Abstract
While both Homologous recombination (HR) and Non Homologous End Joining (NHEJ) can repair DNA double Strand Breaks (DSB), the mechanisms by which one or other of these pathways is chosen remain unclear. Here we show that transcriptionally active chromatin is preferentially repaired by HR. Using chromatin immunoprecipitation-sequencing (ChIP-seq), to analyse repair of multiple DSBs induced throughout the human genome, we identify an “HR-prone” subset of DSBs that recruit the HR protein RAD51, undergo resection and rely on RAD51 for efficient repair. These DSBs are located in actively transcribed genes, and targeted to HR repair via the transcription-elongation associated histone mark, histone H3 lysine 36 trimethylation (H3K36me3). In agreement, depletion of SETD2, the main H3K36 tri-methyltransferase, severely impedes HR at such DSBs. Our study thereby demonstrates a primary role of the chromatin context, in which a break occurs, in DSB repair.
Keywords: DSB repair, NHEJ, HR, chromatin, transcription, ChIP-seq, H3K36me3
Introduction
Cells have developed homologous recombination (HR) and non-homologous end-joining (NHEJ) to repair highly toxic DNA double-strand breaks (DSBs) 1. In contrast to HR, which involves extensive DNA end resection and utilizes an intact copy of the damaged locus, NHEJ ligates broken DSB ends with no or limited processing. Although mostly faithful, both mechanisms can be sources of genomic instability. For example, HR can trigger amplification and depletion of repetitive sequences and can yield loss of heterozygosity (LOH) when the homologous chromosome is employed as the template rather than the sister chromatid. On the other hand, NHEJ can induce point mutations and small deletions, especially when repair is performed by the backup error prone NHEJ pathway (Alt-NHEJ). Considering the wide range of DNA products that can arise from these different repair pathways the choice between NHEJ and HR represents a central aspect of DSB repair.
While cell cycle phase contributes to this choice, these pathways coexist in S and G2 cells 2-4, implying that other factors participate in this decision. Several repair proteins can orient repair towards one or the other pathway. For example, critical roles have been assigned to the Ku heterodimer, whose binding inhibits resection and channels repair towards NHEJ 5, to 53BP1/RIF1 that promotes NHEJ at least in part via counteracting BRCA1 functions 6-12, and to CtIP, MRE11 and Exo1 which promote resection and thus HR 13,14. However, how these factors are targeted to DSBs to achieve their functions is largely unknown.
An appealing hypothesis, supported by recent studies is that the chromatin context where a DSB occurs could contribute to this decision. For example, mono and dimethylation of histone H4K20 15-19, as well as acetylation of H4K16 by Tip60 20, are involved in the recruitment/anchoring of 53BP1 which is proposed to counteract HR 21. Since these histone modifications are not evenly distributed on the genome, this raises the hypothesis that chromatin may play a role in the repair pathway choice. In addition, recent studies have provided very interesting insights regarding the differences of DSB repair between heterochromatin and euchromatin 2,3,22.
Previous studies have benefited from the development of techniques going beyond classical damage induction methods, such as focused lasers and sequence specific DSB(s) inducible cell systems (which greatly increased the resolution of molecular studies on DSBs, by permitting the use of ChIP 23,24). However while these techniques helped to identify new chromatin players targeted to DSBs 25,26 as well as their kinetics of recruitment, they hindered any investigations of the influence of pre-existing chromatin structure on repair. Indeed, on one hand, lasers induce DNA damage along tracks in the nucleus in a random fashion regarding the genomic position. On the other hand, the available sequence–specific DSB inducible systems either trigger one single break (the HO, I-SceI; and FokI systems 24,27,28), or several DSBs but mainly located in ribosomal DNA (the I-PpoI system 23), which preclude the comparison of repair events occurring at different genomic locations.
Recently, we developed stable human cell lines expressing a restriction enzyme, targeting an 8 bp recognition sequence, fused to a modified oestrogen receptor ligand binding domain (henceforth called DIvA for DSB Inducible via AsiSI) 29-31. Treating such cells with 4-hydroxytamoxifen (4OHT) triggers nuclear localisation of the AsiSI enzyme and the ensuing rapid induction of approximately 150 sequence-specific DSBs, dispersed across the genome 31. This system provides a unique opportunity to simultaneously study at a molecular level, the repair events that transpire at many different DSBs located within various chromatin states.
Using ChIP-seq approaches, we found here that distinct DSBs induced across the genome are not necessarily repaired by the same pathway, and that transcriptionally active, H3K36me3 enriched, chromatin is preferentially repaired by homologous recombination, thereby pointing out a critical role of pre-existing chromatin in addressing the DSB repair pathway at sites of breaks.
Results
Differential binding of RAD51 and XRCC4 at distinct DSBs
To investigate how chromatin context might influence DSB repair pathway choice, we performed chromatin immunoprecipitation followed by massively-parallel sequencing (ChIP-seq) in 4OHT treated DIvA cells with antibodies against RAD51 and XRCC4, core components of the HR and NHEJ machineries, respectively. A control Ser-139 phosphorylated histone H2AX (γH2AX) ChIP-seq was also performed to evaluate the efficiency of DSB induction at each AsiSI site, as γH2AX levels reflect the extent of DSB induction 30. As expected, γH2AX showed a typical pattern, with wide spreading away from the DSBs (~1-2 Mb) and a clear drop at the break sites themselves, presumably reflecting reduced nucleosome occupancy (~1 kb) (Supplementary Fig. 1a-b). ChIP-seq analyses indicated clear recruitments of XRCC4 and RAD51 to AsiSI sites (Fig. 1). Notably, while XRCC4 localization was highly focused around the break point, RAD51 spread further away (~2-4 kb), consistent with its role in binding single-stranded DNA generated by DNA-end resection (Fig. 1a).
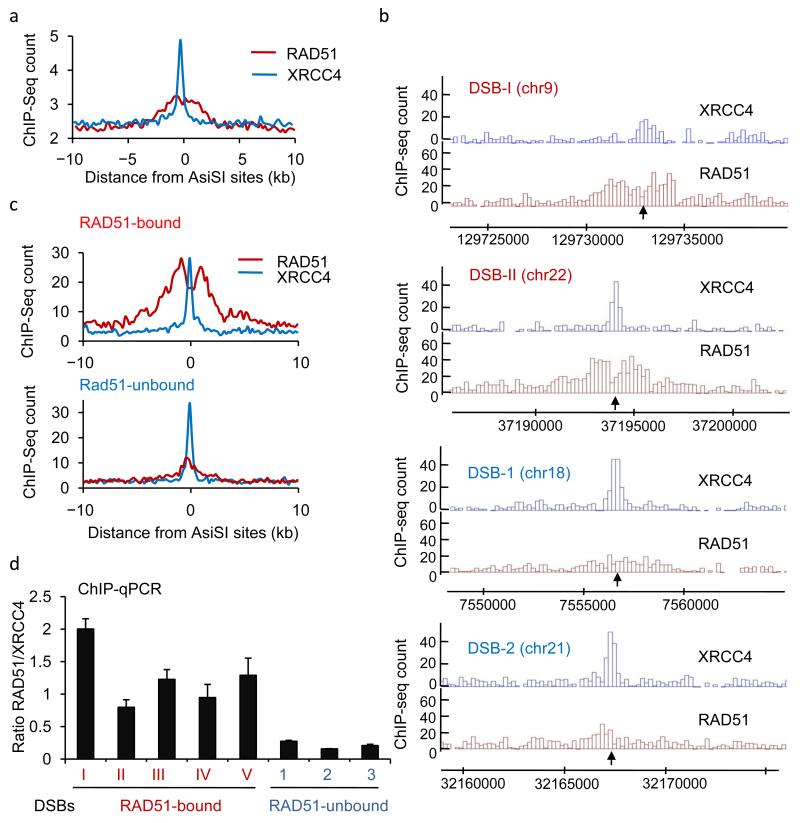
Figure 1. A subset of AsiSI-induced DSBs recruits RAD51.
a. ChIP-seq analyses in DIvA cells after 4OHT treatment (4h), using anti XRCC4 or anti-RAD51 antibodies. The averaged XRCC4 (blue) and RAD51 (red) signals, over a 20kb region flanking annotated AsiSI sites, are shown. b. The profiles of XRCC4 (blue) and RAD51 (red) around four selected AsiSI sites (indicated by arrows) are shown. c. Averaged XRCC4 (blue) and RAD51 (red) signals over 20kb windows and centered at the AsiSI site, are shown for each categories (RAD51-bound or RAD51-unbound subsets). d. ChIP against XRCC4 and RAD51 in 4OHT-treated DIvA cells, analyzed by qPCR. The ratios between the signals observed for XRCC4 and RAD51 are presented for eight AsiSI-induced DSBs, either bound or not by RAD51 in ChIP-seq experiments (RAD51-bound DSBs are labeled with roman numerals, while RAD51-unbound DSBs are labeled with arabic numerals). Mean and s.e.m (technical replicate, n=4) of a representative experiment are shown.
A closer inspection of various AsiSI-induced DSBs revealed that some displayed readily detectable binding of both RAD51 and XRCC4 (Fig. 1b top two panels; note that DSB sites are indicated by arrows). By contrast, other sites displayed little or no discernible RAD51 accrual despite exhibiting robust γH2AX induction and XRCC4 binding (Fig. 1b bottom two panels, and Supplementary Fig. 1c). These findings were confirmed by a second, biological replicate experiment (Supplementary Fig. 2a-b). Based on the respective enrichment of XRCC4 and RAD51, we defined and further characterized two subsets (containing 20 DSBs each) of “RAD51-bound DSBs” and “RAD51-unbound DSBs” (based on the RAD51/XRCC4 ChIP-seq signal ratio, see online Methods). The averaged profiles of XRCC4 and RAD51 on these DSB subsets validated our categorization process (Fig. 1c). To further confirm these data, we determined the relative enrichments of XRCC4 and RAD51 by ChIP followed by real-time quantitative PCR (RT-qPCR) for certain DSBs in each subset. Since our ChIP-seq and ChIP-qPCR analyses revealed that XRCC4 binding was restricted to the DSB, while RAD51 was mainly found further away from the break (Fig. 1a-b, Supplementary Fig. 2c), subsequent XRCC4 and RAD51 ChIP were always analyzed by qPCR at 100 and 800 bp from the DSB, respectively. This analysis by ChIP-qPCR clearly differentiated the two categories, with RAD51-bound DSBs I, II, III, IV and V exhibiting high RAD51/XRCC4 ratios, which contrasted to the substantially lower RAD51/XRCC4 ratios displayed by DSBs 1, 2 and 3 that were categorized as RAD51-unbound (Fig. 1d).
To investigate the influence of the cell cycle phase in RAD51 and XRCC4 recruitment at DSBs, we performed ChIP experiments in G1- or G2-synchronized DIvA cells, after a double thymidine block (Fig. 2a). Consistent with the cell cycle characteristics of HR, we found that RAD51 accumulated at DSBs much more strongly in G2 than in G1, while XRCC4 recruitment was similar in both cell cycle phases (Fig. 2b-c). To exclude a potential bias due to unequal DSB induction at investigated AsiSI annotated sites across the cell cycle, we controlled AsiSI efficiency in synchronized cells, using our previously reported protocol 30,32 based on the ligation of biotinylated double stranded oligonucleotides, cohesive with AsiSI ends, followed by streptavidin purification (“cleavage assay” see online Methods). Importantly, AsiSI-mediated DSB induction was constant throughout the cell cycle (Supplementary Fig. 3a-c).
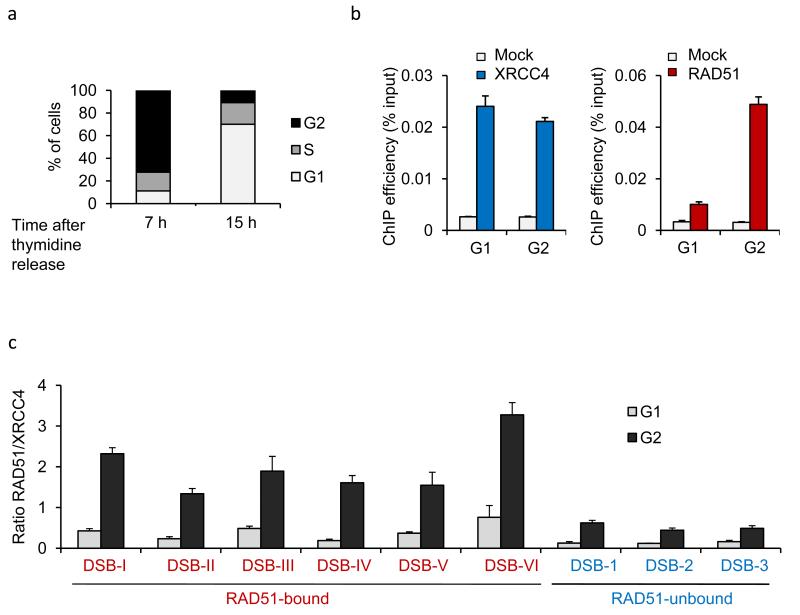
Figure 2. RAD51 recruitment at RAD51-bound DSB occurs mainly in G2.
a. Cell cycle distributions as measured by FACS in synchronized DIvA cells b. ChIP using XRCC4 (blue, left panels) or RAD51 (red, right panels) antibodies in synchronized G1 and G2 DIvA cells. Enrichments in XRCC4 and RAD51 were respectively measured by qPCR at 80 bp and 800 bp from the DSB-I (from our RAD51-bound subset). Mean and s.e.m of a representative experiment is shown. c. The RAD51/XRCC4 ratio obtained by ChIP-qPCR was calculated for various DSBs from the RAD51-unbound subset (named with arabic numerals, in blue) and from the RAD51-bound subset (named with roman numerals, in red) both in G1 and G2 cells. RAD51 recruitment at DSBs occurs preferentially in G2, and on a specific subset of DSBs. Mean and s.e.m (n=4 technical replicates) of a representative experiment are shown.
Therefore, together our data indicate that in G1 phase, XRCC4 recruitment dominated at all DSBs investigated, and that in G2, in addition to XRCC4, RAD51 is also recruited but only at a subset of DSBs. This suggested that in G2, distinct DSBs induced throughout the genome are not necessarily repaired by the same pathway.
RAD51-bound DSBs exhibit HR features
To see whether RAD51 binding correlated with 5′ to 3′ DSB resection, which is characteristic of HR-engaged DSBs, we probed for the presence of ssDNA at RAD51-bound or RAD51-unbound DSBs by using biotinylated double-stranded oligonucleotides with a specific 15-base 3′ end extension that allowed hybridization to the exposed ssDNA (see online Methods). The five RAD51-bound sites investigated indeed bore this hallmark of resection, which contrasted with the three sites investigated that we had defined as RAD51-unbound (Fig. 3a). As shown in Supplementary Fig. 3d, these differences were not due to variations in DSB induction efficiencies.
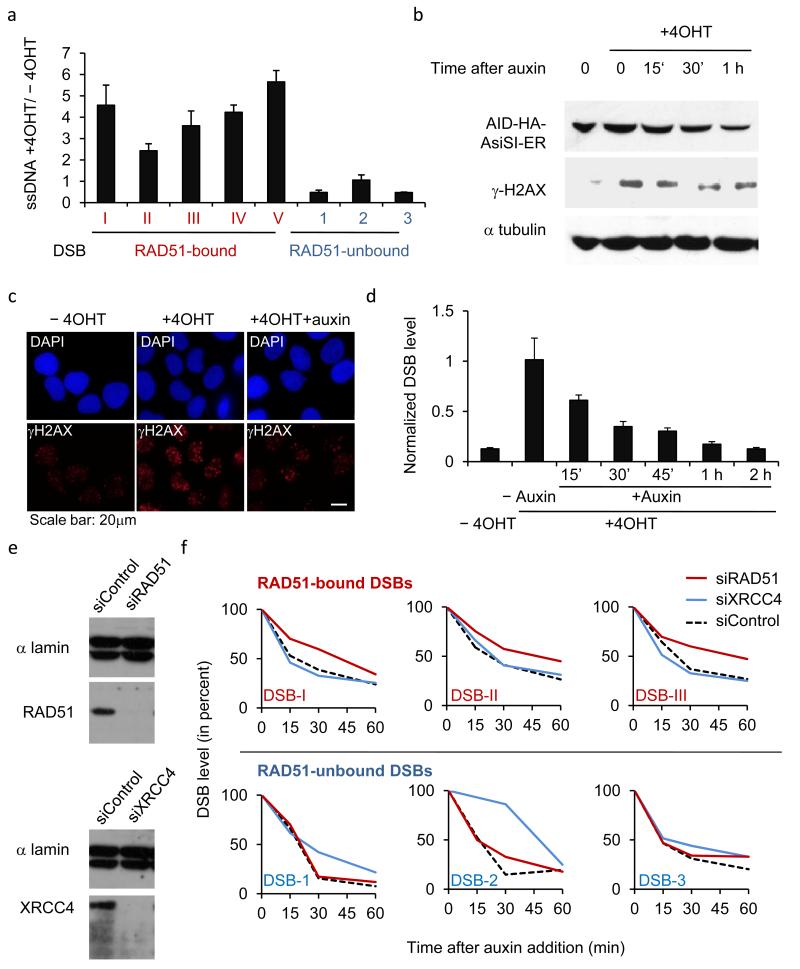
Figure 3. RAD51-bound AsiSI-DSBs are resected and repaired by a RAD51-dependent pathway.
a. ssDNA arising through resection was assessed at each of the eight previously analyzed DSBs. Pull down efficiency was measured by RT-qPCR. Ratios between treated and untreated cells are presented. Mean and s.e.m (n=3, technical replicates) of a representative experiment are shown. b. Western blot analysis of AID-DIvA stably transfected cells induced by 4OHT for 4h and subsequently treated (or not) with auxin for the indicated time. Uncropped images are presented Supplementary Fig.8 c. The presence of γH2AX foci was monitored by immunofluorescence in untreated cells, in 4OHT-induced cells (4h), and in 4OHT-induced cells further incubated with auxin (2h). d. Cleavage assay in AID-DIvA cells, treated as indicated, followed by RT-qPCR close to a cleaved AsiSI site. Normalized pull down efficiencies from a representative experiment are shown. e. Western blot analyses of AID-DIvA cells transfected with a control siRNA (Control), siRNA against RAD51 (left panel) or against XRCC4 (right panel). Uncropped images are presented Supplementary Fig.8. f. Cleavage assay in AID-DIvA cells transfected with control, RAD51, or XRCC4 siRNAs. Immunoprecipitated DNA was analyzed close to six DSBs, either RAD51-unbound (indicated in blue, upper panel) or RAD51-bound (indicated in red, lower panel). The percentage of sites that remain broken for each DSB after the indicated times of auxin treatment are presented. A representative experiment is shown.
To investigate whether the RAD51 binding we observed was biologically relevant and reflected HR repair, we next asked whether DSB repair kinetics at such sites was dependent on RAD51 and/or XRCC4. The DIvA system does not permit one to readily study the kinetics of DSB repair, because the enzyme regenerates DSBs at AsiSI sites even after they have been accurately religated. We therefore further refined our system by adding an auxin-inducible degron (AID) to the AsiSI-ER fusion enzyme, thus allowing us to remove it and stop AsiSI activity upon auxin addition 33. The AID-DIvA cells behaved similarly to the DIvA cells regarding kinetics of DSB induction (30,31 and Supplementary Fig. 4a-b). Our analyses revealed that auxin induced the efficient degradation of AID-AsiSI-ER, whose kinetics of loss correlated well with decreases in γH2AX levels (Fig. 3b). Auxin also triggered the rapid disappearance of γH2AX foci, which reflect the presence of DSBs (Fig. 3c and Supplementary Fig. 4c-d). We next analyzed the repair kinetics of an AsiSI-induced DSB by using the cleavage assay described above that uses adaptor ligation and RT-qPCR to measure the presence of DSB ends (30,32, Supplementary Fig. 3, and online methods). This revealed that repair was nearly completed 1 hour after auxin addition (Fig.3d). Repair kinetics were then compared for DSBs that we had designated as RAD51-bound or RAD51-unbound, upon short-interfering RNA (siRNA)-mediated depletion of either XRCC4 or RAD51 (Fig. 3e). Strikingly, RAD51 depletion substantially slowed the repair kinetics of RAD51-bound DSBs (Fig. 3f top panels and Supplementary Fig. 4e left panels) while it had little or no effect on the repair of RAD51-unbound DSBs (Fig. 3f bottom panels and Supplementary Fig 4e right panels). By contrast, XRCC4 depletion influenced the repair of RAD51-unbound DSBs but had little or no effect on the repair of RAD51-bound DSBs (Fig. 3f and Supplementary Fig. 4e).
Collectively, these data substantiated our ChIP data indicating that all DNA DSBs induced across the genome are not repaired equivalently and revealed that individual DSBs are indeed repaired by the pathway whose factor exhibited preferential binding at each site. DSBs defined as RAD51-bound behave as “HR-prone” DSBs since they undergo resection and rely on RAD51 for their effective repair. In contrast, other DSBs do not undergo detectable resection and rely on XRCC4 for repair.
RAD51-bound DSBs lie in transcriptionally active chromatin
The above findings strongly suggested that genomic or epigenomic features influence DSB repair pathway choice in human cells. To identify such features, we compared our ChIP-seq data with publicly available epigenomic datasets (ENCODE project 34; see online Methods), particularly focusing on histone marks generally associated with transcriptionally active or inactive regions. Interestingly, compared to our experimentally characterized set of RAD51-unbound DSB sites, the RAD51-bound DSBs we identified were reproducibly enriched in chromatin marks associated with active transcription, such as histone H3 trimethylated on lysine 36 (H3K36me3) or acetylated on lysine 9 (H3K9ac) (Supplementary Fig. 5a, top and middle panels). By contrast, repressive chromatin marks (such as histone H3 trimethylated on lysine 9 (H3K9me3) or on lysine 27 (H3K27me3)) were higher on RAD51-unbound sites than on RAD51 enriched sites (Supplementary Fig. 5a, bottom panels). These findings therefore highlighted a potential connection between a locus being transcribed and its propensity to engage in HR. To investigate any association between active transcription and HR-repair, we profiled, by ChIP-Seq in undamaged DIvA cells, the serine 2-CTD phosphorylated form of RNA Polymerase II (PolII-S2P) that is associated with transcriptional elongation. As expected, PolII-S2P spread along the transcribed units, reflecting active transcription (Supplementary Fig. 5b). While almost all the DSBs from our two subsets were located either within or proximal (<1000 bp) to a gene (Supplementary Table 1), the genes close to RAD51-bound DSBs exhibited considerably higher PolII-S2P levels than genes located near RAD51-unbound DSBs, either when taken individually or collectively (Fig. 4a and b, respectively). Such a tendency of RAD51-bound DSBs to be located in actively transcribed regions was also seen when all DSBs were taken into account (Supplementary Fig. 5c). Furthermore, the two RAD51-bound AsiSI sites that turned out not to be in close proximity to an annotated gene (Supplementary Table 1a), were actually located in PolII-S2P enriched regions (Supplementary Fig. 5d), implying that these DSBs in fact lie within transcribed loci. Collectively, the above data strongly suggested that active transcription is a feature that helps target a particular locus for HR repair.
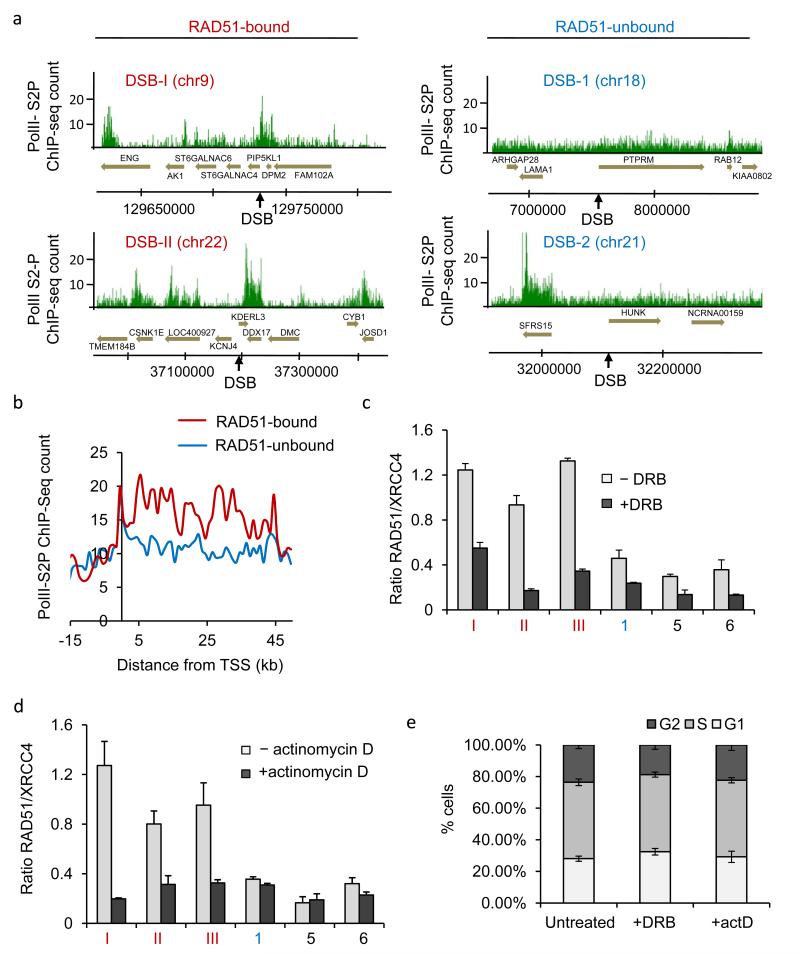
Figure 4. RAD51-bound DSBs lie within transcribed units.
a. PolII-S2P ChIP-seq in untreated DIvA cells. PolII-S2P enrichment is shown around the four AsiSI-induced DSBs presented Fig. 1b. RAD51-bound DSBs are indicated in red and RAD51-unbound DSBs in blue. Positions are in bp. b. The genes associated with each DSB from the RAD51-bound and RAD51-unbound subsets were used to calculate the average PolII-S2P enrichments around the transcription start site. c. XRCC4 and RAD51 ChIP in 4OHT-treated DIvA incubated with DRB (or not). The RAD51/XRCC4 ratios for three RAD51-bound DSBs (DSB-I, -II and -III), one RAD51-unbound DSB (DSB-1), and two DSBs located far from any gene (DSB-5, and -6) are shown. Mean and s.e.m (n=4, technical replicates) of a representative experiment are shown d. Same as c, except that cells were treated with Actinomycin D or not. e. Cell cycle distribution measured by EdU and PI staining and FACS analysis, in untreated, DRB, or Actinomycin D treated DIvA cells, as indicated. Mean and s.e.m of percent of cells in each phase are shown (n=3, biological replicates).
Transcription dependent RAD51 recruitment
To test whether transcription was involved in RAD51 recruitment, we first inhibited transcription by using 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB) or actinomycin D. A ChIP performed against PolII-S2P confirmed that transcription was indeed impaired under these conditions (Supplementary Fig. 6a-b). We studied XRCC4 and RAD51 recruitment at selected DSBs by ChIP followed by RT-qPCR. We included in our analysis, two DSBs arbitrarily chosen among the cleaved AsiSI sites (DSB-5 and -6), located far from any genes. As expected, these DSBs behaved as RAD51-unbound DSBs, showing a low RAD51/XRCC4 ratio (Fig. 4c-d) with a low RAD51 recruitment (Supplementary Fig. 6c). Strikingly, transcription inhibition led to a strong decrease in the RAD51/XRCC4 ratio at RAD51-bound sites, while it induced much less pronounced changes at other AsiSI-induced DSBs studied (Fig. 4c-d). These changes in RAD51/XRCC4 ratio were due to a sharp decrease of RAD51 binding upon transcription inhibition (Supplementary Fig. 6c) while XRCC4 recruitment remained unchanged. Importantly, the effects of transcriptional inhibition on RAD51-bound sites did not reflect appreciable changes in cell cycle distributions (Fig. 4e), and AsiSI cutting efficiencies, either in asynchronous cells (Supplementary Fig. 6d), but also across cell cycle (Supplementary Fig. 6e), excluding a potential bias due to the influence of cell cycle on this observed RAD51 binding impairment. Furthermore, transcription inhibition did not alter substantially the abundance of XRCC4 and RAD51 (data not shown).
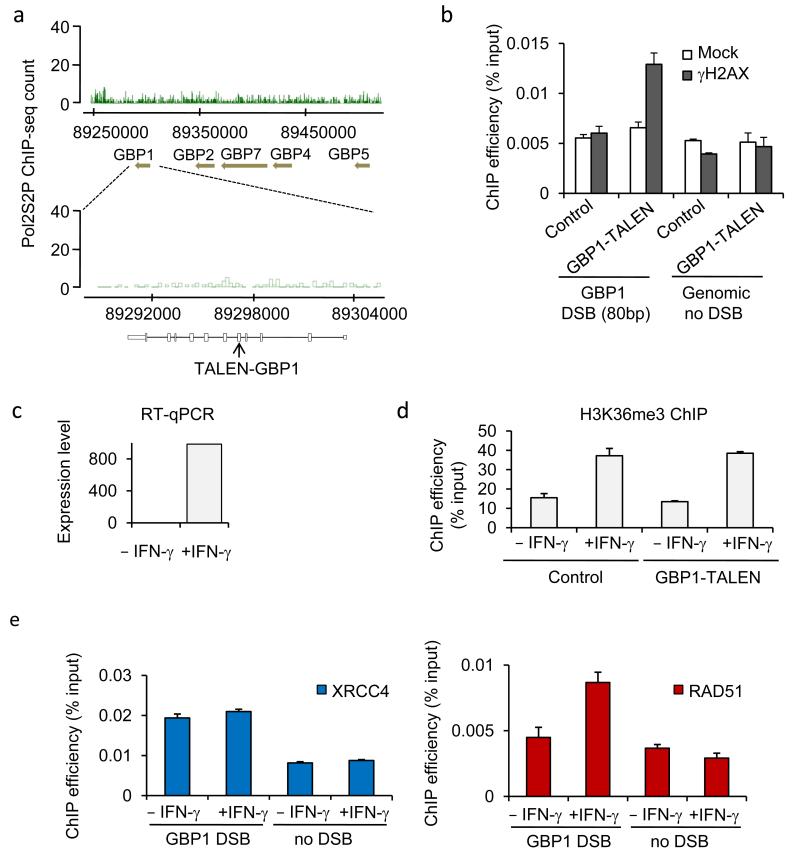
To test the converse – that transcription activation could enhance RAD51 targeting – we employed a transcription activator-like effector nuclease (TALEN) 35 to generate a DSB within an inactive gene that can be easily turned on. We chose to use a GBP1-TALEN to induce a DSB in the interferon-γ responsive gene GBP1 that upon basal condition showed very little enrichment of PolII-S2P in DIvA cells (Fig. 5a). Upon GBP1-TALEN transfection, a DSB was indeed induced within GBP1 as demonstrated by γH2AX induction, detected by ChIP specifically at the vicinity of the DSB (Fig. 5b). Importantly, IFN-γ treatment induced GBP1 activation, as detected by both increased GBP1 mRNA levels (Fig. 5c), and increased H3K36me3 levels on the GBP1 gene body (Fig. 5d). We next analyzed RAD51 and XRCC4 recruitment in TALEN transfected cells, either pretreated with IFN-γ (GBP1 thus being transcriptionally active) or not (GBP1 thus being transcriptionally silent). IFN-γ treatment substantially increased RAD51 binding to the TALEN-induced-DSB, without modifying XRCC4 levels (Fig. 5e). Collectively, these data therefore supported a model in which transcriptionally active loci display preferential RAD51 recruitment and thus HR repair.
Figure 5. Transcriptional activation of an unexpressed gene leads to repair by HR.
a. PolII-S2P profile obtained in DIvA cells is shown on the GBP cluster of gene, located on chromosome 1. A TALEN pair was ordered to specifically induce a DSB in the GBP1 gene, part of the GBP cluster located on chromosome 1. b. γH2AX ChIP in DIvA cells transfected either with pCDNA3 or with the GBP1-TALEN plasmids. Enrichment was measured by qPCR at 80bp from the expected DSB, or on a control genomic region. ChIP efficiency (expressed as % of input) of a representative experiment is shown. c. Levels of GBP1 mRNA measured by reverse transcription followed by qPCR, in DIvA cells treated (or not) with IFN-γ as indicated. cDNA levels were normalized to ribosomal protein P0 cDNA levels. d. H3K36me3 ChIP in control or TALEN transfected cells subjected (or not) to an IFN-γ treatment. H3K36me3 levels were analyzed by qPCR on the GBP1 gene. Mean and s.e.m (n=4, technical replicates) of a representative experiment are shown. e. XRCC4 (blue) and RAD51 (red) ChIP in GBP1-TALEN transfected cells, treated or not with γ-IFN. Enrichments of XRCC4 and RAD51 were measured by qPCR respectively at 80bp and 800bp from the TALEN-induced DSB. Mean and s.e.m (n=4, technical replicates) of a representative experiment are shown.
RAD51 recruitment depends on a H3K36me3-LEDGF (p75) axis
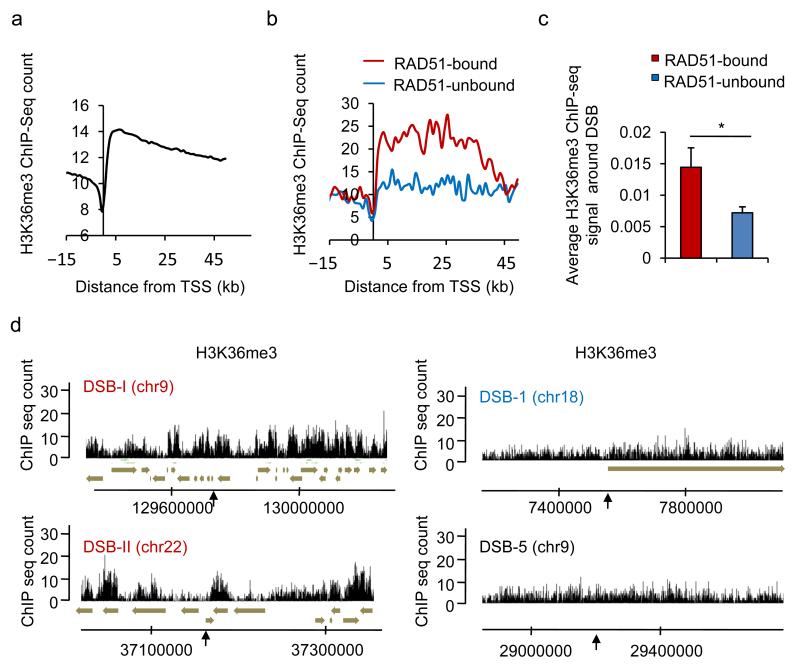
A recent study identified LEDGF(p75) as a protein that facilitates binding of the HR- and resection-promoting factor CtIP on damaged chromatin, likely via a direct interaction between LEDGF and the trimethylated form of H3K36 (H3K36me3)36. Since this histone mark is also associated with transcriptional elongation and is highly enriched on active genes 37,38, we investigated its potential role in directing RAD51 to transcribed loci. First we performed a genome-wide mapping of H3K36me3 distribution in untreated DIvA cells (Fig. 6). As expected, H3K36me3 was strongly enriched on gene bodies, validating our ChIP-seq data (Fig. 6a). Notably, on average, the genes close to RAD51-bound DSBs exhibited much higher H3K36me3 levels than those located near RAD51-unbound DSBs (Fig. 6b). In addition, the regions surrounding RAD51-bound DSBs were significantly enriched in H3K36me3 compared to loci that encompassed RAD51-unbound DSBs (Fig. 6c, and see examples Fig. 6d). Thus our H3K36me3 profiling in DIvA cells revealed that RAD51-bound DSBs are located in chromatin regions enriched for H3K36me3 which is consistent with our previous finding indicating a preferential recruitment of RAD51 to transcriptionally active loci.
Figure 6. H3K36 me3 enrichment on chromatin correlates with the use of HR.
a. H3K36me3 ChIP-seq in untreated DIvA cells. The average H3K36me3 signal was calculated around transcriptional start site (TSS) of all annotated genes (RefSeq, hg18). b. The genes associated with each DSB from the RAD51-bound and RAD51-unbound subsets (respectively red and blue lines) were used to calculate the average H3K36me3 enrichments around the TSS. c. H3K36me3 average signal calculated over a 4kb window centered on the DSB, for each category are shown, together with the s.e.m. * p<0.05 (Mann Whitney).d. Profiles of H3K36me3 in undamaged DIvA cells at two RAD51-bound DSBs (DSB-I and DSB-II), one DSB identified in our RAD51-unbound subset (DSB-1), and a DSB located far from any gene (DSB-5).
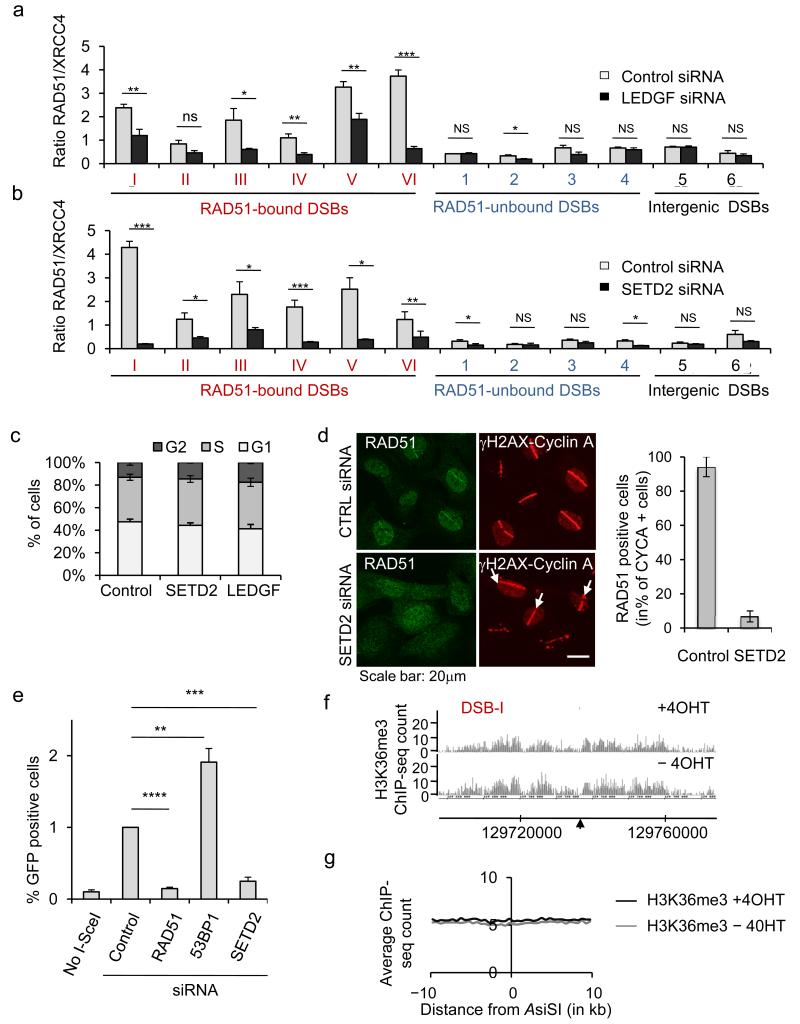
In light of these observations, we investigated whether LEDGF and H3K36me3 participate in targeting RAD51 and HR repair to actively transcribed loci. Depletion of LEDGF by siRNA (Supplementary Fig. 7a) did not affect H3K36me3 levels (as detected by ChIP, Supplementary Fig. 7b) but led to a clear decrease in the RAD51/XRCC4 ratio at all RAD51-bound DSBs studied (Fig. 7a) due to a strong impairment of RAD51 recruitment (Supplementary Fig. 7c). Notably, LEDGF depletion had no or only minor effects on RAD51-unbound DSBs (Fig. 7a, Supplementary Fig. 7c). Such a dramatic effect of LEDGF depletion on RAD51 binding was not due to detectable changes in cell cycle distributions (Fig. 7c) or HR protein levels (data not shown).
Figure 7. The H3K36me3-LEDGF axis targets RAD51 at DSBs induced in active genes.
a. XRCC4 and RAD51 ChIP in 4OHT-treated DIvA cells transfected with control or LEDGF siRNAs. RAD51/XRCC4 ratios were analyzed by RT-qPCR around six RAD51-bound DSBs (DSB-I to-VI), four RAD51-unbound DSBs (DSB-1 to-4) and two DSBs far from any gene (DSB-5 and -6). * p<0.05; ** p<0.01; *** p<0.005; ns non-significant (unpaired Student t-test, two sided). b. Same as in c, except that a siRNA against SETD2 was used. c. Cell cycle distribution measured by EdU and PI staining and FACS analysis, in DIvA cells transfected with siRNA, as indicated. Mean and s.e.m of percent of cells in each phase are shown (n=3, biological replicates). d. U2OS cells, transfected with siRNAs as indicated, were laser micro-irradiated and stained for cyclinA, γH2AX (red) and RAD51 (green). Percentages of cyclin A positive cells, that exhibit RAD51 recruitment at the laser line are shown. e. RG37-GFP-HR reporter cells were transfected with siRNAs as indicated and with a vector expressing I-SceI or not. GFP positive cells were measured by flow cytometry, and expressed relative to the amount of GFP positive cells in control siRNA transfected cells. Mean and s.e.m of independent experiments are shown (n=4, biological replicates). * p<0.05; ** p<0.01; *** p<0.005 (unpaired Student t-test, two sided).f. H3K36me3 ChIP-seq in untreated and 4OHT treated DIvA cells. Profiles obtained around the RAD51-bound DSB-I (indicated by an arrow) are shown. Positions are in bp. g. H3K36me3 signals obtained by ChIP-seq were averaged around all cleaved AsiSI sites.
Similarly we used siRNA to deplete SETD2, the main H3K36 tri-methyltransferase 38, to investigate the consequence of the loss of H3K36me3 on RAD51 binding. SETD2 siRNA led to a strong decrease of SETD2 mRNA levels (Supplementary Fig. 7e) and to an almost complete disappearance of H3K36me3 on chromatin (Supplementary Fig. 7f). Notably SETD2 depletion triggered a strong reduction of RAD51/XRCC4 ratios at RAD51-bound DSBs (Fig. 7b) due to a dramatic decrease of RAD51 recruitment (Supplementary Fig. 7g). As for LEDGF siRNA, SETD2 depletion did not notably modify cell cycle distribution (Fig. 7c) and HR factor level (data not shown). Of note, similar results were observed when using a second siRNA for SETD2 and LEDGF (data not shown). Importantly, both LEDGF and SETD2 depletions led to decreased RAD51 binding without detectably reducing the level of actively transcribing RNA pol II on nearby genes (Supplementary Fig. 7d, h). Taken together, these data therefore indicated a function for this histone mark in RAD51 binding independent from transcription per se.
To further confirm the role of H3K36me3 in RAD51 recruitment at DSBs, we analyzed the effect of SETD2 depletion on RAD51 binding at sites of damage generated by laser micro irradiation. At those laser induced DSBs, SETD2 siRNA also led to a clear decrease of RAD51 recruitment (Fig. 7d).
To investigate whether the loss of RAD51 binding observed upon H3K36me3 removal was associated with HR repair defect, we next used the well characterized DR-GFP reporter assay to analyze the consequences of SETD2 depletion on HR repair 39. As expected depletion of RAD51 by siRNA completely abolished the use of HR to repair the I-SceI induced DSB, while 53BP1 siRNA led to an increased usage of HR. Strikingly, SETD2 depletion almost completely abolished HR repair, to an extent similar to RAD51 depletion (Fig. 7e).
Thus, altogether our data indicate that SETD2 depletion not only impaired RAD51 binding at AsiSI DSBs and laser induced damages sites, but also severely decreased HR repair at an I-SceI induced DSB.
To assess the potential role for de novo H3K36me3 deposition occurring during repair, which could have explained the effect of SETD2 depletion on HR, we analyzed H3K36me3 distribution by ChIP-seq after 4OHT dependent DSB induction. We did not detect marked induction of H3K36 trimethylation at any DSB sites tested upon 4OHT treatment (either taken individually, Fig. 7f, or collectively Fig. 7g), arguing for a role of pre-existing H3K36me3 in the targeting of RAD51 to DSBs.
Discussion
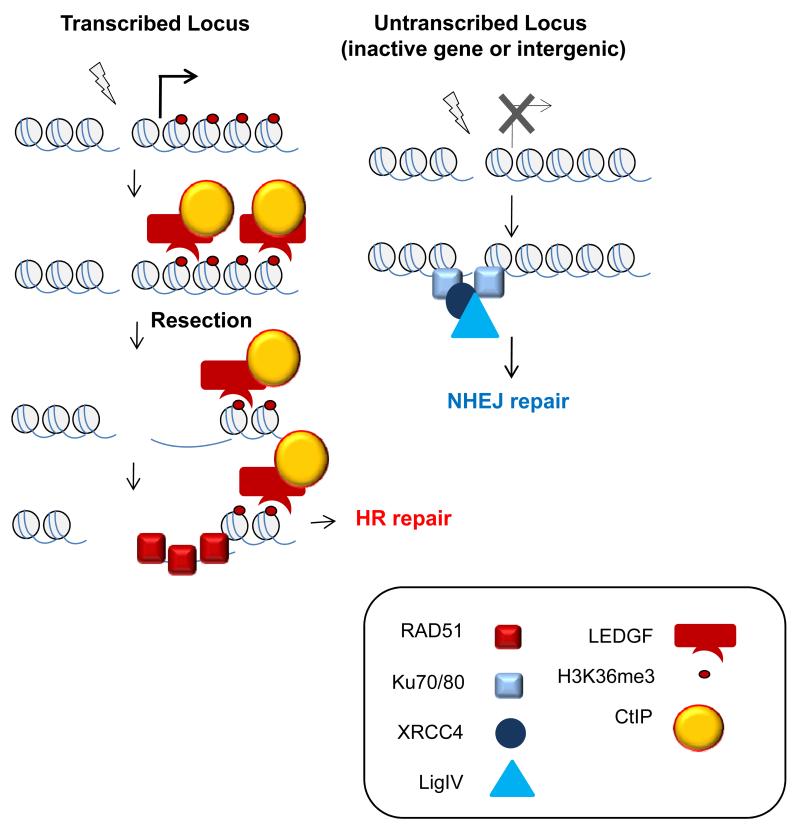
In this study, by using a human cell line (the DIvA cells) expressing a restriction enzyme fused to the ligand binding domain of the oestrogen receptor (AsiSI-ER), together with a ChIP-seq approach, we have found that distinct DSBs induced across the genome are not necessarily repaired by the same pathway. We identified an “HR-prone” subset of DSBs that recruit the HR protein RAD51, undergo resection, and rely on RAD51 for efficient repair. Taken together, our data clearly indicate that the choice of repair pathway at AsiSI-induced DSBs depends, at least in part, on the chromatin context where the break occurs. More specifically, it is influenced by the transcriptional status of the closest nearby gene, through at least one histone mark, H3K36me3, associated with transcriptional elongation. This chromatin mark fosters HR-mediated repair in transcribed genes, most likely through being recognized by LEDGF, which then can promote CtIP recruitment to initiate resection at nearby DSB sites, thereby leading to RAD51 loading and repair by HR (Figure 8).
Figure 8. Targeting of HR at transcriptionally active loci.
The trimethylation of histone H3 on the lysine 36 (H3K36me3), a mark associated with transcriptional elongation, would be recognized by LEDGF, itself interacting with CtIP, thus bringing to the site of break a resection promoting factor. This would subsequently trigger RAD51 binding and HR repair on actively transcribed genes. DSBs occurring within inactive genes or intergenic regions would be unable to recruit a resection factor thus leading to repair by NHEJ.
Repair of AsiSI induced DSB across the cell cycle
Importantly we have also addressed the influence of the cell cycle on AsiSI induced DSB repair. In G1, as expected, we could not detect RAD51 binding while XRCC4 recruitment predominated at all investigated DSBs, either located in transcriptionally active or inactive chromatin. At this cell cycle phase, while repair of DSBs induced on silent chromatin are very likely repaired by NHEJ, the repair status of the DSBs induced on active chromatin still remains to be investigated. Indeed, even if both H3K36me3 and LEDGF are present in G1 (data not shown), the resection promoting activity of CtIP is tightly controlled by phosphorylation events, at the S/G2 transition 40,41. Therefore, although still present in G1 phase, the H3K36me3-LEDGF pathway may not be able to trigger CtIP dependent resection, thus leading to NHEJ repair, at DSBs, induced both in H3K36me3 rich or poor loci.
However, our data also indicate that even though XRCC4 is recruited at DSBs associated with active loci, it may be inefficient in G1 since depletion of XRCC4 did not impair repair of these DSBs. One appealing hypothesis is that DSBs induced in active loci may only be repaired by HR in the subsequent S phase. Of note, a recent study showed that some DSBs induced in late G1, are repaired by HR, as cells progress to S phase 42. Such HR repair at transcribed loci may be initiated in G1: indeed in G0/G1 synchronized cells, I-PpoI dependent DSBs induced in transcriptionally active genes, and not in intergenic regions, are found to undergo ‘chromosome kissing’ (a process believed to be linked to homology search) 43. More investigations are clearly required to address the fate of these DSBs in G1, i.e, to state whether they are left unrepaired until S phase, repaired by NHEJ or repaired by an alternative pathway.
In G2, we could both identify DSBs able and unable to recruit RAD51, in agreement with previous reports, appraising to only about 15% the DSBs repaired by HR in G2 2,42. We show here that chromatin structure that pre-exists at the site of break is one of the major factors that influence the use of HR or NHEJ in G2 (see next section).
Repair in heterochromatin versus in euchromatin
We found that the DSBs located in silent loci and intergenic regions were still unable to recruit RAD51 in G2, while DSBs induced in active chromatin, rich in H3K36me3, were channeled to HR repair.
This conclusion is seemingly at-odds with studies reporting that heterochromatin (generally considered to be devoid of active genes) is repaired by a mechanism involving DNA synthesis by an Artemis-dependent HR pathway 2,3,44, while euchromatin is mainly repaired by a DNA synthesis independent pathway, likely NHEJ. Our work in DIvA cells did not allow us at that point to investigate repair in heterochromatin since AsiSI does not induce DSBs in these regions (likely due to its dense structure, and/or its highly methylated status 30). We note however that beyond its enrichment on active genes, H3K36me3 is also enriched in heterochromatin 45, and that although still poorly characterized, RNA production has been shown to be a feature of constitutive heterochromatin 46-48. Consequently, as for DSBs induced in active genes, DNA breaks occurring within certain heterochromatic regions could also be targeted to HR repair through H3K36me3-dependent mechanisms. Concerning repair in euchromatin we found that actively transcribed and H3K36me3 enriched loci are repaired by HR, while H3K36me3 depleted regions are repaired by NHEJ. This is in agreement with the studies from Jeggo’s and Lobrich’s groups, since H3K36me3 enriched loci represent only a minor fraction (about 5%) of the genome 49, the vast majority of euchromatin being composed of intergenic regions and silent genes.
Pre-existing chromatin contributes to repair pathway choice
Our study demonstrates a critical role of the pre-existing chromatin context on the decision to use HR or NHEJ to repair a DSB induced in the human genome. Notably, changing the transcriptional status of a locus prior DSB induction modified the pathway used for repair: a DSB induced in an inactive gene switched repair from a RAD51 independent to a RAD51 dependent pathway upon transcriptional activation (Fig. 5).
We identified a histone mark, H3K36me3, as essential to recruit RAD51 through LEDGF, a protein that possesses a PWWP domain. This specific module is involved in H3K36me3 recognition 36,50,51 and was found to interact with CtIP 36. Additional H3K36me3 binding factors are likely at work since LEDGF depletion led to a less dramatic effect than SETD2 depletion on RAD51 recruitment at DSBs. Importantly, LEDGF-CtIP anchoring occurs on H3K36me3 that pre-exists on chromatin. Indeed, our ChIP-seq data obtained in 4OHT treated DIvA cells indicated that H3K36me3 is not increased after DSB induction, thus arguing for a role of pre-existing H3K36me3 in RAD51 binding (Fig. 7f-g). Of note, a study recently conducted in the lab of Tim Humphrey also identified SETD2 as required for HR repair in mammalian cells (Pfister S, Humphrey T.C, personal communication).
Interestingly, other studies already suggested a role of pre-existing chromatin in NHEJ versus HR choice. Indeed, mono and di-methylation of histone H4 on lysine 20 serves as an anchoring module for 53BP1, known to promote NHEJ. While their induction at DSBs are still controversial, their involvement in 53BP1 stabilization at the site of damage are well documented 15-19. In addition, a recent study revealed a role of the acetylation of the lysine 16 on histone H4 (H4K16ac), one of the histone marks associated with transcriptional activity, in counteracting 53BP1 binding to H4K20me2. Similarly to our findings, transcriptional activation of a locus prior to DSB induction led to an increased recruitment of BRCA1 and a reduction of 53BP1 binding attributed to the higher H4K16 acetylation levels triggered by transcription 20. How exactly the H3K36me3-LEDGF-CtIP axis cooperates with H4K16ac-Tip60-BRCA1 to tightly regulate HR at DSBs will require further investigation, although our experiments using HR-DRGFP reporter assay suggest that both pathways act independently to promote resection (data not shown).
In conclusion, our study has revealed that DSBs do not all behave in the same manner across the genome and that these differences depend upon whether or not the breaks occur in a transcribed locus. These data underline the extraordinary potential of chromatin in regulating genome stability: beyond the choice between C-NHEJ and HR, explored in this study, one can easily imagine that it could also control the use of Alt-NHEJ, unequal HR, SSA, and other less characterized repair pathways, in order to adapt the repair mechanism to the type and function of the locus to repair. This chromatin dependent control could help to minimize the risks associated with repair events and play a critical role in maintaining genome integrity.
Online Methods
AID-DIvA cell generation
The AsiSI-ER fusion was cloned into the pAID1.1-N vector (BioROIS13), Cloning was performed using a modified E.coli strain (AsiSI-met), provided by New England Biolabs. The AID-AsiSI-ER plasmid was transfected into U20S cells by using the Cell Line Nucleofactor kit V (Amaxa) and selection was performed by using 800 μg/mL G418.
Cell culture
DIvA (AsiSI-ER-U20S),AID-DIvA (AID-AsiSI-ER-U20S) and RG37 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with antibiotics, 10% FCS (InVitrogen) and either 1 μg/mL puromycin (DIvA cells) or 800 μg/mL G418 (AID-DIvA cells) at 37°C under a humidified atmosphere with 5% CO2. For AsiSI-dependant DSB induction, cells were treated with 300 nM 4OHT (Sigma; H7904) for 4h. When indicated, 4OHT treated cells were washed 3 times in pre-warmed PBS and further incubated with 500 μg/mL auxin (Sigma; I5148). Pol II was inhibited with 5μM Actinomycin D (Sigma; A9415) for 10 h, or 100μM of DRB (Sigma; D1916) for 7 h. siRNA transfections were performed with the Cell Line Nucleofactor kit V (Amaxa) according to the manufacturer’s instructions. Sequences for siRNAs are displayed in Supplementary Table 2. For cell synchronization, cells were incubated with 2mM thymidine for 18 h, released for 11 h and subjected to the second thymidine treatment for 18 h. G1 cells were collected after 15 h of release, while G2 cells were collected after 7 h.
TALEN-dependant DSB induction
A TALEN pair targeting the following sequence TCCTCACCTGATGAGAATGA gaatgaggttgaGGATTCAGCTGACTTTGTGA in the GBP1 gene, was ordered at http://www.talenlibrary.net/ (H114918). 5.106 U20S cells were transiently transfected with 12.5 μg of each of the two TALENs by the calcium phosphate co-precipitation method, for 24 h and γ-IFN treatment (InVivogen, rhifn-g) was performed for 8 h at 50ng/mL where indicated.
I-SceI induced DSB HR assay
105 RG37 cells, stably transfected with an I-SceI based HR- GFP substrate (kind gift of B. Lopez, Institut Gustave Roussy) were transfected with siRNA (10nM final) (INTERFERin, PolyPlusTransfection #409-10), and transfected, 24h later, with 1μg of I-SceI vector (jetPEI, PolyPlusTransfection #101-10) After 72h cells were collected and GFP positive cells were analyzed by flow cytometry.
Western Blot
Western blot analysis was performed using NuPAGE Tris-acetate 3-8% gels and reagents (Invitrogen) according to the manufacturer’s indications. Briefly, cells were rinsed twice with ice-cold PBS and then lysed in the appropriate lysis buffer with sample reducing agent (Invitrogen). Liquid transfer of resolved proteins was performed onto PVDF membranes (Invitrogen). After 1 h block in 5% non-fat dry milk / 0.5% PBS-Tween, membranes were incubated overnight with the following primary antibodies: anti-HA (HA-11, Babco, 1/2000), anti-γH2AX (Cell Signaling, 2577s, 1/500), anti-RAD51 (Santa Cruz, SC-8349, 1/200), anti-XRCC4 (Abcam, ab145, 1/1000), anti-lamin A-C (Santa Cruz, H110, 1/500) and anti-α-tubulin (Sigma, DM1A, 1/100000). Validations for these antibodies are available on manufacturers’ websites. Horseradish peroxidase-coupled secondary antibodies were from Amersham Biosciences, and chemiluminescence Lumilight reagent was from Roche Diagnostic. Original images of blots used in this study can be found in Supplementary Figure 8.
Chromatin immunoprecipitation
ChIP assays were carried out according to the protocol described in 30 with the following modifications. 200 μg of chromatin was immunoprecipitated by using 2 μg of anti-γH2AX (Epitomics, 2212-S), anti-XRCC4 (Abcam, ab145), anti-RAD51 (Santa Cruz, SC-8349) or without antibody (mock), all these antibodies were validated in for ChIP in previous studies 30-31. 50 μg of chromatin was used for the PolII-S2P immunoprecipitation (Abcam, ab5095, 2 μg), and 10 μg of chromatin was used for H3K36me3 ChIP (Abcam, ab9050, 2 μg), both antibodies were validated in ChIP by the providers. Immunoprecipitated DNA and input DNA were analyzed in triplicate by RT-qPCR (primer sequences are provided Supplementary Table 3). Since RAD51 recruitment often showed a significant decrease at the immediate vicinity of the DSB (Fig. 1a-b, Supplementary Fig. 2c), primer pairs used to measure XRCC4 enrichment were located at less than 100bp from the DSB while those to assess RAD51 recruitment were located at 800bp. IP efficiency was calculated as percent of input DNA immunoprecipitated. Data are shown either as ratio of RAD51/XRCC4 signals, or as ChIP efficiency, as indicated. For ChIP-seq, sequencing libraries were prepared by using 10 ng of purified DNA (averaged size 250-300 bp), and subjected to high throughput sequencing (single read) by the Genomic Core facility (EMBL, Heidelberg, for XRCC4 and RAD51), by the BGI institute (γH2AX, PolII-S2P and H3K36me3), using a HiSeq 2000 sequencing system or by Genomics core facility at the Cancer Research Institute (CRI), Cambridge, UK using the Illumina Genome Analyzer 2. After quality filtering, the number of uniquely mapped sequencing reads (aligned to hg18) was counted over 200 bp windows.
Cleavage assay/resection assay
The full procedure for the cleavage assay has been previously described 30,32. Briefly, a biotinylated double-stranded oligonucleotide, cohesive with AsiSI sites, was ligated in vitro to genomic DNA after break induction. T4 ligase was heat inactivated at 65°C for 10 min, and DNA was fragmented by EcoRI digestion at 37°C for 2 h followed by heat inactivation at 70°C for 20 min. After a preclearing step, DNA was pulled down with streptavidin beads (Sigma) at 4°C overnight, and then washed 5 times in RIPA buffer and twice in TE. Beads were resuspended in 100 μL of water and digested with HindIII at 37°C for 4 h. After phenol/chloroform purification and precipitation, DNA was resuspended in 100 μL of water. The resection assay is based on the cleavage efficiency assay, but a longer cohesive extremity (15 bp) allows to specifically pull-down resected ends. Specific biotinylated double strand oligonucleotides were thus designed for each analyzed DSB (Supplementary Table 4). For both assays, precipitated DNA was quantified for each site by RT-qPCR using primers described in Supplementary Table 3.
Immunofluorescence and quantification
Detailed methods for immunofluorescence have already been described in 30. γH2AX foci were quantified with a Thermoscientific Cellomics ArrayScan VTI HCS Reader. Laser micro-irradiation was essentially carried out as previously described 13,52.. Cells were plated on glass-bottomed dishes (Willco-Wells) and pre-sensitised for 24 h with 10 μM BrdU before exposing them to a UV-A laser beam using a confocal inverted microscope (Olympus FluoView 1000), a 405-nm laser diode (6 mW, SIM scanner) and a 60× UPlanSApo/1.35 oil objective. Laser settings of 0.2 mW output (50 scans) restricted the generated DNA damage to the laser path in a pre-sensitisation-dependent manner without noticeable cytotoxicity. After a recovery time of 30-50 min cells were co-stained with antibodies against RAD51 (Santa Cruz sc-8349), γH2AX (Millipore 05-636) and cyclin A (BD Biosciences 611268).
“RAD51-bound” and “RAD51-unbound” categories design
We previously reported that only a subset of annotated AsiSI sites over the genome are efficiently cleaved by the AsiSI-ER fusion protein in cells, mostly due to their methylation status, and that efficient cleavage was always associated with a wide γH2AX domain, showing a clear drop in close vicinity to the break30. A subset of the 100 most cleaved AsiSI sites in the genome was therefore determined based on γH2AX enrichment over a 20kb window and γH2AX depletion on 1000 bp around the DSB. This subset was further sorted based on the RAD51/XRCC4 ratio (A 1000bp window surrounding the DSB was taken into account for XRCC4, while a 4kb window was used for RAD51) to identify two categories. The 20 best sites either in favor of XRCC4 or RAD51 were selected (Supplementary Table 1), and used for Fig. 4, 6 and Supplementary Fig.5a
Average profiles around DSBs and TSS
To plot data with respect to DSBs, AsiSI site positions were retrieved from the human genome (hg18). ChIP-seq counts were retrieved for 20kb around each of these DSBs and averaged using a 200 bp window. In order to plot data with respect to transcription start sites (TSS), for each genes associated with the studied DSBs (i.e. located not farther than 1000bp from the AsiSI site), transcript positions and orientations were obtained from the refFlat table from UCSC (hg18) at http://genome.ucsc.edu. Unique genes were taken into account and ChIP-seq counts were averaged using a 1000bp window.
Correlation with epigenomic features
For each of the twenty DSBs of the two subsets, the averaged signal of various histone marks and proteins obtained by ChIP-seq and available from the ENCODE project (http://genome.ucsc.edu/ENCODE/34), was calculated over a 4000 bp window centered on the DSBs. Datasets used for Supplementary Fig. 5a were generated by the Broad Institute (Bernstein), or the USC (Snyder) as indicated. Since some of these data are only available aligned against hg19, AsiSI positions from the two HR-prone and non-HR-prone subsets were converted to hg19 coordinates using the lift over tool (UCSC).
Supplementary Material
Acknowledgements
We thank NEB Biolabs for providing AsiSI genomic DNA. We thank B.Lopez, Institut de Cancérologie Gustave-Roussy, for RG37-HR I-SceI GFP cells. We thank V. Benes and the Solexa team at the EMBL Genomic Core Facility, the Beijing Genomic Institute (BGI), and the Genomics core facility at the Cancer Research Institute (CRI) in Cambridge for high throughput sequencing. We thank the Flow Cytometry platform at LBCMCP-FRBT. FA. is supported by a grant from the Association Contre le Cancer (ARC), SB and EG are supported by grants from the Fondation pour la Recherche Médicale (FRM). Research in the SPJ laboratory is supported by grants from Cancer Research UK (C6/A11226), the European Research Council, the European Community’s Seventh Framework Program (DDResponse) and by core infrastructure funding from Cancer Research UK and the Wellcome Trust. KM was funded by a Wellcome Trust Project grant, CKS by a Return-to-Europe FEBS fellowship and SPJ receives his salary from the University of Cambridge supplemented by Cancer Research UK. PC is supported by a grant from the Association Contre le Cancer (ARC). Funding in GL laboratory was provided by grants from the Association Contre le Cancer (ARC), Agence Nationale pour la Recherche (ANR-09-JCJC-0138), Canceropole Grand Sud Ouest (GSO), and Research Innovation Therapeutic Cancerologie (RITC).
Footnotes
Accession code:
High throughput sequencing data have been deposited to Array Express under accession number E-MTAB-1241.
References
- 1.Hartlerode AJ, Scully R. Mechanisms of double-strand break repair in somatic mammalian cells. Biochem J. 2009;423:157–168. doi: 10.1042/BJ20090942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beucher A, et al. ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J. 2009;28:3413–3427. doi: 10.1038/emboj.2009.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noon AT, et al. 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat Cell Biol. 2010;12:177–184. doi: 10.1038/ncb2017. [DOI] [PubMed] [Google Scholar]
- 4.Shibata A, et al. Factors determining DNA double-strand break repair pathway choice in G2 phase. EMBO J. 2011;30:1079–1092. doi: 10.1038/emboj.2011.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fattah F, et al. Ku regulates the non-homologous end joining pathway choice of DNA double-strand break repair in human somatic cells. PLoS Genet. 2010;6:e1000855. doi: 10.1371/journal.pgen.1000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bothmer A, et al. Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol Cell. 2010;42:319–329. doi: 10.1016/j.molcel.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouwman P, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol. 2010;17:688–695. doi: 10.1038/nsmb.1831. d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bunting SF, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapman JR, et al. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol Cell. 2013;49:858–871. doi: 10.1016/j.molcel.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Virgilio M, et al. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science. 2013;339:711–715. doi: 10.1126/science.1230624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Escribano-Diaz C, et al. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol Cell. 2013;49:872–883. doi: 10.1016/j.molcel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 12.Zimmermann M, Lottersberger F, Buonomo SB, Sfeir A, de Lange T. 53BP1 regulates DSB repair using Rif1 to control 5′ end resection. Science. 2013;339:700–704. doi: 10.1126/science.1231573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sartori AA, et al. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shibata A, et al. DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities. Mol Cell. 2014;53:7–18. doi: 10.1016/j.molcel.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Botuyan MV, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartlerode AJ, et al. Impact of histone H4 lysine 20 methylation on 53BP1 responses to chromosomal double strand breaks. PloS one. 2012;7:e49211. doi: 10.1371/journal.pone.0049211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oda H, et al. Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Mol Cell. 2010;40:364–376. doi: 10.1016/j.molcel.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pei H, et al. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature. 2011;470:124–128. doi: 10.1038/nature09658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanders SL, et al. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell. 2004;119:603–614. doi: 10.1016/j.cell.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 20.Tang J, et al. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat Struct Mol Biol. 2013;20:317–325. doi: 10.1038/nsmb.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.FitzGerald JE, Grenon M, Lowndes NF. 53BP1: function and mechanisms of focal recruitment. Biochem Soc Trans. 2009;37:897–904. doi: 10.1042/BST0370897. [DOI] [PubMed] [Google Scholar]
- 22.Chiolo I, et al. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell. 2011;144:732–744. doi: 10.1016/j.cell.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berkovich E, Monnat RJ, Jr., Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol. 2007;9:683–690. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- 24.Rouet P, Smih F, Jasin M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc Natl Acad Sci U S A. 1994;91:6064–6068. doi: 10.1073/pnas.91.13.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller KM, Jackson SP. Histone marks: repairing DNA breaks within the context of chromatin. Biochem Soc Trans. 2012;40:370–376. doi: 10.1042/BST20110747. [DOI] [PubMed] [Google Scholar]
- 26.Soria G, Polo SE, Almouzni G. Prime, repair, restore: the active role of chromatin in the DNA damage response. Mol Cell. 2012;46:722–734. doi: 10.1016/j.molcel.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 27.Wolner B, van Komen S, Sung P, Peterson CL. Recruitment of the recombinational repair machinery to a DNA double-strand break in yeast. Mol Cell. 2003;12:221–232. doi: 10.1016/s1097-2765(03)00242-9. [DOI] [PubMed] [Google Scholar]
- 28.Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010;141:970–981. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caron P, et al. Cohesin protects genes against gammaH2AX Induced by DNA double-strand breaks. PLoS Genet. 2012;8:e1002460. doi: 10.1371/journal.pgen.1002460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iacovoni JS, et al. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 2010;29:1446–1457. doi: 10.1038/emboj.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Massip L, Caron P, Iacovoni JS, Trouche D, Legube G. Deciphering the chromatin landscape induced around DNA double strand breaks. Cell Cycle. 2010;9:2963–2972. doi: 10.4161/cc.9.15.12412. [DOI] [PubMed] [Google Scholar]
- 32.Chailleux C, et al. Quantifying DNA double-strand breaks induced by site-specific endonucleases in living cells by ligation-mediated purification. Nature protocols. 2014;9:517–528. doi: 10.1038/nprot.2014.031. [DOI] [PubMed] [Google Scholar]
- 33.Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- 34.A user’s guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim Y, et al. A library of TAL effector nucleases spanning the human genome. Nat Biotechnol. 2013;31:251–258. doi: 10.1038/nbt.2517. [DOI] [PubMed] [Google Scholar]
- 36.Daugaard M, et al. LEDGF (p75) promotes DNA-end resection and homologous recombination. Nat Struct Mol Biol. 2012;19:803–810. doi: 10.1038/nsmb.2314. [DOI] [PubMed] [Google Scholar]
- 37.Butler JS, Dent SY. Chromatin ‘resetting’ during transcription elongation: a central role for methylated H3K36. Nat Struct Mol Biol. 2012;19:863–864. doi: 10.1038/nsmb.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edmunds JW, Mahadevan LC, Clayton AL. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J. 2008;27:406–420. doi: 10.1038/sj.emboj.7601967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dumay A, et al. Bax and Bid, two proapoptotic Bcl-2 family members, inhibit homologous recombination, independently of apoptosis regulation. Oncogene. 2006;25:3196–3205. doi: 10.1038/sj.onc.1209344. [DOI] [PubMed] [Google Scholar]
- 40.Huertas P, Jackson SP. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. The Journal of biological chemistry. 2009;284:9558–9565. doi: 10.1074/jbc.M808906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu X, Chen J. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Molecular and cellular biology. 2004;24:9478–9486. doi: 10.1128/MCB.24.21.9478-9486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karanam K, Kafri R, Loewer A, Lahav G. Quantitative live cell imaging reveals a gradual shift between DNA repair mechanisms and a maximal use of HR in mid S phase. Mol Cell. 2012;47:320–329. doi: 10.1016/j.molcel.2012.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gandhi M, et al. Homologous chromosomes make contact at the sites of double-strand breaks in genes in somatic G0/G1-phase human cells. Proc Natl Acad Sci U S A. 2012;109:9454–9459. doi: 10.1073/pnas.1205759109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goodarzi AA, Jeggo P, Lobrich M. The influence of heterochromatin on DNA double strand break repair: Getting the strong, silent type to relax. DNA Repair (Amst) 2010;9:1273–1282. doi: 10.1016/j.dnarep.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 45.Chantalat S, et al. Histone H3 trimethylation at lysine 36 is associated with constitutive and facultative heterochromatin. Genome Res. 2011;21:1426–1437. doi: 10.1101/gr.118091.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Finnegan DJ. Oogenesis: active heterochromatin. Curr Biol. 2011;21:R630–632. doi: 10.1016/j.cub.2011.07.029. [DOI] [PubMed] [Google Scholar]
- 47.Lejeune E, Allshire RC. Common ground: small RNA programming and chromatin modifications. Curr Opin Cell Biol. 2011;23:258–265. doi: 10.1016/j.ceb.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 48.Probst AV, Almouzni G. Heterochromatin establishment in the context of genome-wide epigenetic reprogramming. Trends Genet. 2011;27:177–185. doi: 10.1016/j.tig.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 49.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dhayalan A, et al. The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. The Journal of biological chemistry. 2010;285:26114–26120. doi: 10.1074/jbc.M109.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vezzoli A, et al. Molecular basis of histone H3K36me3 recognition by the PWWP domain of Brpf1. Nat Struct Mol Biol. 2010;17:617–619. doi: 10.1038/nsmb.1797. [DOI] [PubMed] [Google Scholar]
- 52.Miller KM, et al. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol. 2010;17:1144–1151. doi: 10.1038/nsmb.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.