Summary
Through their association with a kleisin subunit (Scc1), cohesin’s Smc1 and Smc3 subunits are thought to form tripartite rings that mediate sister chromatid cohesion. Unlike the structure of Smc1/Smc3 and Smc1/Scc1 interfaces, that of Smc3/Scc1 is not known. Disconnection of this interface is thought to release cohesin from chromosomes in a process regulated by acetylation. We show here that the N-terminal domain (NTD) of yeast Scc1 contains two α helices, forming a four helix bundle with the coiled coil emerging from Smc3’s ATPase head. Mutations affecting this interaction compromise cohesin’s association with chromosomes. The interface is far from Smc3 residues whose acetylation prevents cohesin’s dissociation from chromosomes. Cohesin complexes holding chromatids together in vivo do indeed have the configuration of hetero-trimeric rings and sister DNAs are entrapped within these.
Introduction
A vital member of the Smc/kleisin family is the eukaryotic cohesin complex, which confers sister chromatid cohesion, facilitates the repair of double strand breaks, and modulates the structure and transcription of interphase chromatin (1, 2). Cohesin contains a dimer of two related Smc proteins, Smc1 and Smc3, whose association with an α-kleisin subunit (Scc1/Rad21) has the potential to form an extended tripartite ring (3) within which sister DNAs could be entrapped (4). It has been suggested that the cohesin ring has separate DNA entry and exit gates, located at the Smc1/Smc3 “hinge” (5) and Smc3/kleisin interfaces, respectively (6). To understand how the latter’s disconnection by the regulatory subunits Wapl, Pds5, and Scc3 releases cohesin from chromatin and how Smc3 acetylation locks rings shut (6), we have determined the structure of the Smc3-kleisin interface.
Structure of the Smc3-Scc1 interface
Because in vivo photo-crosslinking experiments(7) showed that Scc1’s N-terminal domain (NTD) binds to the coiled coil emerging from Smc3’s ATPase head (Figs. S1 and S2), we co-expressed in E. coli Scc1’s first 115 residues (Scc1-N) with a version of the Smc3 ATPase head containing a 75-residue long section of its coiled coil (Smc3hdCC). This yielded a complex suitable for X-ray crystallography (Fig. S2D). Diffraction data were obtained to a resolution of 3.3 Å from crystals grown in the presence of ATPγS and their structure solved by a combination of molecular replacement and SeMet SAD phasing. The structure (Fig. 1A) reveals nearly one third of Smc3’s coiled coil including a pronounced and highly conserved kinked region between L991 and F999 (Fig. S3C). The structure of the Smc3 ATPase domain is most closely related to that of Smc1 of all structures currently deposited in the PDB (RMSD ~ 2.2 Å, Fig. 1B).
Figure 1. Crystal structure of the Smc3hdCC:Scc1-N complex.
(A) The coiled coil segment of Smc3 (blue) is interrupted by a ‘kink’. The NTD of Scc1, Scc1-N (green), binds to the coiled coil segment of Smc3, leading to a four-stranded helical arrangement. Inset: aberrant homodimer formation of Smc3 head domains in the crystals.
(B) A superposition of the Smc3hdCC:Scc1-N crystal structure (blue, green; this work) with Smc1hdCC:Scc1-C (red, yellow; PDB 1W1W) reveals that in addition to the ATPase fold, the position of the coiled coil segments is conserved. Crucially, Scc1 binding is completely different for Smc3 and Smc1.
(C) Sequence conservation of Scc1’s NTD.
(D) ATP binding leads to sandwich dimer formation of the head domains of Smc1 and Smc3, closing the ring temporarily. According to the ring model, Scc1 more permanently bridges the two head domains, which can be released through separase-mediated cleavage of Scc1 or in a separase-independent pathway through opening of the Smc3:Scc1 gate. Scc1 contains many more residues in the middle domain. Separase cleavage sites, Pds5 (7) and Scc3 (Brunet et al., in press) binding sited highlighted.
(E) Detail of the KKD strand whose acetylation by Eco1 reduces separase-independent cohesin release. It is far away from the nucleotide binding site on the head domain but the acetylation state may influence the nucleotide binding site through the helix containing R61.
Scc1’s NTD is folded into three helices α1, α2, and α3 (Figs. 1AC). The most prominent is the 34-residue α3 (R69-M102), which forms a long helical bundle with Smc3’s coiled coil. The C-terminal end of α3 almost reaches the coiled coil’s pronounced kink while the N-terminal end extends to Smc3’s ATPase head. Helices α2 and α3 together form a compact four helix bundle with Smc3’s coiled coil and there is no contact with the ATPase head itself. The very different manner by which N- and C-terminal domains interact with Smc ATPase heads (Fig. 1B) means that the path of Scc1 central domain’s polypeptide is complex and possibly influenced by its association with Pds5 and Scc3. Sequences responsible for recruiting Pds5 are situated between H124 and L138 (7), close to the top of α3 (Fig. 1D). This proximity is striking because Pds5 has a key role in releasing cohesin from chromatin, presumably by dissociating the Scc1/Smc3 interface, as well as shutting off this process during S phase by promoting Smc3K113 acetylation.
Conservation of the Scc1-N/Smc3 interface
The entire surface of Smc3’s coiled coil facing Scc1’s α3 is highly conserved while the opposing surface is not (Fig. 2A). The face of Scc1’s α3 that contacts Smc3’s coiled coil is similarly conserved (Figs. 1C and S4AD). Scc1’s α2 helix is in general less conserved than α3. In a B. subtilis complex (8), ScpA sequences corresponding to the N-terminal half of Scc1 α3 form a three helix bundle with the Smc coiled coil similar to that observed between Scc1-N and Smc3 but the structure of the rest of ScpA’s NTD differs substantially from that of Scc1 (Fig. S4B). Several of the characteristics of α3 are found in β- and γ-kleisins from condensin. Thus, hydrophobic residues like L75, Y82, L89, and L97, that seem to have a role in contacting Smc3’s coiled coils, are present at the same positions within β-kleisins (condensin II) and for the most part also γ-kleisins (condensin I) (Fig. S4D). The equivalent residues in B. subtilis ScpA are also hydrophobic and have a similar juxtaposition to their Smc partner (8).
Figure 2. Testing the Smc3-kleisin crystal structure.
(A) Conservation of Smc3’s coiled coil. The surface associated with Scc1 is highly conserved but the solvent-exposed side is not. (B) Thiol-specific cross-linking (bBBr) between α2 and α3 of Scc1-N and Smc3’s coiled coil (CC) after immuno-precipitation of Scc1-HA6. Cross-linking specific to K48C-K1032C was observed in cells expressing C56S (K19796, K19769, K19727, K19732, K19764, K23102, K23103). All mutations were functional and all observed cross-links were dependent on a pair of cysteine substitutions. (C) Scc1-N α2 and α3 helices (green), Smc3 coiled coil (Smc3CC, blue), and substituted residues (yellow).
Testing the structure using thiol-specific cross-linking
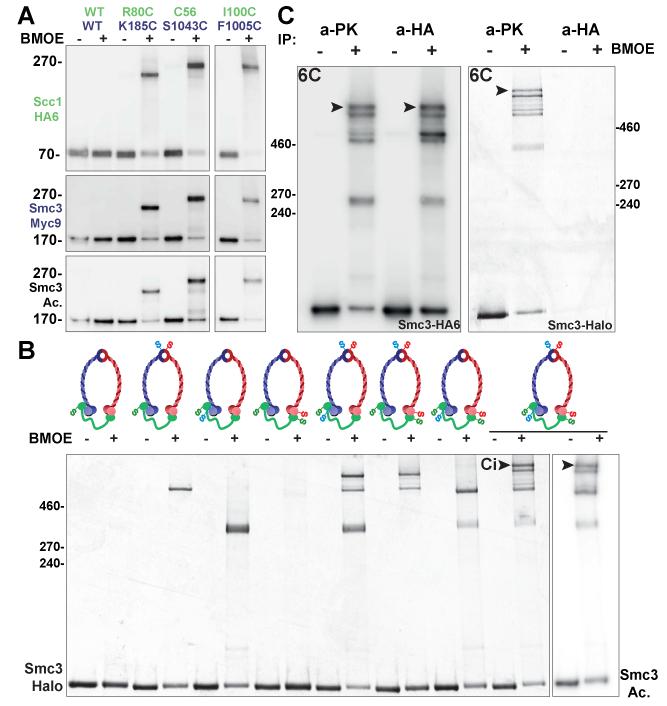
We created a series of pairwise cysteine substitutions within Scc1 and Smc3 and measured the efficiency with which these are cross-linked using the homo-bifunctional sulfhydryl active reagent dibromobimane (bBBr) (9). Treatment of native cohesin complexes following immuno-precipitation revealed the expected pattern of cross-linking between cysteines within Smc3’s coiled coil and cysteines within Scc1’s α3 (Fig. 2BC). This included cross-linking between Scc1I100C and Smc3F1005C, which confirms that Scc1’s α3 extends further up the Smc3 coiled coil than that observed for ScpA (Fig. S4B). We also observed efficient cross-linking between Scc1K48C and Smc3K1032C, confirming the observed juxtaposition of α2 with Smc3 coiled coil. In contrast, cross-linking occurred rarely if at all between residues predicted to be non-adjacent (Fig S4E). Importantly, cysteine pairs involving α2 and α3 can be cross-linked in vivo and the cross-linked species were acetylated (Fig.4A), implying that the interactions revealed by the Smc3/kleisin structure actually exist in complexes engaged in holding sister DNAs together.
Figure 4. Cohesin forms heterotrimeric rings in vivo.
(A) In vivo thiol-specific cross-linking (BMOE) between α2 and α3 of Scc1-N and the Smc3 coiled coil (CC) followed by immuno-precipitation of Scc1-HA6. In all shifted bands Smc3 is acetylated (K19796, K19732, K19764, K23103). (B) In vivo thiol-specific cross-linking of the three Smc1/Smc3/Scc1 interfaces followed by immuno-precipitation of Scc1-PK6 and observation of TMR fluorescence associated with Smc3-Halo. A circular form (Ci) only appears when all three interfaces are linked (black arrowheads). Western blotting using an antibody specific for acetylated K113 shows that Ci is acetylated (last lane) (strains K22013-K22020). (C) Cohesin forms heterotrimeric rings but not higher order complexes. Halo-tagged Ci (black arrowhead) associated with PK- and HA-tagged proteins from Scc1-PK6/Scc1-PK6, Smc3-HA6/Smc3-Halo diploids after in vivo cross-linking. No Smc3-Halo is present in form Ci molecules that had been immuno-precipitated with HA antibodies, implying that they are circular trimers, not tetramers (K22590).
Due to the presence of prolines (10), it is unlikely that α3 from bacterial kleisins extends as far up the coiled coil as is the case for Scc1. This difference between the cohesin and bacterial structures may therefore be genuine. To assess the N-terminal differences, we designed cysteine pairs, namely ScpAE39C-SmcH1043C and ScpAL42C-SmcQ1039C, which should be cross-linked if ScpA adopts the conformation observed in the ScpA/Smc crystal, and an alternative pair, E39C-Q1020C that should be cross-linked only if they adopted the Smc3/Scc1 conformation. Our observation that in B. subtilis bis-maleimidoethane (BMOE) induces efficient cross-linking in vivo between the latter but not former pairs suggests that ScpA binds to the Smc coiled coil by forming a four helix bundle similar to that formed by Scc1 and Smc3 (Fig. S4F).
Interaction between Scc1’s NTD and Smc3’s coiled coil is essential
The adverse effects of mutations within Scc1’s NTD that affect its association with Smc3 (11) are explained by the structure. Three highly conserved leucine residues, namely L68, L75, and L89, line the surface of α3 that faces Smc3’s coiled coil (Fig. 3A). In each case, substitution by lysine causes lethality, reduces association between Scc1 and Smc3 (11), and abrogates (in the case of L75K and L89K) cohesin’s association with centromeres (12). As expected, Scc1L89K abolished cross-linking between S1043C-C56 cysteine pairs (Fig. 3C). To address whether these defects arise from defective interaction with Smc3’s coiled coil, we created a series of mutations intended to alter its surface without affecting coiled coil formation per se. Lethality was caused by three mutations, namely L1019R, I1026R and L1029R, that replaced hydrophobic side chains with charged ones on the surface of the coiled coil facing Scc1’s α2 and α3 (Fig. 3B and Fig. S5A). None of these mutations altered Smc3 levels or prevented Scc1’s association with Smc1/Smc3 heterodimers in vivo (Fig. S5B), presumably because association between Scc1’s CTD and Smc1’s ATPase head recruits the kleisin to heterodimers even when interaction between its NTD and Smc3 is compromised (11). Smc3L1029R also disrupted interaction with Scc1-N when co-expressed in E. coli (Fig. S5F). In yeast cells, the same mutation reduced UV-induced crosslinking between Smc3A181bpa and Scc1 (Fig. S5D) as well as cross-linking between Scc1Q76C and Smc3A181C (Fig. S5C). Disruption of the hydrophobic nature of Smc3’s coiled coil by Smc3L1019R, I1026R and L1029R abrogated cohesin’s ability to associate with centromeric DNAs in vivo and prevented acetylation of Smc3 by Eco1 (Fig. 3D and Fig. S5E).
Figure 3. Functional analysis of the Smc3-kleisin structure.
(A) Essential Scc1-N residues (11) situated within the Smc3-kleisin interface. (B) Conserved residues in Scc1-N (D92, Y82) and Smc3CC (R1015, L1019, I1026, L1029) whose mutation disrupts interaction and causes lethality. (C) Abolition of Scc1-N/Smc3CC cross-linking by Scc1L89K (K19796, K23144, K23145) and Smc3L1029R (K23146, K23128, K23129) as measured by cross-linking C56-S1043C with bBBr following immuno-precipitation of Scc1-HA6. Western blots of Smc3-myc9 and Scc1-HA6 are shown. (D) Diploid cells expressing ectopic WT or mutant Smc3-GFP with white arrowhead pointing at the cohesin peri-centromeric barrel structure absent in the case of L1029R and R61Q (K23107-23109). (E) Scc1Y82A is lethal at 32 °C (K699, K16296, K23105-6). (F) A summary of mutations created and characterized. Lethal (red) and temperature sensitive (brown) mutations, both in Scc1 and Smc3, were found. Note the viability of Scc1D95A, K99D, K99A, and D95A K99A mutants (K23111-23117).
Introduction of charged residues into a hydrophobic interface precludes evaluation of the role of individual residues. For a more nuanced analysis, we focused on Y82, a conspicuous feature of α-, β-kleisins and γ-kleisins (Fig. S4D). Tetrad analysis revealed that Scc1Y82A caused slow growth at 30 °C and lethality at 32 °C (Fig. 3E and Fig. S6A). Scc1Y82I had no obvious effect on proliferation but was synthetic lethal with a temperature sensitive allele of ECO1 (Eco1G211D). We conclude that insertion at this position in Smc3’s coiled coil of a large aromatic residue has an important role in stabilizing its association with Scc1’s NTD.
Though crucial, these hydrophobic interactions are insufficient. For example, lethality is also caused by substitution with glutamic acid of the highly conserved Smc3R1015 (Fig. 3B), which interacts with the equally conserved D92 within Scc1’s α3. Contrary to previous observations, which were made on a version of Scc1 that was doubly tagged and contained a TEV protease cleavage site, D92K is in fact not lethal per se, though it does cause slow growth at 30 °C and lethality at 37 °C when Smc3 is tagged at its C-terminus by PK3 (Fig. S6C). Lastly, the finding that lethality is also caused by Scc1A47K (11), which alters a highly conserved alanine within Scc1’s α2, confirms the importance of α2 (Fig. S4A).
The KKD strand within the Smc3 ATPase head
The Smc3 ATPase heads have an irregular β strand at the top of their N-terminal lobe that contains an invariant aspartic acid residue (D114) next to a pair of highly conserved lysine residues (K112 and K113) whose acetylation by Eco1 is essential for sister chromatid cohesion (13, 14). Our structure also reveals an adjacent α helix (R58-L64) whose base abuts the nucleotide and thereby potentially links the latter to the KKD strand (Fig. 1E). The KKD strand, as well as other highly conserved Smc3-specific residues (S75, R107, and G110) in its vicinity (15), has a role in releasing cohesin from chromatin in a process dependent on cohesin’s Wapl, Pds5, and Scc3 regulatory subunits. Acetylation of K112 and K113 by Eco1 neutralizes this activity and thereby stabilizes cohesin’s association with chromatin. Given that release is thought to involve transient dissociation of Scc1’s NTD from Smc3, thereby creating a gate through which DNA exits the ring, it is striking that the KKD strand is situated some distance (minimal 25 Å) from the part of Smc3 that binds Scc1, namely its coiled coil.
To address whether cohesin containing Smc3R61Q loads onto chromosomes, we compared the distribution of GFP-tagged Smc3R61Q and wild type Smc3 in living cells carrying an untagged endogenous Smc3 gene. Smc3R61Q-GFP failed to accumulate at kinetochores or to form peri-centromeric barrels during G2/M, implying the mutant protein cannot load onto chromosomes, at least in the vicinity of centromeres (Fig. 3D). R61 is close to K112 and K113, whose acetylation by Eco1 not only blocks release but also very possibly blocks cohesin’s ability to engage in a loading reaction capable of producing cohesion (16). It is conceivable therefore that R61 has some role in relaying the state of modification of the KKD strand to Smc3’s nucleotide binding pocket. Because the Smc3K112Q K113Q double mutation is also lethal and reduces cohesin’s loading onto chromosomes (13, 17), it is possible that in their unmodified form K112 and K113 have a role in promoting cohesin loading and that they do so by influencing Smc3’s ATPase activity in a manner involving R61. Thus, the KKD strand might be concerned with Scc2/4-mediated loading of cohesin onto chromosomes as well as Wapl-mediated release.
Cohesin trimers hold sister DNAs together in live cells
To address whether simultaneous interaction of the three Smc/kleisin interfaces actually creates rings in vivo, living cells expressing Smc1-myc9, Scc1-PK6, and Smc3-Halo were incubated with BMOE to cross-link one, two, or three interfaces. Scc1-PK6 was then immuno-precipitated. In the presence of the tetramethyl-rhodamine (TMR) ligand, Smc3-Halo becomes labelled and fluorescent (Fig 4B) and was visualized following SDS-PAGE by scanning gels at 545 nm. Smc1-myc9 and Scc1-PK6 were detected by Western blotting (Fig. S7A). Dimeric molecules were created if cysteine pairs were present at single interfaces and trimers when present at two (Fig. 4B). Cross-linking cohesin containing cysteines at all three interfaces generated a novel form (Ci) whose electrophoretic mobility was slower than that of all three trimeric forms created by cross-linking at merely two out of the three interfaces. This form presumably arises from the creation of circular molecules due to the simultaneous cross-linking of all three interfaces of tripartite rings. The fraction of linear and circular trimers was roughly consistent with cross-linking at each of the three interfaces occurring independently (Fig. S7C). Similar results were obtained by cross-linking with bBBr after immuno-precipitation (Fig. S7B). Thus, most cohesin inside living yeast cells has the form of a heterotrimeric ring. Western blotting using antibodies specific for acetylated Smc3 (17) showed that the Ci form was acetylated to a degree similar to that of molecules that had not been chemically circularized (Fig. 4B, last lane). Importantly, acetylation was detected in cross-linked species created in vivo by several different cysteine pairs (Fig. 4A). Because cohesion is mediated only by acetylated complexes (13, 14), these data suggest that the cohesin complexes responsible for holding sister DNAs together are also circular Smc1/Smc3/Scc1 heterotrimers.
It has been suggested that Scc1 links the ATPase heads of different Smc1/Smc3 heterodimers, thereby creating dimeric rings or multimeric chains (18). According to this scenario, the Ci form could conceivably be a tetramer containing Smc subunits from two Smc1/3 heterodimers. To address whether Ci contains more than one Smc3 molecule, we compared the amount of Halo-tagged Ci associated with PK- and HA-tagged proteins from Scc1-PK6/Scc1-PK6, Smc3-HA6/Smc3-Halo diploids after in vivo cross-linking. This showed that little or no Smc3-Halo is present in Ci molecules that had been immuno-precipitated with HA antibodies, implying that they are circular heterotrimers and not tetramers or multimers (Fig. 4C). We used a similar method, albeit without cross-linking, to show that acetylated wild type Smc3 cannot be co-immuno-preciptiated by a myc-tagged version when extracts were prepared from Smc3/Smc3-myc18 diploids (Fig. S7E).
Sister DNAs are entrapped by cohesin trimers
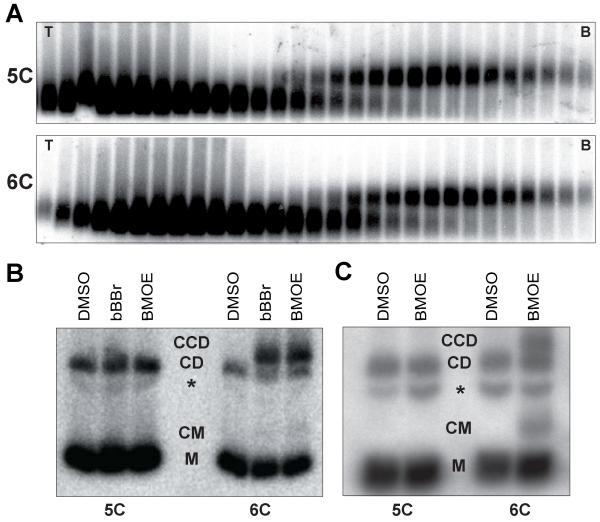
To test whether cross-linking all three interfaces of the cohesin ring entraps sister DNAs in a covalent topological embrace, we isolated dimers of a 2.3 kb circular minichromosome (Fig. 5A), treated them with bBBr or BMOE, and subjected them to gel electrophoresis following denaturation (19). In the absence of crosslinker, most minichromosome DNAs detected by Southern blotting migrated as closed circular monomers (M, Fig. 5B) while 10-20% migrated as monomeric nicked circles or catenated supercoils (CD). With cysteine pairs at all three interfaces, both cross-linkers caused the appearance of dimers (CCD) that migrate more slowly than catenated supercoils (9) (Fig. 5B). No such dimers were formed when minichromosomes were isolated from a strain lacking a single one of the six “interface” cysteines (Fig. 5B). We estimated that the fraction of monomeric DNAs converted to dimers by cross-linking was about 25%, which is similar to the efficiency of cohesin’s chemical circularization in vivo (Fig. 4B) and in vitro (Fig. S7B). Crucially, sister (CCD) as well as monomeric (CM) DNAs were catenated in this manner by chemically circularized cohesin when living yeast cells were treated with BMOE (Fig. 5C), indicating that cohesin rings entrap sister DNAs in vivo
Figure 5. Heterotrimeric cohesin rings trap sister DNAs both in vitro and in vivo.
(A) Dimers of a 2.3 kb circular minichromosome were isolated using sucrose gradients (T=Top, B=Bottom) from strains containing five (5C) or six (6C) cysteines within the Smc1/Smc3/Scc1 interfaces (K20300, K20279). (B) Electrophoresis of dimers denatured with SDS following isolation from sucrose gradients and cross-linking with bBBr or BMOE. DNAs detected by Southern blotting. (C) Electrophoresis of Scc1-PK6 immuno-precipitated DNAs denatured with SDS after in vivo BMOE crosslinking of cycling cells. DNAs detected by Southern blotting. Monomeric circles (M), catenated monomers (CM), nicked DNA (*), catenated dimers (CD)and cohesin catenated dimers (CCD) (K20280, K20279).
Discussion
Our crystal structure of an Smc3/Scc1 complex provides a mechanism by which a single Scc1 polypeptide links the ATPase heads of Smc1/Smc3 heterodimers, creating a heterotrimeric ring structure. Using thiol-specific chemical cross-linking we demonstrate that cohesin complexes holding sister chromatids together in vivo do indeed have the configuration of heterotrimeric rings and that sister minichromosome DNAs are entrapped within these in vivo. The interactions between Smc1/3 hinges, Smc1ATPase/Scc1-C, and Smc3CC/Scc1-N are all very stable (4, 20, 21). Intact cohesin rings are likely therefore to be extremely durable in the absence of specific mechanisms to disrupt them. Such a feature is desirable for a complex that must hold sister DNAs together for extended periods of time, which may last for several decades in the case of human oocytes.
Two mechanisms are known to remove cohesin from chromosomes. Best understood is cleavage of the central domain of Scc1 (22) and its meiotic counterpart Rec8 (23) by separase, an event that triggers sister chromatid disjunction at the onset of anaphase. The simplest explanation for this phenomenon is that the three interactions that create tripartite cohesin rings are both necessary and sufficient to entrap sister DNAs and separase merely destroys topological entrapment. The second mechanism is separase independent, occurs throughout the cell cycle, and is especially active during prophase when most cohesin is removed from chromosome arms (24, 25). The releasing activity responsible for this phenomenon is associated with cohesin itself, involves its Wapl (26, 27), Pds5, and Scc3 subunits (15), and is blocked by fusion of Smc3 to Scc1.Our finding that Smc3 R61 possibly links the KKD strand to the Smc3’s ATP binding pocket raises the possibility that acetylation, which blocks releasing activity, may directly regulate ATPase activity and vice versa.
Sequence comparisons suggest that most if not all eukaryotic Smc/kleisin complexes have a configuration similar to that of cohesin’s ring. Because all three interfaces of cohesin’s ring and those of bacterial Smc/kleisin complexes are structurally homologous, this class of complex must have been present in the last common ancestor of all living organisms and may be an indispensable feature of DNA genomes.
Supplementary Material
Acknowledgements
TGG is supported by an EMBO long-term fellowship (ALTF 2008-127) and the John Fell Fund (132/108). The work was funded by the Medical Research Council (U10518432 to JL, C573/A11625 to JSC), the Wellcome Trust (095514/Z/11/Z to JL, 091859/Z/10/Z to KN), and Cancer Research UK (C573/A 12386 to KN).
References
- 1.Peters JM, Tedeschi A, Schmitz J. The cohesin complex and its roles in chromosome biology. Genes Dev. 2008;22:3089–3114. doi: 10.1101/gad.1724308. [DOI] [PubMed] [Google Scholar]
- 2.Nasmyth K. Cohesin: a catenase with separate entry and exit gates? Nat Cell Biol. 2011;13:1170–1177. doi: 10.1038/ncb2349. [DOI] [PubMed] [Google Scholar]
- 3.Gruber S, Haering CH, Nasmyth K. Chromosomal cohesin forms a ring. Cell. 2003;112:765–777. doi: 10.1016/s0092-8674(03)00162-4. [DOI] [PubMed] [Google Scholar]
- 4.Haering CH, Lowe J, Hochwagen A, Nasmyth K. Molecular Architecture of SMC Proteins and the Yeast Cohesin Complex. Mol Cell. 2002;9:773–788. doi: 10.1016/s1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- 5.Gruber S, Arumugam P, Katou Y, Kuglitsch D, Helmhart W, Shirahige K, Nasmyth K. Evidence that loading of cohesin onto chromosomes involves opening of its SMC hinge. Cell. 2006;127:523–537. doi: 10.1016/j.cell.2006.08.048. [DOI] [PubMed] [Google Scholar]
- 6.Chan KL, Roig MB, Hu B, Beckouet F, Metson J, Nasmyth K. Cohesin’s DNA Exit Gate Is Distinct from Its Entrance Gate and Is Regulated by Acetylation. Cell. 2012;150:961–974. doi: 10.1016/j.cell.2012.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan KL, Gligoris T, Upcher W, Kato Y, Shirahige K, Nasmyth K, Beckouet F. Pds5 promotes and protects cohesin acetylation. Proc Natl Acad Sci U S A. 2013;110:13020–13025. doi: 10.1073/pnas.1306900110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burmann F, Shin HC, Basquin J, Soh YM, Gimenez-Oya V, Kim YG, Oh BH, Gruber S. An asymmetric SMC-kleisin bridge in prokaryotic condensin. Nat Struct Mol Biol. 2013;20:371–379. doi: 10.1038/nsmb.2488. [DOI] [PubMed] [Google Scholar]
- 9.Haering CH, Farcas A, Arumugam P, Metson J, Nasmyth K. The cohesin ring concatenates sister DNAs. Nature. 2008;454:297–301. doi: 10.1038/nature07098. [DOI] [PubMed] [Google Scholar]
- 10.Schleiffer A, Kaitna S, Maurer-Stroh S, Glotzer M, Nasmyth K, Eisenhaber F. Kleisins: a superfamily of bacterial and eukaryotic SMC protein partners. Mol Cell. 2003;11:571–575. doi: 10.1016/s1097-2765(03)00108-4. [DOI] [PubMed] [Google Scholar]
- 11.Arumugam P, Nishino T, Haering CH, Gruber S, Nasmyth K. Cohesin’s ATPase activity is stimulated by the C-terminal Winged-Helix domain of its kleisin subunit. Curr Biol. 2006;16:1998–2008. doi: 10.1016/j.cub.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 12.Hu B, Itoh T, Mishra A, Katoh Y, Chan KL, Upcher W, Godlee C, Roig MB, Shirahige K, Nasmyth K. ATP hydrolysis is required for relocating cohesin from sites occupied by its Scc2/4 loading complex. Curr Biol. 2011;21:12–24. doi: 10.1016/j.cub.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Unal E, Heidinger-Pauli JM, Kim W, Guacci V, Onn I, Gygi SP, Koshland DE. A molecular determinant for the establishment of sister chromatid cohesion. Science. 2008;321:566–569. doi: 10.1126/science.1157880. [DOI] [PubMed] [Google Scholar]
- 14.Ben-Shahar TR, Heeger S, Lehane C, East P, Flynn H, Skehel M, Uhlmann F. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science. 2008;321:563–566. doi: 10.1126/science.1157774. [DOI] [PubMed] [Google Scholar]
- 15.Rowland BD, Roig MB, Nishino T, Kurze A, Uluocak P, Mishra A, Beckouet F, Underwood P, Metson J, Imre R, Mechtler K, Katis VL, Nasmyth K. Building sister chromatid cohesion: smc3 acetylation counteracts an antiestablishment activity. Mol Cell. 2009;33:763–774. doi: 10.1016/j.molcel.2009.02.028. [DOI] [PubMed] [Google Scholar]
- 16.Beckouet F, Hu B, Roig MB, Sutani T, Komata M, Uluocak P, Katis VL, Shirahige K, Nasmyth K. An Smc3 acetylation cycle is essential for establishment of sister chromatid cohesion. Mol Cell. 2010;39:689–699. doi: 10.1016/j.molcel.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sutani T, Kawaguchi T, Kanno R, Itoh T, Shirahige K. Budding yeast Wpl1(Rad61)-Pds5 complex counteracts sister chromatid cohesion-establishing reaction. Curr Biol. 2009;19:492–497. doi: 10.1016/j.cub.2009.01.062. [DOI] [PubMed] [Google Scholar]
- 18.Huang CE, Milutinovich M, Koshland D. Rings, bracelet or snaps: fashionable alternatives for Smc complexes. Philos Trans R Soc Lond B Biol Sci. 2005;360:537–542. doi: 10.1098/rstb.2004.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivanov D, Nasmyth K. A physical assay for sister chromatid cohesion in vitro. Mol Cell. 2007;27:300–310. doi: 10.1016/j.molcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 20.Haering CH, Schoffnegger D, Nishino T, Helmhart W, Nasmyth K, Lowe J. Structure and stability of cohesin’s Smc1-kleisin interaction. Mol Cell. 2004;15:951–964. doi: 10.1016/j.molcel.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 21.Kurze A, Michie KA, Dixon SE, Mishra A, Itoh T, Khalid S, Strmecki L, Shirahige K, Haering CH, Lowe J, Nasmyth K. A positively charged channel within the Smc1/Smc3 hinge required for sister chromatid cohesion. EMBO J. 2011;30:364–378. doi: 10.1038/emboj.2010.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uhlmann F, Lottspeich F, Nasmyth K. Sister chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1p. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- 23.Buonomo SB, Clyne RK, Fuchs J, Loidl J, Uhlmann F, Nasmyth K. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell. 2000;103:387–398. doi: 10.1016/s0092-8674(00)00131-8. [DOI] [PubMed] [Google Scholar]
- 24.Losada A, Hirano M, Hirano T. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev. 1998;12:1986–1997. doi: 10.1101/gad.12.13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sumara I, Vorlaufer E, Gieffers C, Peters BH, Peters J-M. Characterization of vertebrate cohesin complexes and their regulation in prophase. J. Cell Biol. 2000;151:749–762. doi: 10.1083/jcb.151.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gandhi R, Gillespie PJ, Hirano T. Human Wapl is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Curr Biol. 2006;16:2406–2417. doi: 10.1016/j.cub.2006.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kueng S, Hegemann B, Peters BH, Lipp JJ, Schleiffer A, Mechtler K, Peters JM. Wapl controls the dynamic association of cohesin with chromatin. Cell. 2006;127:955–967. doi: 10.1016/j.cell.2006.09.040. [DOI] [PubMed] [Google Scholar]
- 28.Chen HT, Warfield L, Hahn S. The positions of TFIIF and TFIIE in the RNA polymerase II transcription preinitiation complex. Nat Struct Mol Biol. 2007;14:696–703. doi: 10.1038/nsmb1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farcas AM, Uluocak P, Helmhart W, Nasmyth K. Cohesin’s Concatenation of Sister DNAs Maintains Their Intertwining. Molecular Cell. 2011;44:97–107. doi: 10.1016/j.molcel.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stock D, Perisic O, Lowe J. Robotic nanolitre protein crystallisation at the MRC Laboratory of Molecular Biology. Prog Biophys Mol Biol. 2005;88:311–327. doi: 10.1016/j.pbiomolbio.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 31.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr D Biol Crystallogr. 2010;66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turk D. MAIN software for density averaging, model building, structure refinement and validation. Acta Crystallogr D Biol Crystallogr. 2013;69:1342–1357. doi: 10.1107/S0907444913008408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.