ml with PBS. To collect as many cells as possible, rinse the plates with PBS and use the rinsing solution to top up the conical tubes.
8. Centrifuge the cell suspensions at 420g for 5 min at room temperature to obtain cell pellets at the bottom of the conical tubes. Discard the supernatants. If the pellets are to be used immediately, transfer the tubes containing the cell pellets to an ice bucket. Alternatively, please store them as detailed below.
■ PAUSE POINT: The cell pellets can be stored at −80 °C for several months.
Cell lysis and RNase digestion ● TIMING: 45 min–1 h
9. (Optional) If the cell pellets have been stored at −80 °C, see Step 8, thaw the cells by placing the tubes in the palm of your hand, but place them on ice before thawing is complete.? TROUBLESHOOTING
10. Resuspend each cell pellet in 1 ml of ice-cold cell lysis buffer and transfer the resulting suspension to 2-ml Eppendorf tubes.
11. Homogenize the cell pellets by carefully pipetting up and down, e.g., with a 200-μl pipette tip.
12. Centrifuge the suspensions prepared in Step 11 at 2,300g for 5 min at 4 °C. The pellet that forms at the bottom of the tube will be composed of cell nuclei, which will not be pure at this point. Please note that the mentioned lack of purity is not a problem at this stage.
13. Transfer the supernatants (the cytoplasmic fraction) to new tubes. Make sure to transfer the entire amount of the cytoplasmic fraction.
14. Resuspend each nuclei pellet in 500 μl of cell lysis buffer containing 200 μg/ml RNase A and incubate for 15 min at 37 °C; next, put the tubes on ice.
▲ CRITICAL STEP: Do not add RNase A to the buffer if the ChEP sample is to be used to study chromatin-associated RNAs or proteins that associate with chromatin in an RNA-dependent manner.
15. While the nuclei are being digested, make sure that cytoplasmic fractions from cells grown in light and heavy medium have the same volume, and compare their protein concentrations by the Bradford assay (a relative quantification is sufficient; producing a standard curve is not required).
16. Resuspend RNase-digested nuclei well by careful pipetting, and then combine equal amounts of nuclei suspensions from light and heavy SILAC samples on the basis of the protein content of the cytoplasmic fractions. Naturally, this approach is based on the assumption that the concentration of cytoplasmic extracts reflects the concentration of nuclei suspensions, which we found to be correct (data not shown).
17. Centrifuge the pooled nuclei suspension from Step 16 at 2,300g for 10 min at 4 °C. Discard the supernatant and proceed immediately to Step 18.
Chromatin enrichment ● TIMING: 4 h
18. Resuspend the pellet containing the nuclei in 500 μl of SDS buffer using hydrophobic 200-μl pipette tips, and then incubate the suspension for 10 min at room temperature.
▲ CRITICAL STEP: Nuclei will lyse quickly, and the solution will become very viscous owing to the release of chromatin. The use of hydrophobic pipette tips is recommended to avoid retaining a substantial amount of this sticky solution inside the tip. It is crucial that the sample be completely resuspended at this stage, as any remaining clumps will not usually be dissolved at later stages of the protocol. Homogenize the mixture carefully by pipetting up and down slowly, but do not pipette rapidly or vortex the sample in order to avoid mechanical shearing of DNA.
19. Add 1.5 ml of urea buffer and mix thoroughly by inverting the tube several times.
20. Centrifuge the tube at 16,100g for 30 min at 25 °C (or at room temperature, but do not cool the centrifuge).
21. Discard the supernatant. Remove the supernatant carefully, as the pellet will be completely transparent. The pellet is relatively compact, so the supernatant can be removed easily without disturbing it. Note that the pellet will be bigger than the nuclei pellet obtained in Step 17.? TROUBLESHOOTING
22. To wash out contaminants, resuspend the pellet using a hydrophobic tip in 500 μl of SDS buffer; next, add 1.5 ml of urea buffer and mix by inverting the tube several times. Spin down the tube at 16,100g for 25 min, at 25 °C (or at room temperature, but do not cool the centrifuge). Discard the supernatant.
23. To wash out urea, resuspend the pellet using a hydrophobic tip in 500 μl of SDS buffer; add another 1.5 ml of SDS buffer and mix by inverting the tube several times. Spin down the tube at 16,100g for 25 min, at 25 °C (or at room temperature, but do not cool the centrifuge). Discard the supernatant.
24. Cover the pellet with 0.2–0.5 ml of storage buffer and flick the tube a few times to dislodge the pellet from the bottom of the tube. Keep the tube on ice.
▲ CRITICAL STEP: Do not try to resuspend the pellet at this stage, as in the absence of SDS it would get stuck in the pipette tip. The use of less storage buffer will give a more concentrated chromatin sample, which may be desirable for certain downstream applications.
25. Sonicate the tube in a mixture of ice and water for 15 min, in alternating 30-s ‘on’ and 30-s ‘off’ intervals, at the high-intensity setting.
26. Spin down the tube at 16,100g for 30 min at 4 °C.
27. Transfer the supernatant containing sheared, cross-linked chromatin to a new tube. Note that there may not be a pellet at this stage, as Step 25 generally solubilizes the entire sample efficiently.? TROUBLESHOOTING
28. Perform a Bradford or BCA assay to determine the protein yield.
▲ CRITICAL STEP: Components of the storage buffer, or residual traces of SDS or urea from the purification, can interfere with a Bradford or BCA assay. Consult the instructions of your protein assay kit regarding reagent compatibility, as samples may have to be diluted or precipitated to comply with certain protein assay formats. We have successfully used both Bradford and BCA assays to determine protein yield at this point.
29. Add 4× SDS-PAGE loading buffer to a final concentration of 1× and incubate the mixture for 30 min at 98 °C to reverse the formation of cross-links, opening the lid several times to allow the formaldehyde to evaporate.
30. Spin down briefly to collect all of the liquid at the bottom of the tube (e.g., 16,100g for 15 s at room temperature) and store the sample at −80 °C.
■ PAUSE POINT: The sample can be stored at −80 °C for several months.
Proteomic analysis ● TIMING: 24–96 h
31. Digest the protein mixtures into peptides using the protocols described by Shevchenko et al.49 (in-gel trypsin digestion) or by Wiśniewski et al.7 (in-solution trypsin digestion).
▲ CRITICAL STEP: ChEP-based chromatin fractions contain residual SDS, which needs to be removed before mass spectrometry is carried out. Both of the mentioned protocols for trypsin digestion remove SDS efficiently, and they are equally well suited for this step.
32. Desalt the peptide mixtures obtained in Step 31, as described by Rappsilber et al.50, 51, and perform LC-MS/MS.
33. Process MS raw data in MaxQuant software (see Cox et al.52 for a step-by-step guide on how to use MaxQuant).
Data analysis ● TIMING: 2 h
34. A P value for the detection of significant outlier ratios can be calculated, for example using significance B48 in Perseus software.
35. A typical SILAC experiment reveals a number of significant, reproducible outliers, but these outliers often include proteins without expected chromatin function. Interphase chromatin probabilities (see the supplementary material in Kustatscher et al.1) can focus ChEP results and other data sets on likely chromatin proteins. For example, a chromatin probability of 0.8 means that 8 of 10 reference proteins with this value have a reported chromatin-based function. The chromatin probability of significant outliers can therefore be helpful in selecting candidates for follow-up studies1, 25.
TROUBLESHOOTING
Step 9
Cell types such as IMR-90 and WI-38 primary human fibroblasts and chicken DT40 cells appear to be relatively fragile and can sometimes become lysed upon thawing of cross-linked, frozen cell pellets. This problem can be avoided by scaling up the experiment to include more cells. Alternatively, one may directly proceed to the pelleting of chromatin at Step 18, omitting nuclei isolation and RNase digestion, which could potentially provide a (suboptimal) sample for proteomic analysis.
Step 21
If no pellet is observed at this step, it could be due to having applied conditions that led to inadequate cross-linking. To avoid ineffective cross-linking, suitable strategies may include, for example, not using chilled PBS to prepare the 1% (wt/vol) formaldehyde solution, but making sure that it is warmed up in a water bath beforehand. An alternative explanation to the lack of formation of the gelatinous chromatin pellet could be an insufficient amount of starting material. Make sure to follow the guidelines for the amount of starting material provided in the Reagents section. A third possibility is that the solutions referred to in Step 19 were not mixed properly. Chromatin floating on top of a urea cushion would not precipitate under the conditions used in this protocol. Make sure to mix solutions as indicated at Step 19 by inverting the tube several times (e.g., ten times).
Step 27
A very small opaque pellet, if any, should be expected at this step. If there is still a transparent, gelatinous pellet, sonication was not sufficient to solubilize all chromatin, indicating that either sonication or cross-linking conditions were not completely successful or that too much starting material was used. Solve this problem by repeating Steps 25–27.
TIMING
The timings given are for a single quantitative experiment. Processing multiple samples simultaneously will not extend the timing markedly, as long as sufficient space in the centrifuge is available to handle samples in parallel.
Steps 1–8, formaldehyde cross-linking and cell harvesting: 45 min
Steps 9–17, cell lysis and RNase digestion: 45 min–1 h
Steps 18–30, chromatin enrichment: 4 h
Steps 31–33, proteomic analysis: 24–96 h
Steps 34 and 35, data analysis: 2 h
ANTICIPATED RESULTS
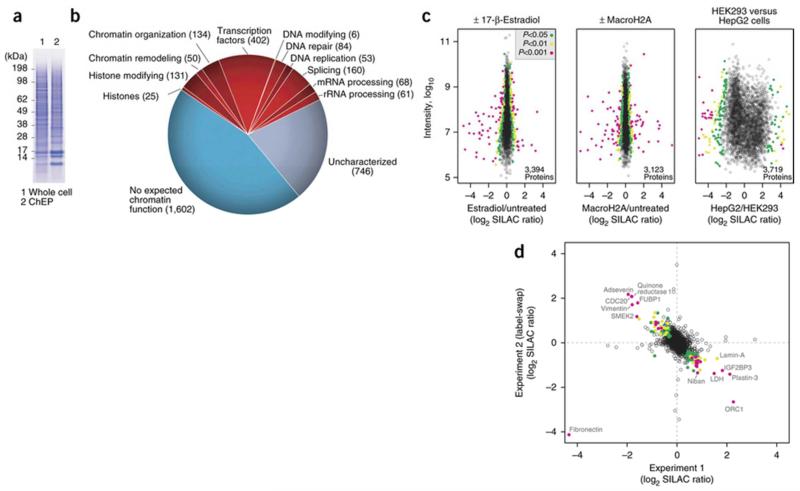
The expected yield of a ChEP preparation from two confluent 150-cm2 plates of cells is in the range of 500 μg of protein. The enrichment of histones should be evident by SDS-PAGE, thus indicating that the procedure worked (Fig. 3a). The number of proteins identified in these samples depends on the scale of the proteomic analysis. Functional protein categories that are typically observed following ChEP are shown in Figure 3b. Note that there can be considerable difference between ChEP fractions isolated from different cell types. This discrepancy reflects not only the cell type–specific chromatin association of transcription factors and readers or modifiers of epigenetic marks, but potentially also the amount of non-chromatin-associated proteins in a cell. ChEP-isolated chromatin from HepG2 cells, for example, is highly enriched in proteins without chromatin-related functions, compared with chromatin isolated from HEK293 cells (Fig. 3c). This difference probably reflects the large number of metabolic enzymes present in liver cells. Quantitative differences in chromatin-associated proteins assessed by ChEP are generally highly reproducible (Fig. 3d). Nevertheless, biological replicate experiments are necessary to ensure reproducibility of individual quantifications before selecting candidates for follow-up studies.
Figure 3.
Typical results from ChEP-based chromatin proteomics. (a) SDS-PAGE showing a ChEP fraction in comparison with a whole-cell extract. Note the enrichment of histones in ChEP, as evident from the histone bands running below 17 kDa. (b) Proteomic analysis of a ChEP sample. Overall, 3,522 proteins were identified and annotated on the basis of UniProt and literature evidence. The number of proteins per category is shown in parentheses. (c) Representative examples of results from SILAC experiments comparing chromatin isolated from different biological conditions. Left: MCF-7 cells treated with 17-β estradiol. Middle: HeLa cells induced to express histone variant macroH2A (ref. 54). Right: comparison of chromatin from wild-type HEK293 and HepG2 cells. Significant outliers were determined with Perseus 1.4.1.3 based on significance B48 and color-coded in green, yellow and magenta according to the level of significance (see inset for different P value thresholds). (d) Quantification reproducibility when the present protocol was applied to obtain ChEP-isolated chromatin from cells with and without Cdk activity1. Outliers that changed significantly in both experiments, compared with the main distribution of protein ratios, are color-coded as in c. Note that the label-swap replicate helps to identify fibronectin as an unlabeled external contaminant. Adapted from Kustatscher et al.1, © 2014 The Authors.
Supplementary Material
ACKNOWLEDGMENTS
We thank F. de Lima Alves and L. Peil for their assistance with mass spectrometric analyses and N. Hegarat and H. Hochegger for testing this protocol on chicken DT40 cells. The Wellcome Trust generously supported this work through a Senior Research Fellowship to J.R. (084229), two Wellcome Trust Centre Core Grants (077707 and 092076) and an instrument grant (091020). G.K. was supported by a Federation of European Biochemical Societies (FEBS) long-term fellowship.
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
Competing financial interests: The authors declare no competing financial interests.
REFERENCES
- 1.Kustatscher G, et al. Proteomics of a fuzzy organelle: interphase chromatin. EMBO J. 2014;33:648–664. doi: 10.1002/embj.201387614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Solomon MJ, Larsen PL, Varshavsky A. Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell. 1988;53:937–947. doi: 10.1016/s0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- 3.Ong SE, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 4.Ishihama Y, et al. Quantitative mouse brain proteomics using culture-derived isotope tags as internal standards. Nat. Biotechnol. 2005;23:617–621. doi: 10.1038/nbt1086. [DOI] [PubMed] [Google Scholar]
- 5.Geiger T, Cox J, Ostasiewicz P, Wiśniewski JR, Mann M. Super-SILAC mix for quantitative proteomics of human tumor tissue. Nat. Methods. 2010;7:383–385. doi: 10.1038/nmeth.1446. [DOI] [PubMed] [Google Scholar]
- 6.Geiger T, et al. Use of stable isotope labeling by amino acids in cell culture as a spike-in standard in quantitative proteomics. Nat. Protoc. 2011;6:147–157. doi: 10.1038/nprot.2010.192. [DOI] [PubMed] [Google Scholar]
- 7.Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat. Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 8.Kohwi-Shigematsu T, deBelle I, Dickinson LA, Galande S, Kohwi Y. Identification of base-unpairing region-binding proteins and characterization of their in vivo binding sequences. Methods Cell Biol. 1998;53:323–354. doi: 10.1016/s0091-679x(08)60885-7. [DOI] [PubMed] [Google Scholar]
- 9.Lewis CD, Laemmli UK. Higher order metaphase chromosome structure: evidence for metalloprotein interactions. Cell. 1982;29:171–181. doi: 10.1016/0092-8674(82)90101-5. [DOI] [PubMed] [Google Scholar]
- 10.Ohta S, et al. The protein composition of mitotic chromosomes determined using multiclassifier combinatorial proteomics. Cell. 2010;142:810–821. doi: 10.1016/j.cell.2010.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kadonaga JT, Tjian R. Affinity purification of sequence-specific DNA binding proteins. Proc. Natl. Acad. Sci. USA. 1986;83:5889–5893. doi: 10.1073/pnas.83.16.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Himeda CL, et al. Quantitative proteomic identification of six4 as the Trex-binding factor in the muscle creatine kinase enhancer. Mol. Cell. Biol. 2004;24:2132–2143. doi: 10.1128/MCB.24.5.2132-2143.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mittler G, Butter F, Mann M. A SILAC-based DNA protein interaction screen that identifies candidate binding proteins to functional DNA elements. Genome Res. 2009;19:284–293. doi: 10.1101/gr.081711.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoshino A, Fujii H. Insertional chromatin immunoprecipitation: a method for isolating specific genomic regions. J. Biosci. Bioeng. 2009;108:446–449. doi: 10.1016/j.jbiosc.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 15.Akiyoshi B, Nelson CR, Ranish JA, Biggins S. Quantitative proteomic analysis of purified yeast kinetochores identifies a PP1 regulatory subunit. Genes Dev. 2009;23:2887–2899. doi: 10.1101/gad.1865909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujita T, Fujii H. Direct identification of insulator components by insertional chromatin immunoprecipitation. PLoS ONE. 2011;6:e26109. doi: 10.1371/journal.pone.0026109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agelopoulos M, McKay DJ, Mann RS. Developmental regulation of chromatin conformation by Hox proteins in Drosophila. Cell Rep. 2012;1:350–359. doi: 10.1016/j.celrep.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Byrum SD, Raman A, Taverna SD, Tackett AJ. ChAP-MS: a method for identification of proteins and histone posttranslational modifications at a single genomic locus. Cell Rep. 2012;2:198–205. doi: 10.1016/j.celrep.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pourfarzad F, et al. Locus-specific proteomics by TChP: targeted chromatin purification. Cell Rep. 2013;4:589–600. doi: 10.1016/j.celrep.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 20.Fujita T, Fujii H. Efficient isolation of specific genomic regions and identification of associated proteins by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP) using CRISPR. Biochem. Biophys. Res. Commun. 2013;439:132–136. doi: 10.1016/j.bbrc.2013.08.013. [DOI] [PubMed] [Google Scholar]
- 21.Fujita T, et al. Identification of telomere-associated molecules by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP) Sci. Rep. 2013;3:3171. doi: 10.1038/srep03171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Déjardin J, Kingston RE. Purification of proteins associated with specific genomic loci. Cell. 2009;136:175–186. doi: 10.1016/j.cell.2008.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kliszczak AE, Rainey MD, Harhen B, Boisvert FM, Santocanale C. DNA mediated chromatin pull-down for the study of chromatin replication. Sci. Rep. 2011;1:95. doi: 10.1038/srep00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sirbu BM, Couch FB, Cortez D. Monitoring the spatiotemporal dynamics of proteins at replication forks and in assembled chromatin using isolation of proteins on nascent DNA. Nat. Protoc. 2012;7:594–605. doi: 10.1038/nprot.2012.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alabert C, et al. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat. Cell Biol. 2014;16:281–293. doi: 10.1038/ncb2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lambert JP, Mitchell L, Rudner A, Baetz K, Figeys D. A novel proteomics approach for the discovery of chromatin-associated protein networks. Mol. Cell. Proteomics. 2009;8:870–882. doi: 10.1074/mcp.M800447-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang CI, et al. Chromatin proteins captured by ChIP-mass spectrometry are linked to dosage compensation in Drosophila. Nat. Struct. Mol. Biol. 2013;20:202–209. doi: 10.1038/nsmb.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soldi M, Bonaldi T. The proteomic investigation of chromatin functional domains reveals novel synergisms among distinct heterochromatin components. Mol. Cell. Proteomics. 2013;12:764–780. doi: 10.1074/mcp.M112.024307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Villar-Garea A, Imhof A. The analysis of histone modifications. Biochim. Biophys. Acta. 2006;1764:1932–1939. doi: 10.1016/j.bbapap.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 30.Vermeulen M, et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 31.Vermeulen M, et al. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010;142:967–980. doi: 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 32.Bartke T, et al. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010;143:470–484. doi: 10.1016/j.cell.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nikolov M, et al. Chromatin affinity purification and quantitative mass spectrometry defining the interactome of histone modification patterns. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M110.005371. M110.005371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spruijt CG, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 35.Shiio Y, et al. Quantitative proteomic analysis of chromatin-associated factors. J. Am. Soc. Mass Spectrom. 2003;14:696–703. doi: 10.1016/S1044-0305(03)00204-6. [DOI] [PubMed] [Google Scholar]
- 36.Kubota T, Hiraga S, Yamada K, Lamond AI, Donaldson AD. Quantitative proteomic analysis of chromatin reveals that Ctf18 acts in the DNA replication checkpoint. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M110.005561. M110.005561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim DR, Gidvani RD, Ingalls BP, Duncker BP, McConkey BJ. Differential chromatin proteomics of the MMS-induced DNA damage response in yeast. Proteome Sci. 2011;9:62. doi: 10.1186/1477-5956-9-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Monte E, et al. Systems proteomics of cardiac chromatin identifies nucleolin as a regulator of growth and cellular plasticity in cardiomyocytes. Am. J. Physiol. Heart Circ. Physiol. 2013;305:H1624–H1638. doi: 10.1152/ajpheart.00529.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khoudoli GA, et al. Temporal profiling of the chromatin proteome reveals system-wide responses to replication inhibition. Curr. Biol. 2008;18:838–843. doi: 10.1016/j.cub.2008.04.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chou DM, et al. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc. Natl. Acad. Sci. USA. 2010;107:18475–18480. doi: 10.1073/pnas.1012946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Franklin S, et al. Quantitative analysis of the chromatin proteome in disease reveals remodeling principles and identifies high-mobility group protein B2 as a regulator of hypertrophic growth. Mol. Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.014258. M111.014258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaguni LS, Lehman IR. Eukaryotic DNA polymerase-primase: structure, mechanism and function. Biochim. Biophys. Acta. 1988;950:87–101. doi: 10.1016/0167-4781(88)90001-2. [DOI] [PubMed] [Google Scholar]
- 43.Fujita N, et al. MTA3, a Mi-2/NuRD complex subunit, regulates an invasive growth pathway in breast cancer. Cell. 2003;113:207–219. doi: 10.1016/s0092-8674(03)00234-4. [DOI] [PubMed] [Google Scholar]
- 44.Nowak DE, Tian B, Brasier AR. Two-step cross-linking method for identification of NF-κB gene network by chromatin immunoprecipitation. Biotechniques. 2005;39:715–725. doi: 10.2144/000112014. [DOI] [PubMed] [Google Scholar]
- 45.Zeng PY, Vakoc CR, Chen ZC, Blobel GA, Berger SL. In vivo dual cross-linking for identification of indirect DNA-associated proteins by chromatin immunoprecipitation. Biotechniques. 2006;41:694. doi: 10.2144/000112297. [DOI] [PubMed] [Google Scholar]
- 46.Choi NM, Boss JM. Multiple histone methyl and acetyltransferase complex components bind the HLA-DRA gene. PLoS ONE. 2012;7:e37554. doi: 10.1371/journal.pone.0037554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ong SE, Mann M. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC) Nat. Protoc. 2006;1:2650–2660. doi: 10.1038/nprot.2006.427. [DOI] [PubMed] [Google Scholar]
- 48.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 49.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 50.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 2003;75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 51.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, prefractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 52.Cox J, et al. A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 2009;4:698–705. doi: 10.1038/nprot.2009.36. [DOI] [PubMed] [Google Scholar]
- 53.Spitzer M, Wildenhain J, Rappsilber J, Tyers M. BoxPlotR: a web tool for generation of box plots. Nat. Methods. 2014;11:121–122. doi: 10.1038/nmeth.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Timinszky G, et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat. Struct. Mol. Biol. 2009;16:923–929. doi: 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.